This study addresses the role of somatic mtDNA mutations in determining cellular phenotype. Wallace and colleagues find a strong selection for mutant mtDNAs during mouse L-cell propagation, with recombination leading to multiple mitochondrial lineages in different cell lines. The mitochondrial defects increase ROS production, providing cells with a proliferative advantage. This work has implications not only for cancer cells, but also for iPS cells and their use and propagation.
Keywords: mtDNA, mutation, recombination, OXPHOS, cell proliferation, mouse L cells
Abstract
The role of mitochondrial DNA (mtDNA) mutations and mtDNA recombination in cancer cell proliferation and developmental biology remains controversial. While analyzing the mtDNAs of several mouse L cell lines, we discovered that every cell line harbored multiple mtDNA mutants. These included four missense mutations, two frameshift mutations, and one tRNA homopolymer expansion. The LA9 cell lines lacked wild-type mtDNAs but harbored a heteroplasmic mixture of mtDNAs, each with a different combination of these variants. We isolated each of the mtDNAs in a separate cybrid cell line. This permitted determination of the linkage phase of each mtDNA and its physiological characteristics. All of the polypeptide mutations inhibited their oxidative phosphorylation (OXPHOS) complexes. However, they also increased mitochondrial reactive oxygen species (ROS) production, and the level of ROS production was proportional to the cellular proliferation rate. By comparing the mtDNA haplotypes of the different cell lines, we were able to reconstruct the mtDNA mutational history of the L–L929 cell line. This revealed that every heteroplasmic L-cell line harbored a mtDNA that had been generated by intracellular mtDNA homologous recombination. Therefore, deleterious mtDNA mutations that increase ROS production can provide a proliferative advantage to cancer or stem cells, and optimal combinations of mutant loci can be generated through recombination.
The importance of understanding the origin of mitochondrial DNA (mtDNA) genotypes and their role in determining a cellular phenotype in vivo and in vitro is becoming increasingly urgent given that somatic mtDNA mutations have been found to accumulate in degenerative diseases (Coskun et al. 2004, 2010; Bender et al. 2006; Kraytsberg et al. 2006), aging (Wallace and Fan 2009), and cancer (Horton et al. 1996; Petros et al. 2005; Wallace 2005; Brandon et al. 2006; Gasparre et al. 2007; Park et al. 2009). Each mammalian cell contains hundreds to thousands of mtDNAs, and when a mtDNA mutation arises, it creates an intracellular mixture of mtDNAs, a state known as heteroplasmy (Wallace 2007). When heteroplasmic cells replicate, the mutant and normal mtDNAs can be segregated toward homoplasmy, either pure wild-type or mutant mtDNAs (Yoneda et al. 1992; Bourgeron et al. 1993; Dunbar et al. 1995; Turner et al. 2005). Heteroplasmic mtDNAs can complement each other in trans (Oliver and Wallace 1982), and severe mtDNA mutations can contribute to tumorigenesis in the heteroplasmic state (Park et al. 2009). Therefore, certain combinations of mtDNA mutant loci may complement each other and provide an advantage to the cell. Homologous recombination of mtDNAs within a cell could accentuate these effects by permitting reassortment of loci to achieve optimal linkage combinations.
Although mtDNA recombination has been reported in a variety of contexts and experimental systems (D'Aurelio et al. 2004; Kraytsberg et al. 2004; Piganeau et al. 2004; Tsaousis et al. 2005; Bacman et al. 2009), the occurrence of homologous recombination between mtDNAs within heteroplasmic cells continues to be controversial. One of the main concerns raised about previous mtDNA recombination reports is that they used the PCR and chain termination sequencing in mixed mtDNA samples, which might have generated spurious “recombinants” during DNA amplification. To prove the existence of intermolecular mtDNA recombination, it will be necessary to identify a cellular system that is heteroplasmic for two parental molecules that have several different loci and then demonstrate recombinant forms in that same cell without resorting to nonnatural DNA amplification.
The mammalian mtDNA encodes the 12S and 16S rRNAs and 22 tRNAs for mitochondrial protein synthesis, plus 13 essential oxidative phosphorylation (OXPHOS) polypeptides: ND1, ND2, ND3, ND4L, ND4, ND5, and ND6 of the 45 polypeptides of complex I; cytochrome b of the 11 polypeptides of complex III; COI, COII, and COIII of the 13 polypeptides of complex IV; and ATP6 and ATP8 of the ∼15 polypeptides of complex V. The mitochondrion generates cellular energy by oxidizing reducing equivalents (electrons) produced by glycolysis, the tricarboxylic acid cycle, and fatty acid oxidation via the successive transfer of electrons down the electron transport chain (ETC) from NADH dehydrogenase (complex I) to coenzyme Q, complex III, cytochrome c (cytc), cytc oxidase (complex IV), and oxygen to generate water. As electrons traverse complexes I, III, and IV, protons are pumped out across the mitochondrial inner membrane, creating an electrochemical gradient that is used by the ATP synthase to generate ATP. As a by-product of respiration, the mitochondrion generates reactive oxygen species (ROS), primarily from complexes I and III. At moderate levels, ROS is an important signal transduction system and a potent stimulator of cellular proliferation (Burdon 1995; Lander 1997; Wallace and Fan 2010; Wallace et al. 2010). However, at excessive levels, ROS can lead to toxicity and death.
To further elucidate the role of mtDNA mutations in cell proliferation and obtain a definitive system for studying homologous recombination, we analyzed the genetics of mtDNA mutations in cultured mouse L cells (Bunn et al. 1974, 1977; Wallace et al. 1976; Trounce et al. 1996). We now provide evidence that mtDNA mutations associated with increased ROS production impart increased proliferative potential to cells and, as a consequence, can potentially be enriched during prolonged propagation. This fosters the long-term maintenance of heteroplasmy, which we found leads to homologous recombination.
Results
L cells contain multiple heteroplasmic mutations
In the process of isolating novel mtDNA mutations in mouse LA9 cells by selection with ETC inhibitors, several homoplasmic polypeptide mutations were repeatedly recovered. These mutations included a C-to-T transition at nucleotide 4794 in the ND2 gene of complex I that changed amino acid 294 from Thr to Ile [ND2 4794T], a T-to-C transition at nucleotide 6589 in the COI gene of complex IV that changed amino acid 421 from Val to Ala [COI 6589C], a T-to-C transition at nucleotide 12,048 in the ND5 gene of complex I that changed amino acid 103 from Phe to Leu [ND5 12048C], and a C insertion in a run of 6Cs at nucleotides 13,880–13,885 in the ND6 gene of complex I that changed the reading frame starting at amino acid 63 and leading to a stop codon at amino acid 79 [ND6 13885insC]. These same mutations had been previously identified: The [ND2 4794T] and [ND5 12048C] mutations were reported in LA9 cells by Enriquez's group in 2003 (Bayona-Bafaluy et al. 2003a) (referred to as LA9E2002), the [COI 6589C] and [ND6 13885insC] mutations were reported in L929 cells also from Enriquez's group in 2003 (Acin-Perez et al. 2003; Bayona-Bafaluy et al. 2003a) (referred to as L929E2002), and the [ND6 13885insC] mutation was reported in an LA9-derived cell line, A4, from Attardi's group in 1998 (Bai and Attardi 1998).
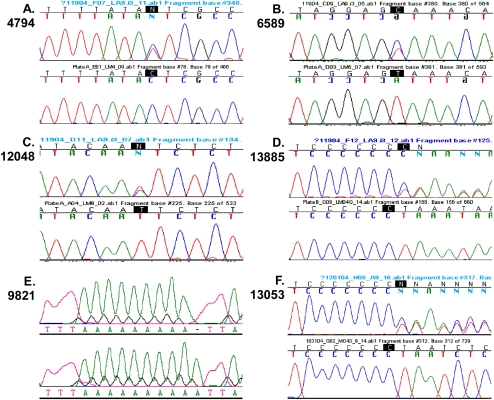
To determine the origin of these mutations, we sequenced the full mtDNAs of our LA9 cell line, which was purchased from American Type Culture Collection (ATCC no. CCL-1.4) in 1998 (referred to as LA9W1998). LA9W1998 was found to contain all four of the above polypeptide mutations—[ND2 4794T], [COI 6589C], [ND5 12048C], and [ND6 13885insC]—in the heteroplasmic state. The cell line was also heteroplasmic for either 9As or 10As of the homopolymer A track starting at nucleotide 9821 in the tRNAArg gene, referred to as [tRNAArg 9821(9A)] or [tRNAArg 9821(10A)], respectively (Fig. 1A–E; Supplemental Fig. S1). Since C3H/He mice have 9As in the tRNAArg gene at nucleotide 9821 (Bayona-Bafaluy et al. 2003a) and C3H mice were the progenitors of mouse L cells (Earle 1943; Sanford et al. 1948), we can define the 9A allele as wild type and the 10A allele as mutant.
Figure 1.
Sequence profiles of the mtDNA mutations in LA9 and LMTK− cells. A–F show six heteroplasmic mtDNA polypeptide mutations. These include ND2 C4794T (A), COI T6589C (B), ND5 T12048C (C), ND6 13885insC (D), [tRNAArg 9821(9A)] versus [tRNAArg 9821(10A)] (E), and ND5 13053insC (F) found in LA9W1998 and/or LA9W2002. In E, the tRNAArg 9821 locus 9As is a wild-type allele and 10As is a mutant. This locus is heteroplasmic in LA9W1998, homoplasmic wild type in LA9W2002 (top; 9As), and homoplasmic mutant in LMTK− (bottom; 10As). With the exception of tRNAArg 9821, the top trace is mutant, and the bottom is wild type. The wild-type genotype for C3H and 129 mice is [ND2 4794C]-[COI 6589T]-[tRNAArg 9821(9A)]-[ND5 12048T]-[ND5 13050(7C)]-[ND6 13885insC].
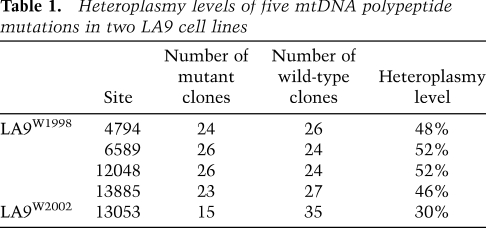
To determine the level of heteroplasmy of the four polypeptide mutations, we cloned the mtDNA surrounding each mutation and sequenced multiple clones for each site. The heteroplasmy levels of the four mutations were 48% for [ND2 4794T], 52% for [COI 6589C], 52% for [ND5 12048C], and 46% for [ND6 13885insC] (Table 1).
Table 1.
Heteroplasmy levels of five mtDNA polypeptide mutations in two LA9 cell lines
To determine whether this surprising array of endogenous mtDNA mutations was a general feature of LA9 cells, we purchased a second LA9 cell line from ATCC (referred to as LA9W2002). The mtDNA of LA9W2002 was sequenced and found to contain the same four heteroplasmic polypeptide mutations found in LA9W1998. In addition, LA9W2002 also contained another mutation, a C insertion in a string of 7Cs at nucleotides 13,047–13,053 in the ND5 gene that changed the reading frame at amino acid 438, introducing a stop codon at position 439 [ND5 13053insC] (Fig. 1F). By cloning and sequencing, the heteroplasmy level of the [ND5 13053insC] allele was determined to be ∼30% (Table 1). This cell line was homoplasmic for the wild-type [tRNAArg 9821(9A)] allele.
The mtDNA of another L-cell derivative, LMTK− (ATCC no. CCL-1.3), was also sequenced. It did not contain any of the LA9 mtDNA polypeptide mutations (Fig. 1A–D), but was homoplasmic for the [tRNAArg 9821(10A)] allele (Fig. 1E; Supplemental Fig. S1).
Isolation of homoplasmic clones from LA9W1998 and LA9W2002 cells
To further clarify the mtDNA genotypes of LA9W1998 and LA9W2002 cells, both cell lines were subjected to ethidium bromide (EtBr)-mediated depopulation–repopulation to segregate each mtDNA type to homoplasmy. A total of 56 and 61 homoplasmic clonal cell lines were obtained and analyzed from LA9W1998 and LA9W2002, respectively.
Listing only the mutant loci found in the mtDNAs isolated from LA9W1998, 28 clones (50%) were homoplasmic for a mtDNA with linked [ND2 4794T]-[ND5 12048C] alleles, 23 (41%) were homoplasmic for linked [COI 6589C]-[tRNAArg 9821(10A)]-[ND6 13885insC] alleles, five (9%) were homoplasmic for linked [COI 6589C]-[tRNAArg 9821(10A)]-[ND5 12048C]- alleles, and none were homoplasmic for wild-type mtDNA (Table 2).
Table 2.
mtDNA composition of three LA9 and one L929 cell lines
From LA9W2002, 19 (31%) clones were homoplasmic for a mtDNA with linked [ND2 4794T]-[ND5 12048C] alleles, 27 (44%) were homoplasmic for linked [COI 6589C]-[ND6 13885insC] alleles, and 15 (25%) were homoplasmic for linked [ND2 4794T]-[ND5 12048C]-[ND5 13053insC] alleles. Again, none of the mtDNAs were wild type (Table 2).
From this data, we calculated the heteroplasmy levels of all six mutations in the two cell lines. LA9W1998 was calculated to harbor 50% of [ND2 4794T], 50% of [COI 6589C], 59% of [ND5 12048C], 41% of [ND6 13885insC], and 50% of [tRNAArg 9821 (10A)], which is close to the polypeptide mutation percentages that we determined by direct cloning of the LA9W1998 mtDNAs (Table 1). Similarly, the heteroplasmy levels of the mutations in LA9W2002 was calculated to be 55% of [ND2 4794T], 44% of [COI 6589C], 55% of [ND5 12048C], 44% of [ND6 13885insC], and 25% of [ND5 13053insC]. Thus, in both cell lines, 100% of the mtDNAs harbored missense mutations, and ∼50% (41%–69%) of the mtDNAs harbored frameshift mutations.
ETC defects in homoplasmic mutant cybrids
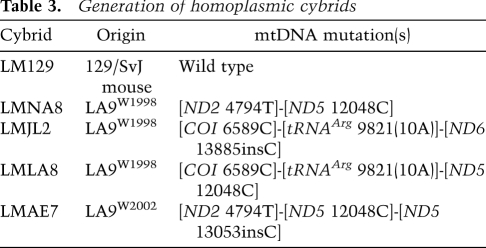
To determine the functional significance of each of the mtDNA types isolated from the LA9W1998 and LA9W2002 cell lines, we enucleated cells harboring each of the four different EtBr-generated mutant mtDNA haplotypes (Table 3) and transferred their mtDNAs into the uniform nuclear background of our mouse LMTK−-derived cell line lacking mtDNA (ρo), LMEB4 (Trounce et al. 1996). The resulting transmitochondrial cybrids were LMNA8 ([ND2 4794T]-[ND5 12048C]), LMJL2 {[COI 6589C]-[tRNAArg 9821(10A)]-[ND6 13885insC]}, LMLA8 {[COI 6589C]-[tRNAArg 9821(10A)]-[ND5 12048C]}, and LMAE7 ([ND2 4794T]-[ND5 12048C]-[ND5 13053insC]). Mitochondria from 129/SvJ mouse brain synaptosomes were also transferred into LMEB4 to make a control cybrid, LM129, whose mtDNA is wild type for all mutant alleles, including [tRNAArg 9821(9A)] (Table 3).
Table 3.
Generation of homoplasmic cybrids
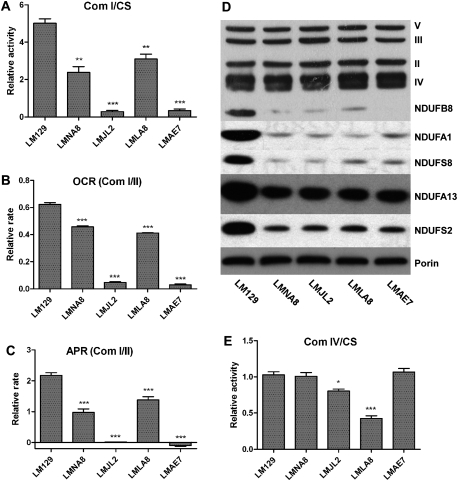
In the two mutant cybrids containing complex I frameshift mutations, LMJL2 and LMAE7, complex I activity and complex I-dependent O2 consumption were both reduced >90%, and complex I-dependent ATP production was virtually undetectable (Fig. 2A–C). In the two cybrids containing complex I missense mutations, LMNA8 and LMLA8, complex I activity, complex I-dependent O2 consumption, and complex I-dependent ATP production were all reduced to 50%–70% of the control levels (Fig. 2A–C).
Figure 2.
Reduced complex I and complex IV function in mutant cybrids. (A) Com (OXPHOS complex activity). Reduced complex I activity (normalized to citrate synthase activity) in all four homoplasmic cybrids: LMNA8 ([ND2 4794T]-[ ND5 12048C]), LMJL2 ([COI 6589C]-[tRNAArg 9821(10A)]-[ND6 13885insC]), LMLA8 ([COI 6589C]-[tRNAArg 9821(10A)]-[ND5 12048C]), and LMAE7 ([ND2 4794T]-[ND5 12048C]-[ND5 13053insC]) relative to the wild-type mtDNA LM129 cybrid. n = 6; (**) P < 0.01; (***) P < 0.001. (B) OCR (Oxygen consumption rate). Reduced complex I-dependent oxygen consumption rate (normalized to complex II-dependent oxygen consumption rate) in all four mutant cybrids. n = 6; (***) P < 0.001. (C) APR (ATP production rate). Reduced complex I-dependent ATP production (normalized to complex II-dependent ATP production) in all four mutant cybrids. n = 6; (***) P < 0.001. (D) Reduced levels of assembled complex I subunits, including subunits of the membrane subcomplex (NDUFB8 [20-kD], NDUFS8 [23-kD], and NDUFA1 [MWFE]), as well as subunits of the peripheral subcomplex (NDUFS2 [49 kD] and NDUFA13 [Grim19]). Subunits of other ETC complexes (II, III, IV, and V) remained unaffected. Porin was used as the loading control. (E) Reduced complex IV activity (normalized to citrate synthase activity) in mutant cybrids containing the COI T6589C mutation (LMJL2 and LMLA8). n = 6; (*) P < 0.05; (***) P < 0.001.
Western blot analysis of the cybrid OXPHOS proteins showed dramatic reduction of assembled complex I subunits in all four complex I mutant cybrids. The polypeptides from the peripheral subcomplex domain, which include the nuclear DNA (nDNA)-encoded NDUFS2 (49 kD) and NDUFA13 (Grim 19) subunits, were reduced 50%–70%, whereas the polypeptides of the membrane subcomplex domain, which encompasses the mtDNA-encoded subunits plus the nDNA-encoded NDUFB8 (20 kD), NDUFA1 (MWFE), and NDUFS8 (23 kD) subunits, were reduced >90% (Fig. 2D). Although the reduction in levels of complex I enzyme activities were different between the frameshift and missense cybrids, the changes in assembled complex I subunits were similar in all four cybrids (Fig. 2A–D). This suggests that the residual complex I enzyme complexes in the missense mutation cells must be enzymatically active, implying that the ND2 C4794T and the ND5 T12048C mutations may compromise complex I assembly but not activity.
The two cybrids containing the COI T6589C missense mutation, LMJL2 and LMLA8, also showed significant reduction in complex IV activity (Fig. 2E). Moreover, LMJL2, where the COI mutation was linked to the complex I ND6 frameshift mutation, had much higher complex IV activity than did LMLA8, where the COI mutation was linked to a less-severe complex I ND5 missense mutation. This supports our previous observation that severe complex I defects can be accompanied by a compensatory induction of complex IV (Fan et al. 2008). Even though the specific activity was reduced, the assembly of complex IV was normal (Fig. 2D, top panel, “IV”). Therefore, the COI T6589C mutation must affect enzymatic function but not assembly (Acin-Perez et al. 2003). The activity (Supplemental Fig. S3) and assembly (Fig. 2D) of complex II and complex III were not altered.
Mutant mtDNAs increased ROS production and stimulated cell proliferation
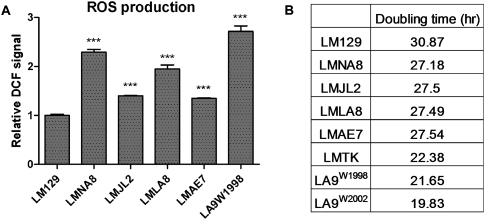
We next performed H2DCFDA staining to determine whether mitochondrial ROS production was impacted by the complex I and complex IV defects (Wallace 2001; Petros et al. 2005; Fan et al. 2008; Ishikawa et al. 2008). All mutant cybrids showed significantly increased ROS production (Fig. 3A). The two cybrids containing the ND5 and ND6 frameshift mutations LMJL2 and LMAE7 had an ∼40% increase relative to the LM129 control cell line. The two cybrids harboring the complex I and complex IV missense mutations, LMNA8 and LMLA8, had an ∼100% increase over LM129. Finally, the multiply heteroplasmic parental cell line LA9W1998 had the highest ROS production, an ∼170% increase over that of the LM129 control (Fig. 3A).
Figure 3.
Increased ROS production and cellular proliferation in mutant cybrids and cells. (A) All four mutant cybrids and LA9W1998 have higher ROS production than the wild-type LM129; the bars report cell line values divided by the LM129 value. n = 6; (***) P < 0.001. (B) Doubling times calculated from growth curves of the control LM129 cybrid; the homoplasmic mutant mtDNA cybrids LMNA8, LMJL2, LMLA8, and LMAE7 (see Table 3); and the mature L-cell lines LMTK−, LA9W1998, and LA9W2002.
These results are consistent with the proposal that most of the mitochondrial matrix O2·− is generated by mitochondrial complex I (Liu et al. 2002). When complex I function is severely impaired, as in LMJL2 and LMAE7 frameshift mutants, ROS production from complex I would be inhibited. However, when complex I is partially inhibited, as is seen for the LMLA8 and LMNA8 cybrids, electron flowthrough complex I is only retarded, and ROS production is accentuated.
Cybrids harboring the COI missense mutation also showed increased ROS production. Presumably, the complex IV blockage increased electron density throughout the upstream ETC, specifically in complex I, CoQ, and complex III. Complex III is the other important source of O2·− after complex I (Muller et al. 2004). Increased ETC electron density should increase ROS production from both complex I and complex III. Apparently, heteroplasmic combinations of a complex I frameshift mutation and complex I and complex IV missense mutations can act synergistically to result in the maximum ROS production, as seen for LA9W1998.
Since ROS has been proposed to be a potent stimulator of cellular proliferation (Burdon 1995; Lander 1997; Wallace and Fan 2010; Wallace et al. 2010), we hypothesized that those mtDNA mutations that increased ROS production would increase the proliferation rate of the mutant cells. To test this hypothesis, we measured the growth rate of all cybrids and the parental LA9 and LMTK− cell lines. Without exception, all mutant cybrids had significantly higher growth rates than the LM129 cybrid with wild-type mtDNA (Fig. 3B). Moreover, the three established cell lines LMTK−, LA9W1998, and LA9W2002 grew even faster than any of the homoplasmic mutant cybrids. Therefore, mtDNA mutations that increase mitochondrial ROS production result in increased cell proliferation.
Stimulation of cell proliferation in culture would provide a selective advantage to cells lines harboring mtDNA mutations. Consequently, the established cell lines seem to have evolved mtDNA genotypes that maximized their cell proliferation rate.
Evolution of mtDNA genotype by mutation and recombination
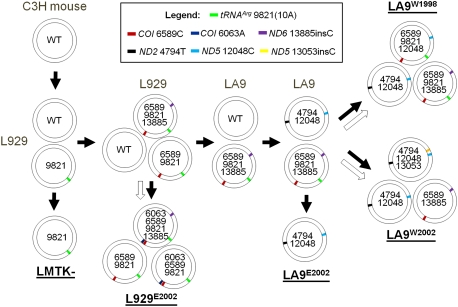
To evaluate the origin and evolution of the various cell mutants, we combined our data on the mtDNA mutation haplotypes found in LA9W1998 and LA9W2002 with those reported by Enriquez and colleagues (Acin-Perez et al. 2003) for L-cell lines LA9E2002 and L929E2002 (Fig. 4).
Figure 4.
Reconstruction of the L-cell lineage mtDNA mutational and recombinational history. Each double circle represents one type of mtDNA, with the numbers inside being the mutation sites that differ from the wild-type allele. The relative position of the mutations on the mtDNA circle is indicated by the colored sector, and the colors are defined in the figure. Filled arrows indicate direction of the evolution of the L-cell genotypes. Open arrows indicate stages at which intra-mtDNA recombination must have occurred.
In this analysis, we assumed that each of the identified mutants arose in L cells once. This follows from the fact that the recurrence of missense mutations is exceedingly rare and alteration of homoplasmic length mutations was not seen in over a hundred clones derived from LA9W2002 and LMTK− cells
Since the ancestral L-cell line (Sanford et al. 1948; Kit et al. 1963; Littlefield 1964) was derived from subcutaneous tissue of a C3H male mouse (Earle 1943), the initial mtDNA must have lacked all of the polypeptide mutations and was [tRNAArg 9821(9A)] (Bayona-Bafaluy et al. 2003a). The initial culture was treated with 20-methylcholanthrene, resulting in the L- and L929 cell lines (Sanford et al. 1948). In the early 1960s, thymidine kinase (TK)-negative and hypoxanthine phosphoribosyltransferase (HPRT)-negative cell lines were isolated from the L–L929 cell lineage to give rise to LMTK− (Kit et al. 1963) and LA9 cells (Littlefield 1964), respectively.
The LMTK− mtDNA is homoplasmic for the [tRNAArg 9821(10A)] allele. Therefore, the L- and L929 cell lineage must have acquired and been heteroplasmic for the [tRNAArg 9821(10A)] allele prior to the origin of LMTK− (Fig. 4).
The L929 line must have next acquired the [COI 6589C] missense mutation on a mtDNA harboring the [tRNAArg 9821(10A)] allele, since this [COI 6589C]-[tRNAArg 9821 (10A)] mtDNA was found in L929E2002. Subsequently, the [ND6 13885insC] frameshift mutation must have arisen on one of the [COI 6589C]-[tRNAArg 9821(10A)] mtDNAs, generating the [COI 6589C]-[tRNAArg 9821(10A)]-[ND6 13885insC] mtDNA found in LA9W1998 (Fig. 4).
The main trunk L929 cell lineage then harbored three mtDNAs: a wild-type mtDNA, a [COI 6589C]-[tRNAArg 9821(10A)] mtDNA, and a [COI 6589C]-[tRNAArg 9821(10A)]-[ND6 13885insC] mtDNA.
The L929E2002 cell line branched from this trunk L929 cell lineage by first acquiring a C-to-A transversion mutation in the COI gene [COI 6063A] (Acin-Perez et al. 2003; Bayona-Bafaluy et al. 2003a) on one of the [COI 6589C]-[tRNAArg 9821(10A)]-[ND6 13885insC] mtDNAs. This created the [COI 6063A]-[COI 6589C]-[tRNAArg 9821(10A)]-[ND6 13885insC] mtDNA found in L929E2002 (Fig. 4).
At this point in the L929E2002 lineage, a recombination event must have occurred between a [COI 6063A]-[COI 6589C]-[tRNAArg 9821(10A)]-[ND6 13885insC] mtDNA and a wild-type mtDNA that separated the [COI 6063A]-[COI 6589C]-[tRNAArg 9821(10A)] mutations from the [ND6 13885insC] mutation, thus creating the [COI 6063A]-[COI 6589C]-[tRNAArg 9821(10A)] mtDNA (Fig. 4). The reciprocal recombinant mtDNA product containing the [ND6 13885insC] allele and the remaining wild-type mtDNAs must then have been lost, resulting in the final L929E2002 genotype (Fig. 4).
Returning to the trunk L929–LA9 lineage, propagation of L929 cells after 1964 must have led to the loss of the [COI 6589C]-[tRNAArg 9821(10A)] mtDNA and the retention of only the wild-type and the [COI 6589C]-[tRNAArg 9821(10A)]-[ND6 13885insC] mtDNAs.
Subsequently, two new mtDNA mutations, [ND2 4794T] and [ND5 12048C], arose within one of the wild-type mtDNAs to generate the [ND2 4794T]-[ND5 12048C] molecule found in LA9E2002, LA9W1998, and LA9W2002 cells. Presumably, the [ND2 4794T]-[ND5 12048C] molecule could partially complement the severe complex I defect caused by the [ND6 13885insC] frameshift mutation in the [COI 6589C]-[tRNAArg 9821(10A)]-[ND6 13885insC] mtDNA, thus permitting the loss of the remaining wild-type mtDNAs in the LA9 trunk lineage (Fig. 4).
One sublineage of LA9 subsequently lost the [COI 6589C]-[tRNAArg 9821(10A)]-[ND6 13885insC] mtDNA. This generated the LA9E2002 cell line, which is homoplasmic for the [ND2 4794T]-[ND5 12048C] mtDNA (Fig. 4).
In the progenitor of the LA9W2002 cell line, a frameshift mutation ([ND5 13035insC]) arose within one of the [ND2 4794T]-[ND5 12048C] mtDNAs, generating the [ND2 4794T]-[ND5 12048C]-[ND5 13053insC] mtDNA. The progenitor of the LA9W2002 cell line must have also retained the [ND2 4794T]-[ND5 12048C] mtDNA and the [COI 6589C]-[tRNAArg 9821(10A)]-[ND6 13885insC] mtDNA from LA9, resulting in a triple heteroplasmy. However, in the final LA9W2002 lineage, the [tRNAArg 9821(10A)] allele in the [COI 6589C]-[tRNAArg 9821(10A)]-[ND6 13885insC] mtDNA was exchanged for the wild-type [tRNAArg 9821(9A)] allele, presumably by recombination between the [COI 6589C]-[tRNAArg 9821(10A)]-[ND6 13885insC] mtDNA and either the [ND2 4794T]- [ND5 12048C] or the [ND2 4794T]-[ ND5 12048C]-[ND5 13035insC] mtDNAs (Fig. 4).
The LA9W1998 derivative of LA9 retained the [COI 6589C]-[tRNAArg 9821(10A)]-[ND6 13885insC] and [ND2 4794T]-[ND5 12048C] mtDNAs. However, it also contains a new mtDNA: [COI T6589C]-[tRNAArg 9821(10A)]-[ND5 12048C]. This latter molecule must have arisen from a homologous recombination between the [COI 6589C]-[tRNAArg 9821(10A)]-[ND6 13885insC] and the [ND2 4794T]-[ND5 12048C] mtDNAs (Fig. 4). Consistent with the relatively recent occurrence of this recombination event, the recombinant [COI 6589C]-[tRNAArg 9821(10A)]-[ND5 12048C] mtDNA is present at the much lower frequency, 9%, than its parental mtDNAs: [ND2 4794T]-[ND5 12048C] at 50%, and [COI 6589C]-[tRNAArg 9821(10A)]-[ND6 13885insC] at 41%. This recombination event is particularly important, since it involved the reassortment of missense mutations. Hence, it was unlikely that this “recombination” event could have been the result of a reversion mutation in a homopolymeric nucleotide sequence (Fig. 4).
Discussion
During the evolution of the LMTK−, L929E2002, LA9E2002, LA9W1998, and LA9W2002 cell lines, the L–L929–LA9 cell lineage acquired seven de novo gene mutations: four missense mutations, two frameshift mutations, and one tRNA homopolymer A insertion mutation. Each of these mutations became enriched to a significant degree over the thousands of other mtDNAs in the various cell lines to the point that all wild-type mtDNAs were lost, and every mtDNA in all five established cell lines harbors at least one of these mutations. Therefore, these mutations must have been enriched by selection. The fact that all six polypeptide mutations affect complexes I and IV and that complex I mutations have frequently been observed in established cell lines (Bai et al. 1997, 2001; Bai and Attardi 1998; Acin-Perez et al. 2003) suggests that specific types of mtDNA OXPHOS mutations can be advantageous to cells proliferating in vitro.
To determine the basis for the selective enrichment of the mutant mtDNAs, we isolated each mtDNA in a separate cybrid cell line and then analyzed their mitochondrial physiology. Since all of the mutations inhibited OXPHOS, reduced mitochondrial ATP production does not seem to be a major factor in determining the proliferative potential of the L-cell lines. Generally, L cells are grown in high-glucose medium. Hence, they presumably generate sufficient ATP from glycolysis.
While mitochondrial OXPHOS function did not correlate with L-cell growth, mitochondrial ROS production did. Cybrid LM129 with wild-type mtDNAs had the lowest ROS production and grew the slowest, all of the homoplasmic cybrid lines with various LA9 mutant mtDNAs had higher ROS production and grew faster, and the heteroplasmic parental cell line LA9W1998 with three mutant mtDNAs had the highest ROS production rate and grew the fastest.
Presumably, then, during prolonged culture, the L–L929–LA9 cell lines evolved a mtDNA genotype that had high ROS production, thus driving maximum growth rate. For the L929E2002, LA9W1998, and LA9W2002 cell lines, this optimal mitochondrial genotype stabilized as a heteroplasmic mixture of mtDNA missense and frameshift mutations. That the frameshift mtDNA mutants would remain heteroplasmic is consistent with the report that cells homoplasmic for an ND5 frameshift mutation grew less rapidly and had a lower neoplastic potential than heteroplasmic cells (Park et al. 2009). Hence, it follows that combinations of missense and frameshift mutant mtDNAs may generate the maximum ROS production and growth stimulation.
LMTK− stabilized with a homoplasmic mtDNA harboring only the [tRNAArg 9821 (10A)] mutation. This mutation has also been associated with increased ROS production (Moreno-Loshuertos et al. 2006).
The evolution of multiple heteroplasmic mtDNA mutations in the mouse L929–LA9 cell lineage provided the perfect situation to foster mtDNA homologous recombination. Our elucidation of the individual mtDNA haplotypes, together with knowledge of the founding mtDNA genotype (the “common” mtDNA haplotype shared by 129 and C3H mice) (Ferris et al. 1982; Bayona-Bafaluy et al. 2003a), permitted us to reconstruct the series of events that led to the final L-cell line mtDNA genotypes. From this reconstruction, we were able to identify a different recombinant mtDNA in each of the three heteroplasmic cell lines examined: L929E2002, LA9W1998, and LA9W2002. Therefore, this proves that heteroplasmic mtDNAs within cells can and do undergo homologous recombination and that recombinant mtDNA haplotypes can incorporate combinations of alleles that result in their selective enrichment within proliferating cells.
These observations have profound implications for cancer biology and stem cell therapeutics. For cancer cells, the acquisition of mtDNA mutations and their reassortment by recombination could generate mtDNA genotypes that have a significant effect on the proliferative potential of the cancer cell (Petros et al. 2005). Moreover, cancer cells heteroplasmic for multiple different mtDNA haplotypes can rapidly adjust to varying growth environments by simply shifting the percentage of their various mtDNAs.
For stem cell therapeutics, current proposals envision conversion of adult patient fibroblasts into induced pluripotent stem (iPS) cells, followed by differentiating the iPS cells into cells to be used to repair the damaged tissue. However, as we age, we accumulate mutations in the resident mtDNAs (Corral-Debrinski et al. 1992; Michikawa et al. 1999; Murdock et al. 1999), and the level of somatic mtDNA mutations is systemically elevated in certain neurodegenerative diseases (Coskun et al. 2003, 2010). Therefore, iPS cells generated from adult tissues may already contain mutant mtDNAs that could become reassorted by recombination during dedifferentiation and culturing. Those haplotypes that impart the highest proliferative potential would then be selectively enriched. As a result, iPS cell-derived tissues may carry with them the seeds for premature aging, imperfect differentiation, and neoplastic transformation.
Materials and methods
Cell culture
All mouse fibroblast cell lines were cultured in DMEM with high glucose (4.5 g/L) and L-glutamine supplemented with 10% FBS, 1 mM pyruvate, and 50 μg/mL uridine. Cultures were held in a humidified 5% CO2 and 95% air incubator at 37°C (Trounce et al. 1996).
For growth rate analysis, cells were plated at the concentration of 50,000 cells per well in six-well plates. Cells were trypsinized and counted every day for 6 d. The doubling time was calculated using exponential regression analysis by the method of Doubling Time (Roth V. 2006, http://www.doubling-time.com/compute.php).
mtDNA segregation
Heteroplasmic LA9 or A9 cells were cultured in the above medium supplemented with 1 μg/mL EtBr for 2 wk (King 1996). mtDNA-depleted single colonies were then isolated, expanded, and analyzed for segregation to homoplasmy.
Cybrid generation
To generated cybrid lines LMNA8, LMJL2, and LMLA8 derived from homoplasmic subclones of LA9W1998, the clonal lines were enucleated by treatment with cytochalasin B and cytoplasts isolated using Ficoll step gradients (Table 3). To generate the cybrid line LMAE7 derived from a homoplasmic clone of LA9W2002, the clonal line was chemically enucleated by treatment with 0.5 μg/mL actinomycin D for 12 h (Table 3; Bayona-Bafaluy et al. 2003b). The cytoplasts or actinomycin D-treated cells were then mixed with 2 × 106 freshly harvested mtDNA-deficient (ρ0) LMEB4 cells (Trounce et al. 1996). To generate the wild-type cybrid line, synaptosomes from a 1-mo-old 129/SvJ mouse were isolated using Ficoll gradient centrifugation (Sligh et al. 2000) and fused to the LMEB4 ρ0 cells (Trounce et al. 1996). The mixtures were centrifuged at 3000g for 5 min, and the pellet was overlaid with 1 mL of 50% PEG (w/v) and 10% DMSO in phosphate buffered saline (PBS) (Sigma, P-7306) for 1 min. The pellet was then gently resuspended in 20 mL of medium and plated. The cybrids were selected with 50 μg/mL bromodeoxyuridine in medium without pyruvate and uridine for 3 wk and cloned (Trounce et al. 1996).
To confirm that the nuclei of the cybrid lines were derived from LMEB4, the karyotypes of LMJL2 and LMAE7, representing the two enucleation procedures used, were prepared and stained with Hoechst 33258. This gave chromosome numbers of 39.80 ± 0.45 and 39.60 ± 0.89, respectively, and both cell lines harbored the characteristic LMTK− marker chromosome. The mtDNA genotypes were confirmed by sequencing the mtDNAs. To confirm that the cell lines had the same relative levels of mtDNA and mitochondria, the mtDNA/nDNA ratios were determined by real-time PCR using the mtDNA cytochrome b primers (forward, 5′-CATTTATTATCGCGGCCCTA-3′; and reverse, 5′-TGTTGGGTTGTTTGATCCTG-3′) and the nDNA glucagon primers (forward, 5′-CAGGGCCATCTCAGAACC-3′; and reverse, 5′-GCTATTGGAAAGCCTCTTGC-3′) (Supplemental Fig. S3). The relative levels of mitochondrial protein were determined by Western blot comparing porin to actin (Supplemental Fig. S3). The mtDNA/nDNA ratios and mitochondrial protein levels were the same for all cybrids within experimental error.
mtDNA isolation and amplification
Whole genomic DNA was isolated from 2 × 106 cultured cells using the Puregene DNA isolation kit (Gentra Systems). Eleven pairs of primers were used to amplify overlapping fragments covering the whole-mouse mtDNA (Supplemental Table S1).
mtDNA sequencing
A total of 66 primers were used to sequence the whole-mouse mtDNA (Supplemental Table S1). Dye terminator sequencing was performed using BigDye terminator version 3.1 (Applied Biosystems). The sequencing results were analyzed using Sequencher 4.5 software.
mtDNA cloning sequencing
Five pairs of primers (Supplemental Table S1) were used to amplify ∼500-nucleotide (nt) fragments surrounding the five polypeptide mutations found in LA9 and A9 cells. The fragments were cloned into Zero Blunt TOPO cloning vectors (Invitrogen). Bacteria colonies were picked, lysed, and PCR-amplified for target mtDNA fragments, which were next purified and sequenced. By calculating the bacteria colonies containing mutant alleles and wild-type alleles, the heteroplasmic levels in the original LA9 and A9 cells were obtained. A total of 50 clones per mutation were sequenced to give a resolution of 2%.
Mitochondria isolation
Two billion cells were collected and washed with 20 mL of isolation buffer (H buffer: 210 mM mannitol, 70 mM sucrose, 1 mM EGTA, 0.5% BSA [fatty acid free], and 5 mM HEPES at pH 7.2), and resuspended in 4 mL of H buffer. Digitonin (100 mg/mL DMSO) was added drop-wise to a final concentration of 0.15 mg/mL and mixed by swirling for 1 min. Then, 10 mL of H buffer was added, and the cell suspension was centrifuged at 3000g for 5 min. The pellet was resuspended in 5 mL of H buffer, transferred into a Dounce homogenizer, and disrupted with six passes at 300 rpm. Ten milliliters of H buffer was added, and the cells were centrifuged at 700g for 5 min. The supernatant was collected and centrifuged at 10,000g for 20 min, and the mitochondria pellet was resuspended in 200 μL of H buffer. Twenty microliters of this suspension was further centrifuged, and the pellet was used to determine the protein concentration (Trounce et al. 1996).
Polarographic assay of mitochondrial respiration
O2 consumption was measured in freshly isolated mitochondria using the Hansatech Instruments Oxygraph system in 1 mL of respiration buffer (225 mM mannitol, 75 mM sucrose, 10 mM KCl, 10 mM Tris-HCl, 5 mM KH2PO4 at pH 7.2) at 30°C. Subsequently, 0.3–1 mg of mitochondria and 5 μL of each of complex I substrates pyruvate (1 M) and malate (1 M) or complex II substrate succinate (1 M) were then added. Three 5-μL aliquots of ADP (25 mM) were added to assess state III versus state IV respiration. Finally, 5 μL of DNP or FCCP (10 mM) was added to determine the uncoupled respiration rate (Trounce et al. 1996). Complex I- versus complex II-dependent respiration rate was compared between wild-type and mutant cybrids.
ATP production
A kinetic measurement of ATP synthesis was performed using the luciferase–luciferin system in digitonin-permeabilized cells (Milakovic and Johnson 2005). Ten million cells were harvested, washed with PBS, and resuspended in 1 mL of H buffer. Ten microliters of 2 mg/mL digitonin was added to the cell suspension and swirled for 1 min. Ten microliters of H buffer was then added to stop permeabilization, and the cell suspension was centrifuged at 1000g for 3 min and resuspend in 1 mL of buffer A (150 mM KCl, 25 mM Tris-HCl, 2 mM EDTA, 0.1% BSA, 10 mM potassium phosphate, 0.1 mM MgCl2 at pH 7.4). One-hundred-sixty microliters of this cell suspension was mixed with 10 μL of freshly prepared buffer B (0.5 M Tris-acetate at pH 7.75, 0.8 mM luciferin, 20 μg/mL luciferase), 5 μL of 6 mM P1,P5-Di(adenosine), 5 μL of 4 mM ADP, and 5 μL each of 1 M pyruvate and malate or succinate in the presence or absence of 2 μL of 0.2 mg/mL oligomycin. The mixed reaction was measured for light emission using a NOVOStar fluorimeter (BMG Labtech). Complex I- versus complex II-dependent ATP production rates were compared between wild-type and mutant cybrids.
ROS production
ROS production in mouse fibroblasts was measured by monitoring the oxidation of carboxy-H2DCFDA (Invitrogen) in 96-well plates. Two million cells per well were incubated in 200 μL of phenol red-free DMEM containing 8 μM carboxy-H2DCFDA for 15–30 min at 37°C and washed once in warm medium. The cellular ROS-induced fluorescence was monitored for 1 h at 37°C in a NOVOStar fluorimeter using 485-nm excitation and 530-nm emission.
Mitochondrial enzymatic assays
The activity of mitochondrial ETC complexes and citrate synthase was measured spectrophotometrically using freeze–thaw-disrupted mitochondria in a SpectraMAX Plus spectrophotometer (Molecular Device) (Trounce et al. 1996).
Complex I activity was measured as the rate of NADH oxidation at 340 nm in a reaction containing 80 μL of water, 100 μL of 2× buffer (500 mM sucrose, 2 mM EDTA, 100 mM Tris-HCl at pH 7.4), 2 μL of 10 mM decylubiquinone (DB; Sigma), 0.5 μL of 2 M KCN, and 10 μL of 1 mg/mL mitochondria and incubated for 5 min at 30°C. Ten microliters of 1 mM NADH was added to initiate the reaction, and absorption at 340 nm was measured for 5 min. A parallel experiment was performed in the presence of 1 μL of 1 mg/mL rotenone to determine the rotenone-insensitive activity.
Complex II+III activity was measured as the rate of cytc reduction at 550 nm in a reaction containing 110 μL of water, 80 μL of 100 mM potassium phosphate buffer (pH 7.4), 4 μL of 1 M succinate, 1 μL of 0.1 M EDTA, 0.5 μL of 2M KCN, and 6 μL of 1 mM cytc. Five microliters of 1 mg/mL mitochondria was added, and the absorption was monitored at 550 nm for 2 min.
Complex IV activity was measured as the rate of cytc oxidation at 550 nm in a reaction containing 170 μL of water, 20 μL of 100 mM potassium phosphate buffer (pH 7.4), and 10 μL of 1 mM-reduced cytc. Five microliters of 0.1 mg/mL mitochondria was added, and the absorption at 550 nm was monitored for 2 min. Reduced cytc was prepared by adding 2 μL of 1 M dithiothreitol (DTT) to 1 mL of 1 mM cytc and incubating for 15 min.
Citrate synthase activity was monitored as the reduction of 5,5′-dithio-bis (2-nitrobenzoic acid) (DTNB) at 412 nm in a reaction containing 160 μL of water, 20 μL of 1 M Tris-HCl (pH 8.0), 10 μL of 6 mM acetyl-CoA, and 2 μL of 10 mM DTNB. Five microliters of 1 mg/mL mitochondria was added, and the absorption at 412 nm was monitored for 2 min.
Western blot
Mitochondrial proteins (10 μg) were resolved by SDS-PAGE and transferred to Immobilon-P (0.2 μm) membranes. Mitoprofile antibody cocktail for rodent OXPHOS proteins, NDUFB8, and porin antibodies were from Mitosciences. Actin antibody was from Sigma. NDUFS8 and NDUFS2 antibodies were provided by Professor Joel Lunardi and NDUFA1 antibody was provided by Professor Immo Scheffler.
Statistical analysis
Two-tailed unpaired t-test was performed to analyze the data in Figures 2 and 3.
Acknowledgments
This work was supported by National Institutes of Health grants R01 NS211328, AG24373, DK73691, AG16573, and NS41850. CIRM Comprehensive Grant RC1-00353-1 was awarded to D.C.W. D.C.W. and W.F. contributed to the design of the study and writing of this manuscript. W.F. performed the majority of the experimental work. C.S.L. contributed to the mtDNA sequencing analysis and cellular proliferation assay. P.P. contributed to the Western blotting analysis. V.P. contributed to the discussion and writing of the manuscript.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.175802.111.
References
- Acin-Perez R, Bayona-Bafaluy MP, Bueno M, Machicado C, Fernandez-Silva P, Perez-Martos A, Montoya J, Lopez-Perez MJ, Sancho J, Enriquez JA 2003. An intragenic suppressor in the cytochrome c oxidase I gene of mouse mitochondrial DNA. Hum Mol Genet 12: 329–339 [DOI] [PubMed] [Google Scholar]
- Bacman SR, Williams SL, Moraes CT 2009. Intra- and inter-molecular recombination of mitochondrial DNA after in vivo induction of multiple double-strand breaks. Nucleic Acids Res 37: 4218–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Attardi G 1998. The mtDNA-encoded ND6 subunit of mitochondrial NADH dehydrogenase is essential for the assembly of the membrane arm and the respiratory function of the enzyme. EMBO J 17: 4848–4858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai U, Seidman MD, Hinojosa R, Quirk WS 1997. Mitochondrial DNA deletions associated with aging and possibly presbycusis: A human archival temporal bone study. Am J Otol 18: 449–453 [PubMed] [Google Scholar]
- Bai Y, Hajek P, Chomyn A, Chan E, Seo BB, Matsuno-Yagi A, Yagi T, Attardi G 2001. Lack of complex I activity in human cells carrying a mutation in mtDNA-encoded ND4 subunit is corrected by the Saccharomyces cerevisiae NADH-quinone oxidoreductase (NDI1) gene. J Biol Chem 276: 38808–38813 [DOI] [PubMed] [Google Scholar]
- Bayona-Bafaluy MP, Acin-Perez R, Mullikin JC, Park JS, Moreno-Loshuertos R, Hu P, Perez-Martos A, Fernandez-Silva P, Bai Y, Enriquez JA 2003a. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res 31: 5349–5355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayona-Bafaluy MP, Manfredi G, Moraes CT 2003b. A chemical enucleation method for the transfer of mitochondrial DNA to rho(0) cells. Nucleic Acids Res 31: e98 doi: 10.1093/nar/gng100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, Betts J, Klopstock T, et al. 2006. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet 38: 515–517 [DOI] [PubMed] [Google Scholar]
- Bourgeron T, Chretien D, Rotig A, Munnich A, Rustin P 1993. Fate and expression of the deleted mitochondrial DNA differ between human heteroplasmic skin fibroblast and Epstein-Barr virus-transformed lymphocyte cultures. J Biol Chem 268: 19369–19376 [PubMed] [Google Scholar]
- Brandon M, Baldi P, Wallace DC 2006. Mitochondrial mutations in cancer. Oncogene 25: 4647–4662 [DOI] [PubMed] [Google Scholar]
- Bunn CL, Wallace DC, Eisenstadt JM 1974. Cytoplasmic inheritance of chloramphenicol resistance in mouse tissue culture cells. Proc Natl Acad Sci 71: 1681–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunn CL, Wallace DC, Eisenstadt JM 1977. Mitotic segregation of cytoplasmic determinants for chloramphenicol resistance in mammalian cells. I: Fusions with mouse cell lines. Somatic Cell Genet 3: 71–92 [DOI] [PubMed] [Google Scholar]
- Burdon RH 1995. Superoxide and hydrogen peroxide in relation to mammalian cell proliferation. Free Radic Biol Med 18: 775–794 [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, Beal MF, Wallace DC 1992. Mitochondrial DNA deletions in human brain: Regional variability and increase with advanced age. Nat Genet 2: 324–329 [DOI] [PubMed] [Google Scholar]
- Coskun PE, Ruiz-Pesini EE, Wallace DC 2003. Control region mtDNA variants: Longevity, climatic adaptation and a forensic conundrum. Proc Natl Acad Sci 100: 2174–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun PE, Beal MF, Wallace DC 2004. Alzheimer's brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc Natl Acad Sci 101: 10726–10731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun PE, Wyrembak J, Derbereva O, Melkonian G, Doran E, Lott IT, Head E, Cotman CW, Wallace DC 2010. Systemic mitochondrial dysfunction and the etiology of Alzheimer's disease and down syndrome dementia. J Alzheimers Dis 20: S293–S310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aurelio M, Gajewski CD, Lin MT, Mauck WM, Shao LZ, Lenaz G, Moraes CT, Manfredi G 2004. Heterologous mitochondrial DNA recombination in human cells. Hum Mol Genet 13: 3171–3179 [DOI] [PubMed] [Google Scholar]
- Dunbar DR, Moonie PA, Jacobs HT, Holt IJ 1995. Different cellular backgrounds confer a marked advantage to either mutant or wild-type mitochondrial genomes. Proc Natl Acad Sci 92: 6562–6566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earle WR 1943. Production of malignancy in vitro. IV. The mouse fibroblase cultures and changes seen in the living cells. J Natl Cancer Inst 4: 165–212 [Google Scholar]
- Fan W, Waymire K, Narula N, Li P, Rocher C, Coskun PE, Vannan MA, Narula J, MacGregor GR, Wallace DC 2008. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science 319: 958–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris SD, Sage RD, Wilson AC 1982. Evidence from mtDNA sequences that common laboratory strains of inbred mice are descended from a single female. Nature 295: 163–165 [DOI] [PubMed] [Google Scholar]
- Gasparre G, Porcelli AM, Bonora E, Pennisi LF, Toller M, Iommarini L, Ghelli A, Moretti M, Betts CM, Martinelli GN, et al. 2007. Disruptive mitochondrial DNA mutations in complex I subunits are markers of oncocytic phenotype in thyroid tumors. Proc Natl Acad Sci 104: 9001–9006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton TM, Petros JA, Heddi A, Shoffner J, Kaufman AE, Graham SD Jr, Gramlich T, Wallace DC 1996. Novel mitochondrial DNA deletion found in a renal cell carcinoma. Genes Chromosomes Cancer 15: 95–101 [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J 2008. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science 320: 661–664 [DOI] [PubMed] [Google Scholar]
- King MP 1996. Use of ethidium bromide to manipulate ratio of mutated and wild-type mitochondrial DNA in cultured cells. Methods Enzymol 264: 339–344 [DOI] [PubMed] [Google Scholar]
- Kit S, Dubbs DR, Pielarskik LJ, Hsu TS 1963. Deletion of thymidine kinase activity from L cells resistant to bromodeoxyuridine. Exp Cell Res 31: 297–312 [DOI] [PubMed] [Google Scholar]
- Kraytsberg Y, Schwartz M, Brown TA, Ebralidse K, Kunz WS, Clayton DA, Vissing J, Khrapko K 2004. Recombination of human mitochondrial DNA. Science 304: 981. [DOI] [PubMed] [Google Scholar]
- Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K 2006. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet 38: 518–520 [DOI] [PubMed] [Google Scholar]
- Lander HM 1997. An essential role for free radicals and derived species in signal transduction. FASEB J 11: 118–124 [PubMed] [Google Scholar]
- Littlefield JW 1964. Three degrees of guanylic acid-inosinic acid phosphorylase deficiency in mouse fibroblasts. Nature 203: 1142–1144 [DOI] [PubMed] [Google Scholar]
- Liu Y, Fiskum G, Schubert D 2002. Generation of reactive oxygen species by the mitochondrial electron transport chain. J Neurochem 80: 780–787 [DOI] [PubMed] [Google Scholar]
- Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G 1999. Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science 286: 774–779 [DOI] [PubMed] [Google Scholar]
- Milakovic T, Johnson GV 2005. Mitochondrial respiration and ATP production are significantly impaired in striatal cells expressing mutant huntingtin. J Biol Chem 280: 30773–30782 [DOI] [PubMed] [Google Scholar]
- Moreno-Loshuertos R, Acin-Perez R, Fernandez-Silva P, Movilla N, Perez-Martos A, Rodriguez de Cordoba S, Gallardo ME, Enriquez JA 2006. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat Genet 38: 1261–1268 [DOI] [PubMed] [Google Scholar]
- Muller FL, Liu Y, Van Remmen H 2004. Complex III releases superoxide to both sides of the inner mitochondrial membrane. J Biol Chem 279: 49064–49073 [DOI] [PubMed] [Google Scholar]
- Murdock DG, Christacos N, Melov S, Wallace DC 1999. A PNA-clamping based method of detection of point mutations in mtDNA. Abstract #2627. Am J Hum Genet 65: A463 [Google Scholar]
- Oliver NA, Wallace DC 1982. Assignment of two mitochondrially synthesized polypeptides to human mitochondrial DNA and their use in the study of intracellular mitochondrial interaction. Mol Cell Biol 2: 30–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Sharma LK, Li H, Xiang R, Holstein D, Wu J, Lechleiter J, Naylor SL, Deng JJ, Lu J, et al. 2009. A heteroplasmic, not homoplasmic, mitochondrial DNA mutation promotes tumorigenesis via alteration in reactive oxygen species generation and apoptosis. Hum Mol Genet 18: 1578–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, et al. 2005. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci 102: 719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piganeau G, Gardner M, Eyre-Walker A 2004. A broad survey of recombination in animal mitochondria. Mol Biol Evol 21: 2319–2325 [DOI] [PubMed] [Google Scholar]
- Sanford KK, Earle WR, Likely GD 1948. The growth in vitro of single isolated tissue cells. J Natl Cancer Inst 9: 229–246 [PubMed] [Google Scholar]
- Sligh JE, Levy SE, Waymire KG, Allard P, Dillehay DL, Nusinowitz S, Heckenlively JR, MacGregor GR, Wallace DC 2000. Maternal germ-line transmission of mutant mtDNAs from embryonic stem cell-derived chimeric mice. Proc Natl Acad Sci 97: 14461–14466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trounce IA, Kim YL, Jun AS, Wallace DC 1996. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol 264: 484–509 [DOI] [PubMed] [Google Scholar]
- Tsaousis AD, Martin DP, Ladoukakis ED, Posada D, Zouros E 2005. Widespread recombination in published animal mtDNA sequences. Mol Biol Evol 22: 925–933 [DOI] [PubMed] [Google Scholar]
- Turner CJ, Granycome C, Hurst R, Pohler E, Juhola MK, Juhola MI, Jacobs HT, Sutherland L, Holt IJ 2005. Systematic segregation to mutant mitochondrial DNA and accompanying loss of mitochondrial DNA in human NT2 teratocarcinoma cybrids. Genetics 170: 1879–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC 2001. A mitochondrial paradigm for degenerative diseases and ageing. Novartis Found Symp 235: 247–263 [DOI] [PubMed] [Google Scholar]
- Wallace DC 2005. Mitochondria and cancer: Warburg address. Cold Spring Harb Symp Quant Biol 70: 363–374 [DOI] [PubMed] [Google Scholar]
- Wallace DC 2007. Why do we have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu Rev Biochem 76: 781–821 [DOI] [PubMed] [Google Scholar]
- Wallace DC, Fan W 2009. The pathophysiology of mitochondrial disease as modeled in the mouse. Genes Dev 23: 1714–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Fan W 2010. Energetics, epigenetics, mitochondrial genetics. Mitochondrion 10: 12–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Pollack Y, Bunn CL, Eisenstadt JM 1976. Cytoplasmic inheritance in mammalian tissue culture cells. In Vitro 12: 758–776 [DOI] [PubMed] [Google Scholar]
- Wallace DC, Fan W, Procaccio V 2010. Mitochondrial energetics and therapeutics. Annu Rev Pathol 5: 297–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda M, Chomyn A, Martinuzzi A, Hurko O, Attardi G 1992. Marked replicative advantage of human mtDNA carrying a point mutation that causes the MELAS encephalomyopathy. Proc Natl Acad Sci 89: 11164–11168 [DOI] [PMC free article] [PubMed] [Google Scholar]