Chromatin domains display a constant “near-random walk” movement within the nucleus. Gasser and colleagues now investigate the molecular cause of such movement. Using high-precision single-particle tracking in live cells, they find that targeting of the INO80 chromosomal remodeler enhances locus movement in a manner dependent on its ATPase activity but independent of active transcription. The study suggests that changes in nucleosome density influences increased chromatin movement.
Keywords: chromatin remodeling, nuclear organization, transcription, VP16, Ino80, fluorescence microscopy, homologous recombination
Abstract
Chromatin in the interphase nucleus moves in a constrained random walk. Despite extensive study, the molecular causes of such movement and its impact on DNA-based reactions are unclear. Using high-precision live fluorescence microscopy in budding yeast, we quantified the movement of tagged chromosomal loci to which transcriptional activators or nucleosome remodeling complexes were targeted. We found that local binding of the transcriptional activator VP16, but not of the Gal4 acidic domain, enhances chromatin mobility. The increase in movement did not correlate strictly with RNA polymerase II (PolII) elongation, but could be phenocopied by targeting the INO80 remodeler to the locus. Enhanced chromatin mobility required Ino80's ATPase activity. Consistently, the INO80-dependent remodeling of nucleosomes upon transcriptional activation of the endogenous PHO5 promoter enhanced chromatin movement locally. Finally, increased mobility at a double-strand break was also shown to depend in part on the INO80 complex. This correlated with increased rates of spontaneous gene conversion. We propose that local chromatin remodeling and nucleosome eviction increase large-scale chromatin movements by enhancing the flexibility of the chromatin fiber.
DNA-based transactions such as transcription, replication, and repair take place in distinct nuclear subcompartments. Transcriptional silencing is frequently associated with the nuclear envelope or occurs near nucleoli (Towbin et al. 2009), whereas the activation of tissue-specific genes correlates with a shift of the relevant genes away from the nuclear periphery (Egecioglu and Brickner 2011). In contrast, genes activated under conditions of nutrient or temperature stress move to nuclear pores when they are induced (Taddei 2007). Finally, some types of damage—namely, irreparable DNA double-strand breaks (DSBs), collapsed replication forks, and eroded telomeres—relocate to nuclear pores to be processed for repair, unlike DSBs that can be repaired by recombination with a homologous template (for review, see Nagai et al. 2010). For these relocalization events to occur, whether at a promoter, a replication fork, or a DSB, chromatin must be mobile.
Rapid time-lapse fluorescence microscopy of GFP-LacI-tagged genomic loci has shown that individual chromosomal domains move constantly in a near-random walk within a restrained volume of the nucleus (Hubner and Spector 2010). The measured radius of constraint for the movement of an average chromosomal locus (∼0.6 μm) was roughly similar in every species investigated, although mobility was also shown to be influenced by local chromatin context (Marshall et al. 1997; Heun et al. 2001; Vazquez et al. 2001; Chubb et al. 2002; Gartenberg et al. 2004). For instance, lacO arrays inserted near budding yeast centromeres or telomeres, which are tethered to the nuclear envelope through protein–protein interactions, move within radii of 0.3–0.4 μm, which is significantly less than the 0.6 μm measured for loci in the middle of chromosomal arms (Marshall et al. 1997; Heun et al. 2001; Gartenberg et al. 2004). The binding of the repressive SIR complex in budding yeast also leads to the anchoring of silent loci to the inner nuclear envelope through Esc1 or Mps3, which also restricts locus movement (Gartenberg et al. 2004; Taddei et al. 2004; Bupp et al. 2007). Whereas it is obvious how the tethering of chromatin to an immobile structural element might limit movement, little is known about the forces that accentuate the movement of an untethered locus to allow its relocalization.
Chromatin movement is not always a “random walk” type of motion. In the case of strongly induced transcriptional activation in a repetitive chromosomal array in cultured mammalian cells, directional movement could be observed, and nonrandom movement was scored during Drosophila spermatocyte differentiation (Vazquez et al. 2001; Chuang et al. 2006). Similarly, the targeting of the viral transactivator VP16 to a telomere moved it away from the nuclear envelope (Taddei et al. 2006). The observation that chromatin movement in yeast is sensitive both to glucose levels in the medium and intracellular levels of ATP also argued for active or non-Brownian modes of movement (Heun et al. 2001). Consistently, movement is suppressed by the addition of inhibitors such as sodium azide or carbonyl cyanide chlorophenyl hydrazine, which lower intracellular ATP concentrations by collapsing membrane potentials (Marshall et al. 1997; Heun et al. 2001; Gartenberg et al. 2004; Hubner and Spector 2010). While this suggests that chromatin movement requires ATP-dependent processes, to date the enzymes that contribute to chromatin mobility remain unknown.
The basic unit of chromatin, the nucleosome, is formed from 147 base pairs (bp) of DNA tightly wrapped around eight core histones. When transcription and repair enzymes act on their DNA substrates, nucleosomes must be shifted and, in some cases, removed or replaced (Flaus and Owen-Hughes 2004; Clapier and Cairns 2009). This is achieved primarily by ATP-dependent nucleosome remodelers, the founding member of which was the Snf2/Swi2 complex of yeast (Winston and Carlson 1992). Although the recruitment of transactivators triggers the unfolding of heterochromatin created by repetitive arrays (Tumbar and Belmont 2001; Carpenter et al. 2005), it has not been documented whether local changes in chromatin structure induced by nucleosome remodeling can alter the freedom of movement of the chromatin fiber.
Nucleosome remodelers influence transcription and DNA repair by modulating nucleosome position and altering accessibility for DNA-binding factors (Flaus and Owen-Hughes 2004; Clapier and Cairns 2009). Indeed, the recruitment of remodelers profoundly affects both transcription and the repair of DSBs (for reviews, see van Attikum and Gasser 2005; Hargreaves and Crabtree 2011). The SWI/SNF and INO80 complexes, like all known nucleosome remodeling complexes, contain a large, catalytic subunit with ATPase activity (Snf2 and Ino80, respectively). In complex with eight to 15 other subunits, these macromolecular machines translocate along DNA and redistribute nucleosomes (Clapier and Cairns 2009). Intriguingly, often more than one remodeler, as well as histone tail modifiers, are recruited to a promoter or DSB (Neely et al. 1999; Barbaric et al. 2007; van Attikum et al. 2007). For example, the histone acetyltransferase Gcn5 is recruited to promoters as part of the SAGA complex to ensure full transcriptional activation (Grant et al. 1997; Teng et al. 2002; Huisinga and Pugh 2004), and the HAT-containing NuA4 complex is one of the first enzymes recruited to DSBs (Downs et al. 2004). Histone tail modification and altered nucleosome positioning may cooperate to alter local chromatin compaction (Segal and Widom 2009), yet it has not been explored whether they modulate long-range chromatin mobility.
Remodeling complexes can read specific histone marks, which allows them to carry out their remodeling tasks. Of particular interest in this respect is the INO80 remodeler. At DSBs, the INO80 complex is specifically recruited by C-terminally phosphorylated H2A (γH2A), where it evicts nucleosomes to facilitate end-resection (for review, see van Attikum and Gasser 2005). This may be achieved by the replacement or removal of nucleosomes containing the histone H2A variant H2A.Z (Htz1 in yeast), which is deposited by the related SWR1 nucleosome remodeling complex (Mizuguchi et al. 2004; Papamichos-Chronakis et al. 2011).
Combining our interest in nucleosome remodelers with the subnuclear dynamics of chromatin, we explored the possibility that remodelers contribute to chromatin mobility. We quantified chromatin movement using rapid time-lapse microscopy of living yeast cells and tracked the movement of GFP-tagged loci in the presence or absence of a targeted chromatin modifier. Whereas transcriptional elongation and the targeting of either the Gal4 activation domain (GAD), acetyltransferase Gcn5, or remodeler Snf2 had little effect on chromatin movement, the binding of the transcriptional activator VP16 or the INO80 complex did. The VP16 effect itself depends on an intact INO80 complex for its action. Finally, we found that enhanced chromatin mobility correlates with increased rates of ectopic recombination, suggesting a role for chromatin movement in genome stability.
Results
Measuring mobility of chromatin in live cells
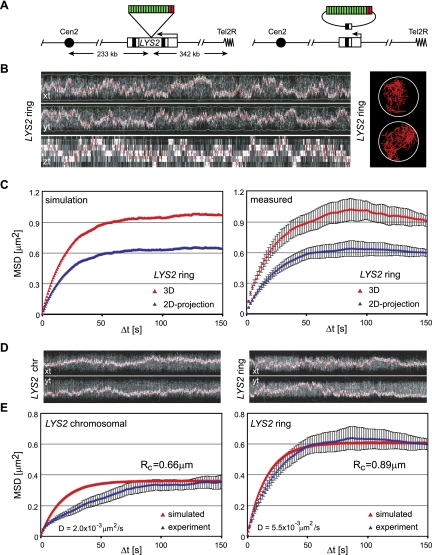
To measure the mobility of chromosomal loci, we used the lacO/LacI system in which an array of lac operators (lacO) inserted at the locus of interest binds GFP-LacI and creates a fluorescent spot that is detectable above a weak background of free GFP-LacI. This tagging method is coupled with three-dimensional (3D) time-lapse microscopy to measure and compare the dynamic movement of chromosomal loci (Marshall et al. 1997). The strains also express a GFP-Nup49 fusion, which allows a frame-by-frame alignment of nuclear centers to eliminate extraneous translational movement (Heun et al. 2001). For each locus monitored, we quantify the movement of the tagged chromosomal site and extract basic parameters that include the number of large steps (i.e., movement in one direction exceeding 500 nm within a 10.5-sec window) (Heun et al. 2001), the diffusion coefficient (D), and the radius of constraint (RC). The latter two parameters are derived from a mean square displacement (MSD) analysis, which consists of measuring the square of the distance a particle travels between two images separated by an increasing time interval. For instance, the MSD value at the 1.5-sec interval is the mean of all distances (squared) that occurred between every pair of frames separated by 1.5 sec. The mean value of the squared displacements for every time interval between 1.5 and 150 sec is then plotted. The MSD plot of a particle moving with a random walk in a constrained spherical volume has the characteristic curve of an exponential approach to the plateau (Meister et al. 2010). The initial slope of the curve is proportional to D (Berg 1993), and the plateau reached is proportional to the square of RC.
There are three major challenges in the analysis of chromatin movement by time-lapse fluorescence microscopy. The first is light-induced damage, the second is the limited resolution in z (along the optical axis), and the third stems from the very rapid movement of chromatin. Whereas deconvolution will overcome part of the resolution problem in z, this usually requires oversampling for increased accuracy, which leads to higher phototoxicity. To circumvent these problems, we took very rapid and limited sampling in z (seven to eight focal slices at ∼1-μsec dwell time per pixel) and tracked locus movement in a two-dimensional (2D) projection of the planes. To demonstrate the accuracy of this method, we first present a comparative study in which we simulated the movement of a particle in 3D and analyzed the resulting tracks both as 3D and as projected 2D movies.
MSD analysis of the simulated particle moving randomly inside a spherical confinement revealed that the projection of the trajectory leads to a reduction in both the initial slope and the plateau of the MSD curve by a factor of 2/3 (see the Supplemental Material). The analysis of simulated random walks confirmed that this relationship holds for the full MSD curve (Fig. 1C). This theoretical analysis suggests that for isotropic movement inside a spherical confinement, the MSD of the 3D trajectory can be fully recovered from the analysis of projected trajectories. To confirm that this method is also applicable to tracks acquired by live fluorescence microscopy, we analyzed the actual movement of an excised lacO-tagged chromatin ring in both 3D and 2D projections. We used the LYS2 locus located 342 kb from the telomere on the right arm of Chr II, which was tagged by the insertion of lacO repeats and flanking sites of recognition for R recombinase (Fig. 1A,B; Gartenberg et al. 2004). The excised 16.5-kb episome moves freely through the nucleoplasm and could be tracked in 3D stacks of images over 7.5 min at 1.5-sec intervals. As expected, the MSD curves reached a higher plateau and had a steeper slope when analyzed in 3D than the MSD curves derived from the 2D projections (Fig. 1C). In agreement with the simulations, the values were systematically 1.5-fold higher than those obtained from projected movies. This similarity between measured and simulated track analysis convinced us that no essential information about the diffusion coefficient (D) or radius of constraint (RC) was lost by analyzing 2D projected movies, and, in fact, error was reduced.
Figure 1.
Validation of chromatin movement analysis. (A) Map of the LYS2 locus on chromosome II. Site-specific recombination leads to excision of parts of the LYS2 gene and the lacO repeats (green boxes), resulting in an extrachromosomal ring of 16.5 kb. (B) Kymograph of a representative cell tracked over 7.5 min in 3D (left) and representative 2D projected traces (right). (C) The left panel shows simulated MSD plots generated using a constrained random walk model, which uses the diffusion coefficient measured experimentally (step size = 1 nm; RC = 900 nm) in both 3D (red) and the same simulations projected in 2D (blue). The right panel shows MSD curves from the same six 3D movies (7.5-min each) determined in 3D (red) or on a 2D projection (blue). The strain used was GA2627 after expression of the recombinase. The error bars represent the standard error of the mean. The agreement between the measured and simulated data for both 3D and 2D projection data, along with the smaller error in the latter, validates use of the 2D projection for analysis. (D) Representative kymographs of tracking data from typical time-lapse capture of the chromosomal LYS2 locus (left) and the excised ring (right). (E) MSD analysis of experimental values (blue) for the chromosomal LYS2 locus (eight movies of 7.5-min each in GA2627) (left) and the excised ring (six movies of 7.5-min each after expression of the recombinase in GA2627) (right) overlaid onto in silico simulated curves (red). The movement of the chromosomal LYS2, unlike the excised ring, does not fit a random walk model, reflecting constraint by the chromosomal fiber. “D” indicates diffusion coefficient, and “RC” indicates radius of constraint.
This method allowed us to rapidly and accurately quantify the movement of tagged loci with limited sampling and reduced phototoxicity and mathematically generate accurate 3D values for both D and RC. To ensure that the imaging conditions do not generate checkpoint-activating DNA damage, we used phase contrast microscopy to score the timely progression of the imaged cell through a subsequent mitosis and G1 phase. We could not detect any arrest or delay in cell cycle progression within 4 h after imaging in our conditions (Materials Methods).
Equipped with a robust method for measuring locus mobility, we first examined whether the movement of a genomic locus also conforms to a constrained random walk model. We simulated random walks using the diffusion coefficient and radius of constraint obtained from measurements of the chromosomal LYS2 locus and then compared these with the movements monitored by microscopy. The chromosomal LYS2 locus moves in a nuclear volume 2.5-fold smaller than that of the chromatin ring (RC = 0.66 μm and 0.89 μm, respectively) (Fig. 1D,E). The excised chromatin diffuses in all directions 2.5-fold faster than its chromosomal counterpart (Dring = 5.5 × 10−3 μm2/sec vs. Dchromo = 2.0 × 10−3 μm2/sec) (Fig. 1D,E). Whereas the experimental MSD plot of the excised chromatin ring agrees perfectly with a simulated random walk, that of the chromosomal locus does not (Fig. 1E). At short time intervals, the slope of the curve is lower, meaning that diffusion is less than expected for a random walk. We conclude that the contiguity of the chromosomal fiber limits the volume of the nucleus explored and constrains its rate of diffusion within that space. A similar conclusion was drawn for a centromere-proximal locus tracked by Marshall et al. (1997) with a different sampling resolution, although in that case, restricted movement may have been imposed by the tethering of the centromere by microtubules (Heun et al. 2001).
To rule out that our imaging technologies themselves introduce systematic errors, we compared chromatin movement at a tagged locus captured with two significantly different imaging instruments: either a scanning laser confocal microscope with minimal pinhole or a spinning disc confocal microscope, each equipped with a hyperfine piezo. We found no significant difference for the movement determined for ATG2 on the two different instruments (Supplemental Fig. S1). This highlights the reliability of both the tracking and analysis methods.
Local recruitment of VP16 increases chromatin mobility
If one considers DNA movement in terms of polymer chain dynamics, restrictions on chromatin mobility can be explained as a function of a fiber's persistence length and mass density (Kratky and Porod 1949), which are a function of both the compaction of the nucleosomal fiber and its inherent flexibility (Bystricky et al. 2004). Protein–protein interactions may also impact chromatin mobility. Moreover, in the nucleus of actively growing cells, where transcription, replication, and repair occur, it is likely that chromatin movement is influenced by the ATP-dependent machinery involved in these processes. For example, it has been amply demonstrated that the targeting of the viral transactivator VP16 can provoke the relocalization of tagged chromosomal loci to the interior of the nucleus in both mammalian cells and yeast (Chuang et al. 2006; Taddei et al. 2006), as it triggers the unfolding of the targeted transgene array (Tumbar and Belmont 2001; Carpenter et al. 2005). However, given that the mammalian studies are performed on arrays that cover many megabases of DNA, it is difficult to extrapolate this chromatin unfolding effect to single-gene mobility.
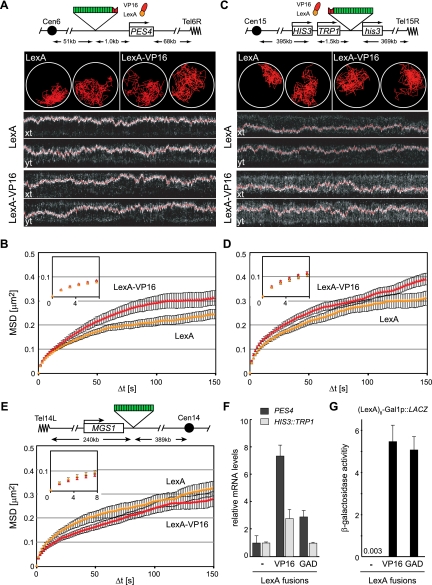
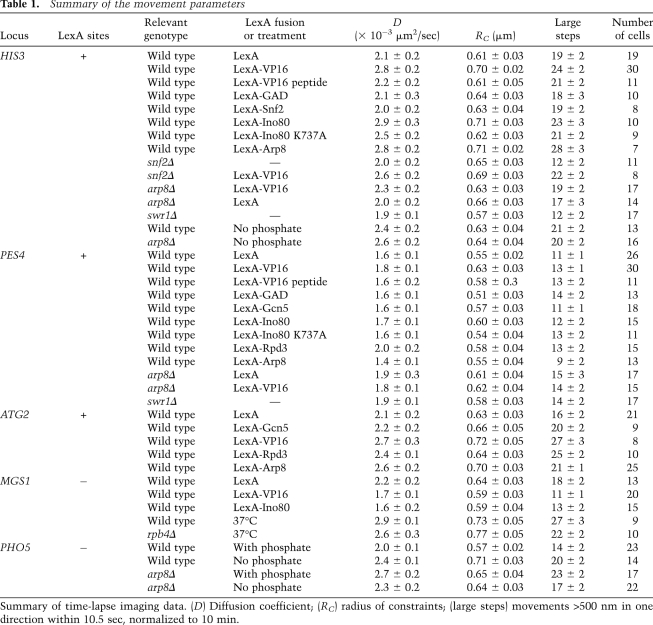
To see whether such gross movements also entail local changes in mobility, we examined the effects of VP16 binding on the mobility of a single locus in yeast by targeting a LexA-VP16 fusion to a genomic locus tagged with a lacO array and four LexA-binding sites. Three different loci were used, distal from a centromere or telomere, to rule out effects of chromosomal anchorage sites (HIS3, PES4, or ATG2, respectively) (Fig. 2; Supplemental Fig. S2). When locus mobility was monitored in the presence of LexA alone, the three loci showed somewhat different degrees of constrained diffusion, most likely reflecting differences in their chromosomal contexts. The least mobile is PES4 (D = 1.6 ± 0.1 × 10−3 μm2/sec) (Fig. 2A,B). It is centrally located on the short right arm of Chr VI, only ∼60 kb away from CEN6 and TEL6R, while HIS3 and ATG2 are more mobile (D = 2.3 ± 0.2 × 10−3 μm2/sec and 2.1 ± 0.2 μm2/sec, respectively) (Fig. 2C,D; Supplemental Fig. S2A,B) and are located in the middle of two of the longest chromosomal arms in yeast (XIV-L and XV-R).
Figure 2.
LexA-VP16 targeting increases chromatin movement of two independent loci. (A) Using the methodology of Figure 1C, we tracked the movement of the PES4 locus in GA1461 and GA4500, captured using a spinning disc confocal microscope. The locus contains four LexA-binding sites (red) and a lacO array (green). Two representative traces and kymographs (2D projections) over a 5-min time-lapse movie showing the marked locus along the X-axis and Y-axis are provided. (B) MSD analysis of movement of the PES4 locus after targeting LexA alone (yellow) or the LexA-VP16 activation domain (red). The inset shows the first five time points (7.5 sec) from which the diffusion coefficient is calculated. Error bars for MSD plots correspond to the standard error. Quantitative details from the analysis are found in Table 1. (C) As in A, but monitoring the LexA- and lacO-tagged locus HIS3 (in GA3441). Two representative traces and kymograph representations of cells showing the tracked locus along the X-axis and Y-axis. (D) MSD analysis of HIS3 in GA3441 after expression of LexA alone (yellow) or the LexA-VP16 activation domain (red). (E) Map and MSD analysis of the MGS1 locus in GA1590, which does not contain LexA-binding sites, after expressing LexA alone (yellow) or LexA-VP16 (red). (F) Relative transcript levels of PES4 (black) and TRP1 (inserted at the HIS3 locus [light gray]) quantified by real-time RT–PCR from total mRNA and normalized to ACT1 after targeting LexA, LexA-VP16, or LexA-GAD. The error bars represent the standard error of three independent RNA preparations. (G) β-Galactosidase reporter assay on strains carrying pSH18-34, which contains the LacZ gene preceded by eight LexA-binding sites and the core promoter of GAL1, during coexpression of LexA, LexA-VP16, or LexA-GAD.
Importantly, the binding of the LexA fusion of the acidic domain of VP16 (amino acids 413–490) at the tagged HIS3 locus significantly increased the diffusion coefficient of this site (from 2.1 ± 0.2 × 10−3 μm2/sec to 2.8 ± 0.3 × 10−3 μm2/sec; P = 0.001) and the radius of constraint (from 610 nm to 700 nm) (Fig. 2D; Table 1) over the values for LexA alone. Similar increases could be scored at PES4 and ATG2; however, at the former locus, the increase in D was not significant (P = 0.07) (Fig. 2B; Table 1; Supplemental Fig. S2B). As a further parameter of movement, we compared large steps, which are also seen to increase upon the tethering of LexA-VP16 to both HIS3 and ATG2 but not at PES4 (P = 0.01, 0.003, and 0.11, respectively). All changes in mobility reflect the action of VP16 in cis, since the expression of LexA-VP16 in a strain lacking the LexA sites had only minor effects, and these went in the opposite direction (Fig. 2E; Table 1). We conclude that local recruitment of VP16 increases chromatin diffusion and enlarges the radius of constraint.
Table 1.
Summary of the movement parameters
Transcriptional elongation is not sufficient for enhanced movement
Since this activating domain of VP16 can strongly induce transcription (Sadowski et al. 1988), we asked whether transcription rates were higher from the VP16-bound loci. Indeed, at the two loci monitored, HIS3 and PES4, LexA-VP16 targeting increased the level of mRNA from the nearest promoter by 2.8-fold and 7.5-fold, as compared with targeting LexA alone (P < 0.03 for both loci) (Fig. 2F). To see whether transcriptional activators generally increase movement, we tested whether the targeting of the yeast GAD fused to LexA would similarly increase locus mobility. Interestingly, we found that the movement of the tagged HIS3 and PES4 loci was unchanged (Table 1; Supplemental Fig. S3), even though the targeting LexA-GAD 1 kb upstream of the PES4 promoter increased mRNA levels by nearly threefold (P = 0.008) over that scored by LexA expression alone. As previously documented, the Gal4 domain did not increase transcription when targeted 3′ of the TRP1 gene at the HIS3 locus (Fig. 2F), although we could confirm that LexA-GAD and LexA-VP16 are equally potent activators of transcription when targeted upstream of a β-galactosidase reporter (Fig. 2G). Given that LexA-GAD and LexA-VP16 both activate transcription, but only one significantly increases chromatin movement, it is unlikely that transcription itself is sufficient to drive increased locus mobility.
To test directly whether transcriptional elongation influences chromatin movement, we inhibited RNA polymerase II (PolII) elongation by genetic means and tracked the mobility of the MGS1 locus. This active gene, located on the left arm of Chr XIV, was tagged with a lacO array but no LexA-binding sites in both wild-type and rpb4Δ backgrounds (Supplemental Fig. S4A). Loss of Rpb4 renders RNA PolII transcription elongation sensitive to elevated temperature, such that polymerase elongation fails at 37°C (Woychik and Young 1989). Within 45 min of the temperature switch, 96% of transcripts have dropped by fourfold or more (Miyao et al. 2001). In our hands, the radius of constraint of the MGS1 locus was indistinguishable in rpb4Δ and RPB4+ cells at restrictive temperature (Table 1; Supplemental Fig. S4B). Thus, whereas VP16-induced gene transcription coincides with transcriptional activation, RNA PolII elongation per se is not sufficient to ensure enhanced chromatin mobility.
Movement is largely independent of Gcn5, Rpd3, and Snf2
To explore what other changes provoked by VP16 might alter chromatin movement, we monitored the effects of other proteins that VP16 recruits to activate a promoter (Berger et al. 1992). One of these is Gcn5, the acetyltransferase of the SAGA complex (Grant et al. 1997), which appears to assist transcriptional activation by opening nucleosomal structure. We expressed a well-characterized LexA-Gcn5 fusion protein that has demonstrated transactivation potential (Marcus et al. 1994; Candau et al. 1997; Kadosh and Struhl 1997) in strains carrying a lacO array and four LexA-binding sites at the ATG2 or PES4 loci (Supplemental Fig. S5A,B). LexA-Gcn5 did not significantly increase the diffusion coefficient (P > 0.43 at both loci), the number of large steps (P > 0.37 at both loci), or the radius of constraint over the values obtained with LexA alone (Table 1; Supplemental Fig. S5C,D). Similar results were obtained upon expression of LexA-Rpd3, the deacetylase that prevents recruitment of SAGA to promoters (Deckert and Struhl 2002). LexA-Rpd3 caused no change in the diffusion coefficients (P > 0.12 for both loci) or in the radii of constraint for either tagged locus (Table 1; Supplemental Fig. S5E,F). It did increase slightly the number of large steps at HIS3 (P = 0.01) but not at ATG2 (P = 0.23). Given that two robust histone-modifying enzymes failed to alter chromatin mobility at two targeted loci, it seems unlikely that acetylation by Gcn5 accounts for the effects of LexA-VP16 on chromatin movement.
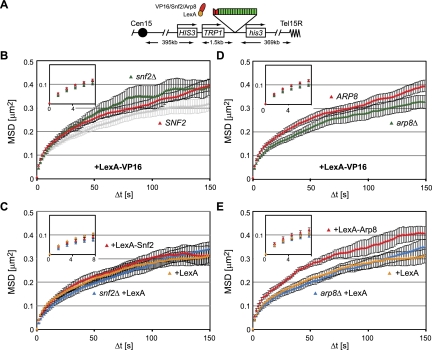
In addition to SAGA, VP16 recruits two nucleosome remodelers to activate transcription: the SWI/SNF complex and the INO80 complex (Neely et al. 1999; Shen et al. 2000). The SWI/SNF complex remodels nucleosomes in vitro mainly through a sliding mechanism (Kassabov et al. 2003), but in vivo it also evicts nucleosomes, possibly depending on genomic context (Boeger et al. 2004; Schwabish and Struhl. 2007). If we target LexA-VP16 to the tagged HIS3 locus in a strain lacking Snf2 (snf2Δ) and track the mobility of the tagged locus, we do not compromise the ability of LexA-VP16 to increase the diffusion coefficient, the radius of constraint, or the number of large steps (Fig. 3A,B; Table 1). This suggests that VP16 uses a SWI/SNF-independent mechanism to increase chromatin movement. We further tested the effects of targeting the SWI/SNF complex to the tagged HIS3 locus through a LexA-Snf2 fusion. The expression of a functional LexA-Snf2 construct that is able to suppress the growth defects of a snf2Δ strain (Laurent et al. 1991), however, did not increase the radius of constraint for the HIS3 locus movement (Fig. 3C; Table 1). Taken together, these results suggest that the effects of VP16 on locus mobility do not stem from the recruitment of the SWI/SNF remodeling complex.
Figure 3.
Snf2 and Arp8 have differential effects on chromatin movement. (A) Map of the HIS3 locus with lacO-binding sites (green) and LexA-binding sites (red). (B) MSD plots from time-lapse imaging of the HIS3 locus in SNF2 (GA3441 [red]) and snf2Δ (GA3444 [green]) cells expressing LexA-VP16, and SNF2 (GA3441) cells expressing LexA alone (light gray) performed and analyzed as in Figure 2, A and B. (C) SNF2 cells (GA3441) expressing either LexA (yellow) or LexA-Snf2 (red) along with snf2Δ (GA3444 [blue]) expressing LexA alone. (D) MSD plots obtained as in Figure 2, A and B, showing ARP8 (GA3441 [red]) and arp8Δ (GA6447 [green]) expressing LexA-VP16 (plasmid no. 2007). (E) MSD analysis of ARP8 cells (GA3441) expressing either LexA (yellow) or LexA-Arp8 (red) along with arp8Δ cells (GA6447) expressing LexA alone (blue).
Targeting the INO80 complex to chromatin increases its movement
VP16 can also recruit the INO80 complex to remodel nucleosomes in vitro (Shen et al. 2000). Given that the deletion of ino80 is lethal in most yeast backgrounds, we tested whether LexA-VP16 acts through the recruitment of INO80 by monitoring locus mobility in a strain lacking the nonessential gene ARP8. Arp8 binds directly to the Ino80 catalytic subunit and is required for the remodeling activity of the complex (Shen et al. 2000, 2003). Indeed, the targeting of LexA-VP16 to the tagged HIS3 locus in an arp8Δ background showed a small but significant reduction in mobility as compared with LexA-VP16 binding in a wild-type strain (Fig. 3D). The radius of constraint of arp8Δ was 630 nm upon targeting of VP16 versus 700 nm in the ARP8 cells. Similarly, D was decreased in arp8Δ cells expressing LexA-VP16, from 2.3 ± 0.2 × 10−3 μm2/sec to 2.8 ± 2 × 10−3 μm2/sec in ARP8 cells (P = 0.04), and the number of large steps fell significantly (P = 0.03). In addition, the targeting of LexA-VP16 to PES4 failed to increase mobility of the locus in arp8 cells (Supplemental Fig. S6A,B).
To confirm that Arp8 was indeed directly implicated in the VP16-induced increase in mobility, we next created a LexA-Arp8 fusion and targeted this to the same lacO- and LexA-tagged HIS3 locus. The expression of LexA-Arp8 led to a striking increase in the radius of constraint, comparable with that induced by targeting the LexA-VP16 fusion (Fig. 3E). The diffusion coefficient was also significantly increased (P = 0.01), as was the number of large steps (P = 0.004) (Table 1). An increase of equal dimension was detected upon the targeting of LexA-Arp8 to the ATG2 locus on Chr XIV (Supplemental Fig. S6C,D). Like the snf2 deletion, the basal-level movement of HIS3 was not affected by arp8 deletion (Fig. 3C,E).
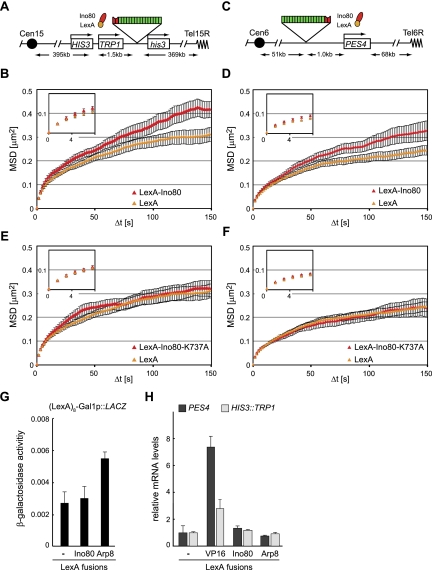
To confirm that the effect of LexA-Arp8 targeting indeed reflects the action of the INO80 complex, we next generated a LexA-Ino80 fusion, which was expressed in strains bearing the tagged HIS3 and PES4 loci (Fig. 4). Upon targeting LexA-Ino80 to the tagged HIS3 locus, D increases from 2.1 ± 0.2 × 10−3 μm2/sec to 2.9 ± 0.3 × 10−3 μm2/sec (P = 0.004), as compared with targeting LexA alone. At the same time, the RC increased from 610 to 710 nm (Fig. 4A,B; Table 1). For unknown reasons, the number of large steps in the presence of LexA-Ino80 was highly variable and was therefore not statistically different from the control (P = 0.07). Like LexA-VP16, LexA-Ino80 was also effective at a second locus: The radius of constraint of LexA-tagged PES4 increased upon LexA-Ino80 expression, although increases in the diffusion coefficient (P = 0.24) and large step number (P = 0.27) (Fig. 4C,D; Table 1) were not significant. The increase in mobility was not an indirect effect of LexA-Ino80 expression, since the mobility of a site lacking the LexA-binding sites (MGS1) did not increase in strains expressing the fusion construct (Table 1; Supplemental Fig. S4C). Rather, the diffusion coefficient of MGS1 decreased slightly (P = 0.02), as did the radius of constraint, possibly due to slightly negative effects in trans (Supplemental Fig. S4C).
Figure 4.
INO80 tethering promotes chromatin movement and requires the ATPase activity of the complex. (A) Map of the HIS3 locus in GA3441. (B) MSD analysis presented as in Figure 2, A and B, showing the dynamics of the HIS3 locus in GA3441 expressing LexA alone (yellow) or LexA-Ino80 (red). (C) Map of the tagged PES4 locus in GA1461. (D) Tracking and resulting MSD plots of the PES4 locus in GA1461 cells expressing LexA alone (yellow) or LexA-Ino80 (red), performed as in Figure 2, A and B. (E,F) Tracking and resulting MSD curves of the HIS3 locus in GA3441 (E) or the PES4 locus in GA1461 (F) upon targeting of LexA alone (yellow) or LexA-Ino80K737A (red), a mutant incapable of binding ATP. (G) β-Galactosidase reporter assay as in Figure 2G. LexA, LexA-Ino80, or LexA-Arp8 are expressed in cells carrying a reporter construct of the LacZ gene preceded by eight LexA-binding sites and the GAL1 core promoter (pSH18-34). (H) Relative transcript levels of TRP1 (inserted at the HIS3 locus) in GA3441 (light gray) and of PES4 in GA1461 (black) upon targeting of LexA, LexA-VP16, LexA-Ino80, or LexA-Arp8. The expression levels were normalized to ACT1.
Finally, we expressed a mutant form of Ino80, Ino80K737A, fused to LexA to test whether the nucleosome remodeling activity of Ino80 was essential for its effect on chromatin movement. The K737A mutation alters the ATP-binding pocket, and although it still permits the complex to assemble, the mutant complex is incapable of remodeling nucleosomes (Shen et al. 2000). Targeting LexA-Ino80K737A to both HIS3 and PES4 gave MSD results indistinguishable from those in cells expressing LexA alone (PES4: P = 0.45 for D and P = 0.1 for large steps; HIS3: P = 0.10 for D and P = 0.07 for large steps) (Fig. 4E,F; Table 1). In conclusion, the binding of INO80 to a locus is able to enhance the radius of constraint of its movement in a manner dependent on the effects of its remodeling activity in cis. Moreover, the effect of LexA-VP16 targeting is dependent on an intact INO80 complex.
We argued above that transcriptional elongation is not sufficient to ensure increased chromatin mobility, yet it remained of interest to test whether the targeting of LexA-Arp8 and LexA-Ino80 had effects similar to LexA-VP16 on the transcription of the nearest promoter. To this end, we measured transcript levels after targeting LexA-Ino80 and LexA-Arp8 to either a β-galactosidase reporter (Fig. 4G) or the tagged loci at which we monitored movement (HIS3 or PES4) (Fig. 4H). Although there is an increase in transcription upon LexA-Arp8 targeting to the β-galactosidase reporter, this was not observed for the endogenous promoters at which we scored increases in chromatin mobility (Fig. 4H). Moreover, the targeting of LexA-Ino80 itself had no effect on either mRNA or β-galactosidase levels. This argues that the contribution of the INO80 remodeling activity to chromatin movement most likely reflects the changes in chromatin structure and not the activity of RNA polymerase. INO80 has been recently shown to evict the histone variant Htz1 (Papamichos-Chronakis et al. 2011), which is deposited by the SWR1 complex (Mizuguchi et al. 2004). We therefore tested whether deletion of swr1 would increase chromatin mobility, mimicking the effect of INO80 targeting. This, however, was not the case at two tagged loci (Supplemental Fig. S7).
The activation of PHO5 by INO80 coincides with increase in locus mobility
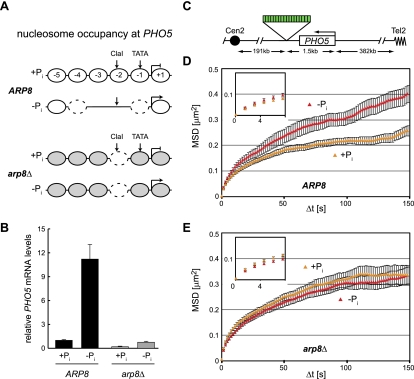
The implication from our targeting experiments is that the remodeling of nucleosomes by INO80 in promoters should lead to both gene induction and an enhanced mobility of the locus. This increase may reflect an increased flexibility of the chromatin fiber, which might be generated by the eviction or shifting of nucleosomes. To test whether a known instance of gene activation by the INO80 complex correlates with increased movement, we chose to study the PHO5 locus. Activation of the PHO5 promoter by low phosphate (Pi) requires Snf2, Gcn5, and INO80, which contribute to the opening of the promoter (Steger et al. 2003; Barbaric et al. 2007). As indicated in Figure 5A, the PHO5 promoter is occupied by five well-positioned nucleosomes, at least one of which is displaced upon gene induction in an INO80-dependent manner (Steger et al. 2003; Barbaric et al. 2007). While we do not rule out a role for SWI/SNF, we note that in an arp8 mutant, the key nucleosome at −2 is not efficiently displaced, and the promoter assumes a slightly altered conformation that is between an inactive and activated state independent of phosphate levels (Steger et al. 2003).
Figure 5.
The mobility of the PHO5 locus depends on Arp8. (A) Representation of the nucleosome occupancy at the PHO5 promoter in the presence and absence of phosphate (Pi) in ARP8 cells as summarized by Ertel et al. (2010). In arp8Δ cells, the ClaI site is partially accessible with and without phosphate, as shown by Steger et al. (2003). The nucleosomes in gray have not been assayed for stability in this mutant. (B) PHO5 mRNA levels in the presence or absence of phosphate in ARP8 (GA7333 [black]) and arp8Δ (GA7347 [gray]) strains. The PHO5 levels were normalized to ACT1, and the repressed conditions in the ARP8 cells were arbitrarily set to 1 for easy comparisons. (C) Map of the PHO5 locus where we inserted a LacO array for monitoring movement in GA7333 and GA7347. (D,E) MSD plots as in Figure 2B showing the movement of the PHO5 locus with (yellow) and without (red) phosphate in ARP8 cells (GA7333) (D) and arp8Δ cells (GA7347) (E).
We tagged the PHO5 locus by inserting a lacO array 3′ of the gene, ∼1.5 kb from the transcription start site (Fig. 5C). We then scored the mobility of the locus under conditions of induction (no Pi medium) and repression (high Pi medium). Under low-phosphate conditions, the PHO5 locus is induced >10-fold in an INO80-dependent manner (cf. arp8Δ in Fig. 5B). We scored the mobility of the locus as performed above for the other tagged loci and found that the radius of constraint of the PHO5 gene is significantly increased in medium lacking Pi, which induces transcription (Fig. 5C). Consistent with previously demonstrated dependency on Arp8 and functional INO80, neither transcription nor mobility are increased on low-Pi medium in the arp8Δ strain (Fig. 5B,E). Intriguingly, the mobility of PHO5 in the strain lacking Arp8 is slightly higher than that of the repressed promoter in ARP8+ cells, consistent with an intermediate level of occupancy at nucleosome −2 in the arp8 mutant (Steger et al. 2003). There is no change in movement at an unrelated promoter that is insensitive to Arp8 and phosphate levels (Supplemental Fig. S8). We conclude that INO80-dependent promoter remodeling correlates well with changes in chromatin mobility at an endogenous locus.
INO80 is needed for the increased mobility at an induced DSB
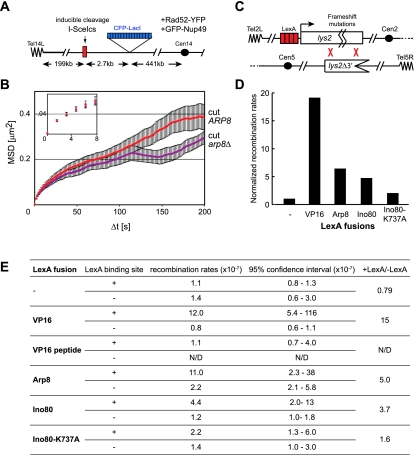
The INO80 complex not only alters nucleosome organization at promoters, but is also recruited to DSBs, where it facilitates the eviction of nucleosomes and end-resection (for review, see van Attikum and Gasser 2005). During the repair of a DSB by homologous recombination (HR), a break must find and pair with its template before recombination can occur and thus must be able to move within the nucleus (Gehlen et al. 2011). We recently showed in yeast that a locus bearing a cleavage site for an inducible endonuclease, I-SceI (Plessis et al. 1992), increases its mobility upon induction of a DSB. Given that a homology search is rate-limiting for the repair of DSBs by HR (Wilson et al. 1994), we examined whether the recruitment of the INO80 complex to this lesion contributes to the observed increase in chromatin movement.
To examine the role of INO80 in the enhanced mobility of a lacO-tagged DSB, we quantified the movement of an I-SceI-induced DSB in both wild-type (ARP8) and arp8Δ backgrounds. The endonuclease I-SceI has no cleavage consensus in the native yeast genome and thus generates only one lesion at the consensus site inserted on Chr XIV–L, which we marked with a lacO array. By inducing I-SceI in a galactose-containing medium for 30 min, we could score the presence of a persistent cut by the binding of Rad52-YFP. Tracking this fluorescent focus at the resected DSB allows us to compare the mobility of a cleaved locus in the presence or absence of a functional INO80 complex. The strain further carried GFP-Nup49 to ensure accurate alignment by subtraction of translational movement.
Upon DSB induction, the radius of constraint of the cleaved I-SceI site increases such that the broken locus can scan nearly 50% of the nuclear volume within minutes (RC cut = 0.70 μm). We compared the mobility of the Rad52-YFP focus in wild-type and arp8Δ cells and indeed scored a drop in the mobility of the DSB in the arp8Δ cells (RC = 0.61 ± 0.03 μm for arp8Δ) (Fig. 6A,B). This suggests that DSB mobility is at least in part dependent on a functional INO80 complex.
LexA-Ino80 and LexA-VP16 promote HR
To see whether the enhanced movement is relevant to the rate of DSB repair by HR, we used a well-established assay for the repair of spontaneous lesions by ectopic gene conversion (GC) events (Freedman and Jinks-Robertson 2002). In this assay, two differently mutated copies of the LYS2 gene are inserted on Chr II and Chr V, one with frameshift mutations that render it nonfunctional, and the second bearing a 3′ truncation. In the promoter of the frameshifted allele, we added four LexA-binding sites to which we targeted modulators of chromatin mobility (Fig. 6C; Nagai et al. 2008). Spontaneous GC between the two sites is rare (10−7), but because it restores a functional copy of the LYS2 gene that allows cells to survive on medium lacking lysine, GC events can be readily quantified. Single crossovers are eliminated, as they would lead to dicentric chromosomes and cell lethality. Recombination rates were calculated using a fluctuation test for the number of Lys+ colonies compared with the total number of cells plated, using at least eight independent transformants for each test (see the Materials and Methods).
Figure 6.
Targeting VP16 and INO80 to a recombination substrate increases the rates of GC. (A) An I-SceI cut site (I-SceIcs) was inserted at the ZWF1 locus on Chr XIV (see the Materials and Methods). The locus is marked by a lacO/CFP-LacI system, and Rad52-YFP is recruited to sites of I-SceI-induced cleavage. The endonuclease I-SceI is under the control of the GAL1 promoter. (B). The movement of the Rad52-YFP focus in arp8Δ mutant cells (GA6318 [violet curve]) is partially diminished in comparison with that of ARP8+ cells (GA6208 [red curve]). Tracking and MSD were performed as in Figure 2B. (C) Map of the relevant sites for GC by HR in GA3232. Sequences were inserted in the lys2 gene on Chr II, generating frameshifts in all reading frames (Freedman and Jinks-Robertson 2002). LexA-binding sites (red) are inserted upstream of the gene (Nagai et al. 2008). The inactive lys2 copy on Chr V carries a 3′ truncation. Recombination by GC restores a functional LYS2, allowing growth on medium lacking lysine. LexA constructs driven by the Tet-off promoter allowed the induction only during the 3-d period of growth before selection of cells on plates lacking lysine. (D) Lys+ colonies were scored for GA3232 transformed with plasmids expressing the indicated LexA fusion (at least eight independent transformants for each construct). Recombination rates for strains carrying the LexA-binding sites were divided by those of strains without the binding sites (GA3208) and normalized to that of cells expressing LexA alone. (E) Details of the recombination rates, including the 95% confidence interval.
We first analyzed the effect of targeting LexA-VP16, LexA-Ino80, or LexA-Arp8 to the frameshifted allele by scoring the frequency of appearance of spontaneous Lys+ colonies. We placed all of the LexA constructs for this assay under control of doxycyline (Tet-off promoter) so that there would be no negative effects due to long-term expression during growth. When induced and targeted, LexA-VP16 increased GC rates by nearly 20-fold and had little effect when the tracked locus lacked LexA-binding sites (Fig. 6D,E). LexA-Ino80 and LexA-Arp8 expression improved GC rates by roughly fivefold (Fig. 6D,E). In contrast, in an isogenic strain lacking LexA operators, expression of LexA-VP16, LexA-Ino80, or LexA-Arp8 had little or no impact on spontaneous GC rates (Fig. 6E). The effect of Ino80 targeting was dependent on the catalytic activity of Ino80 because the targeting LexA-Ino80K737A did not significantly increase GC rates (Fig. 6D,E). We conclude that targeting VP16 or the INO80 complex can spontaneously enhance rates of HR in yeast.
Whereas this may indicate that mobility enhances recombination rates, it was previously reported that an increase in transcription also can increase GC rates (Freedman and Jinks-Robertson 2002). To evaluate whether the effect of LexA-VP16 was due to an increase in mobility or transcription, we compared its effects with that of LexA fused to the minimal 11-amino-acid VP16 transactivating peptide (DALDDFDLDML) (Seipel et al. 1992). Intriguingly, this peptide can activate transcription both in a β-galactosidase reporter assay (Seipel et al. 1992; data not shown) and slightly when targeted to the lacO-tagged HIS3 and PES4 loci, yet it had no detectable effect on chromatin mobility (Supplemental Fig. S9). Consistently, when tested for its effects on recombination at the LYS2 locus, we found no effect on the efficiency of GC (Fig. 6E). This supports our conclusion that increased chromatin mobility correlates positively with increased rates of ectopic HR.
Discussion
The movement of genomic loci is associated with a wide range of DNA transactions, including transcription (Chuang et al. 2006), DNA replication (Kitamura et al. 2006), and DNA repair (for review, see Nagai et al. 2010), yet the forces behind chromatin movement are unclear. Using methodology that provides highly reproducible quantitation of single-locus tracking, we show here that the action of the INO80 nucleosome remodeling complex enhances the subnuclear movement of chromosomal loci. This is manifested as an enlarged radius of constraint within which a near-random walk motion occurs. In some cases, the diffusion coefficient and frequency of large steps also increase. We correlate enhanced movement with the removal of nucleosomes, a well-documented phenomenon that occurs in an INO80-dependent manner at the PHO5 promoter, which shows enhanced movement upon activation. We can also trigger increased movement by targeting the viral transactivator VP16, which recruits both the SWI/SNF and INO80 remodelers to its binding site. The VP16-associated increase in movement was significantly reduced in a strain lacking Arp8, an INO80 subunit that is essential for nucleosome remodeling (Shen et al. 2003), but not in a strain lacking Snf2. Moreover, the targeting of Gcn5 histone acetyltransferase had no impact on long-range chromatin dynamics.
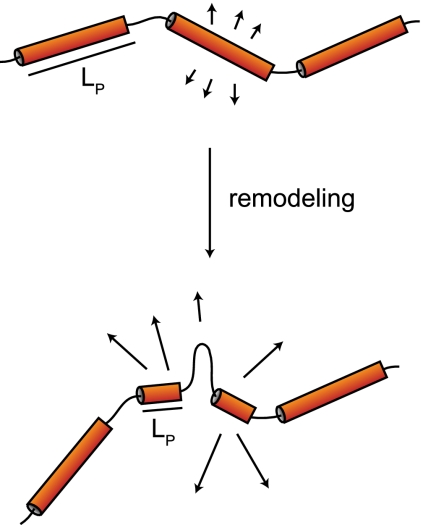
We did not find a consistent correlation between enhanced chromatin movement and transcriptional activation. For instance, targeting of the Gal4 transcription factor increases transcription without increasing long-range mobility, while the targeting of Ino80 or Arp8 enhances mobility without increasing transcription. Finally, we found that a reduction in RNA PolII elongation achieved by inactivation of its Rpb4 subunit does not alter chromatin mobility. Collectively, these results suggest that although enhanced movement can coincide with transcription in some cases, transcription is not the sole source of chromatin dynamics. Moreover, RNA PolII elongation itself does not drive the movement we score. Rather, we propose that it is the alteration of the local nucleosome organization that leads to altered dynamics by changing the folding of chromatin and its persistence length (Fig. 7).
Figure 7.
Model of how chromatin remodeling leads to enhanced movement. Folded chromatin domains, represented by stiff tubes, have rather large persistent length (LP) values compared with the flexible unfolded regions of chromatin. Upon local nucleosome remodeling and/or eviction, the persistent length is reduced, creating an extra flexible linker that may exhibit freer movement.
It is unclear how increased movement would facilitate events associated with transcription, but, by definition, increased movement would favor exploration of the nuclear volume, which could contribute to a homology search during recombination-mediated DSB repair (Gehlen et al. 2011). Consistently, we demonstrate a correlation between increased movement and enhanced rates of GC through HR with an ectopic donor site. Although other explanations can account for more frequent recombination events, we show that a fluorescently tagged locus does increase its mobility upon induction of a DSB in an INO80-dependent manner, consistent with the well-documented recruitment of this remodeler to DSBs (for review, see van Attikum and Gasser 2005). We propose that a homology search is at least one nuclear event that is favored by enhanced chromatin mobility, given that the contact between appropriate sequences is the rate-limiting step in HR (Wilson et al. 1994; Gehlen et al. 2011).
The mechanism of chromatin movement
Analysis of the PHO5 promoter allows us to propose a mechanism through which INO80 influences chromatin mobility. At the PHO5 promoter, gene activation by a lack of phosphate depends on INO80 and coincides with the alteration of four well-positioned nucleosomes immediately upstream of the start site. Restriction enzyme accessibility is increased in an Arp8 (Ino80)-dependent manner on this promoter, although SWI/SNF is also important for PHO5 induction (Steger et al. 2003; Barbaric et al. 2007). We speculate that the removal of nucleosomes by INO80 alters the higher-order packing of the locus and hence the persistence length of the chromatin structure. The statistical properties of polymer chains (Kratky and Porod 1949) suggest that the flexibility of a fiber influences its radius of movement. Therefore, a local disruption of chromatin packaging that would shorten the persistence length could contribute to the increase in locus mobility (Fig. 7).
Our targeting results, which show a dependence on the INO80 ATPase activity for enhanced mobility, are fully consistent with this model. We note that both INO80 and SWI/SNF contain a related ATPase subunit, actin, and actin-related proteins, but the INO80 complex contains RuvB-like helicase subunits and several INO80-specific subunits (Ies1 to Ies6) as well. The two remodelers have very different binding distributions through the genome, with INO80 mapping to replication origins, tRNA genes, and a subset of RNA PolII promoters (Shimada et al. 2008). Snf2 is present at high levels in ribosomal protein genes (Shivaswamy and Iyer 2008). Recent work has argued that INO80 preferentially exchanges Htz1-containing nucleosomes for canonical H2A/H2B dimers (Papamichos-Chronakis et al. 2011), while SWI/SNF does not. INO80 was also shown to facilitate nucleosome removal at DSBs, in contrast to the closely related remodeler SWR1 (for review, see van Attikum and Gasser 2005). Since specific Htz1 occupancy does not alter mobility, we conclude that general INO80-dependent nucleosome displacement is likely to be the mechanism that alters chromatin movement.
Alternative mechanisms influencing chromatin movement
Other scenarios may account for alterations in chromatin movement, such as the removal or establishment of reversible protein–protein interactions, or the alteration of torsional stress through removal of a nucleosome or polymerase movement. The simplest alternative mechanism would be that INO80 promotes movement by removing a proteinaceous anchor. While there is no evidence that INO80 evicts anything other than nucleosomes, we cannot at present rule this out. Indeed, previous work has shown that VP16-triggered changes at promoters can relocate a locus away from the nuclear envelope, apparently by disrupting the anchoring mechanism (Tumbar and Belmont 2001; Chuang et al. 2006; Taddei et al. 2006). None of the loci tracked in our study are associated with the nuclear envelope, although we do note differences in the sensitivity of some loci to LexA-Arp8 targeting (e.g., PES4), suggesting that local context can also affect the ability of INO80 to stimulate mobility.
In mammalian cells, VP16-induced directional movement of large arrays has been shown to depend on nuclear myosin and its interaction with actin (Chuang et al. 2006). Such data provide an attractive model to explain increases in directional movement: Actin might polymerize to provide a scaffold onto which a cargo (i.e., chromatin), possibly in complex with myosin, might move. This would suggest that actin plays a dynamic role in chromatin movement (for review, see Dion et al. 2010), yet nuclear actin does not seem to form filaments in somatic nuclei (Gieni and Hendzel 2009), and modulators of filamentous actin (e.g., latrunculin A) do not affect the mobility observed here (data not shown).
Finally, a further alternative for changing chromatin mobility may involve changes in DNA supercoiling. Nucleosomes wrap DNA in a negative supercoil, relieving torsional stress, and therefore the removal of a nucleosome alters the linking number of torsionally constrained domains (Germond et al. 1975). The “spring”-like movements detected in Drosophila nuclei (Vazquez et al. 2001) were proposed to stem from rapid changes in torsional stress. Given that many remodelers are capable of inducing supercoiling in closed circular DNA molecules (Havas et al. 2000), it is possible that this also alters higher-order chromatin structure. We note, however, that SWI/SNF, which can promote supercoiling in vitro, does not alter chromatin mobility in our hands (Havas et al. 2000).
We cannot exclude that other mechanisms also affect chromatin movement, but our data clearly implicate the activity of the INO80 remodeler in the dynamics documented here. Moreover, the elevated chromatin mobility observed upon PHO5 induction coincides with Ino80-dependent nucleosome displacement. Importantly, the loss of Arp8 impairs activation and leads to mobility that is in between the repressed and active states. INO80 may evict nucleosomes at other promoters as well and may help remove nucleosomes at DSBs (for review, see van Attikum and Gasser 2005). In all of these instances, enhanced chromatin mobility is scored, suggesting that INO80-induced remodeling provides at least one molecular mechanism behind the spatial dynamics of the genome.
Materials and methods
Yeast strains and growth conditions for microscopy experiments
Genotypes of the yeast strains and plasmids are in Supplemental Tables S1 and S2, respectively. For microscopy, yeast was grown in minimal synthetic medium appropriately supplemented. Experiments used exponentially growing cell cultures from freshly transformed cells to avoid potential long-term effects of LexA fusions and used two independent cultures. For the PHO5 experiments, cells were incubated 2–4 h in SC medium without phosphate, supplemented with KCl (Formedium). It was not possible to constitutively express VP16 in arp8Δ strains, so a Tet-inducible VP16 construct was used (no. 2007), and cells were grown in the presence of 5 μg/mL doxycycline, which was removed for 1 h to induce VP16 expression. Both constitutive and induced expression of VP16 gave identical results in wild-type cells. For the experiments done at 37°C, the cells were precultured at this temperature for 45 min before being imaged.
Microscopy
Time-lapse fluorescence imaging was performed as described (Meister et al. 2010), unless otherwise stated, using either LSM point scanning confocal or a spinning disc confocal (see below). Cells were mounted in a Ludin chamber (Life Imaging Services) filled with the appropriate minimal medium. For scanning confocal capture, we used a Zeiss LSM510 Axiovert 200M equipped with a Zeiss Plan-Apochromat 100×/NA = 1.4 oil immersion objective to image the cells. The stage of the microscope was equipped with a hyperfine motor HRZ 200, and temperature was maintained at 25°C during acquisition using a temperature-controlled box that surrounded the microscope, unless otherwise specified. The 488-nm laser was used at a tube current of 4.7 A and 25% output. We used an Lp 505 filter and acquired the images with a pinhole radius of 1–1.2 airy units. The detector gain was 930–999, and the amplifier gain was 1–1.5. We used the amplifier offset at levels of 0.2–0.1 V, and the AOTF was set between 0.1 and 2%. Other conditions were as follows: a dwell time of 1.28 μsec per pixel in an 8-bit format with one scanning direction, four average/mean/line, and a restricted region of interest to smaller than 40 × 38 pixels with a zoom of 1.8 to yield a pixel size of 100 nm in XY.
Spinning disc confocal microscopy used an Olympus IX81 with Yokogawa CSU-X1 scan head and equipped with an ASI MS-2000 Z-piezo. We excited the GFP-LacI and GFP-Nup49 with a 491-nm laser set at 30% of the total power (∼75 μW). A Semrock Di01-T488/568-13x15x0.5 dichroic and a Semrock FF01-525/40-25 filter along with a PlanApo 100×/1.45 TIRFM oil objective were used. A 512 × 512-pixel EM-CCD Cascade II camera (Photometrics) acquired the signal, leading to a pixel size of 94 nm. On the LSM microscope, seven or eight focal steps of 300 μm were taken every 1.5 sec for 5–7.5 min at a speed of >200 msec per slice. On the spinning disc microscope, 4 μm were scanned at 300-μm steps with 30-msec exposure for each optical slice. Cell cycle progression was monitored by rebudding of wild-type cells with phase contrast imaging for 4 h after fluorescence acquisition. The tracking of the I-SceI-induced DSB was performed on the spinning disc confocal microscope in a similar manner.
Movie tracking and analysis
The LSM software (Zeiss) was used to project images to one plane, which was exported as 8-bit TIFF images. We used the Huygens Remote Manager (Ponti et al. 2007; http://huygens-rm.org) to deconvolve the images acquired with the spinning disc confocal microscope and Imaris to project them. The SpotTracker plug-in for ImageJ (Sage et al. 2005) was used to track the moving GFP-LacI signal. This plug-in corrects for translational movement of the nucleus by defining the center of the nucleus in each frame based on background nuclear fluorescence or GFP-Nup49. We used the following settings: cone aperture, 5; normalization factor, 80; center constraint, 20–25; movement constraint, 20; active subpixel resolution. A custom-tailored Excel macro was used to calculate the MSD, large steps (defined as displacements >500 nm in 10.5 sec normalized to 10-min movies), RC, and D. We used the slope (m) of the first five time intervals (1.5–7.5 sec) to determine the 3D diffusion coefficient (D) knowing that D = m/2 × d, where d is the number of dimensions (see Supplemental Material). For practical reasons, the radius of constraint (RC) was derived from the maximum MSD value within the first 150 sec. For 2D time-lapse movies, the plateau of the MSD equals 4/5 RC2 (see the Supplemental Material).
Random walk simulations and calculations
Simulations of random walks in a spherical volume were programmed in C++ using reflective boundary conditions. The radius of the sphere was 1000 times bigger than the step size of the walk. To compare the simulation data with experiments, the step length and the time step of the simulated random walks were normalized as follows: First, the size of confinement was chosen such that the plateau of the MSD curve matched the experimentally measured MSD plateau (see also the Supplemental Material). This also determined the step length of the simulated random walks. Second, the time step of the walks was chosen such that the MSD of the simulated walks during 1.5 sec matched the quadratically averaged step size of the measured trajectories. The simulated MSD curves were each calculated from 750,000 independent simulations.
Quantitative real-time PCR and β-galactosidase reporter assay
We extracted RNA from a 5-mL culture of exponentially growing cells using the RNeasy kit (Qiagen) and generated cDNA from 500 ng of total RNA using the Protoscript AMV First Strand cDNA synthesis kit (New England Biolabs). We used an ABI 7500 Fast real-time thermocycler to quantify the cDNA produced essentially as described (Taddei et al. 2006). All experiments were performed in triplicate and normalized to the ACT1 message. Supplemental Table S3 lists primers and TaqMan probes used to quantify the message level at ACT1, TRP1, and PES4, and the PHO5 primers (Wongwisansri and Laybourn 2005). We used Promega GoTaq qPCR master mix to quantify the levels of the PHO5 transcripts and normalized them to the ACT1 locus amplified with the same primers as for the TaqMan approach described above. The β-galactosidase assay was performed as described (Burke et al. 2000) using crude yeast extracts from strains transformed with pSH18-34 and the appropriate LexA fusion.
Ectopic recombination assay
We determined the frequencies of HR as described (Freedman and Jinks-Robertson 2002; Nagai et al. 2008). The lexA fusion proteins that are targeted to the lys2 locus on Chr II are under control of the Tet-off promoter to ensure there is no detriment to growth by extended overexpression. The Tet-off plasmid was derived from pCM190 (Gari et al. 1997), which we subcloned into pRS415. We transformed GA3232 and GA3208 in the presence of doxycyline (expression off) with the appropriate inducible plasmid encoding LexA fusion proteins (Supplemental Table S2). For each fusion tested, at least eight transformants were inoculated into 5-mL cultures and propagated for 3 d without doxycycline (expression ON). The cells were then plated onto SC-lysine to select recombinants. The recombination rate was determined using the median method and the FALCOR Web tool (Hall et al. 2009).
Statistics and error propagation
The P-values presented for the comparison between diffusion coefficients and large steps were derived from one-tailed Student's t-tests. In the case of mRNA level comparisons, we used a two-tailed t-test. P-values of 0.05 or less were considered statistically significant. To calculate the error on the RC value, we propagated the error on the maximum MSD value using e = 5/(8RC)p, where e is the standard error on RC, and p is the standard error on the MSD value.
Acknowledgments
We thank members of the Gasser laboratory from the University of Geneva and at FMI for helpful discussions and critical reading of the manuscript. We are grateful to David Shore for the GAD vector, Kevin Struhl for the LexA-Rpd3 plasmid, Marian Carlson for the LexA-Snf2 plasmid, Shelley Berger for pLexA-Gcn5, Marc Gartenberg for pRINT, Ulrich Laemmli for pGBD-Ino80K737A and pGBD-Ino80, and Jonas Dorn for help in RC calculations. We thank Thierry Laroche and the Facility for Advanced Imaging and Microscopy (FAIM) at the FMI for excellent technical support. V.D. is supported by a Post-doctoral Research Fellowship from the Terry Fox Foundation (award no. 19759). Research in S.M.G.'s laboratory is supported by the Novartis Research Foundation, the Swiss National Science Foundation, and the SystemsX initiative.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.176156.111.
References
- Barbaric S, Luckenbach T, Schmid A, Blaschke D, Horz W, Korber P 2007. Redundancy of chromatin remodeling pathways for the induction of the yeast PHO5 promoter in vivo. J Biol Chem 282: 27610–27621 [DOI] [PubMed] [Google Scholar]
- Berg HC. 1993. Random walks in biology. Princeton University Press, Princeton, NJ. [Google Scholar]
- Berger SL, Pina B, Silverman N, Marcus GA, Agapite J, Regier JL, Triezenberg SJ, Guarente L 1992. Genetic isolation of ADA2: A potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70: 251–265 [DOI] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD 2004. Romoval of promoter nucleosomes by disassembly rather than sliding in vivo. Mol Cell 14: 667–673 [DOI] [PubMed] [Google Scholar]
- Bupp JM, Martin AE, Stensrud ES, Jaspersen SL 2007. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1–UNC-84 domain protein Mps3. J Cell Biol 179: 845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D, Dawson D, Stearns T 2000. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Bystricky K, Heun P, Gehlen L, Langowski J, Gasser SM 2004. Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc Natl Acad Sci 101: 16495–16500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candau R, Zhou JX, Allis CD, Berger SL 1997. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J 16: 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Memedula S, Plutz MJ, Belmont AS 2005. Common effects of acidic activators on large-scale chromatin structure and transcription. Mol Cell Biol 25: 958–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS 2006. Long-range directional movement of an interphase chromosome site. Curr Biol 16: 825–831 [DOI] [PubMed] [Google Scholar]
- Chubb JR, Boyle S, Perry P, Bickmore WA 2002. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol 12: 439–445 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Deckert J, Struhl K 2002. Targeted recruitment of Rpd3 histone deacetylase represses transcription by inhibiting recruitment of Swi/Snf, SAGA, and TATA binding protein. Mol Cell Biol 22: 6458–6470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion V, Shimada K, Gasser SM 2010. Actin-related proteins in the nucleus: Life beyond chromatin remodelers. Curr Opin Cell Biol 22: 383–391 [DOI] [PubMed] [Google Scholar]
- Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Cote J 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell 16: 979–990 [DOI] [PubMed] [Google Scholar]
- Egecioglu D, Brickner JH 2011. Gene positioning and expression. Curr Opin Cell Biol 23: 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertel F, Dirac-Svejstrup AB, Hertel CB, Blaschke D, Svejstrup JQ, Korber P 2010. In vitro reconstitution of PHO5 promoter chromatin remodeling points to a role for activator-nucleosome competition in vivo. Mol Cell Biol 30: 4060–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Owen-Hughes T 2004. Mechanisms for ATP-dependent chromatin remodelling: Farewell to the tuna-can octamer? Curr Opin Genet Dev 14: 165–173 [DOI] [PubMed] [Google Scholar]
- Freedman JA, Jinks-Robertson S 2002. Genetic requirements for spontaneous and transcription-stimulated mitotic recombination in Saccharomyces cerevisiae. Genetics 162: 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari E, Piedrafita L, Aldea M, Herrero E 1997. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13: 837–848 [DOI] [PubMed] [Google Scholar]
- Gartenberg MR, Neumann FR, Laroche T, Blaszczyk M, Gasser SM 2004. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell 119: 955–967 [DOI] [PubMed] [Google Scholar]
- Gehlen L, Gasser SM, Dion V 2011. How broken DNA finds its template for repair: A computational approach. Prog Theor Phys 191: 20–29 [Google Scholar]
- Germond JE, Hirt B, Oudet P, Gross-Bellark M, Chambon P 1975. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci 72: 1843–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieni RS, Hendzel MJ 2009. Actin dynamics and functions in the interphase nucleus: Moving toward an understanding of nuclear polymeric actin. Biochem Cell Biol 87: 283–306 [DOI] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev 11: 1640–1650 [DOI] [PubMed] [Google Scholar]
- Hall BM, Ma CX, Liang P, Singh KK 2009. Fluctuation analysis CalculatOR: A Web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics 25: 1564–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Crabtree GR 2011. ATP-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res 21: 396–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havas K, Flaus A, Phelan M, Kingston R, Wade PA, Lilley DM, Owen-Hughes T 2000. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell 103: 1133–1142 [DOI] [PubMed] [Google Scholar]
- Heun P, Laroche T, Shimada K, Furrer P, Gasser SM 2001. Chromosome dynamics in the yeast interphase nucleus. Science 294: 2181–2186 [DOI] [PubMed] [Google Scholar]
- Hubner MR, Spector DL 2010. Chromatin dynamics. Annu Rev Biophys 39: 471–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell 13: 573–585 [DOI] [PubMed] [Google Scholar]
- Kadosh D, Struhl K 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89: 365–371 [DOI] [PubMed] [Google Scholar]
- Kassabov SR, Zhang B, Persinger J, Bartholomew B 2003. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol Cell 11: 391–403 [DOI] [PubMed] [Google Scholar]
- Kitamura E, Blow JJ, Tanaka TU 2006. Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell 125: 1297–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratky O, Porod G 1949. Röntgenuntersuchung gelöster Fadenmoleküle. Recueil des Travaux Chimiques des Pays-Bas 68: 1106–1123 [Google Scholar]
- Laurent BC, Treitel MA, Carlson M 1991. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc Natl Acad Sci 88: 2687–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus GA, Silverman N, Berger SL, Horiuchi J, Guarente L 1994. Functional similarity and physical association between GCN5 and ADA2: Putative transcriptional adaptors. EMBO J 13: 4807–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, Sedat JW 1997. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol 7: 930–939 [DOI] [PubMed] [Google Scholar]
- Meister P, Gehlen LR, Varela E, Kalck V, Gasser SM 2010. Visualizing yeast chromosomes and nuclear architecture. Methods Enzymol 470: 535–567 [DOI] [PubMed] [Google Scholar]
- Miyao T, Barnett JD, Woychik NA 2001. Deletion of the RNA polymerase subunit RPB4 acts as a global, not stress-specific, shut-off switch for RNA polymerase II transcription at high temperatures. J Biol Chem 276: 46408–46413 [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348 [DOI] [PubMed] [Google Scholar]
- Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela E, Hediger F, Gasser SM, Krogan NJ 2008. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322: 597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S, Heun P, Gasser SM 2010. Roles for nuclear organization in the maintenance of genome stability. Epigenomics 2: 289–305 [DOI] [PubMed] [Google Scholar]
- Neely KE, Hassan AH, Wallberg AE, Steger DJ, Cairns BR, Wright AP, Workman JL 1999. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol Cell 4: 649–655 [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL 2011. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 144: 200–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessis A, Perrin A, Haber JE, Dujon B 1992. Site-specific recombination determined by I-SceI, a mitochondrial group I intron-encoded endonuclease expressed in the yeast nucleus. Genetics 130: 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponti A, Gulati A, Bäker V, Schwarb P 2007. Huygens Remote Manager: A Web interface for high-volume batch deconvolution. Imaging & Microscopy 9: 57–58 [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M 1988. GAL4-VP16 is an unusually potent transcriptional activator. Nature 335: 563–564 [DOI] [PubMed] [Google Scholar]
- Sage D, Neumann FR, Hediger F, Gasser SM, Unser M 2005. Automatic tracking of individual fluorescence particles: Application to the study of chromosome dynamics. IEEE Trans Image Process 14: 1372–1383 [DOI] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K 2007. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol Cell Biol 27: 6987–6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Widom J 2009. What controls nucleosome positions? Trends Genet 25: 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipel K, Georgiev O, Schaffner W 1992. Different activation domains stimulate transcription from remote (‘enhancer’) and proximal (‘promoter’) positions. EMBO J 11: 4961–4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, Wu C 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406: 541–544 [DOI] [PubMed] [Google Scholar]
- Shen X, Ranallo R, Choi E, Wu C 2003. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol Cell 12: 147–155 [DOI] [PubMed] [Google Scholar]
- Shimada K, Oma Y, Schleker T, Kugou K, Ohta K, Harata M, Gasser SM 2008. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr Biol 18: 566–575 [DOI] [PubMed] [Google Scholar]
- Shivaswamy S, Iyer VR 2008. Stress-dependent dynamics of global chromatin remodeling in yeast: Dual role for SWI/SNF in the heat shock stress response. Mol Cell Biol 28: 2221–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Haswell ES, Miller AL, Wente SR, O'Shea EK 2003. Regulation of chromatin remodeling by inositol polyphosphates. Science 299: 114–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A 2007. Active genes at the nuclear pore complex. Curr Opin Cell Biol 19: 305–310 [DOI] [PubMed] [Google Scholar]
- Taddei A, Hediger F, Neumann FR, Bauer C, Gasser SM 2004. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J 23: 1301–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM 2006. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441: 774–778 [DOI] [PubMed] [Google Scholar]
- Teng Y, Yu Y, Waters R 2002. The Saccharomyces cerevisiae histone acetyltransferase Gcn5 has a role in the photoreactivation and nucleotide excision repair of UV-induced cyclobutane pyrimidine dimers in the MFA2 gene. J Mol Biol 316: 489–499 [DOI] [PubMed] [Google Scholar]
- Towbin BD, Meister P, Gasser SM 2009. The nuclear envelope-a scaffold for silencing? Curr Opin Genet Dev 00: 180–186 [DOI] [PubMed] [Google Scholar]
- Tumbar T, Belmont AS 2001. Interphase movements of a DNA chromosome region modulated by VP16 transcriptional activator. Nat Cell Biol 3: 134–139 [DOI] [PubMed] [Google Scholar]
- van Attikum H, Gasser SM 2005. ATP-dependent chromatin remodeling and DNA double-strand break repair. Cell Cycle 4: 1011–1014 [DOI] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Gasser SM 2007. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J 26: 4113–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez J, Belmont AS, Sedat JW 2001. Multiple regimes of constrained chromosome motion are regulated in the interphase Drosophila nucleus. Curr Biol 11: 1227–1239 [DOI] [PubMed] [Google Scholar]
- Wilson JH, Leung WY, Bosco G, Dieu D, Haber JE 1994. The frequency of gene targeting in yeast depends on the number of target copies. Proc Natl Acad Sci 91: 177–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Carlson M 1992. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet 8: 387–391 [DOI] [PubMed] [Google Scholar]
- Wongwisansri S, Laybourn PJ 2005. Disruption of histone deacetylase gene RPD3 accelerates PHO5 activation kinetics through inappropriate Pho84p recycling. Eukaryot Cell 4: 1387–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woychik NA, Young RA 1989. RNA polymerase II subunit RPB4 is essential for high- and low-temperature yeast cell growth. Mol Cell Biol 9: 2854–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]