In this study, Orkin and colleagues uncover a mechanism of selectively affecting leukemic stem cells by regulating the DNA methyltransferase Dnmt1. Dnmt1 conditional knockout blocks AML development, and its haploinsufficiency specifically delays progression of leukemogenesis due to derepression of a gene expression program. This study has significant therapeutic implications for the treatment of AML.
Keywords: DNA methylation, leukemia stem cell, Dnmt1, bivalent chromatin
Abstract
Epigenetic mechanisms regulating leukemia stem cells (LSCs) are an attractive target for therapy of blood cancers. Here, we report that conditional knockout of the DNA methyltransferase Dnmt1 blocked development of leukemia, and haploinsufficiency of Dnmt1 was sufficient to delay progression of leukemogenesis and impair LSC self-renewal without altering normal hematopoiesis. Haploinsufficiency of Dnmt1 resulted in tumor suppressor gene derepression associated with reduced DNA methylation and bivalent chromatin marks. These results suggest that LSCs depend on not only active expression of leukemogenic programs, but also DNA methylation-mediated silencing of bivalent domains to enforce transcriptional repression.
Epigenetic changes play important roles in the development and progression of cancer. Historically, alterations in DNA methylation have been associated with transformation. However, it is now being appreciated that within the complex chromatin network, DNA methylation contributes to and is influenced by histone modifications that also may be disrupted in cancer (Esteller 2008). In a seminal study, Dick and colleagues (Lapidot et al. 1994) showed that human acute myeloid leukemia (AML) cells are organized in a hierarchy that originates from a primitive cell termed a “leukemia stem cell” (LSC). These cells are able to initiate tumorigenesis by undergoing self-renewal and differentiation, processes similar to normal stem cells, whereas the remaining majority of leukemia cells lack these properties. It has been demonstrated that DNA methylation is a shared epigenetic program controlling both normal hematopoietic stem cells (HSCs) and LSCs, but these stem cell programs may rely on different methylation dosage-dependent control mechanisms (Bröske et al. 2009). It remains unclear whether there are unique gene targets of DNA methylation in LSCs and, furthermore, whether specific cooperating epigenetic mechanisms exist to regulate the critical functions of these cells. In that DNA methylation regulates both HSCs and LSCs, issues of specificity are pertinent to assessing whether DNA methylation and/or its gene targets are to be considered for use in directed leukemia therapy.
Here, using a mouse model of MLL-AF9-induced AML (Krivtsov et al. 2006), we found that Dnmt1 is the only DNA methyltransferase with sustained levels of expression in LSCs compared with HSCs and consequently focused on Dnmt1 as a candidate for maintaining DNA methylation patterns in LSCs. Through functional and molecular studies, we examined the specific consequences of Dnmt1 loss in LSCs and demonstrated selectivity in the response of LSCs versus normal HSCs, revealing that a favorable therapeutic index may be achieved by specifically targeting Dnmt1 in AML therapy.
Results and Discussion
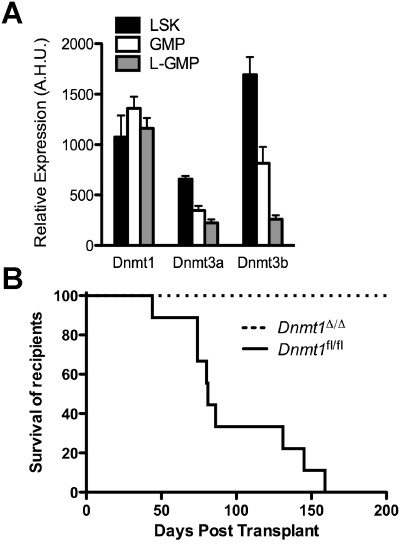
We first examined expression of the DNA methyltransferases Dnmt1, Dnmt3a, and Dnmt3b in MLL-AF9-induced LSCs, defined as leukemic granulocyte–macrophage progenitors (L-GMPs; Lin− IL-7Rα− c-Kit+ Sca-1− CD34+ FcγR+), compared with normal hematopoietic stem/progenitor cell-enriched LSK (Lin− Sca-1+ c-Kit+) cells. We found that expression of Dnmt1 was sustained in L-GMPs versus LSK cells; however, the expression of Dnmt3a and Dnmt3b was reduced in L-GMPs (Fig. 1A). The selective maintenance of Dnmt1 in L-GMPs suggests that DNMT1 may be responsible for maintaining methylation patterns in L-GMPs.
Figure 1.
Deletion of Dnmt1 prevents development of MLL-AF9-induced leukemia. (A) Relative expression of Dnmt1, Dnmt3a, and Dnmt3b in LSK (n = 5), GMP (n = 4), and L-GMP (n = 5) cells. (A.H.U.) Array hybridization units. (B) Kaplan-Meier survival curve of mice transplanted with MLL-AF9-transduced Dnmt1Δ/Δ (n = 6) or control Dnmt1fl/fl (n = 9) cells.
To define the dependence of MLL-AF9-induced AML on Dnmt1, we used a conditional knockout of Dnmt1 (Dnmt1fl/fl) driven by the IFNα-inducible Mx-Cre (Trowbridge et al. 2009). Dnmt1 deletion was induced by polyI–polyC injection prior to isolation of LSK cells, infection with MLL-AF9-IRES-GFP retrovirus, and transplant into recipients. In all experiments, both control and experimental groups received polyI–polyC treatment to control for induced interferon-α expression. While control MLL-AF9-transduced Dnmt1fl/fl cells gave rise to fully penetrant AML with a median recipient survival of 81 d, mice transplanted with MLL-AF9-transduced Dnmt1Δ/Δ cells failed to develop AML (Fig. 1B). Upon sacrifice of Dnmt1Δ/Δ recipients at ∼200 d post-transplant, GFP+ leukemia cells were not detectable by flow cytometry in the bone marrow or spleen (data not shown), suggesting that the initiation of transformation by MLL-AF9 or, alternatively, survival of target cells is dependent on Dnmt1.
To examine the effect of Dnmt1 deletion on the maintenance of pre-existing leukemia, Dnmt1fl/fl Mx-Cre+ or control Dnmt1fl/fl L-GMPs were transplanted into recipients and allowed to establish leukemia over the course of 2 wk before inactivation of Dnmt1 (Supplemental Fig. S1A). Prior to polyI–polyC injection, all recipient mice demonstrated elevated peripheral white blood cell (WBC) counts (>50 × 109 per liter), thrombocytopenia (PLT, <250 × 109 per liter), and >80% GFP+ cells in peripheral blood by FACS analysis (data not shown). At this stage, deletion of Dnmt1 resulted in a significant delay in leukemia progression and doubled the survival time to a median of 64 d compared with control Dnmt1fl/fl with a median survival of 31 d (Supplemental Fig. S1B). Of note, genomic excision PCR revealed that the leukemia outgrowth from Dnmt1fl/fl Mx-Cre+ cells was comprised of only partially excised leukemic clones (Supplemental Fig. S1C), indicating probable selection against Dnmt1Δ/Δ leukemic cells. To investigate this further, L-GMPs were isolated from Dnmt1fl/fl Mx-Cre+ or control leukemias (shown in Supplemental Fig. S1B) and transplanted into secondary recipient mice. Strikingly, secondary recipients transplanted with Dnmt1fl/fl Mx-Cre+ L-GMPs demonstrated a prolonged median survival of 66.5 d compared with those transplanted with control L-GMPs with a median survival of 22.5 d (Supplemental Fig. S1D). Excision PCR of the secondary leukemias revealed the absence of the excised Dnmt1 allele (Supplemental Fig. S1E), further indicating selection against leukemic cells bearing the Dnmt1Δ allele. Together, these results demonstrate a dependence of MLL-AF9-induced AML on Dnmt1 in (1) transformation giving rise to leukemia, (2) maintenance of established leukemia, and (3) re-establishment of leukemia by transplanted L-GMPs.
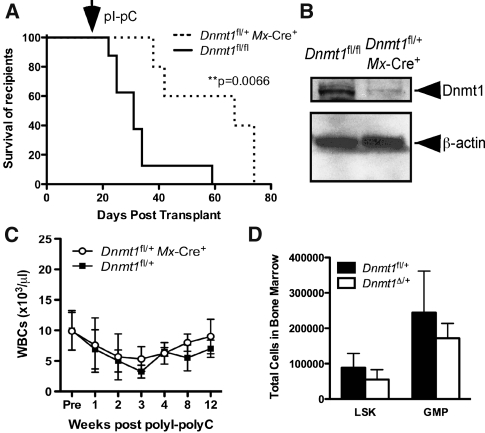
As our results indicated the existence of selection pressure against leukemic cells bearing the Dnmt1Δ allele, we tested the effect of Dnmt1 haploinsufficiency on the maintenance of leukemia using cells with one wild-type and one floxed allele of Dnmt1 for excision by Mx-Cre (Dnmt1fl/+ Mx-Cre+). MLL-AF9-transduced Dnmt1fl/+ Mx-Cre+ and control Dnmt1fl/fl cells were transplanted into recipients to establish leukemia 2 wk prior to injection of polyI–polyC. Importantly, the control Dnmt1fl/fl cells and mice are fully viable (data not shown), indicating that there are no partial defects resulting from the presence of the Dnmt1fl allele. Deletion of one allele of Dnmt1 after establishment of leukemia resulted in a significant delay in leukemia progression and increased survival time to a median of 67 d compared with control with a median survival of 31 d (Fig. 2A). Genomic excision PCR confirmed the genotype of leukemic clones as Dnmt1Δ/+ (Supplemental Fig. S2A). Furthermore, Western blot demonstrated a reduction in DNMT1 protein in Dnmt1Δ/+ leukemic cells (Fig. 2B), demonstrating that Dnmt1 haploinsufficiency is sufficient to delay the progression of MLL-AF9-induced AML.
Figure 2.
Haploinsufficiency of Dnmt1 delays progression of AML but does not alter normal hematopoiesis. (A) Kaplan-Meier survival curve of mice transplanted with MLL-AF9-transduced Dnmt1fl/+ Mx-Cre+ (n = 5) or control Dnmt1fl/fl (n = 8) cells. The arrow denotes the start of the polyI–polyC injections. (B) Western blot demonstrating DNMT1 protein levels in Dnmt1fl/+ Mx-Cre+ or control leukemias. (C) WBC counts obtained from Dnmt1fl/+ Mx-Cre+ or control mice (n ≥ 3), collected before and up to 12 wk after polyI–polyC injection. (D) Total number of LSK and GMP cells in Dnmt1Δ/+ (n = 3) or control (n = 3) mice.
Haploinsufficiency of Dnmt1 may be achieved pharmacologically in a clinical context; however, the therapeutic index of the LSC response compared with the effects on normal HSCs and hematopoiesis is not known. To evaluate the effects of Dnmt1 haploinsufficiency on nonmalignant hematopoiesis, we used Dnmt1fl/+ Mx-Cre+ or control Dnmt1fl/+ mice. As shown in Figure 2C, deletion of one allele of Dnmt1 did not alter significantly the number of WBCs in the peripheral blood compared with control mice up to 12 wk post-injection of polyI–polyC. Evaluation of the multilineage distribution of WBCs in the peripheral blood demonstrated that Dnmt1Δ/+ mice have equivalent distribution of granulocytes, macrophages, and mature B cells and T cells compared with control (Supplemental Fig. S2B). Examination of bone marrow hematopoiesis also revealed an equivalent total number of cells per femur in Dnmt1Δ/+ mice (Supplemental Fig. S2C), multilineage distribution (Supplemental Fig. S2D), and no significant change in numbers of hematopoietic stem/progenitor cells, including the LSK and GMP populations (Fig. 2D), compared with control mice. Together, these studies indicate that Dnmt1 haploinsufficiency fails to perturb nonmalignant hematopoiesis, including GMPs that are a target population for transformation by MLL-AF9 (Cozzio et al. 2003; Krivtsov et al. 2006). Thus, the delayed progression of MLL-AF9-induced AML resulting from Dnmt1 haploinsufficiency is mediated by a Dnmt1-dependent mechanism selective or unique to transformed cells.
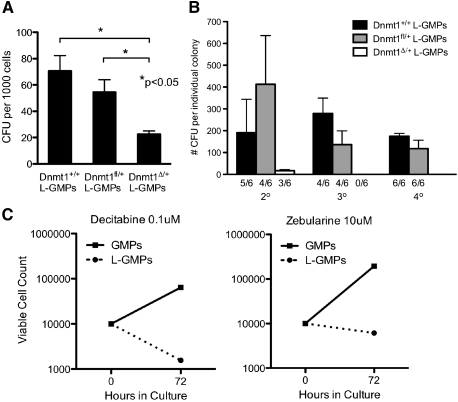
It has been demonstrated that DNA hypomethylation induced by a hypomorphic allele of Dnmt1 can impair self-renewal of MLL-AF9 LSCs (Bröske et al. 2009). As our model of Dnmt1 haploinsufficiency results in expression of DNMT1 to a higher level than the hypomorphic allele (Gaudet et al. 2003), we investigated whether Dnmt1 haploinsufficiency was sufficient to impair the self-renewal and differentiation of L-GMPs. MLL-AF9-transduced Dnmt1Δ/+ or control Dnmt1fl/+ L-GMPs were plated in methylcellulose for serial colony-forming unit (CFU) assays. In the initial round of plating, Dnmt1Δ/+ L-GMPs formed significantly fewer blast colonies compared with control (Dnmt1+/+ or Dnmt1fl/+) L-GMPs (Fig. 3A). This result demonstrates that Dnmt1 haploinsufficiency impairs the survival and/or differentiation of L-GMPs into blast colonies in vitro. In subsequent replatings of single colonies, Dnmt1Δ/+ L-GMPs formed only a small number of secondary colonies and were unable to form tertiary colonies (Fig. 3B). These results demonstrate a severe replating deficiency compared with control L-GMPs, indicating that Dnmt1 haploinsufficiency is sufficient to cause impaired survival, self-renewal, and differentiation of L-GMPs in vitro.
Figure 3.
Haploinsufficiency of Dnmt1 and DNA methylation inhibitors impair the self-renewal and proliferation of L-GMPs. (A) Blast CFUs per 1000 Dnmt1Δ/+ L-GMPs (n = 5) compared with control Dnmt1+/+ (n = 3) or Dnmt1fl/+ (n = 2) after 7 d of culture in methylcellulose. (B) Serial CFU assay of Dnmt1Δ/+ L-GMPs (white bars; n = 6) compared with control Dnmt1+/+ (black bars; n = 6) or Dnmt1fl/+ (gray bars; n = 6). Bars represent average number of CFUs formed from individual replated colonies. Ratios indicate the number of colonies giving rise to secondary or tertiary colonies over total. (C) Growth curve of L-GMPs (dotted line) and GMPs (solid line) treated with 0.1 μM decitabine or 10 μM zebularine for 72 h.
To evaluate whether pharmacological inhibition of DNA methylation can also impair the growth of L-GMPs, we plated equal numbers of GMPs and L-GMPs in liquid culture with two concentrations of the DNA methylation inhibitors decitabine (5-aza-2′-deoxycytidine) and zebularine (Stresemann et al. 2006). After 72 h, viable cell counts revealed that decitabine and zebularine inhibit the expansion of L-GMPs at concentrations that fail to impair the expansion of normal GMPs (Fig. 3C; Supplemental Fig. 3A). The 0.1 μM decitabine treatment, which we observed to be effective in limiting growth of L-GMPs, has been demonstrated to reduce DNA methylation by 20%–30% in human AML cell lines (Hollenbach et al. 2010), and treatment of cancer cells with 10 μM zebularine reduces genomic DNA methylation by ∼20% (Stresemann et al. 2006). Together, these studies demonstrate that reductions in DNA methylation through either Dnmt1 haploinsufficiency or pharmacological inhibition of DNA methylation impair the self-renewal and expansion of L-GMPs. These findings suggest that a favorable therapeutic index targeting L-GMPs may be achieved by specific pharmacological inhibition of Dnmt1.
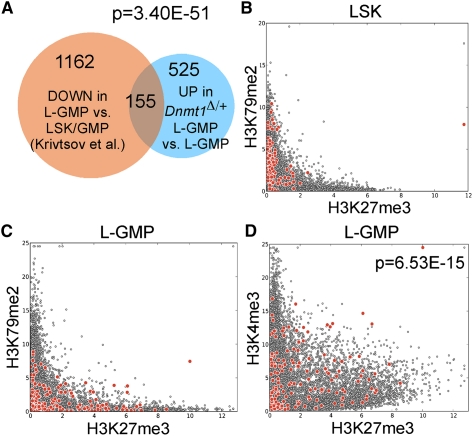
To investigate potential mechanisms underlying the dependence of MLL-AF9 L-GMPs on Dnmt1, we took an unbiased, genome-wide expression-profiling approach. Importantly, we observed an ∼50% reduction in Dnmt1 transcripts in Dnmt1Δ/+ L-GMPs and did not observe altered expression of Dnmt3a or Dnmt3b (Supplemental Fig. S3A). In addition, we did not observe significant changes in expression of Mll1, Hoxa9, Meis1, Myb, Hmgb3, or Cbx5 in Dnmt1Δ/+ L-GMPs (Supplemental Fig. S3A), demonstrating that deficiencies in these mediators of leukemogenesis are unlikely to account for the impaired function of Dnmt1Δ/+ L-GMPs. As DNA methylation has historically been associated with transcriptional repression, we focused our analysis on genes that were up-regulated or derepressed as a consequence of Dnmt1 haploinsufficiency. Approximately 30% of genes up-regulated more than twofold in Dnmt1Δ/+ L-GMPs were not up-regulated in Dnmt1Δ/+ bulk leukemia cells (Supplemental Fig. S3B), suggesting that a significant complement of genes were specifically derepressed in Dnmt1Δ/+ L-GMPs. Interestingly, the majority of these genes were not significantly altered in expression in Dnmt1fl/fl Mx-Cre+ HSCs (Supplemental Fig. S3C) or myeloid progenitors (Trowbridge et al. 2009; Supplemental Fig. S3D), suggesting that derepression of this gene set is unique to L-GMPs. We hypothesized that there may be a direct relationship between genes repressed by MLL-AF9 transformation in L-GMPs and genes derepressed in Dnmt1Δ/+ L-GMPs. We observed a significant overlap of 155 genes that were down-regulated in MLL-AF9 L-GMPs versus LSK and GMP cells (Krivtsov et al. 2006) and also up-regulated (or derepressed) in Dnmt1Δ/+ versus control L-GMPs (Fig. 4A). These 155 genes represent strong candidates for regulation by DNA methylation in L-GMPs. Querying against the CancerGenes database (Higgins et al. 2007) revealed that many of the 155 genes are categorized as “cancer-related” (36%) and a significant number are implicated as tumor suppressor genes (12%). To gain further insight into the cellular functions of the 155-gene set derepressed in Dnmt1Δ/+ L-GMPs, we queried the MSigDB databank (Subramanian et al. 2005) for other conditions or treatments that could result in induction of a significant number of the same 155 genes. Within the top five gene sets (Supplemental Fig. S3E), we identified an enrichment of EZH2 (Enhancer of Zeste homolog 2) target genes. As part of Polycomb-repressive complex 2 (PRC2), EZH2 catalyzes the addition of methyl groups to histone H3 at Lys 27 (H3K27) and is associated with transcriptional repression. Enrichment of this EZH2 target gene set suggests that polycomb-mediated repression may cooperate with DNA methylation-mediated silencing of target genes in L-GMPs.
Figure 4.
Dnmt1 haploinsufficiency results in derepression of genes normally silenced and marked by bivalent chromatin domains in L-GMPs. (A) Venn diagram demonstrating the overlap between 1317 genes repressed in L-GMPs (orange circle) (Krivtsov et al. 2006) and 680 genes derepressed in Dnmt1Δ/+ L-GMPs (blue circle). The 155-gene overlap represents our “derepressed in Dnmt1Δ/+ L-GMPs” signature. (B) Graphical representation of genes marked by H3K27me3 and H3K79me2 in LSK cells. Our 155-gene signature is highlighted (red circles) compared with genome-wide (gray circles). (C) Graphical representation of genes marked by H3K27me3 and H3K79me2 in L-GMPs. (D) Graphical representation of genes marked by H3K27me3 and H3K4me3 in L-GMPs.
To further explore the relationship between DNA methylation and histone modifications in L-GMPs, we examined H3K27me3, H3K4me3, and H3K79me2 of our 155-gene set in L-GMPs as well as LSK cells (Bernt et al. 2011). H3K4me3 and H3K79me2 are associated with transcriptional activation, with DOT1L-mediated H3K79me2 specifically required for the MLL-AF9-associated oncogenic program (Bernt et al. 2011; Jo et al. 2011; Nguyen et al. 2011). Furthermore, colocalizations of H3K27me3 and H3K4me3 have been termed “bivalent” chromatin domains that mark developmental genes in embryonic stem cells (Bernstein et al. 2006) and are associated with tumor suppressor genes in cancer (Ohm et al. 2007). These bivalent chromatin domains are considered to mark silent genes poised for transcriptional activation. In LSK cells, our 155-gene set was predominantly marked by H3K79me2 but not H3K27me3 (Fig. 4B, red circles), consistent with active transcription in hematopoietic stem/progenitor cells. In L-GMPs, however, many of these genes were marked by H3K27me3 and H3K4me3 but not H3K79me2 (Fig. 4C,D, red circles), these genes being in a bivalent chromatin state in L-GMPs. Bivalent chromatin domains were significantly enriched in the 155-gene set (Fig. 4D, red circles) compared with genome-wide (Fig. 4D, gray circles), suggesting that DNA methylation may serve to enforce transcriptional repression of these bivalent domains.
We also examined the occupancy of these regions by MLL and MLL-AF9 proteins. Shown as a positive control, the region targeted by MLL-AF9 in the HoxA cluster is associated with maintenance of H3K4me3 in L-GMPs versus LSK cells, and enriched H3K79me2 is associated with MLL-AF9 occupancy (Supplemental Fig. S4A). In our 155-gene set, we observed predominant occupancy of the wild-type MLL protein (87 out of 155 genes; 56%) but a near absence of MLL-AF9 occupancy (eight out of 155 genes; 5%). Occupancy of wild-type MLL at these loci was associated with maintenance of H3K4me3 in L-GMPs compared with LSK cells, reduced H3K79me2, and enrichment of H3K27me3 (Supplemental Fig. S4B–D). These results suggest that the 155-gene set that is repressed in association with the presence of bivalent chromatin and DNA methylation is bound by MLL but not MLL-AF9 in L-GMPs.
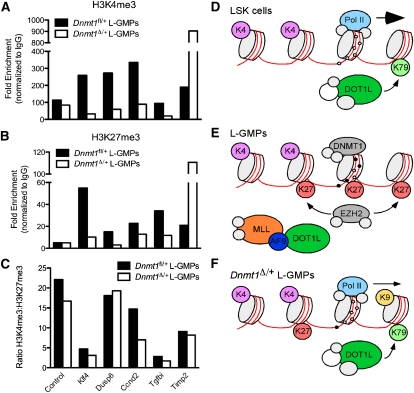
To characterize alterations in DNA methylation and histone modifications induced by Dnmt1 haploinsufficiency, we focused on a subset of tumor suppressor genes (Klf4, Dusp6, Ccnd2, Tgfbi, and Timp2) expressed in LSK cells and repressed in L-GMPs (Supplemental Fig. S6A). As anticipated, these genes were derepressed upon haploinsufficiency of Dnmt1 (Supplemental Fig. S6B). Consistent with derepression, we observed increased occupancy of RNA polymerase II (Pol II) at the proximal promoter regions of these genes (Supplemental Fig. S6C). In Dnmt1 haploinsufficient L-GMPs, the 5′ regulatory regions of Ccnd2 and Timp2 were largely unmethylated (Supplemental Fig. S6D,E). Examining changes in histone modifications, H3K4me3 and H3K27me3 were reduced at the Klf4, Dusp6, Ccnd2, and Tgfbi loci but enriched at the Timp2 locus in Dnmt1Δ/+ L-GMPs (Fig. 5A,B). Importantly, despite different levels of H3K4me3 and H3K27me3, the ratio of H3K4me3:H3K27me3 remained consistent at the majority of these loci (Fig. 5C), indicating that these bivalent domains were not resolved into monovalent H3K4me3 active domains. A similar observation has been reported in cancer cell lines treated with DNA methylation inhibitors, in which up-regulation of silenced genes was not associated with resolution of bivalent domains (Rodriguez et al. 2008). We also assessed the presence of other transcriptional activation-associated histone marks (H3K9ac and H3K79me2). We observed enrichment in both H3K9ac and H3K79me2 in the proximal promoter regions in Dnmt1Δ/+ L-GMPs compared with control (Supplemental Fig. S6F,G). These data suggest that haploinsufficiency of Dnmt1 in L-GMPs results in reduced DNA methylation and levels of bivalent chromatin marks at these tumor suppressor genes and increased levels of H3K79me2, occupancy of RNA Pol II, and activation of transcription.
Figure 5.
Haploinsufficiency of Dnmt1 results in reduced bivalent chromatin marks in L-GMPs. (A,B) Enrichment of histone marks H3K4me3 (A) and H3K27me3 (B) at the Ccnd2, Tgfbi, Dusp6, Timp2, and Klf4 loci in Dnmt1Δ/+ and control Dnmt1fl/+ L-GMPs. (C) Ratio of H3K4me3:H3K27me3 in Dnmt1Δ/+ and control L-GMPs. (D) In LSK cells, transcription is initiated by Pol II at promoters lacking DNA methylation and marked by H3K4me3. DOT1L-mediated H3K79me2 is distributed broadly across promoter and ORF regions. (E) In L-GMPs, bivalent chromatin domains are established by the addition of H3K27me3. DNA methylation acts to enforce transcriptional repression of these loci. (F) Upon haploinsufficiency of Dnmt1, these loci lose DNA methylation, have reduced bivalent chromatin marks, and gain H3K79me2. This allows Pol II binding, deposition of H3K9ac, and active transcription.
We propose a model to account for these observations. In normal LSK cells, these tumor suppressor genes are marked by H3K4me3 and DOT1L-mediated H3K79me2 in association with active transcription (Fig. 5D). Upon transformation by MLL-AF9, these genes remain marked by H3K4me3 but also gain the polycomb-mediated H3K27me3 mark to establish bivalent chromatin and DNMT1-mediated DNA methylation that enforces transcriptional repression (Fig. 5E). Our observation of wild-type MLL binding at a subset of these loci with DNA methylation (Supplemental Fig. S5B–D) is not consistent with the known function of MLL in selectively binding unmethylated DNA (Birke et al. 2002). We speculate that this observation is a consequence of heterogeneity within the L-GMP population and the true co-occurrence of protein occupancy and methylation marks will be resolved with the development of technology to evaluate these at the single-cell or clonal resolution. In Dnmt1 haploinsufficient L-GMPs, reductions in DNA methylation, H3K4me3, and H3K27me3 allow the establishment of H3K79me2 at these loci, presumably mediated by a DOT1L-containing complex, and activation of transcription (Fig. 5F). Together, these findings suggest that L-GMPs depend on not only active expression of leukemogenic programs, but also reversible, DNA methylation-mediated silencing of bivalent chromatin domains to enforce transcriptional repression of genes involved in tumor suppression, hematopoietic lineage restriction, and apoptosis.
In MLL-AF9-induced AML, it has been previously demonstrated that a hypomorphic allele of Dnmt1 (Dnmt1−/chip) impaired the self-renewal of LSCs (Bröske et al. 2009). Importantly, this level of DNMT1 expression also significantly impaired normal hematopoiesis and HSC function (Bröske et al. 2009), leading to the hypothesis that both LSCs and HSCs are similarly dependent on moderate levels of DNMT1. Here, we demonstrate that haploinsufficiency of Dnmt1 significantly impairs leukemogenesis and LSC self-renewal without altering normal hematopoiesis. Similarly in intestinal tract tumors, Dnmt1 haploinsufficiency was found to suppress tumor development (Laird et al. 1995), suggesting that a broad spectrum of tumor types may have enhanced dependence on Dnmt1. Two DNA methylation inhibitors, azacitidine and decitabine, have been FDA-approved for clinical use in the United States. In recent preclinical and clinical studies, therapeutic doses of decitabine were demonstrated to induce terminal differentiation of AML cells without altering normal HSC self-renewal (Negrotto et al. 2011), and azacitidine was found to have an acceptable safety profile and could substantially prevent or delay hematologic relapse in MDS and AML following HSC transplant (Platzbecker et al. 2011). Despite these successes, the high-dose cytotoxic effects and chemical instability of these DNA methylation inhibitors have prompted the development of additional strategies to target DNA methylation (Fathi and Abdel-Wahab 2011). Our work suggests that LSCs and leukemias have an increased dependence on or “addiction” to DNA methylation specifically mediated by Dnmt1 and that direct targeting of Dnmt1 will generate a favorable therapeutic index.
Materials and methods
Mice
Dnmt1fl/fl Mx1-Cre mice (Trowbridge et al. 2009) were administered polyI–polyC (InVivoGen) at 25 mg/kg per injection (Hock et al. 2004). Deletion of Dnmt1 was confirmed by PCR genotyping (Jackson-Grusby et al. 2001). Our Institutional Animal Care and Use Committee approved all experiments.
Generation of transformed cells and leukemia
Retroviral supernatant was made as described (Krivtsov et al. 2006). Bone marrow LSK cells were transduced for 2 d, then sorted GFP+ cells were transplanted into sublethally irradiated (600 rad) recipients at 1 × 104 or 1 × 105 cells per mouse. For secondary transplants, 500 GFP+ L-GMPs were injected.
Western blot
Whole lysates from spleens of leukemic mice were separated by SDS-PAGE. Western blot was performed using monoclonal anti-Dnmt1 (Active Motif) or β-actin (Abcam).
Peripheral blood and bone marrow analysis
Peripheral blood differential counts were taken using an AcT10 (Beckman Coulter). For analysis of LSK and GMP populations, lineage depletion of bone marrow cells and FACS were performed as described (Trowbridge et al. 2009).
CFU assays
GFP+-transduced leukemia cells or L-GMPs were plated in MethoCult GF M3534 (StemCell Technologies) at 1000 or 5000 cells per plate and replated weekly at 1000 cells per plate. Colonies were scored on day 7.
In vitro culture with DNA methylation inhibitors
GMPs and L-GMPs (1 × 104) were seeded in IMDM with 20% FCS, 20 ng/mL mSCF, 10 ng/mL mIL-6, 10 ng/mL mIL-3 (all Peprotech), 100 U/mL penicillin/streptomycin, and dilutions of decitabine and zebularine (Tocris). Viable cell counts were taken following 72 h of culture.
Chromatin immunoprecipitaion (ChIP)-quantitative PCR (qPCR)
L-GMPs (1 × 106) were processed for ChIP as previously described (Krivtsov et al. 2008) using antibodies against H3K4me3 (Abcam), H3K27me3 (Millipore), H3K79me2 (Abcam), H3K9ac (Millipore), and RNA Pol II (Millipore).
Statistical analyses
Statistical analysis of survival curves was performed using the log-rank (Mantel-Cox) test, bar graphs using the unpaired t-test or one-way ANOVA with Tukey's post-test, Venn diagrams by calculating the hypergeometric probability of the overlapping region, and enrichment of bivalent chromatin domains using ratios comparing the observed overlap and expected overlap.
Accession numbers
Raw data were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession numbers GSE31626 and GSE34261.
Acknowledgments
We thank members of the Orkin and Armstrong laboratories for helpful discussion, and John Daley, Suzan Lazo-Kallanian, and Rob Smith of the DFCI HemNeo Flow Facility. J.J.T. was supported by a fellowship from the Leukemia and Lymphoma Society, the Children's Leukemia Research Foundation, and ASH Scholar award. This work was supported by NIH U01 grant CA105423 (to S.H.O.), GO grant RC2 CA148222 (to S.H.O. and S.A.A.), and the HSCI blood program (S.H.O. and S.A.A.). S.H.O. is an Investigator of the HHMI.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.184341.111.
References
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, et al. 2011. MLL-rearranged Leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20: 66–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birke M, Schreiner S, Garcia-Cuellar MP, Mahr K, Titgemeyer F, Slany RK 2002. The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic Acids Res 30: 958–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröske A-M, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, et al. 2009. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet 41: 1207–1215 [DOI] [PubMed] [Google Scholar]
- Cozzio A, Passegué E, Ayton PM, Karsunky H, Cleary ML, Weissman IL 2003. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev 17: 3029–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M 2008. Epigenetics in cancer. N Engl J Med 358: 1148–1159 [DOI] [PubMed] [Google Scholar]
- Fathi AT, Abdel-Wahab O 2011. Mutations in epigenetic modifiers in myeloid malignancies and the prospect of novel epigenetic-targeted therapy. Adv Hematol 2012: 469–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R 2003. Induction of tumors in mice by genomic hypomethylation. Science 300: 489–492 [DOI] [PubMed] [Google Scholar]
- Higgins ME, Claremont M, Major JE, Sander C, Lash AE 2007. CancerGenes: A gene selection resource for cancer genome projects. Nucleic Acids Res 35: D721–D726 doi: 10.1093/nar/gkl811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock H, Hamblen MJ, Rooke HM, Schindler JW, Saleque S, Fujiwara Y, Orkin SH 2004. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature 431: 1002–1007 [DOI] [PubMed] [Google Scholar]
- Hollenbach PW, Nguyen AN, Brady H, Williams M, Ning Y, Richard N, Krushel L, Aukerman SL, Heise C, MacBeth KJ 2010. A comparison of azacitidine and decitabine activities in acute myeloid leukemia cell lines. PLoS ONE 5: e9001 doi: 10.1371/journal.pone.0009001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, et al. 2001. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet 27: 31–39 [DOI] [PubMed] [Google Scholar]
- Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL 2011. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood 117: 4759–4768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al. 2006. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 442: 818–822 [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al. 2008. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell 14: 355–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird PW, Jackson-Grusby L, Fazeli A, Dickinson SL, Jung WE, Li E, Weinberg RA, Jaenisch R 1995. Suppression of intestinal neoplasia by DNA hypomethylation. Cell 81: 197–205 [DOI] [PubMed] [Google Scholar]
- Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE 1994. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 367: 645–648 [DOI] [PubMed] [Google Scholar]
- Negrotto S, Ng KP, Jankowska AM, Bodo J, Gopalan B, Guinta K, Mulloy JC, Hsi E, Maciejewski J, Saunthararajah Y 2011. CpG methylation patterns and decitabine treatment response in acute myeloid leukemia cells and normal hematopoietic precursors. Leukemia doi: 10.1038/leu.2011.1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, Taranova O, He J, Zhang Y 2011. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood 117: 6912–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. 2007. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 39: 237–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platzbecker U, Wermke M, Radke J, Oelschlaegel U, Seltmann F, Kiani A, Klut IM, Knoth H, Rollig C, Schetelig J, et al. 2011. Azacitidine for treatment of imminent relapse in MDS or AML patients after allogeneic HSCT: Results of the RELAZA trial. Leukemia doi: 10.1038/leu.2011.1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, Muñoz M, Vives L, Frangou CG, Groudine M, Peinado MA 2008. Bivalent domains enforce transcriptional memory of DNA methylated genes in cancer cells. Proc Natl Acad Sci 105: 19809–19814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stresemann C, Brueckner B, Musch T, Stopper H, Lyko F 2006. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res 66: 2794–2800 [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowbridge JJ, Snow JW, Kim J, Orkin SH 2009. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell 5: 442–449 [DOI] [PMC free article] [PubMed] [Google Scholar]