E2F proteins are key drivers of cell proliferation. Dyson and colleagues uncover a novel mechanism of E2F regulation. Pumilio is identified to enhance the ability of miRNAs to control their specific E2F targets. Pumilio and miRNAs thus cooperate to limit E2F levels. Importantly, this regulation is abrogated in tumor cells: Bladder cancer cells adopt multiple mechanisms to circumvent such Pumilio/miRNA regulation, for example, by shortening E2F 3′ UTRs to eliminate regulation via the Pumilio regulatory element.
Keywords: cancer, E2F3, Pumilio, miRNA
Abstract
E2F transcription factors are important regulators of cell proliferation and are frequently dysregulated in human malignancies. To identify novel regulators of E2F function, we used Drosophila as a model system to screen for mutations that modify phenotypes caused by reduced levels of dE2F1. This screen identified components of the Pumilio translational repressor complex (Pumilio, Nanos, and Brain tumor) as suppressors of dE2F1-RNAi phenotypes. Subsequent experiments provided evidence that Pumilio complexes repress dE2F1 levels and that this mechanism of post-transcriptional regulation is conserved in human cells. The human Pumilio homologs Pum 1 and Pum 2 repress the translation of E2F3 by binding to the E2F3 3′ untranslated region (UTR) and also enhance the activity of multiple E2F3 targeting microRNAs (miRNAs). E2F3 is an oncogene with strong proliferative potential and is regularly dysregulated or overexpressed in cancer. Interestingly, Pumilio/miRNA-mediated regulation of E2F3 is circumvented in cancer cells in several different ways. Bladder carcinomas selectively down-regulate miRNAs that cooperate with Pumilio to target E2F3, and multiple tumor cell lines shorten the 3′ end of the E2F3 mRNA, removing the Pumilio regulatory elements. These studies suggest that Pumilio–miRNA repression of E2F3 translation provides an important level of E2F regulation that is frequently abrogated in cancer cells.
E2F proteins are important regulators of cellular proliferation and apoptosis. The mammalian family of E2F proteins contains eight members (E2F1–8). E2F1–3 are often described as transcriptional activators, while E2F4–8 are thought to primarily act as transcriptional repressors (for reviews, see Iaquinta and Lees 2007; Chen et al. 2009; Lammens et al. 2009). Dysregulation of E2F activity, either by the amplification of activator E2Fs (E2F3) (Feber et al. 2004) or through the inactivation of repressor mechanisms (pRb mutation/CDK activation) (Hanahan and Weinberg 2000; Sherr 1995), is a frequent oncogenic event in human tumorigenesis.
E2F activity is regulated at multiple levels. However, the size of the mammalian family of E2F proteins and the potential for functional redundancy between related family members has made it difficult to identify new mechanisms of E2F regulation by examining changes in overall E2F activity. The use of simpler metazoan systems such as Caenorhabditis elegans and Drosophila that have fewer E2F family members has provided important insights into the functions of E2F and has contributed to our understanding of the processes regulating the cell cycle and apoptosis. In Drosophila, dE2F1 is the sole activator E2F and is necessary for the expression of a large number of genes that are essential for DNA replication, cell cycle progression, and DNA damage responses (Duronio and O'Farrell 1994; Duronio et al. 1995; Dimova et al. 2003). To identify novel regulators of E2F activity in vivo, we used Drosophila as a model system to conduct genome-wide screens for mutations that modify phenotypes caused by tissue-specific depletion of dE2F1. From these screens, we uncovered an important and unexpected interaction between dE2F1 and the Pumilio translational repressor.
The Pumilio complex is a multisubunit repressor containing a PUF RNA-binding protein, Pumilio (Pum) (Murata and Wharton 1995); a Zinc finger RNA-binding protein, Nanos (Nos) (Barker et al. 1992; Sonoda and Wharton 1999); and a Trim-NHL protein, brain tumor (Brat) (Sonoda and Wharton 2001). The Puf proteins (of which Pumilio is the founding member) are a family of essential RNA-binding proteins that are conserved throughout the plant and animal kingdoms and provide post-transcriptional regulation of their targets (for review, see Quenault et al. 2011). Puf proteins regulate important developmental and cell cycle processes and are critical for stem cell maintenance and pluripotency in worms, flies, mice, and humans (Lin and Spradling 1997; Crittenden et al. 2002; Moore et al. 2003; Spassov and Jurecic 2003). In Drosophila, the RNA-binding proteins of the Pumilio complex, Pumilio and Nanos, bind to target mRNAs via distinct motifs (GUUGU Nanos regulatory element [NRE] and UGUAXAUA Pumilio regulatory element [PRE]) within the 3′ untranslated region (UTR) (Murata and Wharton 1995). The Pumilio complex inhibits translation via a number of mechanisms, including deadenylation (Goldstrohm et al. 2007) and decapping (Coller and Parker 2005), or by recruiting competitors of the translation initiation machinery to the target mRNA (Chagnovich and Lehmann 2001).
Two Pumilio (Pum 1 and Pum 2) and three Nanos (Nos 1–3) proteins have been identified in humans (Spassov and Jurecic 2002, Moore et al. 2003). The human Pumilio proteins have functions that are similar to their Drosophila homologs and have important roles in regulating male germ cell development (Moore et al. 2003; Ginter-Matuszewska et al. 2009), synaptic function (Vessey et al. 2010), and cell cycle re-entry following quiescence (Kedde et al. 2010). RNA pull-down assays (RPAs) have identified transcripts regulated by the endogenous human Pumilio machinery (Galgano et al. 2008). The mRNAs that associate with Pumilio show an enrichment for microRNA (miRNA) seed sequences (Galgano et al. 2008), and localized cooperation between Pum and miRNAs has previously been identified in C. elegans (Nolde et al. 2007) and human cells (Kedde et al. 2010).
miRNAs are 21- to 23-nucleotide (nt) noncoding RNAs that act as key post-transcriptional regulators and have important roles in the control of various cellular processes. Mature miRNAs associate with Argonaute (Ago) proteins to form RNA-induced silencing complexes (RISCs) (Hutvagner and Zamore 2002). These complexes interact directly with target mRNAs and are guided by sequence complementarity between the miRNA and the target. Interaction between the RISC and target mRNA inhibits gene expression through site-specific cleavage, mRNA degradation, or translation inhibition (for recent review, see Gu and Kay 2010).
Here we show that Drosophila dE2F1 is regulated by the Pumilio complex and that this mechanism of regulation is conserved in the human ortholog E2F3. Pumilio and Nanos also facilitate the ability of multiple miRNAs to regulate the complex 3′ UTR of the E2F3 mRNA, and the importance of this regulation is underscored by changes that occur in human cancer cells. E2F3 is often dysregulated during tumorigenesis. Amplification or elevated expression from the E2F3 locus (6p22) has been identified in breast (Tordai et al. 2008), prostate (Olsson et al. 2007), and lung (Cooper et al. 2006) cancers. Deregulated E2F3 also appears to be an important driver of proliferation in bladder carcinomas, since amplification of E2F3 is particularly common in this type of cancer (Feber et al. 2004; Oeggerli et al. 2004; Olsson et al. 2007; Hurst et al. 2008). Here we show that several miRNAs that have been shown to be selectively down-regulated in bladder carcinomas in clinical studies (Dyrskjot et al. 2009) target seed sequences within the E2F3 3′ UTR and repress E2F3 in a manner that is strongly cooperative with Pumilio. We found that many cancer cell lines circumvent Pumilio-mediated regulation of E2F3 by shortening the 3′ end of the E2F3 transcript, which eliminates the PRE required for Pumilio regulation. Taken together, these findings suggest that a functional cooperation between the Pumilio complex and miRNAs constrains E2F3 levels and that cancer cells consistently select changes that eliminate this regulation.
Results
Pumilio is a novel post-transcriptional regulator of dE2F1
To identify novel regulators of dE2F1, we used tissue-specific Gal4 drivers to allow UAS-RNAi transgenes targeting dE2F1 to be expressed in a temporally and spatially constrained manner during Drosophila development. Using this strategy, Glass Multimer Reporter-Gal4 (GMR-Gal4) transgenes enabled the levels of dE2F1 to be reduced specifically in the developing eye, generating a rough eye phenotype. Patched-Gal4 (ptc-GAL4)-driven expression of UAS-dE2F1 RNAi lowered levels of dE2F1 in the developing wing, and the resulting reduction in cell proliferation caused a narrowing of the intervein region between the L3 and L4 veins in the adult wing (Morris et al. 2008). To identify novel regulators of dE2F1 function, we examined the capacity of Exelixis deficiencies (Parks et al. 2004; Thibault et al. 2004) to modify these dE2F1-RNAi phenotypes (J Ji, A Herr, and N Dyson, unpubl.). By screening a collection of PiggyBac insertional mutants, we identified alleles that enhance or suppress the effects of dE2F1 depletion in both the eye and the wing (Morris et al. 2008; J Ji and A Herr, unpubl.).
Using this approach, we discovered that mutant alleles that affect subunits of the Pumilio translational repressor complex are strong suppressors of dE2F1-RNAi phenotypes. Mutations affecting pumilio (pum), nanos (nos), or brain tumor (brat) suppressed both the GMR-dE2F1-RNAi and ptc-dE2F1-RNAi phenotypes (Fig. 1A; Supplemental Fig. S1). A search of the 3′ UTRs of the Drosophila E2F-Rbf components for putative NREs (GUUGU) and PREs (UGUAXAUA) revealed the presence of a putative PRE and NREs within the 3′ UTR of dE2F1 (Fig. 1B) but not in other E2F/Rbf mRNAs. To test whether the endogenous dE2F1 mRNA is a target of Pumilio regulation, we conducted TAP tag RPAs from transgenic flies expressing the Pum RNA-binding motif (Gerber et al. 2006). dE2F1 transcripts were strongly enriched in Pum RPAs compared with controls (w1118 and TAP-tagged dNeurofibromin-1; gift from James Walker) in a manner that resembled the previously characterized Pum target CG8414 (Fig. 1C; Gerber et al. 2006). In contrast, no other members of the E2F-Rbf family were enriched in the TAP-Pum RPAs, although they all interacted with Poly-A-binding protein (PABP) (Fig. 1C; Supplemental Fig. S1C). These results suggest that there is a specific interaction between dE2F1 and Pumilio.
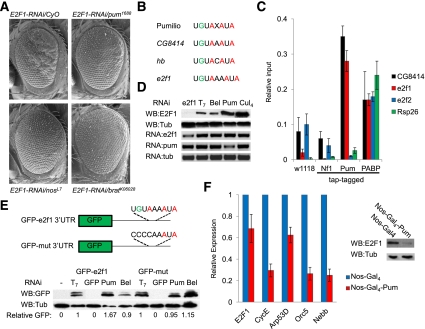
Figure 1.
Pumilio is a novel post-transcriptional regulator of dE2F1. (A) GMR-dE2F1-RNAi (E2F1-RNAi/CyO) causes a rough eye phenotype. Mutations affecting pum (pum1688), nanos (nosL7), or brat (bratK06028) dominantly suppress this phenotype. (B) PRE alignment showing the Pumilio consensus, PREs in validated Pum targets CG8414 and Hunchback (hb), and the putative PRE in dE2F1 (de2f1). (C) RT-qPCR results from tap tag RNA pull-down experiments from normal flies (w1118), tap tag Neurofibramin-1 (Nf1; tap tag negative control), tap tag Pumilio RNA-binding domains, and tap tag Poly-A-binding protein (PABP; positive control) showing the relative levels of CG8414, e2f1, e2f2, and Rsp26 (negative control) transcripts that are associated. (Relative input) Input levels set at 1. (D) Western blots of dE2F1 and Tubulin (Tub) from Drosophila SL2 cells depleted with either e2f1, T7, Bel, Pum, or Cul4. RT-qPCR data of e2f1, pumilio (pum), and tubulin (tub) demonstrating transcript levels upon knockdown. (E) Schematic of GFP constructs containing de2f1's 3′ UTR with the sequence of the putative PRE (GFP-e2f1-3′ UTR) or a mutated PRE sequence (GFP-mut-3′ UTR). Western blots of GFP and Tubulin (Tub) from cells depleted of T7, GFP, Pum, or Bel. Relative GFP readings comparing the lower GFP band and Tubulin. GFP readings from T7 depletion set to 1. (F) RT-qPCR results of e2f1, cycE, arp53D, orc5, and nebb from dissected ovaries from Nos-Gal4/CyO or Nos-Gal4-UAS-Pumilio (Nos-Gal4-Pum) flies. Westerns blots from these same ovaries of dE2F1 (E2F1) and Tubulin (Tub).
To assess the influence of Pumilio on the levels of dE2F1 expression, RNAi was used to deplete Pum from Drosophila S2 cells (Supplemental Fig. S2A). Knockdown of Pumilio elevated the levels of dE2F1 protein without changing transcript levels (Fig. 1D) and also increased expression of the E2F target genes (Supplemental Fig. S2B). Depletion of Cullin4, a known regulator of dE2F1 ubiquitination and degradation (Shibutani et al. 2008), similarly increased dE2F1 levels; in contrast, RNAi of belle (bel), an RNA-binding protein that has previously been reported not to regulate dE2F1 (Ambrus et al. 2007), had no effect (Fig. 1D). These data suggest that Pumilio may be a novel regulator of dE2F1 translation. As Rbf1 is a known repressor of transcription of dE2F1-regulated genes, we tested the prediction that Pumilio regulation of dE2F1 is distinct from regulation by Rbf1. To do this, we depleted Pumilio, Rbf1, or both (Supplemental Fig. S2C) and measured the effect on E2F target gene expression. Knocking down either Pumilio or Rbf1 elevated E2F target gene expression. The codepletion of both proteins had an additive effect on E2F target gene expression, suggesting that Pumilio and Rbf1 act in parallel to suppress the activity of dE2F1 via separate mechanisms (Supplemental Fig. S2D), although due to the fact that there is residual protein remaining after RNAi, we cannot completely rule out the possibility that they act cooperatively. Taken together, these results suggest that Pumilio and Rbf provide different mechanisms of dE2F1 regulation and that the cumulative activity of both proteins is required to constrain dE2F1 activity.
To determine whether Pumilio regulation of dE2F1 is dependent on the putative PRE, we generated a GFP reporter fused to the 3′ UTR of dE2F1 (GFP-dE2F1-3′ UTR). To test whether the putative PRE is functional, we mutated the sequence encoding the PRE (UGUAXXAUA) to give a nonfunctional element (CCCCXXAUA; GFP-mut-3′ UTR). GFP-dE2F1-3′ UTR and GFP-mut-3′ UTR expression constructs were transfected into Drosophila S2 cells and treated with dsRNA targeting Pumilio, Belle, or T7 (control). Depletion of Pum increased the level of GFP-dE2F1-3′ UTR translation compared with both the Bel and T7 controls; however, this effect was abolished when the PRE was mutated (GFP-mut) (Fig. 1E). In addition, disruption of the PRE (GFP-mut) elevated the expression of GFP in cells treated with control dsRNAs (T7 or bel) (Fig. 1E), suggesting the putative PRE is functional. To confirm that Pumilio regulates dE2F1 in vivo, we overexpressed Pum in the Drosophila ovary using the Nos-Gal4 driver. Elevated levels of Pumilio strongly reduced the levels of endogenous dE2F1 and decreased the expression of dE2F target genes (CycE, Orc5, and Nebb) (Fig. 1F). Together, these results strongly suggest that Pumilio is an important regulator of endogenous dE2F1 activity in Drosophila.
Pumilio regulation of activator E2F levels is conserved in human cells
To ask whether Pum regulation of E2F translation might be conserved in humans, we searched the 3′ UTRs of the human activator E2Fs (E2F1–3) for putative NRE and PRE sequences. Potential NRE/PREs were found within the 3′ UTRs of E2F2 and E2F3 (Fig. 2A) but not E2F1 (or the pocket proteins [pRb, p107, and p130]). To test whether Pumilio regulation of these activator E2Fs is conserved in human cells, we conducted RPAs using antibodies against endogenous Pum 1 and Pum 2 from primary human fibroblasts (IMR90). E2F3 transcripts were strongly enriched in immunoprecipitates of both Pum proteins (Fig. 2B) as compared with E2F1 or pRb mRNAs, suggesting that Pumilio regulation of activator E2F levels is conserved. The level of E2F2 mRNA in these cells was below the level of detection prior to RPA analysis and was not studied further. To evaluate the role of the endogenous Pum proteins in regulating the levels of E2F3, we depleted Pum from primary human fibroblasts (IMR90) using shRNAs specific to each Pumilio protein (cell cycle profiles) (Supplemental Fig. S3A). Knockdown of Pum 1 or Pum 2 specifically increased the levels of E2F3 (Fig. 2C; data not shown). The levels of E2F1 and the pocket proteins were not increased (Supplemental Fig. S3A–C), suggesting that Pumilio regulation is specific to E2F3.
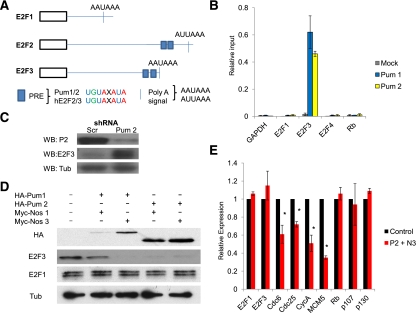
Figure 2.
Pumilio regulation of activator E2F levels is conserved in human cells. (A) Schematic of the human activator E2Fs (E2F1–3) demonstrating the relative positions and sequences of the PRE and distal polyadenylation sites (Poly-A signals). (B) RT-qPCR results of gapdh (GAPDH), e2f1 (E2F1), e2f3 (E2F3), e2f4 (E2F4), and retinoblastoma (Rb) from RPAs from IMR90 cells using human Pum 1 and Pum 2 antibodies compared with HA controls (Mock). Note that insufficient e2f2 transcript was detected in IMR90 cells, so it was excluded from further analysis. (Relative input) Input levels set at 1. (C) Western blots of Pumilio 2 (Pum 2), E2F3, and Tubulin (Tub) from IMR90s infected with lentiviral shRNA targeting Scrambled controls (Scr) or Pumilio 2 (Pum 2). (D) Western blots of HA, E2F3, E2F1, and Tub from primary human fibroblast (IMR90s) transfected with HA-Pum and Myc-nos overexpression constructs. (E) RT-qPCR results for the E2F target genes (e2f1, e2f3, cdc6, cdc25, cycA, and mcm5) and the pocket proteins (rb, p107, and p130) from IMR90s overexpressing Pum 2 and Nos 3. (*) P < 0.01.
To evaluate the functionality of the Pum and Nos proteins on E2F3, IMR90 cells were transfected with different combinations of Pum and Nos overexpression constructs. Elevated levels of Pum and Nos dramatically diminished the levels of E2F3 protein (compared with E2F1) (Fig. 2D), and this reduction in E2F3 was accompanied by decreased expression of E2F target genes (Fig. 2E; Supplemental Fig. S3D) or E2F-responsive reporter constructs (E2F4-Luciferase) (Supplemental Fig. S3E). None of the E2F target genes analyzed in these experiments contain PREs. The suppression of target gene expression by Pum/Nos expression was rescued by the cotransfection of an E2F3 overexpression construct that lacked the 3′ UTR sequence (Supplemental Fig. S3D). We conclude from these results that Pumilio regulation of activator E2F activity is conserved between Drosophila and (nontransformed) human cells.
The E2F3 3′ UTR contains two PREs
To identify the specific elements that mediate Pumilio regulation of E2F3, we characterized the PREs in the 3′ UTR of its mRNA. Sequence analysis suggested that the E2F3 3′ UTR contains two putative PRE sequences at the 3′ distal end of the transcript. To determine whether these elements are functional, we cloned the 3′ UTR containing both motifs downstream from Luciferase and used site-directed mutagenesis to mutate the motifs (which we designate as A and B, respectively) from the canonical sequence (UGUAXAUA) to an inactive form (UCCAXAUA) (Fig. 3A). We then tested the capacity of the putative PREs to regulate the translation of Luciferase in TCCSUP bladder carcinoma cells. Previously published constructs containing the E2F1 3′ UTR (Sylvestre et al. 2007), which does not contain a PRE, served as a negative control. Mutations compromising all PRE activity (AB) elevated Luciferase's levels compared with the wild-type E2F3 (F3) sequence (Fig. 3B). Mutation of single PRE sequences (A or B) produced only minor increases in Luciferase levels, suggesting that both PREs are functional. To confirm that these changes in Luciferase levels are dependent on Pumilio activity, we depleted Pum using shRNAs. Knockdown of Pumilio significantly increased Luciferase production from all constructs containing an intact PRE (F3, A, and B), while the double PRE mutant (AB) remained unresponsive (Fig. 3C). Pum knockdown had no effect on Luciferase production from control vectors (Gl3 or E2F1 3′ UTR) (Supplemental Fig. S4A). Conversely, ectopic Pum and Nos expression repressed Luciferase production from F3, A, and B but not AB or negative controls (Fig. 3D; Supplemental Fig. S4B). In these experiments, constructs containing a single PRE sequence showed an intermediate sensitivity to Pum levels, suggesting that both PREs are necessary to mediate full suppression (Fig. 3C,D). Ectopic expression of Pum or Nos alone is insufficient to repress E2F3 translation (Supplemental Fig. S4C,D), indicating that both components of the complex are required in stoichiometric concentrations for repression (NRE schematic in Supplemental Fig. S5A). These results show that there are two functional PREs 161 and 482 base pairs (bp) from the distal end of the E2F3 transcript.
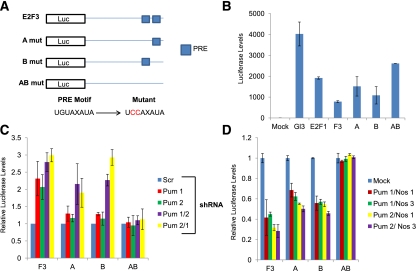
Figure 3.
The E2F3 3′ UTR contains two functional PREs. (A) Schematic of the E2F3 3′ UTR structure: The distal portion of the E2F3 3′ UTR containing both putative PRE sequences was cloned downstream from Luciferase. Site-directed mutagenesis was used to change the encoded PRE sequences from UGUAXAUA to UCCAXAUA within A, B, and AB. (B) Relative Luciferase readings from TCCSUP cells transfected with the empty vector control (Gl3), E2F1 3′ UTR (E2F1), E2F3 3′ UTR containing both PRE sequences (F3), mutation within putative PRE 1 (A), mutation within putative PRE 2 (B), and mutations within both putative PRE sequences (AB). (C) Relative Luciferase readings from TCCSUP cells of transfected F3-, A-, B-, and AB-Luciferase constructs after lentiviral depletion of Scrambled controls (Scr), single Pum proteins (Pum 1 or Pum 2), or both Pum proteins (Pum1/2 or Pum 2/1). Expression of each Luciferase construct infected with Scrambled shRNA was set at 1. (D) Relative Luciferase readings from TCCSUP cells cotransfected with F3-, A-, B-, and AB-Luciferase; Pum; and Nos overexpression constructs. Luciferase expression without Pum cotransfection was set to 1.
PREs enhance miRNA regulation of E2F3
Previous studies have suggested that there may be cross-talk between Pumilio-binding sites and miRNA regulation. Indeed, the 3′ UTR of E2F3 contains a substantial number of putative target sites for miRNA (TargetScan) (Supplemental Fig. S5A), and siRNA-mediated depletion of the miRNA processing enzyme Dicer 1 (Dcr 1) increased the levels of E2F3 protein (Fig. 4A), indicating that at least some of these sites are likely to be functional. To identify miRNAs that impact E2F3 levels, we took advantage of the knowledge that miRNAs are known to be selectively down-regulated in human tumors. E2F3 is an important regulator of proliferation, and gains in E2F3 gene copy number have been identified in a diverse assortment of human cancers. However, E2F3 is a particularly important driver in bladder cancer, and amplifications of the E2F3 locus have been found in a substantial proportion of bladder carcinomas (Veltman et al. 2003; Oeggerli et al. 2004; Olsson et al. 2007). Genomic profiling experiments have identified a set of miRNAs that are down-regulated in bladder carcinomas compared with normal patient tissue (Dyrskjot et al. 2009). Remarkably, E2F3 is a predicted target for three of the five miRNAs that show a greater than twofold decrease in levels in bladder carcinomas compared with normal tissue and is a predicted target for three of the four miRNAs that decrease in expression with increasing tumor grade (TargetScan) (Supplemental Fig. S5B). These observations suggest that the process of bladder carcinogenesis selects for the down-regulation of miRNAs that target E2F3.
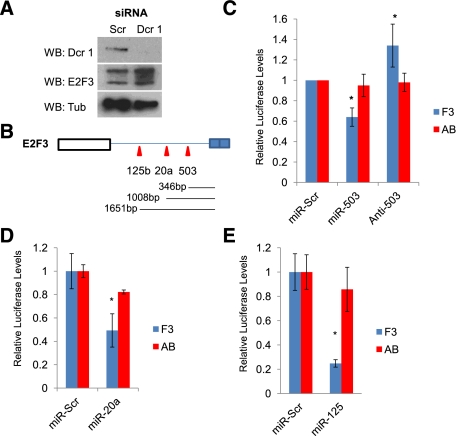
Figure 4.
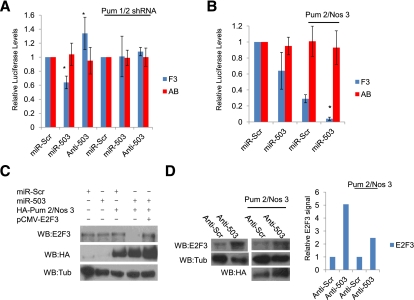
miRNA regulation of E2F3 is enhanced by the PREs. (A) Western blots of Dcr-1, E2F3, and Tub from TCCSUP cells depleted with a pool of Scrambled siRNAs (Scr) or Dicer-1 siRNAs (Dcr 1). (B) Schematic of the E2F3 3′ UTR outlining the relative position of the seed sequences for miR-503, miR-20a, and miR-125b relative to PRE 1. (C) Relative Luciferase assays from TCCSUP cells transfected the E2F3 or AB constructs, testing the effect of adding miR-503 or the anti-mer inhibitor (anti-503). (*) P < 0.05. (D) Relative Luciferase readings from TCCSUP cells comparing the capacity of miR-20a to regulate F3 and AB translation with scrambled controls (miR-Scr). (*) P = 0.01. (E) Relative Luciferase readings from TCCSUP cells comparing the capacity of miR-125b to regulate F3 and AB translation with scrambled controls (miR-Scr). (*) P < 0.01.
To test whether these miRNAs cooperate with Pum to regulate E2F3, we first focused on miR-503, the miRNA identified in the analysis of bladder carcinomas that had the highest-quality predicted seed sequence within the E2F3 3′ UTR (Supplemental Fig. S5A) and the highest number of shared targets with Pum 1 that were down-regulated in bladder cancer (Supplemental Fig S5C). We transfected cells with both scrambled controls (miR-Scr or anti-Scr), miR-503, or the anti-miR of miR-503 (anti-503) and measured the effect on E2F3-Luciferase expression. Addition of miR-503 repressed translation of F3-Luciferase, confirming that E2F3 is a bona fide miR-503 target (Fig. 4C). In complementary experiments, anti-503 enhanced F3 translation (Fig. 4C). To test whether Pum activity contributes to the capacity of miR-503 to suppress E2F3, we assayed the capacity of miR-503 to regulate translation from Luciferase plasmids containing single PREs or the AB-Luciferase plasmid that lacks a functional PRE. Addition of either miR-503 or anti-503 modifies expression from plasmids containing single PREs (A and B) (Supplemental Fig. S5E) but had little effect on AB-Luciferase levels (Fig. 4C).
To determine whether Pum cooperates solely with miR-503, we examined other miRNAs that contain seed sequences proximal to miR-503. miR-20a and miR-125b were selected for further characterization because they have been linked to E2F regulation (miR-20a) or are down-regulated in bladder cancer (miR-125b). miR-20a has highly conserved binding sites within all three human activator E2Fs (Supplemental Fig. S6A) and has been shown to weakly regulate E2F3 (Sylvestre et al. 2007). To test whether Pum actively cooperates with miR-20a to repress E2F3, we analyzed the effect of miR-20a on the translation of F3- or AB-Luciferase constructs. miR-20a strongly repressed F3 levels but had only weak effects on AB translation (Fig. 4D), indicating that the PREs enhance the repressive capacity of miR-20a on E2F3. Reduced expression of miR-125b has also been linked to the initiation and progression of bladder carcinogenesis (Supplemental Fig. S5B,C; Dyrskjot et al. 2009); we therefore tested its ability to regulate E2F3 and whether this requires the PRE sequence. As seen with miR-503, the addition of miR-125b dramatically repressed F3 expression compared with AB (Fig. 4E). Our findings suggest that PREs are important for miRNA activity on the E2F3 3′ UTR at seed sequences proximal to the PREs.
Pumilio facilitates miRNA regulation of E2F3
Having demonstrated that the PREs are important for miRNA regulation of E2F3, we next asked whether Pumilio is the critical factor acting on these sites. To do this, we depleted both Pumilio proteins (Pum 1/2 shRNA) (Supplemental Fig. S6C,D) from cells before transfecting them with Luciferase constructs and miR-503. Knockdown of Pum strongly suppressed the capacity of miR-503 to repress F3-Luciferase translation; however, it did not affect the translation of AB-Luciferase (Fig. 5A). These findings suggested to us that it may be possible to enhance the repressive effect of miR-503 by elevating the levels of Pumilio. To test this, we overexpressed Pum/Nos and miR-503 and assayed the effect on F3 and AB translation. Consistent with this prediction, cotransfection of Pum/Nos and miR-503 strongly enhanced the repression of F3 compared with either alone (Fig. 5B), while AB remained unresponsive.
Figure 5.
Pumilio facilitates miRNA regulation of E2F3. (A) Relative Luciferase readings comparing the capacity of miR-503 and anti-503 to regulate AB translation in the presence and absence of Pumilio (Pum 1/2 shRNA). In both conditions, Luciferase readings using the scrambled controls (miR-Scr) were set to 1. (*)P < 0.05. (B) Relative Luciferase readings of F3 and AB, testing the effect of Pum 2 and Nos 3 overexpression on the capacity of miR-503 to regulate F3 levels. (*) P = 0.01. (C) Western blots of E2F3, HA, and Tub from TCCSUP cells transfected with levels of Pum 2 and Nos 3 insufficient for repression of E2F3, testing the capacity of miR-503 to enhance Pumilio repression compared with Scrambled controls (miR-Scr). pCMV-E2F3 without a 3′ UTR was included as a control. (D, left panels) Western blots of E2F3, Tub, and HA from TCCSUP cells, testing the effect of the anti-503 on the endogenous Pumilio regulation of E2F3. (Right panels) TCCSUP cells were then transfected with levels of Pum 2 and Nos 3, which only weakly represses E2F3 in combination with either Scrambled anti-mer (anti-Scr) or 503 anti-mer (anti-503), investigating the effect of loss of miR-503 expression on the capacity of exogenous Pumilio to regulate E2F3. Graph measuring the relative E2F3 Western blot signal compared with Tub under each transfection condition (anti-Scr control set to 1).
To determine whether cooperation between Pum and miR-503 regulates the levels of endogenous E2F3 in bladder carcinoma cells, we first titrated the levels of transfected Pum/Nos plasmids to find a concentration that was insufficient to reduce the level of E2F3 and then assayed the effect of adding miR-503. miR-503 alone only weakly reduced the levels of E2F3, but this effect was dramatically enhanced by the addition of exogenous Pum/Nos (Fig. 5C). As expected, the suppression of E2F3 levels by Pum/Nos/miR-503 was rescued by cotransfection of an E2F3 expression construct lacking a 3′ UTR sequence. To test whether miR-503 influences the levels of endogenous E2F3 in bladder cancer cells, we transfected TCCSUP cells with anti-503. Addition of anti-503 increased E2F3 protein levels and also impaired the ability of exogenous Pum to repress E2F3 (Fig. 5D). We conclude that miR-503 is a physiological regulator of E2F3 and that it acts in cooperation with Pumilio.
Taken together, these results show that both the PREs and Pumilio are important for multiple miRNAs to target E2F3 and this regulation affects at least two miRNAs (503 and 125b) whose expression is suppressed in bladder cancers. These results suggest that the silencing of specific sets of miRNAs may enable bladder carcinoma cells to escape Pumilio-mediated regulation of E2F3. miRNA silencing is a common feature of cancer cells, and our findings raise the possibility that the changes in miRNA levels may not only reduce miRNA regulation of their targets but also compromise the capacity of Pumilio to repress translation of specific targets.
Pumilio-mediated regulation of E2F3 in cancer cells
Pumilio is an essential gene, and, to date, genome-wide cancer studies have not identified changes in Pumilio expression level or activity in cancer cells. We focused, therefore, on the more specific question of whether Pumilio-dependent regulation of E2F3 is compromised in cancer cells. As bladder cancer cells represent a physiological cellular context in which the control of E2F3 is particularly meaningful, we used these to examine Pumilio regulation of E2F3.
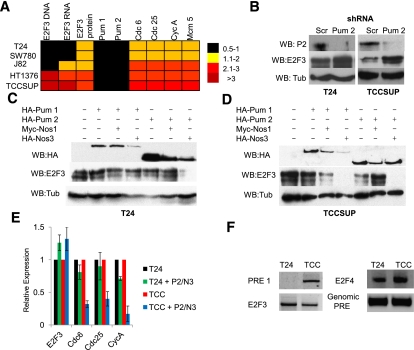
As a first step, we surveyed a panel of bladder cancer cell lines that reflect the variety of E2F3 copy numbers seen in human tumors. Quantification of E2F3 genomic amplification, RNA, and protein levels for these cell lines has been previously published (Feber et al. 2004). Real-time quantitative PCR (RT-qPCR) was used to compare the relative expression levels of E2F1, E2F3, Pum, and various E2F target genes. While the levels of E2F1 and Pum transcripts changed little between cell lines, large differences were evident in the level of E2F3 mRNA and, to a lesser degree, in the expression of E2F target genes (Cdc6, Cdc25, CycA, and Mcm5) (Fig. 6A). Interestingly, E2F3 mRNA correlates poorly with E2F3 protein levels (Fig. 6A). For subsequent experiments, we selected TCCSUP cells to represent one end of the spectrum: These cells have amplified E2F3, a high level of E2F3 mRNA, and a high expression of E2F targets (Feber et al. 2004; Hurst et al. 2008). At the other end of the spectrum, we examined T24 bladder cancer cells. T24 cells lack E2F3 amplification, have much lower levels of E2F3 mRNA, and express the E2F target genes to a lower level than that of TCCSUP cells.
Figure 6.
Bladder carcinomas circumvent Pumilio regulation of E2F3. (A) E2F3 DNA, RNA, and protein levels in bladder cancer cells. RT-qPCR analysis of a panel of bladder carcinoma cell lines (T24, SW780, J82, HT1376, and TCCSUP) showing the expression levels of E2F1, E2F3, Pum 1, Pum 2, and select E2F target genes (cdc6, cdc25, and cycA). Black squares represent 0.5–1.0, yellow squares represent 1.1–2.0, light-red squares represent 2.1–3.0, and dark-red squares represent >3. (B) Western blots of Pumilio 2 (Pum2), E2F3, and Tub from nonamplified T24 cells and in amplified TCCSUP cells depleted of Scrambled controls (Scr) or Pumilio 2 (Pum 2) using specific shRNAs. (C) Western blots of HA, E2F3, and Tub from T24 cells transfected with Pum and Nos overexpression constructs. (D) Western blots of HA, E2F3, and Tub from TCCSUP cells transfected with Pum and Nos overexpression constructs. (E) RT-qPCR results of Pumilio transfections from T24 and TCCSUP cells measuring the effect on E2F target gene expression from e2f3, cdc6, cdc25, and cycA. (F) 3′ RACE results from T24 and TCCSUP cells. Comparing E2F3 transcript levels (E2F3), the PRE 1 region in the E2F3 3′ UTR (PRE 1), the 3′ end of the E2F4 3′ UTR (control), and the genomic PRE 1 sequence for E2F3.
To assess whether Pumilio regulates E2F3 protein levels in bladder carcinoma cells, we manipulated Pum levels. Depletion of Pum 1 or Pum 2 caused a substantial increase in E2F3 levels in TCCSUP cells, but had only minor effects in T24 cells (Fig. 6B; Supplemental Fig. S6). In converse experiments, the ectopic expression of Pum and Nos again had only modest effects in T24 cells (Fig. 6C), but significantly reduced E2F3 protein levels in TCCSUP cells (Fig. 6D). This reduction in E2F3 levels in TCCSUP cells dramatically suppressed the expression of E2F target genes (Fig. 6E). Taken together, both sets of data suggest that the level of E2F3 in TCCSUP cells is sensitive to Pumilio regulation. The amplification of the E2F3 loci in TCCSUP cells appears to produce sufficient mRNA to exceed the regulatory capacity of the endogenous Pumilio protein, since additional Pum/Nos complex significantly reduces the amount of E2F3. To understand why T24 cells are largely insensitive to Pum when compared with both TCCSUP cells and primary human cells, we examined the E2F3 mRNA present in these cells. Analysis of the E2F3 transcript using 3′ rapid amplification of cDNA ends (RACE) and RT-qPCR revealed that the mRNA present in T24 bladder cancer cells has a shortened 3′ UTR that lacks both of the putative PRE sequences (Fig. 6F). In contrast, this region is retained in the E2F3 transcripts expressed in TCCSUP cells. Our findings suggest that 3′ end shortening (seen in T24 cells) and gene amplification (seen in TCCSUP cells) may represent two distinct mechanisms by which E2F3 can escape Pum regulation.
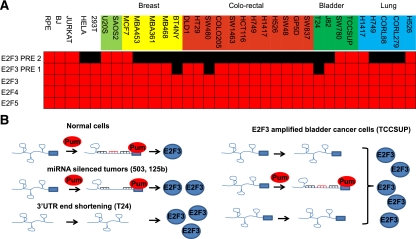
To determine whether 3′ end shortening of the E2F3 transcript was unique to bladder cancers or is a more widespread mechanism to evade Pumilio translational regulation, we expanded the 3′ RACE analysis to include a panel of cancer cell lines. We examined the length of the E2F3 3′ UTR in nontransformed human cells (RPE and BJ) and in cancer cell lines derived from sarcoma, breast, colorectal, bladder, and lung tumors (Fig. 7A). We observed shortening of the E2F3 transcript that removed the distal PRE sequence in the majority of cancer cell lines. A smaller subset of cell lines had shortened the 3′ UTR sufficiently to remove both PREs (BT4NY, SW480, and T24). By way of comparison, no shortening of the 3′ UTRs for E2F4 and E2F5 (two ubiquitously expressed repressor E2Fs) was observed in any of the cells tested. Taken together, these findings suggest that Pumilio regulation of E2F3 is circumvented in many cancer cells by a shortening the 3′ UTR that compromises regulation via the PRE. Given that Pum-binding sites affect the ability of multiple miRNAs to suppress E2F3, this shortening may not only affect regulation by Pum, but also reduce the capacity of miRNA to limit the expression of E2F3.
Discussion
We found that E2F proteins are subject to a regulatory pathway that is conserved between flies and humans, in which the Pumilio repressor complex interacts with defined motifs within the 3′ UTRs of activator E2Fs and, by regulating their translation, limits the synthesis of these key drivers of cell proliferation. The human E2F3 mRNA contains multiple regulatory features within a long 3′ UTR (3.3 kb). These include a large number of miRNA seed sequences, two PRE motifs, two NRE motifs, and several alternative polyadenylation signals. The results described here show that Pumilio plays a critical role in the regulation of E2F3 and has a major impact on the ability of multiple miRNAs to target the 3′ UTR. While previous studies in C. elegans (Nolde et al. 2007) and human primary cells (Kedde et al. 2010) have shown that Puf proteins can influence the effects of specific miRNAs on nearby seed sequences, these results show that in the regulation of E2F3, Pumilio proteins can modulate miRNA regulation within the context of a complex 3′ UTR. Pumilio interaction has been shown to disrupt the secondary folding of RNA stem–loop structures and to open up the surrounding RNA (Kedde et al. 2010). In a similar way, an interaction between the Pumilio complex and the E2F3 transcript may promote conformational changes in the secondary structure of the mRNA, enabling multiple miRNAs to gain access to seed sequences throughout the 3′ UTR and enhancing the repression of E2F3 (Fig. 7B). Additionally, the brat homolog in mice, Trim 32, can increase the activity of miRNAs (including miR-503) by interacting with Argonaute 1 (Schwamborn et al. 2009). This suggests that Pumilio complexes may also influence the recruitment of miRNAs to their targets via Trim 32. We note, however, that Pumilio repressor components were not identified in mass spectrometry analysis of the RISC (Meister et al. 2005). A close relationship between Pumilio regulation and miRNA function has broad implications, as previous in silico studies have noted that Pumilio targets are especially enriched with miRNA seed sequences (Galgano et al. 2008). Potentially, the functional interaction between Pumilio and miRNAs as described here on E2F3 may be a widespread phenomenon.
Figure 7.
3′ End shortening of E2F3 is widespread in cancer cell lines. (A) 3′ RACE analysis of E2F3 (PRE 2, PRE 1, and E2F3), E2F4, and E2F5 from primary cells (RPE and BJ) and representative sarcoma, breast, colorectal, bladder, and lung cancer cell lines. A gene-specific primer and an amplification primer were used in a first-round amplification step. Purified 3′ RACE products were then used in a second round of PCR using a different gene-specific primer. Amplicons were designed to map key regulatory regions within each 3′ UTR, including PREs and miRNA clusters. Red boxes indicate sequence detected in transcripts, and black boxes represent absence sequence. (B) Model of Pum and miRNA regulation of E2F3 translation. In normal cells, the E2F3 3′ UTR is highly structured. Interaction of Pumilio with the PRE sequences facilitates the relaxing of the secondary structure, enabling miRNAs to gain access to their seed sequences and regulate E2F3 protein levels. In tumors that have selectively silenced miRNAs that cooperate with Pumilio (miR-503 or miR-125b), translation control is weakened and more protein is produced. In cells that have shortened their 3′ UTRs (T24), no PRE sequences remain and the E2F3 transcript is rapidly translated, producing elevated levels of E2F3 protein. In cancer cells that amplify the E2F3 locus (TCCSUP), the elevated levels of transcript exceed the capacity of Pumilio regulation, and only a subset of transcripts are regulated. The remainder is rapidly translated.
Activator E2Fs, such as E2F3 and dE2F1, are potent regulators of cell cycle progression, with the capacity to stimulate quiescent cells to proliferate. Cells have evolved multiple tiers of regulation to limit the potentially damaging consequences of inappropriate E2F activation, and these mechanisms are often perturbed in cancer cells. Mammalian E2F activity is the integrated function of a large family of proteins, with different family members playing key roles in specific cellular contexts. Although elevated levels of E2F3 have been found in a variety of human cancers, including breast (Tordai et al. 2008; Bild et al. 2009), prostate (Olsson et al. 2007), and lung (Cooper et al. 2006), the activity of E2F3 is known to be especially important in bladder carcinoma, where amplification of the E2F3 locus (6p22) has been identified as a driver mutation (Feber et al. 2004; Oeggerli et al. 2004; Olsson et al. 2007). Interestingly, genomic studies have shown that tumors that amplify E2F3 often also acquire mutations in RB1 (Hurst et al. 2008), suggesting that there may be continuous selection in these tumors for changes that sequentially elevate E2F3 activity.
There are several types of changes in bladder cancer cells that undermine Pumilio/miRNA regulation of E2F3. Amplification of the E2F3 locus and increased mRNA synthesis may enable E2F3 to overwhelm the effects of Pumilio/miRNA repression. We found that TCCSUP cells, which contain amplified E2F3, remain highly sensitive to elevated levels of Pumilio (Fig. 6D). The PREs within E2F3 are located at the very distal end of the 3′ UTR. 3′ UTR shortening has been linked to accelerated cellular growth rates (Sandberg et al. 2008) and cancer (Mayr and Bartel 2009). We found shortened E2F3 transcripts (lacking PRE sequences) in T24 bladder carcinoma cells that lack E2F3 amplification and in a large number of additional cancer cell lines (Fig. 7A). This suggests that 3′ UTR shortening frequently eliminates Pumilio regulation of E2F3, potentially contributing to elevated E2F3 levels and increased cancer cell proliferation. In contrast, we saw little change in the 3′ UTR of the highly expressed, nononcogenic repressor E2Fs, E2F4 and E2F5.
Previously published clinical studies have also identified a set of miRNAs that consistently show reduced levels of expression in bladder carcinomas (Dyrskjot et al. 2009). Interestingly, E2F3 is a predicted target of many of these miRNAs, and it is striking that several of the miRNAs that are selectively down-regulated in bladder cancers target E2F3 and act in a manner that is strongly dependent on Pumilio (Figs. 4, 5). Together, these observations suggest that the normal regulation of E2F3 is compromised in bladder carcinomas by a combination of changes that include gene amplification, RB1 mutation, 3′ UTR shortening, and miRNA silencing (Fig. 7B). Since individual tumors use multiple avenues to compromise the regulation of E2F3, these alternative mechanisms may provide opportunities for therapeutic intervention in specific subsets of cancer cells. For example, by enhancing Pumilio/miRNA regulation, it may be possible to suppress the activity of E2F3 in bladder carcinomas that have amplified E2F3 and/or mutated RB1. E2F3 protein levels are generally elevated in transformed cells compared with primary cells, even though the changes in mRNA levels are often very modest (Supplemental Fig. S7). Disruption of Pumilio/miRNA regulation is likely to be one contributory factor, but E2F3 may also be subject to additional mechanisms of post-translation deregulation that are altered by oncogenesis. The complete elimination of Pumilio regulation is unlikely to promote tumor cell proliferation. Pum proteins are thought to have hundreds, perhaps more than a thousand, potential targets (Galgano et al. 2008), and Pum genes are essential for cell survival and proliferation (Barker et al. 1992). To date, mutations or changes in Pumilio expression levels have not been identified in cancer genomic studies, but these results raise the possibility that 3′ UTR shortening is one mechanism by which Pumilio targets that have important roles in the control of proliferation and differentiation can circumvent Pumilio/miRNA regulation.
The overall effects of Puf proteins are likely to be narrower than miRNAs. While over half of the human genome contains seed sequences for miRNAs (Lewis et al. 2005; Friedman et al. 2009), the number of identified Pumilio targets is much smaller (Galgano et al. 2008), indicating that only a subset of miRNA targets are likely to be influenced by Pumilio. Pumilio binding may not be advantageous in 3′ UTRs with relaxed secondary structures or high-quality miRNA seed sequences. Alternatively, Pumilio interaction with the 3′ UTR may be necessary to dislodge RNA-binding proteins that inhibit miRNA function (e.g., HuR/Dnd1) (Bhattacharyya et al. 2006; Kedde et al. 2007). Another possibility is that other RNA-binding proteins may cooperate with miRNAs to promote the regulation of other transcripts. Interestingly, seed sequences for miR-20a are found within the 3′ UTRs of all three activator E2Fs (E2F1–3). Although miR-20a regulation of E2F3 is dependent on Pumilio, miR-20a is far more efficient at regulating E2F1 that lacks a PRE (Sylvestre et al. 2007). These findings suggest that cooperation between Pumilio and the miRNA is transcript-specific rather than miRNA-specific.
Bioinformatic studies suggest that the vast majority of Pumilio targets contain miRNA seeds (Galgano et al. 2008), raising the possibility that cooperation with miRNAs may be a general feature of Pumilio-mediated regulation. We speculate that cooperation between Pumilio and miRNA may be especially relevant on key regulators of cell proliferation (such as E2F3 and p27) that need to provide a graded response to multiple types of regulatory inputs. The importance of this control is underscored by the fact that tumor cells that are driven by E2F3 select changes that bypass this regulation to generate the high levels of E2F3 activity that promote tumorigenesis. In human malignancies like these, where a key driver of cell proliferation can be controlled by the cooperative effects of Pumilio and miRNAs, manipulating Pumilio activity may provide a novel opportunity to re-establish/strengthen the regulatory potential of miRNAs and suppress tumor cell proliferation.
Materials and methods
Fly stocks, genetic crosses, and modifier screens
The following stocks were used for these studies: wild-type (w1118) (Bloomington Stock Center), GMR-Gal4 (Bloomington Stock Center), Ptc-Gal4 (Bloomington Stock Center), nos-Gal4 (Bloomington Stock Center), UAS-TAP-PumHD (Gerber et al. 2006), UAS-TAP-Nf1 (kind gift of James Walker), UAS-TAP-PABP (Gerber et al. 2006), and UAS-Pum (Schweers et al. 2002). The UAS-dE2F1 transgenic lines contain a 650-bp (2690–3340) region of the dE2F1 gene within a pWIZ vector (Dimova et al. 2003, Morris et al. 2008). The GMR-Gal4, UAS-dE2F1 RNAi #10 (Morris et al. 2008) and ptc-Gal4, UAS-dE2F1 RNAi #3 (Morris et al. 2008) lines were used to screen against Exelixis deficiencies or PiggyBac mutations for modifiers of the dE2F1-RNAi phenotype. Exelixis PiggyBac insertions that affected the Pumilio complex, which suppressed these phenotypes, were c01494, c05844, and e01497. Mutant alleles or UAS-RNAi constructs used to validate this interaction were as follows: brat (brat18 [Bloomington Stock Center], bratfs1 [Bloomington Stock Center], bratv31333 [Vienna Drosophila RNAi Center {VDRC}]), Pumilio (pum13 [Bloomington Stock Center], pum01688 [Bloomington Stock Center], pumv45815 [VDRC]), and nanos (nosL7 [Bloomington Stock Center], nosv108900 [VDRC], and nosv22693 [VDRC]).
Tap tag RPAs
Wild-type (w1118), nos-Gal4/UAS-TAP-PumHD, nos-Gal4/UAS-TAP-Nf1, and nos-Gal4/UAS-TAP-PABP adult females were grown on standard cornmeal–molasses fly food. The adults were then frozen in liquid nitrogen and stored at −80C. Adult flies (2.5 g) were suspended in 15 mL of buffer 1 (20 mM Tris-HCL at pH 8, 150 mM NaCl, 10 mM EDTA, 0.2% Nonidet P-40, 0.02 mg/mL heparin, 1.5 mM DTT, 1 mM PMSF, 0.5 μg of leupeptin, 20 U/mL DNase I, 10 U/mL RNasin) and ground to a powder with a pestle. The powder was then dounced and centrifuged twice at 10,000g for 10 min. Cleared extracts were then incubated with 500 μL of slurry (50% [v/v]) of IgG-agarose beads for 90 min at 4°C . The beads were then washed once in buffer 1 for 15 min before being washed three more times in buffer 2 (20 mM Tris-HCL at pH 8, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.01% Nonidet P-40, 1 mM DTT, 10 U/mL RNasin). Tap-tagged proteins were removed from the beads by incubating 150 U of AcTEV protease (Invitrogen) for 2 h. RNA was then isolated using TRIzol reagent (Invitrogen), followed by RNeasy (Qiagen) purification as per the manufacturer's instructions (Gerber et al. 2006). Each RNA Tap tag pull-down in Figure 1 was conducted in triplicate, and averages and standard deviations from RT-qPCR experiments were used in this study.
RNAi in Drosophila S2 cells
dsRNA for RNAi experiments was generated using the RiboMax large-scale RNA production system (Promega) following the manufacturer's instruction. Drosophila S2 cells were incubated with 50 μg of dsRNA for 4 d and performed as previously described (Dimova et al. 2003). T7 and belle dsRNA were used as controls in our experiments, as neither effected dE2F1 transcription or translation. All RNAi experiments used in Figure 1 were conducted in triplicate, and averages and standard deviations are displayed in this study.
RT-qPCR
Total RNA or immunoprecipitated RNA was purified using the RNeasy extraction kit (Qiagen). Reverse transcription was performed using TaqMan Reverse Transcription (PE Applied Biosystems) according to the manufacturer's specifications. RT–PCR was performed for 50 cycles using an ABI prism 7900 HD Sequence Detection system. mRNA levels were measured using SYBR Green detection chemistry (Applied Biosystems). Quantification was performed using the comparative ΔCt method as described by the manufacturer. Tubulin, Actin, GAPDH, and Rsp26 were used a control for normalization. Gene-specific primers are available on request. All RT-qPCR experiments used in Figures 1–6 were conducted in biological triplicates and technical duplicates. Graphs representing RT-qPCR data contain averages and standard deviations.
Cell culture, expression constructs, and miRNA
Plasmids used in this study include pAC-GFP (Invitrogen), pAC-GFP-dE2F1-3′ UTR, pAC-GFP-mut-3′ UTR, pDEST-HA-Pum 1, pDEST-HA-Pum 2, pMyc-Nos 1 (kind gift from Frans Van Roy), pDEST-Myc-Nos 3, pCMV-E2F3, pCMV-E2F4-Lucfierase, pGl4 (Promega), pGL3 (Promega), pGL3-E2F1 3′ UTR, pGl3-E2F3 3′ UTR (2527–4940) (F3), pGl3-E2F3 3′ UTR Mut A (2527–4940), pGl3-E2F3 3′ UTR (2527–4940) Mut B, and pGl3-E2F3 3′ UTR (2527–4940) Mut AB. Drosophila S2 cells were transfected for 48 h using FuGene 6 transfection reagent (Roche) as per the manufacturer's instructions. Human cells (IMR90, HeLa, T24, SW780, J82, HT1376, and TCCSUP cells) were transfected for 48 h with FuGene 6 transfection reagent (Roche) and Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. miRNA were transfected using Lipofectamine 2000, miR-503 (Ambion, AM17100), anti-503 (Ambion AM17000), miR-20a (pTMP-20a; kind gift from Pascal Chartand), and miR-125b (Dharmacon, C-300595-03). All transfection experiments in Figures 1–7 were conducted in biological triplicate.
Antibodies
Antibodies used in this study include dE2F1 (polyclonal anti-rabbit; C. Seum), GFP (Santa Cruz Biotechnology, sc-8334), Tubulin (Developmental Studies Hybridoma Bank, E7), Pumilio 1 (Bethyl Laboratories, A300-201A), Pumilio 2 (Bethyl Laboratories, A300-202A), E2F1 (Santa Cruz Biotechnology, sc-193), E2F3 (pg-30), Rb (Santa Cruz Biotechnology, sc-50), p107 (Santa Cruz Biotechnology, sc-318), p130 (Santa Cruz, sc-317), Dicer 1 (Abcam, ab4735), and anti-HA epitope (Covance, 16B12).
RPAs
RNA affinity isolations were performed as described previously (Galgano et al. 2008). HeLa cells were grown to 90% confluency, then washed in 1× PBS and collected by centrifugation at 2000g for 10 min. Cells were then lysed in Polysome lysis buffer (10 mM HEPES-KOH at pH 7, 100 mM KCL, 5 mM MgCl2, 25 mM EDTA, 0.5% Nonidet P-40, 2 mM DTT, 0.2 mg/mL heparin, 50 U/mL RNase OUT [Invitrogen], 50 U/mL Superase IN, 1× complete protease inhibitor tablet [Roche]). This lysate was then spun three times at 14,000g for 10 min, and any cellular debris was removed. Aliquots were then flash-frozen and stored at −80°C.
Fifty microliters of protein G or protein A beads (Amersham) was equilibrated in NT2 buffer (50 mM Tris-HCL at pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.05% Nonidet P-40, 5% BSA [Sigma], 0.02 mg/mL heparin). Twenty microliters of Pum 1 antibody and 50 μg of Pum 2 were then coupled to the beads and incubated for 12 h at 4°C. The beads were then washed three times in NT2 buffer. Twenty milligrams of lysate was then added to the bead-coupled antibody and mixed for 6 h at 4°C. The beads were then used four times in NT2 buffer, and RNP was eluted in SDS-EDTA buffer (50 mM Tris at pH 8, 100 mM NaCl, 10 mM EDTA, 1% SDS). Purified RNA from these pull-downs was analyzed using RT-qPCR. Each RPA was done in duplicate, and the RT-qPCR of each sample was analyzed in triplicate. RT-qPCR data from these experiments are displayed as averages and standard deviations.
3′ RACE and genomic DNA preparations
RNA was extracted as described above and used in 3′ RACE assays in the Invitrogen SuperScript II protocol. One microgram of RNA was used in the first strand cDNA synthesis before 1 μL of RNase H (Invitrogen, 18021-14) was added to remove the remaining RNA. Two microliters of the initial cDNA reaction was then used in a standard PCR reaction using a E2F3 3′ UTR-specific primer (GGCACTCAACTCATATGT) and amplification-specific primer (GGCCACGCGTCGACTAGTAC). DNA from this secondary record of PCR was purified using the PCR purification kit (Qiagen), and 2 μL of this was used in another PCR reaction using the amplification-specific primer and another E2F3 primer (GGAATACTAATAAGTCTT) distal from the original primer. Ten microliters of this PCR reaction was run out on a 2% agarose gel. The RNA used in the 3′ RACE experiments was extracted in duplicate. The 3′ RACE experiments to map the 3′ end of the transcript in Figures 6 and 7 were conducted in biological triplicate and technical duplicate.
Luciferase assays
In E2F4-Luciferase assays, IMR90 cells were transfected in six-well plates with 100 ng of pGl4 and 150 ng of the E2F4-Luciferase constructs. For E2F3-based Luciferase experiments, TCCSUP cells were transfected in six-well plates with 100 ng of pGl4 and 150 ng of pGl-E2F3 (or variations thereof). Unless otherwise stated, Luciferase levels were measured 48 h post-transfection (data are expressed as mean ± SE; n = 3).
Luciferase readings were taken using the Dual-Luciferase Reporter Assay system (Promega) as per the manufacturer's instructions. All Luciferase assays in Figures 3–5 were conducted in biological triplicate and technical duplicate. Luciferase readings in this study are averages and standard deviations of these measurements.
Lentiviral shRNA
The DNA preparation, transfections, and virus preparation methods have been published elsewhere (Pearlberg et al. 2005). LKO.1 shRNA vectors targeting PUM 1 (sh#1, TRCN0000147347, sh#2,, TRCN0000148785; sh#3, TRCN0000148491; sh#4, TRCN0000148263; sh#5, TRCN0000146945), PUM 2 (NM_015317; sh#1, TRCN0000061858; sh#2, TRCN0000061859; sh#3, TRCN0000061860; sh#4, TRCN0000061861; sh#5, TRCN0000061862), NANOS 1 (NM_199461; sh#1, TRCN0000118075; sh#2, TRCN0000118076; sh#3, TRCN0000118072; sh#4, TRCN0000118073; sh#5, TRCN0000118074), and NANOS 2 (NM_001029861; sh#1, TRCN0000118123; sh#2, TRCN0000118122; sh#3, TRCN0000118124; sh#4, TRCN0000118125; sh#5, TRCN0000118126) were obtained from the RNAi Consortium. The effect of each shRNA was tested in triplicate, and representative samples are displayed.
FACS
Flow cytometry was performed as described earlier (Binne et al. 2007; Tschöp et al. 2011). Briefly, cells were labeled for 1 h with BrdU (GE), fixed in 75% ethanol, denatured in 2 M HCl and 0.5% Triton X-100, and neutralized in 0.1 M borate (pH 8.5). Cells were incubated with anti-BrdU antibody (BD; 1:80 in PBS, 0.5% Tween-20, 1% BSA) and secondary anti-mouse FITC-conjugated antibody (1:400; BD). Cells were stained with 5 μg/mL propidium iodide in PBS, 1% BSA, and 250 μg/mL RNase A. Cells labeled with EdU were detected using the Click-iT EdU Flow Cytometry Assay kit (Invitrogen) with modification of the protocol by fixation of cells in 75% ethanol. Cell morphology (cell size and granularity) was determined using flow cytometry on cells fixed in 1% paraformaldehyde/PBS. All samples were analyzed in triplicate using a BD FACSCalibur flow cytometer and CellQuest software.
Acknowledgments
We thank Dr. Spyros Artanavis-Tsakonas for providing fly stocks. We also thank A. Gladden, A. Walker, H. Najafi-Shoushtari, S. Vasudevan, A. Naar, and J. Walker for careful reading of the manuscript, and all members of the Naar and Dyson laboratories for helpful comments and discussions. This work was supported by National Institutes of Health (NIH) grant R01GM053203 to N.J.D. N.J.D. is the Saltonstall Scholar of the Massachusetts General Hospital Cancer Center.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.182568.111.
References
- Ambrus AM, Nicolay BN, Rasheva VI, Suckling RJ, Frolov MV 2007. dE2F2-independent rescue of proliferation in cells lacking an activator dE2F1. Mol Cell Biol 27: 8561–8570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DD, Wang C, Moore J, Dickinson LK, Lehmann R 1992. Pumilio is essential for function but not for distribution of the Drosophila abdominal determinant Nanos. Genes Dev 6: 2312–2326 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125: 1111–1124 [DOI] [PubMed] [Google Scholar]
- Bild AH, Parker JS, Gustafson AM, Acharya CR, Hoadley KA, Anders C, Marcom PK, Carey LA, Potti A, Nevins JR, et al. 2009. An integration of complementary strategies for gene-expression analysis to reveal novel therapeutic opportunities for breast cancer. Breast Cancer Res 11: R55 doi: 10.1186/bcr2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binne UK, Classon MK, Dick FA, Wei W, Rape M, Kaelin WG Jr, Naar AM, Dyson NJ 2007. Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat Cell Biol 9: 225–232 [DOI] [PubMed] [Google Scholar]
- Chagnovich D, Lehmann R 2001. Poly(A)-independent regulation of maternal hunchback translation in the Drosophila embryo. Proc Natl Acad Sci 98: 11359–11364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HZ, Tsai SY, Leone G 2009. Emerging roles of E2Fs in cancer: An exit from cell cycle control. Nat Rev Cancer 9: 785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R 2005. General translational repression by activators of mRNA decapping. Cell 122: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper CS, Nicholson AG, Foster C, Dodson A, Edwards S, Fletcher A, Roe T, Clark J, Joshi A, Norman A, et al. 2006. Nuclear overexpression of the E2F3 transcription factor in human lung cancer. Lung Cancer 54: 155–162 [DOI] [PubMed] [Google Scholar]
- Crittenden SL, Bernstein DS, Bachorik JL, Thompson BE, Gallegos M, Petcherski AG, Moulder G, Barstead R, Wickens M, Kimble J 2002. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417: 660–663 [DOI] [PubMed] [Google Scholar]
- Dimova DK, Stevaux O, Frolov MV, Dyson NJ 2003. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev 17: 2308–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio RJ, O'Farrell PH 1994. Developmental control of a G1-S transcriptional program in Drosophila. Development 120: 1503–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio RJ, O'Farrell PH, Xie JE, Brook A, Dyson N 1995. The transcription factor E2F is required for S phase during Drosophila embryogenesis. Genes Dev 9: 1445–1455 [DOI] [PubMed] [Google Scholar]
- Dyrskjot L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R, Fristrup N, Jensen JL, Andersen CL, Zieger K, et al. 2009. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res 69: 4851–4860 [DOI] [PubMed] [Google Scholar]
- Feber A, Clark J, Goodwin G, Dodson AR, Smith PH, Fletcher A, Edwards S, Flohr P, Falconer A, Roe T, et al. 2004. Amplification and overexpression of E2F3 in human bladder cancer. Oncogene 23: 1627–1630 [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP 2008. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS ONE 3: e3164 doi: 10.1371/journal.pone.0003164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D 2006. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci 103: 4487–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginter-Matuszewska B, Spik A, Rembiszewska A, Koyias C, Kupryjanczyk J, Jaruzelska J 2009. The SNARE-associated component SNAPIN binds PUMILIO2 and NANOS1 proteins in human male germ cells. Mol Hum Reprod 15: 173–179 [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Seay DJ, Hook BA, Wickens M 2007. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J Biol Chem 282: 109–114 [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP 2007. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell 27: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Kay MA 2010. How do miRNAs mediate translational repression? Silence 1: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2000. The hallmarks of cancer. Cell 100: 57–70 [DOI] [PubMed] [Google Scholar]
- Hurst CD, Tomlinson DC, Williams SV, Platt FM, Knowles MA 2008. Inactivation of the Rb pathway and overexpression of both isoforms of E2F3 are obligate events in bladder tumours with 6p22 amplification. Oncogene 27: 2716–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060 [DOI] [PubMed] [Google Scholar]
- Iaquinta PJ, Lees JA 2007. Life and death decisions by the E2F transcription factors. Curr Opin Cell Biol 19: 649–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA, et al. 2007. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131: 1273–1286 [DOI] [PubMed] [Google Scholar]
- Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R 2010. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol 12: 1014–1020 [DOI] [PubMed] [Google Scholar]
- Lammens T, Li J, Leone G, De Veylder L 2009. Atypical E2Fs: New players in the E2F transcription factor family. Trends Cell Biol 19: 111–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Lin H, Spradling AC 1997. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124: 2463–2476 [DOI] [PubMed] [Google Scholar]
- Mayr C, Bartel DP 2009. Widespread shortening of 3′ UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, Tuschl T 2005. Identification of novel Argonaute-associated proteins. Curr Biol 15: 2149–2155 [DOI] [PubMed] [Google Scholar]
- Moore FL, Jaruzelska J, Fox MS, Urano J, Firpo MT, Turek PJ, Dorfman DM, Pera RA 2003. Human Pumilio-2 is expressed in embryonic stem cells and germ cells and interacts with DAZ (deleted in azoospermia) and DAZ-like proteins. Proc Natl Acad Sci 100: 538–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris EJ, Ji JY, Yang F, Di Stefano L, Herr A, Moon NS, Kwon EJ, Haigis KM, Naar AM, Dyson NJ 2008. E2F1 represses β-catenin transcription and is antagonized by both pRB and CDK8. Nature 455: 552–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y, Wharton RP 1995. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell 80: 747–756 [DOI] [PubMed] [Google Scholar]
- Nolde MJ, Saka N, Reinert KL, Slack FJ 2007. The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3′UTR-mediated repression of the let-7 microRNA target gene, hbl-1. Dev Biol 305: 551–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeggerli M, Tomovska S, Schraml P, Calvano-Forte D, Schafroth S, Simon R, Gasser T, Mihatsch MJ, Sauter G 2004. E2F3 amplification and overexpression is associated with invasive tumor growth and rapid tumor cell proliferation in urinary bladder cancer. Oncogene 23: 5616–5623 [DOI] [PubMed] [Google Scholar]
- Olsson AY, Feber A, Edwards S, Te Poele R, Giddings I, Merson S, Cooper CS 2007. Role of E2F3 expression in modulating cellular proliferation rate in human bladder and prostate cancer cells. Oncogene 26: 1028–1037 [DOI] [PubMed] [Google Scholar]
- Parks AL, Cook R, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, et al. 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet 36: 288–292 [DOI] [PubMed] [Google Scholar]
- Pearlberg J, Degot S, Endege W, Park J, Davies J, Gelfand E, Sawyer J, Conery A, Doench J, Li W, et al. 2005. Screens using RNAi and cDNA expression as surrogates for genetics in mammalian tissue culture cells. Cold Spring Harb Symp Quant Biol 70: 449–459 [DOI] [PubMed] [Google Scholar]
- Quenault T, Lithgow T, Traven A 2011. PUF proteins: Repression, activation and mRNA localization. Trends Cell Biol 21: 104–112 [DOI] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwamborn JC, Berezikov E, Knoblich JA 2009. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136: 913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweers BA, Walters KJ, Stern M 2002. The Drosophila melanogaster translational repressor pumilio regulates neuronal excitability. Genetics 161: 1177–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ 1995. Mammalian G1 cyclins and cell cycle progression. Proc Assoc Am Physicians 107: 181–186 [PubMed] [Google Scholar]
- Shibutani ST, de la Cruz AF, Tran V, Turbyfill WJ 3rd, Reis T, Edgar BA, Duronio RJ 2008. Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev Cell 15: 890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP 1999. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev 13: 2704–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP 2001. Drosophila Brain tumor is a translational repressor. Genes Dev 15: 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassov DS, Jurecic R 2002. Cloning and comparative sequence analysis of PUM1 and PUM2 genes, human members of the Pumilio family of RNA-binding proteins. Gene 299: 195–204 [DOI] [PubMed] [Google Scholar]
- Spassov DS, Jurecic R 2003. Mouse Pum1 and Pum2 genes, members of the Pumilio family of RNA-binding proteins, show differential expression in fetal and adult hematopoietic stem cells and progenitors. Blood Cells Mol Dis 30: 55–69 [DOI] [PubMed] [Google Scholar]
- Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P 2007. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem 282: 2135–2143 [DOI] [PubMed] [Google Scholar]
- Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, et al. 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet 36: 283–287 [DOI] [PubMed] [Google Scholar]
- Tordai A, Wang J, Andre F, Liedtke C, Yan K, Sotiriou C, Hortobagyi GN, Symmans WF, Pusztai L 2008. Evaluation of biological pathways involved in chemotherapy response in breast cancer. Breast Cancer Res 10: R37 doi: 10.1186/bcr2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschöp K, Conery AR, Settleman J, Harlow E, Dyson N 2011. A kinase shRNA screen links LATS2 and the pRb tumor suppressor pathway. Genes Dev 25: 814–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman JA, Fridlyand J, Pejavar S, Olshen AB, Korkola JE, DeVries S, Carroll P, Kuo WL, Pinkel D, Albertson D, et al. 2003. Array-based comparative genomic hybridization for genome-wide screening of DNA copy number in bladder tumors. Cancer Res 63: 2872–2880 [PubMed] [Google Scholar]
- Vessey JP, Schoderboeck L, Gingl E, Luzi E, Riefler J, Di Leva F, Karra D, Thomas S, Kiebler MA, Macchi P 2010. Mammalian Pumilio 2 regulates dendrite morphogenesis and synaptic function. Proc Natl Acad Sci 107: 3222–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]