Membrane-anchored metalloproteinases are widely believed to function primarily in the remodeling of the extracellular matrix. Weiss and colleagues investigated the role of one membrane-tethered matrix metalloenzyme (MT1-MMP) in macrophages, which regulate cytokines and inflammation. In this study, the authors demonstrate that macrophages target the extracellular domain of MT1-MMP to the nuclear compartment, where it regulates immune responses through the transcriptional regulation of a novel PI3Kδ/Mi-2/NuRD nucleosome remodeling axis. Thus, the authors provide insight into novel functions for metalloproteinases in controlling macrophage immune responses.
Keywords: MT1-MMP, macrophage, PI3Kδ, immune response, nucleosome remodeling
Abstract
Macrophages play critical roles in events ranging from host defense to obesity and cancer, where they infiltrate affected tissues and orchestrate immune responses in tandem with the remodeling of the extracellular matrix (ECM). Despite the dual roles played by macrophages in inflammation, the functions of macrophage-derived proteinases are typically relegated to tissue-invasive or -degradative events. Here we report that the membrane-tethered matrix metalloenzyme MT1-MMP not only serves as an ECM-directed proteinase, but unexpectedly controls inflammatory gene responses wherein MT1-MMP−/− macrophages mount exaggerated chemokine and cytokine responses to immune stimuli both in vitro and in vivo. MT1-MMP modulates inflammatory responses in a protease-independent fashion in tandem with its trafficking to the nuclear compartment, where it triggers the expression and activation of a phosphoinositide 3-kinase δ (PI3Kδ)/Akt/GSK3β signaling cascade. In turn, MT1-MMP-dependent PI3Kδ activation regulates the immunoregulatory Mi-2/NuRD nucleosome remodeling complex that is responsible for controlling macrophage immune response. These findings identify a novel role for nuclear MT1-MMP as a previously unsuspected transactivator of signaling networks central to macrophage immune responses.
In events ranging from host defense and cancer to the chronic inflammatory disease states that characterize atherosclerosis and obesity, macrophages infiltrate affected interstitial tissues and mobilize proteolytic enzymes whose functions are most commonly linked to the remodeling of the extracellular matrix (ECM) (Martin et al. 2005; Mosser and Edwards 2008; Kessenbrock et al. 2010; Nathan and Ding 2010). However, macrophages also play key roles in orchestrating local immunity by secreting a complex mix of pro- and anti-inflammatory molecules that direct the intensity and duration of host responses (Martin et al. 2005; Mosser and Edwards 2008; Kessenbrock et al. 2010; Nathan and Ding 2010). While inflammatory mediators have long been known to regulate the expression of macrophage proteinases (Martin et al. 2005; Mosser and Edwards 2008; Kessenbrock et al. 2010; Nathan and Ding 2010), the possibility that proteinases themselves directly control immune responses in a cell-autonomous fashion has received little attention.
Recent studies suggest that the membrane-anchored metalloproteinase MT1-MMP controls macrophage invasion by hydrolyzing ECM components and cell surface molecules as well as activating signal transduction cascades that regulate motility and energy metabolism (Cao et al. 2004; Matias-Roman et al. 2005; Sithu et al. 2007; Barbolina and Stack 2008; D'Alessio et al. 2008; Sakamoto and Seiki 2009, 2010; Cougoule et al. 2010; Gonzalo et al. 2010; Van Goethem et al. 2010). However, other related myeloid populations (e.g., neutrophils) readily traffic through host tissues and do not express MT1-MMP (Huber and Weiss 1989; Weiss 1989; Sabeh et al. 2009b), suggesting that macrophages may reserve the proteinase for additional purposes. In an effort to assign definitive roles for MT1-MMP in macrophage function, we compared and contrasted the behavior and gene expression patterns of macrophages recovered from wild-type and MT1-MMP-deficient mice in vitro as well as in vivo. Here we found that macrophage-derived MT1-MMP does not, as often assumed, play a major role in regulating cell trafficking through host tissues. Instead, MT1-MMP acts as a critical transactivator of the gene networks central to the control of macrophage inflammatory responses. Unexpectedly, MT1-MMP modulates the expression of >100 genes, of which almost 20% are directly linked to immune regulation, via a process that operates independently of its proteolytic activity. In the absence of MT1-MMP, macrophages display a proinflammatory phenotype characterized by the inappropriate up-regulation of multiple proinflammatory mediators while down-regulating anti-inflammatory cytokines. Downstream transcriptional targets of MT1-MMP include phosphoinositide 3-kinase δ (PI3Kδ), a key regulator of macrophage immune responses (Fukao and Koyasu 2003; Beurel et al. 2010), whose active transcription is associated with the subcellular trafficking of MT1-MMP to the nuclear compartment. In turn, the MT1-MMP-dependent induction of PI3Kδ expression triggers an Akt/GSK3 signaling cascade that controls the Mi-2/NuRD complex of nucleosome remodeling enzymes that normally serve to limit the expression of macrophage-derived proinflammatory mediators (Ramirez-Carrozzi et al. 2006; Lai et al. 2009; Zhou et al. 2010). Together, these results identify a novel, proteinase-independent role for MT1-MMP as a nuclear trafficking, cell-autonomous regulator of macrophage immune function.
Results
MT1-MMP regulates macrophage-mediated subjacent proteolysis, but not migration or invasion
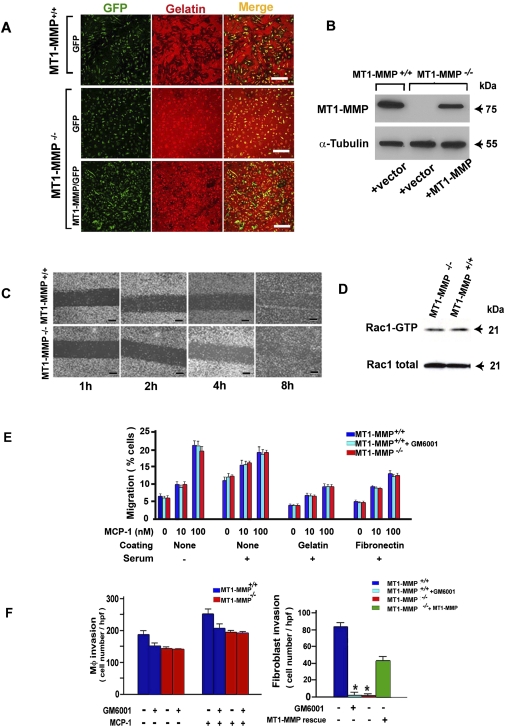
Following a 7-d culture period with M-CSF, mouse bone marrow cells recovered from wild-type (Mmp14+/+) or MT1-MMP-deficient (Mmp14−/−) mice give rise to morphologically identical populations of F4/80-positive macrophages whose expression of other collagenolytic MMPs is left intact (Supplemental Fig. 1). Consistent with earlier studies, wild-type macrophages assemble podosome-like, F-actin- and cortactin-rich punctae on their basal surface that lie atop discrete zones of the degraded subjacent matrix (Fig. 1A; Supplemental Fig. 1; Cougoule et al. 2010). In contrast, although MT1-MMP−/− macrophages are likewise able to form podosome-like structures, proteolytic activity is ablated by both morphologic and quantitative criteria (Fig. 1A; Supplemental Fig. 1). Following transduction with an MT1-MMP lentiviral expression vector, however, proteinase expression approaches wild-type levels, and the proteolytic activity of MT1-MMP−/− macrophages is largely restored (Fig. 1A,B; Supplemental Fig. 1). Nevertheless, despite their marked deficiency in proteolytic activity, MT1-MMP-deficient macrophages do not display significant defects in two-dimensional motile response, or motility-associated changes in Rac activation (Fig. 1C,D). Likewise, MT1-MMP−/− macrophages mount a chemotactic response across either uncoated or ECM-coated filters that is indistinguishable from that observed with wild-type macrophages (cultured in either the absence or presence of the broad-spectrum, synthetic matrix metalloproteinase inhibitor GM6001) (Fig. 1E; Sabeh et al. 2009a).
Figure 1.
MT1-MMP-independent regulation of macrophage trafficking. (A) Confocal laser micrographs of MT1-MMP+/+ and MT1-MMP−/− macrophages cultured atop fluorescently labeled gelatin (red) for 2 d with 10 ng/mL TNF-α. Black areas mark zones of gelatin proteolysis. Macrophages were transduced with either a control (GFP) or MT1-MMP/GFP expression vector (cells; green). Bar, 200 μm. (B) Western blot of endogenous MT1-MMP levels in wild-type macrophages versus MT1-MMP−/− macrophages before or after transduction with a lentiviral MT1-MMP expression vector. (C) Confluent monolayers of MT1-MMP+/+ or MT1-MMP−/− macrophages were disrupted by scratch-wounding, and migration was monitored in serum-containing medium over an 8-h time course. Bar, 200 μm. (D) Representative immunoblot of total Rac1 and Rac1-GTP levels monitored in MT1-MMP+/+ or MT1-MMP−/− macrophages. (E) MT1-MMP+/+ or MT1-MMP−/− macrophage chemotactic responses to MCP-1 were quantified after 24 h with or without 10% serum in the absence or presence of 10 μM GM6001. Filter surfaces were left uncoated or coated with either gelatin or fibronectin. Results are expressed as the mean ± SEM (n = 5). (F) MT1-MMP+/+ or MT1-MMP−/− macrophages (2 × 105) were cultured with 100 ng/mL MCP-1 (left panel), or fibroblasts (5 × 104) were cultured with PDGF-BB (right panel) in the absence or presence of GM6001 atop 3D type I collagen gels for 5 d, and the number of invaded cells was quantified. In the right panel, the defect in invasive potential of MT1-MMP-null fibroblasts is rescued following MT1-MMP rescue. Results shown are the mean ± SEM; (*) P < 0.01; n = 5).
To next assess the roles of MT1-MMP in regulating tissue-invasive activity, wild-type or MT1-MMP-null macrophages were cultured atop three-dimensional (3D) matrices of native type I collagen, the major ECM component of the interstitial matrix (Rowe et al. 2009; Sabeh et al. 2009b), and invasion was monitored in the absence or presence of GM6001. Under these conditions, a modest but not statistically significant trend is observed for decreased macrophage invasion in the presence of the broad-spectrum MMP inhibitor (regardless of whether cell invasion is stimulated by serum alone or serum supplemented with the chemotactic protein MCP-1) (Fig. 1F). Similarly, MT1-MMP−/− macrophages retain an invasive potential comparable with wild-type cells (Fig. 1F). The intact invasive potential of MT1-MMP-deficient macrophages stands in direct contrast to that observed with wild-type fibroblasts treated with GM6001 or fibroblasts recovered from Mmp14−/− mice, which display a complete loss of collagen-invasive activity until such time that MT1-MMP expression is rescued (Fig. 1F).
An unexpected role for MT1-MMP in regulating macrophage proinflammatory gene responses
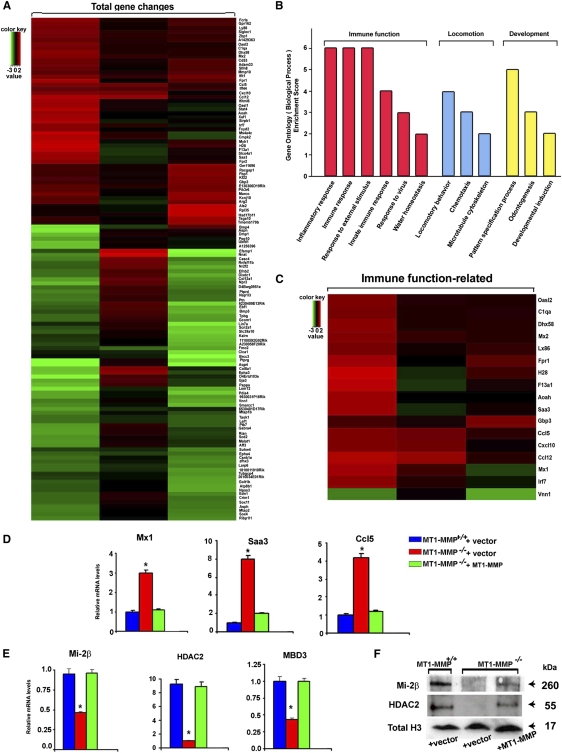
Given the fact that MT1-MMP−/− macrophages display normal motile and invasive responses in vitro, we sought to identify alternate functional roles for macrophage MT1-MMP in an unbiased fashion. Hence, MT1-MMP-null macrophages were transduced with either a control or MT1-MMP lentiviral expression vector, and mRNA recovered for transcriptional profiling. Using a minimum twofold cutoff and a P-value of 0.05, MT1-MMP rescue significantly alters the expression of 130 unique transcripts (Fig. 2A). Unexpectedly, gene ontology (GO) analysis indicates that MT1-MMP governs numerous cellular processes, including those impacting inflammation as well as viral and innate immune functions (Fig. 2B,C). Upon inspection of the profile of altered transcripts, MT1-MMP−/− macrophages display up-regulated levels of multiple inflammatory response regulator genes (e.g., Mx1, Mx2, Saa3, CXCL10, and CCL5) whose markedly enhanced expression is confirmed by quantitative PCR (qPCR) (Fig. 2D). As the expression of each of these proinflammatory mediators has been linked previously to the Mi-2/NuRD axis of ATP-dependent nucleosome remodeling complexes (Ramirez-Carrozzi et al. 2006; Lai et al. 2009), the expression status of a subset of these immunomodulatory genes was next assessed. Indeed, consistent with the fact that Mi-2/NuRD complex activity normally functions to suppress macrophage inflammatory gene responses (Ramirez-Carrozzi et al. 2006; Lai et al. 2009), the heightened proinflammatory response of MT1-MMP-null macrophages correlates with a significant down-regulation of multiple transcripts belonging to the Mi-2/NuRD complex, including Mi-2β, HDAC2, and MBD3 (Fig. 2E; Vignali et al. 2000). Although attempts to target expression of these Mi-2/NuRD components in wild-type macrophages were unsuccessful (data not shown), silencing either Mi-2β or HDAC2 in the J744.1 macrophage cell line similarly up-regulates expression of Mx1, Saa3, or CCL5 (Supplemental Fig. 2). Following transduction of MT1-MMP-null macrophages with an MT1-MMP lentiviral expression vector, transcript and protein levels of the Mi-2/NuRD complex family members return to wild-type levels in tandem with the normalization of Mx1, Saa3, and CCL5 expression (Fig. 2D–F).
Figure 2.
MT1-MMP-dependent control of macrophage immune function-related gene expression. (A–C) Microarray data for three biological replicates of MT1-MMP−/− macrophages transduced with either a lentiviral control or MT1-MMP expression vector. Heat maps of total genes affected (A), GO analysis (B), and genes specifically related to immune function (C) are presented. The key on the top left assigns heat map colors to the absolute gene expression value on a log2 scale. Green and red indicate lower and higher expression, respectively, in control transfected versus MT1-MMP-rescued MT1-MMP−/− macrophages. (D) Transcript expression of late immune response genes was analyzed by qPCR in MT1-MMP+/+ macrophages transduced with a lentiviral control vector or MT1-MMP−/− macrophages transduced with either a lentiviral control vector or full-length MT1-MMP. Results are expressed as the mean ± SEM; (*) P < 0.01; versus MT1-MMP+/+ + vector; n = 5. (E,F) Mi-2/NuRD complex expression at both transcript (E) and protein (F) levels was assessed by qPCR and Western blotting, respectively. MT1-MMP+/+ macrophages were transduced with a lentiviral control vector or MT1-MMP−/− macrophages were transduced with either a lentiviral control vector or full-length MT1-MMP. Results are expressed as the mean ± SEM; (*) P < 0.01; versus MT1-MMP+/+ + vector; n = 5.
Differential regulation of macrophage pro- and anti-inflammatory cytokine production by MT1-MMP
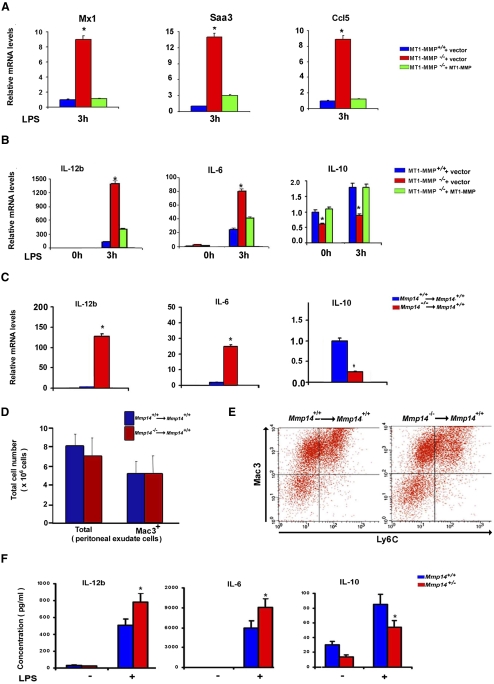
In addition to immune response regulator genes, Mi-2/NuRD complexes also control proinflammatory cytokine responses in macrophages (Weinmann et al. 2001; Ramirez-Carrozzi et al. 2006; Lai et al. 2009). Given the fact that Mmp14−/− mice display a morbid phenotype reminiscent of a chronic inflammatory response (Holmbeck et al. 1999), the immune responses and cytokine profiles of MT1-MMP wild-type and deficient macrophages were monitored in response to LPS challenge in vitro and in vivo. Remarkably, MT1-MMP-null macrophages display not only eightfold to 12-fold increases in Mx1, Saa3, and CCL5 expression relative to wild-type controls, but also exaggerated responses in IL-12b and IL-6 mRNA and protein expression levels (Fig. 3A,B; Supplemental Fig. 3). Following transduction of MT1-MMP-deficient macrophages with the lentiviral MT1-MMP expression vector, both the immune regulatory and cytokine responses to LPS stimulation are attenuated to levels similar to those observed in wild-type macrophages (Fig. 3A,B), confirming that MT1-MMP is responsible for controlling proinflammatory responses. As macrophages committed to a proinflammatory status frequently down-regulate their expression of anti-inflammatory cytokines (Fukao and Koyasu 2003; Martin et al. 2005; Rehani et al. 2009), profiling was extended to include IL-10. Indeed, as predicted, IL-10 expression is decreased significantly in MT1-MMP-null macrophages, but returns to wild-type baseline levels following MT1-MMP rescue (Fig. 3B). Silencing Mi-2β or HDAC2 expression in J744.1 cells likewise derepresses IL-12b and IL-6 expression while inhibiting IL-10 levels (Supplemental Fig. 2).
Figure 3.
MT1-MMP orchestrates Mi-2/NuRD complex-mediated regulation of macrophage pro- and anti-inflammatory cytokines. (A) MT1-MMP+/+ or MT1-MMP−/− macrophages lentivirally transduced with a control vector or full-length MT1-MMP were stimulated with 1 μg/mL LPS for 3 h and assessed for representative inflammatory mediator gene expression by qPCR. Results are expressed as the mean ± SEM; (*) P < 0.01; versus MT1-MMP+/+ vector; n = 5. (B) qPCR analysis of IL-12b, IL-6, and IL-10 mRNA levels in MT1-MMP+/+ macrophages transduced with a lentiviral control vector or MT1-MMP−/− macrophages transduced with either a lentiviral control vector or full-length MT1-MMP. Cells were either cultured alone or stimulated with 1 μg/mL LPS for 3 h. Values shown are the mean ± SEM; n = 5; (*) P < 0.01; versus MT1-MMP+/+ + vector. (C) qPCR analysis of IL-12b, IL-6, or IL-10 mRNA levels in thioglycollate-elicited peritoneal macrophages harvested from Mmp14+/+ or Mmp14−/− bone marrow-transplanted wild-type recipient mice. Results are expressed as the mean ± SEM; n = 3, with three animals in each group; (*) P < 0.01 versus Mmp14+/+ → Mmp14+/+. (D,E) Thioglycollate-elicited peritoneal exudate cells were harvested from MT1-MMP+/+ or MT1-MMP−/− bone marrow-transplanted recipient mice. Peritoneal exudate cells from bone marrow-transplanted recipient mice were analyzed for Mac3, and both total and Mac3-positive cell numbers were determined (D). Results are expressed as the mean ± SEM; n = 3, with three animals in each group. Peritoneal exudate cells were analyzed for Mac3 and Ly6C (E). Representative plots are shown; n = 3, with three animals in each group. (F) IL-12b, IL-6, or IL-10 protein levels were quantified in serum collected from either Mmp14+/+ or Mmp14+/− littermates before or 6 h after a single intraperitoneal injection of LPS (3 μg/kg). Results are expressed as the mean ± SEM; (*) P < 0.05 versus Mmp14+/+; n = 5 for Mmp14+/+ mice, and n = 7 for Mmp14+/− mice.
In vitro, macrophages are routinely differentiated from bone marrow-derived monocytes that have been cultured in the presence of M-CSF and fetal calf serum—conditions that may not recapitulate accurately those encountered in vivo. Hence, to assess the role of MT1-MMP in regulating macrophage function in the in vivo setting, thioglycollate-elicited peritoneal macrophages were recovered from wild-type recipient mice transplanted with MT1-MMP-null bone marrow. After a 6-wk recovery period, at which time peripheral blood monocyte levels are equivalent between the two groups of animals (data not shown), MT1-MMP−/− macrophages recovered from thioglycollate-challenged mice displayed an even more striking increase in IL-6 and IL-12b expression than that observed in vitro, coupled with decreased IL-10 expression relative to Mmp14+/+ marrow-transplanted mice (Fig. 3C). Furthermore, the proinflammatory phenotype of MT1-MMP-deficient macrophages is not associated with defective tissue recruitment, as equivalent numbers of elicited wild-type or MT1-MMP−/− macrophages (derived primarily from circulating blood monocytes) (Ghosn et al. 2010) are recovered from the peritoneal cavity (Fig. 3D). As monocytes and macrophages can be further segregated into activated M1 (Ly6C+) or alternatively activated cells (Ly6C−) (Geissmann et al. 2003), elicited macrophages recovered from Mmp14+/+ or Mmp14−/− bone marrow-transplanted mice were analyzed for Ly6C cell surface expression. Under these conditions, significant differences are not detected between the responses of Mmp14+/+ or Mmp14−/− transplanted mice (Fig. 3E). Finally, while the debilitated status of Mmp14−/− mice precludes attempts to monitor cytokine responses to LPS challenge in the in vivo setting (i.e., even saline-injected mice succumb to this mild stress) (data not shown), MT1-MMP hemizygous (Mmp14+/−) mice are viable, fertile, and phenotypically indistinguishable from wild-type animals (Filippov et al. 2005; Chun et al. 2010). Hence, Mmp14+/+ or Mmp14+/− mice were administered a single dose of LPS, and 6 h later, blood cytokine levels were determined. Despite only a partial reduction in MT1-MMP levels, hemizygous mice likewise display—relative to their littermate controls—significant increases in IL-12b and IL-6 blood levels, coupled with a muted IL-10 response following acute LPS challenge (Fig. 3F).
MT1-MMP regulates Mi-2/NuRD-dependent nucleosome remodeling
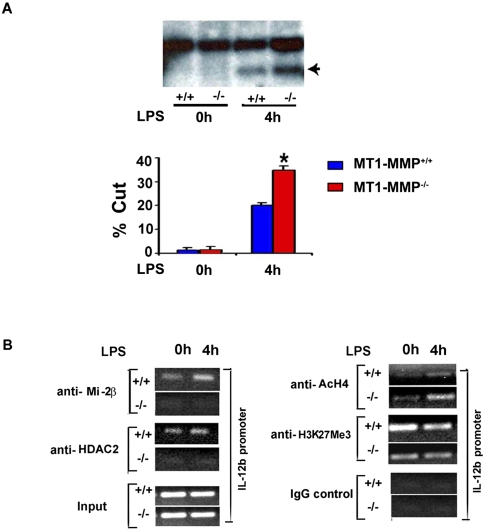
In primary macrophages, LPS activation triggers nucleosome remodeling at the IL-12b promoter region, an activity that can be assessed as a function of increased restriction enzyme accessibility at a single positioned nucleosome (Weinmann et al. 1999; Ramirez-Carrozzi et al. 2006; Zhou et al. 2010). Earlier studies have demonstrated that the exaggerated inflammatory gene responses observed following decreases in Mi-2/NuRD activity correlate with increased exposure of the IL-12b promoter as a consequence of altered nucleosome remodeling (Weinmann et al. 1999; Ramirez-Carrozzi et al. 2006). As shown in Figure 4A, in the absence of LPS, the IL-12b promoter regions of both wild-type and MT1-MMP-null macrophages proved insensitive to the restriction endonuclease SpeI, confirming nucleosome structure-dependent silencing of the region. Following LPS stimulation, however, SpeI access to the IL-12b locus in MT1-MMP−/− macrophages is increased significantly relative to the wild-type control (Fig. 4A). Furthermore, consistent with defects in recruitment of Mi-2/NuRD components to the IL-12b promoter, chromatin immunoprecipitation (ChIP) analysis demonstrates that binding of Mi-2β or HDAC2 to the IL-12b promoter can only be detected in wild-type macrophages (Fig. 4B). As a consequence of the marked loss in HDAC2 recruitment observed in MT1-MMP-null macrophages, total acetylated histone 4 (AcH4) levels are also increased at the IL-12b promoter in these cells (Fig. 4B). Interestingly, the increased AcH4 content at the IL-12b promoter of MT1-MMP-null macrophages also correlates with a decrease in the repressive H3K27Me3 chromatin mark at this locus (Fig. 4B). Taken together, the exaggerated IL-12b induction observed in MT1-MMP−/− macrophages is consistent with the decreased expression of Mi-2/NuRD complex components that consequently allow for increased nucleosome remodeling and histone acetylation at the cytokine promoter.
Figure 4.
Characterization of IL12b promoter regions of MT1-MMP+/+ versus MT1-MMP−/− macrophages. (A) Restriction enzyme accessibility at the IL-12b promoter region was assessed in MT1-MMP+/+ or MT1-MMP−/− macrophages. Cells were either cultured alone or stimulated with 1 μg/mL LPS for 4 h. A representative image of Southern blotting is shown with the percentage of DNA cleavage by SpeI calculated as described in the Materials and Methods. Results are expressed as the mean ± SEM; (*) P < 0.01; versus MT1-MMP+/+; n = 3. (B) ChIP assay in MT1-MMP+/+ or MT1-MMP−/− macrophages. NuRD complex (left column) and histone modification marker expression (right column) associated with the IL12b promoter region were assessed. Results are shown as representative images of semiquantitative PCR.
MT1-MMP-dependent, noncatalytic regulation of macrophage immune function via the transcriptome-centric control of PI3Kδ expression
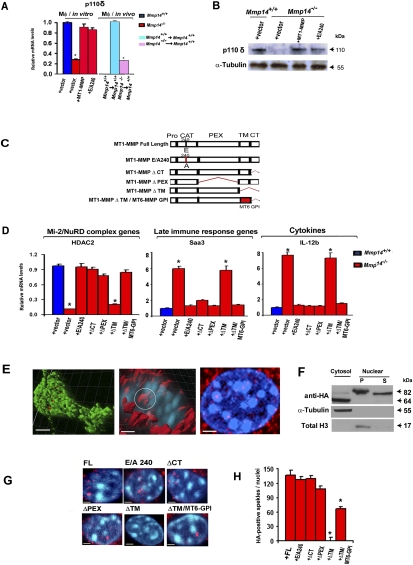
The orchestrated control of pro- and anti-inflammatory responses in myeloid cells is normally dependent on the induction of a PI3Kδ-Akt signaling network that acts to suppress GSK3 activity (Fukao and Koyasu 2003; Martin et al. 2005; Rommel et al. 2007; Papakonstanti et al. 2008; Rehani et al. 2009). As such, we considered the possibility that MT1-MMP−/− macrophages harbor additional, upstream defects in PI3Kδ expression that consequently contribute to their hyperactive, proinflammatory status. Indeed, expression of p110δ mRNA (the catalytic subunit of PI3Kδ) and PI3Kδ protein levels are decreased markedly in MT1-MMP-null macrophages differentiated in vitro as well as those differentiated in vivo from bone marrow-transplanted mice (Fig. 5A,B).
Figure 5.
Nuclear localization of membrane-tethered MT1-MMP regulates macrophage cytokine response by modulating p110δ expression. (A) p110δ mRNA levels were assessed by qPCR in MT1-MMP+/+ bone marrow-derived macrophages transduced with a lentiviral control vector or MT1-MMP−/− macrophages transduced with either a lentiviral control vector, full-length MT1-MMP, or the E/A240 mutant. Alternatively, p110δ mRNA levels were quantified in thioglycollate-elicited peritoneal macrophages harvested from Mmp14+/+ or Mmp14−/− bone marrow-transplanted recipient wild-type mice. Values are expressed as the mean ± SEM; (*) P < 0.01 test versus Mmp14+/+ + vector; (*) P < 0.01 test versus Mmp14+/+ → Mmp14+/+, with n = 5 for in vitro differentiated macrophages, and n = 3 for in vivo differentiated macrophages. (B) Western blot analysis of p110δ in Mmp14+/+ and Mmp14−/− bone marrow-derived macrophages lentivirally transduced with a control vector, full-length MT1-MMP, or the E/A240 mutant. (C) A schematic diagram of full-length MT1-MMP (FL) and its deletion mutants and chimeras. (^) Deleted domains. (D) HDAC2, Saa3, and IL-12b mRNA levels were assessed by qPCR in Mmp14+/+ bone marrow-derived macrophages transduced with a lentiviral control vector, or Mmp14−/− macrophages transduced with either a lentiviral control vector, full-length MT1-MMP, the E/A240 mutant, or domain deletion mutants. Cells were either cultured without LPS for HDAC2 determinations or stimulated with 1 μg/mL LPS for 4 h for Sac3 and IL-12b mRNA levels. Values shown are the mean ± SEM; n = 5; (*) P < 0.01; versus Mmp14+/+ + vector. (E) Nuclear localization of HA-tagged MT1-MMP lentivirally expressed in MT1-MMP+/+ macrophages. Ubiquitous HA staining on the cell surface is colored green (bar, 5 μm), and intracellular HA staining of MT1-MMP is colored red. Representative confocal photomicrographs of MT1-MMP localization are shown as 3D reconstructions using IMARIS software. In the middle panel, a white circle marks MT1-MMP localized within the nucleus (DAPI; blue) (bar, 2 μm). The third panel from the left is a representative Z-section of MT1-MMP-positive intranuclear speckles (bar, 1 μm). (F) Macrophages were fractionated, and HA-tagged MT1-MMP expression in cytosolic (α-tubulin-positive), nuclear-soluble (Mi-2β-positive) (data not shown), or nuclear pellet (total histone 3-positive) compartments was assessed by Western blot analysis with anti-HA antibody. (G) Representative confocal images of intranuclear MT1-MMP mutant localization shown as Z-slice sections. Wild-type macrophages (or MT1-MMP-null) (data not shown) were transduced with epitope-tagged MT1-MMP mutants and stained for MT1-MMP with either anti-HA or anti-His antibody (red). Nuclei are stained with DAPI (blue). Bar, 2 μm. (H) Representative quantitation of MT1-MMP-positive speckles in macrophages transduced with either wild-type or mutant MT1-MMP constructs. Speckle number was quantified in three randomly selected nuclei from a single representative experiment. Values shown are the mean ± SEM; (*) P < 0.05; versus wild-type MT1-MMP-transduced macrophages.
To determine whether MT1-MMP-dependent proteolysis of heretofore unrecognized substrates plays a role in controlling macrophage PI3Kδ expression, wild-type cells were cultured in the presence of GM6001. Unexpectedly, GM6001 did not affect p110δ expression or the cytokine profiles of LPS-stimulated, wild-type macrophages (Supplemental Fig. 4). As these results raise the possibility that MT1-MMP regulates macrophage PI3Kδ expression independently of its proteolytic activity, MT1-MMP-null macrophages were transduced with either wild-type MT1-MMP or a proteolytically inactive form of MT1-MMP harboring an E/A240 point mutation within its catalytic domain (Fig. 5C; Li et al. 2008; Sabeh et al. 2009a). Following expression of either the wild-type or mutant MT1-MMP constructs in null macrophages, p110δ expression increases to wild-type levels (Fig. 5A,B). In tandem fashion, the expression of Mi-2/NuRD complex family members, immune response genes, and cytokines in MT1-MMP−/− macrophages is normalized (Fig. 5D). As MT1-MMP has been proposed to trigger signal transduction cascades by interacting with partner molecules that bind to either the MT1-MMP C-terminal cytosolic tail or the extracellular hemopexin domain (Cao et al. 2004; Matias-Roman et al. 2005; Sithu et al. 2007; Sakamoto and Seiki 2009; Gonzalo et al. 2010), MT1-MMP-null macrophages were next transduced with MT1-MMP mutants wherein either the cytosolic tail or hemopexin domain was deleted (ΔCT or ΔPEX, respectively) (Fig. 5C; Li et al. 2008), and the expression of HDAC2, Saa3, or IL-12b was monitored. Inconsistent with previously described structure/function models that require the participation of the MT1-MMP tail or hemopexin domains, however, expression of either the ΔCT or ΔPEX MT1-MMP mutants effectively rescues the altered NuRD complex, late immune response, and cytokine gene expression profiles of MT1-MMP−/− macrophages (Fig. 5D).
Unlike most MMP family members, MT1-MMP is tethered to the cell surface by a type I transmembrane domain (Barbolina and Stack 2008; Rowe and Weiss 2009). To determine whether the immune regulatory function of MT1-MMP requires its anchorage to the cell surface, MT1-MMP-null macrophages were transduced with a catalytically active but transmembrane-deleted form of MT1-MMP (i.e., ΔTM) (Fig. 5C). Unlike the other MT1-MMP mutants tested, the secreted form of MT1-MMP is unable to rescue the expression of Mi-2/NuRD complex members or the exaggerated expression of immune-response genes or cytokines (Fig. 5D). To rule out the possibility that the MT1-MMP transmembrane domain itself contains embedded motifs that modulate transcriptional activity, MT1-MMP-null macrophages were transduced with a MT1-MMP construct that anchors MT1-MMP to the cell surface via the GPI anchor of MT6-MMP, a more distant member of the MT-MMP family (ΔTM MT1-MMP/MT6·GPI) (Fig. 5C; Li et al. 2008). Even under these conditions, however, the GPI-anchored MT1-MMP chimera maintains the ability to rescue the gene expression profile of MT1-MMP-null macrophages (Fig. 5D). Hence, the hemopexin-free, extracellular domain of membrane-tethered MT1-MMP maintains all of the transcriptional regulatory elements critical to the immune regulatory pathways operative in wild-type macrophages.
A potential link between MT1-MMP nuclear trafficking and PI3Kδ expression
To determine whether MT1-MMP regulates macrophage function in a cell-intrinsic or -extrinsic fashion, wild-type and MT1-MMP−/− macrophages were cocultured in Transwell dishes. Under these conditions, however, neither the hyperimmune responses of the MT1-MMP−/− macrophages nor the normal responses of wild-type macrophages are affected (data not shown). In considering alternate mechanisms by which catalytically inactive MT1-MMP might impact macrophage function, recent studies demonstrate that cell surface-associated macromolecules, ranging from receptor tyrosine kinases to cell adhesion molecules, can traffic to the nuclear compartment, where they regulate transcriptional programs (Lin et al. 2001; Hieda et al. 2008; Huo et al. 2010; Wang et al. 2010a). Indeed, when assessed by confocal microscopy, HA-tagged MT1-MMP is detected in a speckle-like distribution within the macrophage nuclear compartment that is similar to that reported for nuclear-associated receptor tyrosine kinases in other cell types (Fig. 5E; Huo et al. 2010; Lo et al. 2010; Sehat et al. 2010). Consistent with these results, following a high-salt extraction of isolated macrophage nuclei (Sehat et al. 2010), MT1-MMP is detected in both the nuclear pellet (containing proteins tightly bound to DNA and insoluble nuclear membrane proteins) and the membrane-free nucleoplasm fraction (Fig. 5F). Trafficking to the nuclear compartment (as assessed by confocal laser microscopy as well as quantitative analysis of MT1-MMP-positive nuclear speckles) is similarly preserved when either wild-type or MT1-MMP-null macrophages are transduced with catalytically inactive MT1-MMP, MT1-MMPΔCT, MT1-MMPΔPEX, or GPI-anchored MT1-MMP (Fig. 5G,H). In contrast, the transmembrane-deleted, secreted form of MT1-MMP that is incapable of restoring macrophage immune responses does not traffic to the nucleus (Fig. 5G,H).
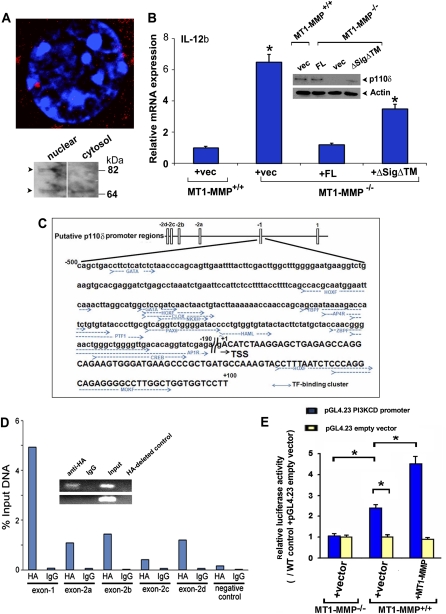
The correlation between MT1-MMP nuclear localization and the rescue of macrophage immune function raises the possibility that MT1-MMP impacts transcriptional programs independently of its normal trafficking to the cell surface. To test this hypothesis, MT1-MMP−/− macrophages were transduced with an MT1-MMP mutant wherein the signal peptide, transmembrane domain, and tail were deleted (i.e., ΔSigΔTM), hence redirecting the proteinase to the cytosolic compartment. Under these conditions, low-level, cytosolic expression of ΔSigΔTM was confirmed by Western blotting, while high and low Mr forms of MT1-MMP (similar to those detected in MT1-MMP−/− macrophages transduced with full-length MT1-MMP) (Fig. 5F) are found in the nuclear compartment (Fig. 6A). Significantly, ΔSigΔTM not only retains nuclear trafficking activity, but also partially restores PI3Kδ expression and IL-12b regulation (Fig. 6B). Given these findings, coupled with a recent precedent wherein a secreted MMP family member displays transcription factor-like activity (Eguchi et al. 2008), we considered the possibility that MT1-MMP might directly regulate PI3Kδ expression. To this end, potential interactions between MT1-MMP and the p110δ promoter were interrogated by quantitative ChIP. Following transduction of wild-type or MT1-MMP-null macrophages with an HA-tagged MT1-MMP expression vector, specific binding interactions between MT1-MMP and the five conserved promoter elements previously identified in PI3KCD were explored (Supplemental Table I; Kok et al. 2009). Under these conditions, primers specific for exon −1 of PI3KCD predominately amplify DNA–protein complexes immunoprecipitated from nuclear lysates with a monospecific anti-HA antibody (Fig. 6C,D). Given that (1) primer controls designed 5 kb downstream from the target region did not amplify the immunoprecipitated DNA–protein complex and (2) no complex was immunoprecipitated when cells were transduced with a control expression vector that did not carry an HA insert (Fig. 6D), these data support a specific binding interaction between nuclear MT1-MMP and the p110δ promoter. To determine whether exon −1/MT1-MMP interactions are sufficient to drive promoter activity, a minimal 0.6-kb fragment of exon −1 was inserted into a PGL4.23 reporter vector and transduced into either MT1-MMP-null, wild-type, or MT1-MMP-transduced wild-type macrophages. As shown in Figure 6E, the reporter activity of this minimal construct can be increased almost fivefold in an MT1-MMP-specific fashion.
Figure 6.
MT1-MMP nuclear routing and interactions with the p110δ promoter. (A) Nuclear localization of ΔSigΔTM in transduced MT1-MMP−/− macrophages as assessed in confocal laser, Z-section micrographs or by Western blot analyses of the macrophage cytosolic and nuclear fractions. (B) IL-12b mRNA expression was determined in control vector transduced MT1-MMP+/+ macrophages or MT1-MMP−/− macrophages transduced with a control vector, full-length MT1-MMP (FL) or ΔSigΔTM following a 4-h incubation with LPS. In tandem, macrophage lysates were prepared from each sample, and p110δ protein levels were assessed by Western blot analysis. (C) Schematic representation of the p110δ promoter regions and the exon −1 region. Transcription factor (TF)-binding clusters are indicated in blue (Kok et al. 2009). (D) Wild-type or MT1-MMP-null macrophages (data not shown) were transduced with a control vector or HA-tagged MT1-MMP, and ChIP of PCR was performed with anti-HA antibody or control IgG. Results are presented as the mean of two experiments. (Inset) A representative image of semiquantitative PCR for the p110δ promoter (exon −1 region; top row) and negative control primers (+5 kb coding region primer; bottom row). Primers specific for the p110δ promoter were designed to amplify −358 to −19 of exon −1 (Kok et al. 2009). (E) Relative luciferase activity of the pGL4.23 and the pGL4.23 PI3KCD promoter in MT1-MMP−/− macrophages, MT1-MMP+/+ macrophages transduced with a control expression vector, or an MT1-MMP expression vector. Results are expressed as the mean ± SEM of five experiments; (*) P < 0.01; versus MT1-MMP+/+ + pGL4.23 empty vector + pcDNA 3.1 control vector.
MT1-MMP modulates macrophage immune function via the PI3Kδ/Akt/GSK3β-dependent control of Mi-2/NuRD activity
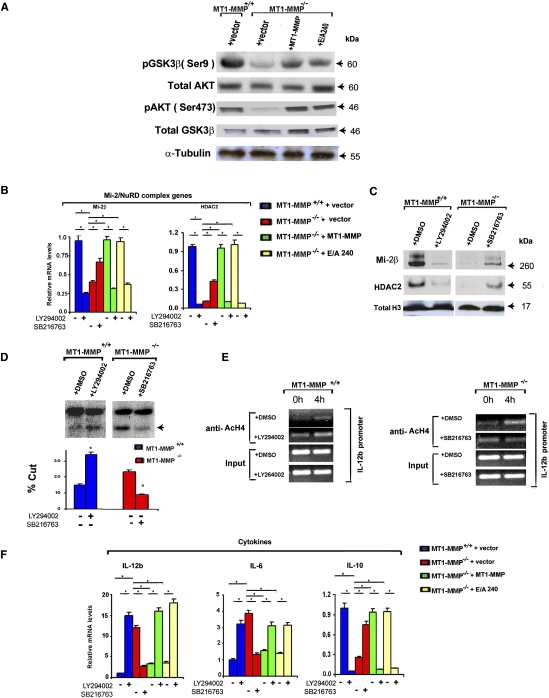
PI3Kδ activity controls macrophage inflammatory responses by triggering the Akt-dependent inactivation of GSK3β activity [i.e., via phosphorylation at Ser 9; pGSK3β(Ser 9)] (Monick et al. 2001; Guha and Mackman 2002; Martin et al. 2005; Beurel et al. 2010). As predicted, and in tandem with the decreased PI3Kδ levels detected in MT1-MMP-null macrophages, Akt(Ser 473) phosphorylation is likewise depressed in association with the near undetectable levels of GSK3β(Ser 9) relative to wild-type macrophages (Fig. 7A). Taken together, the results support a potential linkage between MT1-MMP expression, PI3Kδ activity, and the Mi-2/NuRD complex-dependent regulation of the macrophage inflammatory gene network. Hence, to assess the functional importance of MT1-MMP-induced increases in PI3Kδ activity on the dampening of the proinflammatory phenotype in null macrophages, MT1-MMP-null cells were transduced with either wild-type MT1-MMP or the E/A240 mutant in the absence or presence of the PI3K inhibitor LY294002 (Martin et al. 2005; Papakonstanti et al. 2008). Under these conditions, wild-type macrophages treated with LY294002 phenocopy the proinflammatory status of MT1-MMP−/− macrophages with (1) decreased expression of Mi-2/NuRD complex components at the mRNA and protein levels (Fig. 7B,C), (2) increased nucleosome remodeling at the IL-12b promoter with enhanced levels of AcH4 chromatin marks at the cytokine promoter (Fig. 7D,E), and (3) up-regulated IL-12b and IL-6 expression, coupled with decreases in IL-10 levels and increased late immune response gene expression (Fig. 7F; Supplemental Fig. 4). In similar fashion, the ability of wild-type MT1-MMP or MT1-MMP E/A240 to suppress the proinflammatory phenotype of MT1-MMP-deficient macrophages is blocked completely following PI3Kδ inhibition with either LY294002 (Fig. 7B–F) or the PI3Kδ-specific inhibitor IC877114 (Supplemental Fig. 4; Sadhu et al. 2003). Finally, when the unregulated GSK3β activity detected in MT1-MMP-null cells is targeted with the synthetic GSK3 inhibitor SB216763 (Martin et al. 2005), Mi-2/NuRD complex gene expression increases in tandem with decreased nucleosome remodeling, decreased histone acetylation at the IL-12b promoter, and the return of cytokine and late immune response gene expression to the wild-type baseline (Fig. 7B–F; Supplemental Fig. 3). Hence, MT1-MMP exerts protease-independent control over macrophage immune regulatory responses through its ability to modulate PI3Kδ expression and the subsequent Akt/GSK3β-dependent modulation of the Mi-2/NuRD nucleosome remodeling complex (Supplemental Fig. 5).
Figure 7.
MT1-MMP exerts catalytic activity-independent control over the PI3K/Akt/GSK3β·Mi-2/NuRD network. (A) Western blot analysis of pAKT(Ser 473), total GSK, and pGSK3β(Ser 9) in MT1-MMP+/+ and MT1-MMP−/− macrophages lentivirally transduced with a control vector, full-length MT1-MMP, or the E/A240 mutant. (B,C) MT1-MMP+/+ and MT1-MMP−/− macrophages lentivirally transduced with a control vector, full-length MT1-MMP, or the E/A240 mutant were incubated with or without 20 μM LY294002 or 10 μM SB216763 and assessed for Mi-2β or HDAC2 expression by qPCR (B) or for protein level by Western blotting (C). Results are expressed as the mean ± SEM; (*) P < 0.01. (D) Restriction enzyme accessibility at the IL-12b promoter region. MT1-MMP+/+ and MT1-MMP−/− macrophages were incubated with or without 20 μM LY294002 or 10 μM SB216763, respectively, for 1 h and stimulated with 1 μg/mL LPS for 4 h. A representative image of Southern blotting is shown along with the percent DNA cleavage by SpeI. Results are expressed as the mean ± SEM; (*) P < 0.01; versus MT1-MMP+/+; n = 3. (E) ChIP assay with MT1-MMP+/+ and MT1-MMP−/− macrophages incubated with or without 20 μM LY294002 or 10 μM SB216763, respectively. AcH4 histone marks associated with the IL-12b promoter region in MT1-MMP+/+ (left column) and MT1-MMP−/− (right column) macrophages were assessed. Results are shown as representative images of semiquantitative PCR. (F) MT1-MMP+/+ or MT1-MMP−/− macrophages where lentivirally transduced with a control vector, full-length MT1-MMP, or the E/A240 mutant; incubated with or without 20 μM LY294002 or 10 μM SB216763; and assessed for cytokine expression by qPCR. LPS (1 μg/mL) was added to cell cultures 2 h prior to mRNA collection. Results are expressed as the mean ± SEM; (*) P < 0.01; n = 5.
Discussion
The inception and propagation of chronic inflammatory responses are marked by increased monocyte and macrophage trafficking to affected tissues (Mosser and Edwards 2008; Grivennikov et al. 2010; Nathan and Ding 2010). Armed with an array of proteolytic enzymes, monocytes and macrophages have been assumed to use these effectors as the dominant means by which to affect both tissue-invasive and ECM remodeling phenotypes. Indeed, in nonmyeloid cells such as fibroblasts, endothelial cells, smooth muscle cells, mesenchymal stem/stromal cells, or adipocytes, MT1-MMP predominately acts as a pericellular collagenase that confers cells with the ability to infiltrate and degrade type I collagen-rich tissues (Hotary et al. 2003; Chun et al. 2004; Sabeh et al. 2004, 2009a,b; Filippov et al. 2005; Rowe and Weiss 2009; Lu et al. 2010). Thus, by extension, MT1-MMP has been assumed to play similar roles in myeloid cell populations (Sakamoto and Seiki 2009; Cougoule et al. 2010; Van Goethem et al. 2010). In our study, however, MT1-MMP does not play a required role in supporting macrophage motility, chemotaxis, or collagen-invasive activity in vitro. Moreover, in the in vivo setting, we found that MT1-MMP-deficient macrophages infiltrate host tissues comparably with wild-type cells, a finding consistent with earlier reports that have monitored the normal trafficking of MT1-MMP-null macrophages into inflammatory sites in bone marrow-transplanted mice (Schneider et al. 2008; Xiong et al. 2009). While Seiki and colleagues (Sakamoto and Seiki 2009) have reported on the impaired trafficking of MT1-MMP-null macrophages in vivo, these studies are complicated by the fact that they were performed in Mmp14−/− mice that suffer from multiple—and ultimately fatal—organ defects affecting bone, vascular, skeletal muscle, and adipose tissue functions (Holmbeck et al. 1999; Zhou et al. 2000; Chun et al. 2004, 2006; Ohtake et al. 2006). The fact that MT1-MMP does not play a major role in the tissue-invasive activity in macrophages should not be misconstrued to suggest that the proteinase is unable to participate in other ECM remodeling events. MT1-MMP readily hydrolyzes a wide variety of ECM components ranging from fibronectin and laminin to surface proteoglycans (Barbolina and Stack 2008; Butler et al. 2008; Rowe and Weiss 2009). Nevertheless, while MT1-MMP serves as the dominant pericellular collagenase in a wide range of epithelial or mesenchymal cell populations, macrophages appear to direct the proteinase to alternate purpose.
MT1-MMP-dependent immune regulation
Transcriptional profiling of MT1-MMP-null macrophages revealed an unexpected role for the proteinase in modulating immune responses, affecting multiple proinflammatory gene products that fall under the regulation of both the Mi-2/NuRD nucleosome remodeling complex (Weinmann et al. 2001; Ramirez-Carrozzi et al. 2006; Lai et al. 2009) and the PI3Kδ/Akt/GSK3 axis (Martin et al. 2005; Rehani et al. 2009; Beurel et al. 2010). The molecular mechanisms that inversely link GSK3 activity to the expression of chromatin remodeling complexes require further study (Foster et al. 2006; Trivedi et al. 2007; Zhou et al. 2010), although further defining this pathway will likely prove daunting given the multiplicity of GSK3 substrates (Beurel et al. 2010). Nevertheless, the central role played by increased GSK3 activity in MT1-MMP-null macrophage function is underlined by the complete rescue of immune responses observed following GSK3 inhibition. That is, despite the fact that the expression patterns of >100 transcripts are altered in MT1-MMP-deficient macrophages, inhibiting endogenous GSK3 activity alone normalized expression levels of Mi-2β, HDAC2, and MBD3 in tandem with the correction of both proinflammatory and anti-inflammatory responses to the wild-type baseline. While it is tempting to assign all immune-related effects of the MT1-MMP/PI3Kδ axis to changes in Mi-2/NuRD activity, TNFα, an early immediate response gene whose expression is controlled independently of chromatin remodeling complexes (Ramirez-Carrozzi et al. 2006; Lai et al. 2009; Zhou et al. 2010), is also misregulated in MT1-MMP−/− macrophages via a process that is partially dependent on PI3Kδ and GSK3 activities (Supplemental Fig. 5). As PI3Kδ can exert direct control over TNFα expression as well as secretion (Martin et al. 2005; Low et al. 2010), MT1-MMP likely regulates macrophage immune responses through PI3Kδ pathways that operate both dependently and independently of the Mi-2/NuRD complex.
Are nuclear MT1-MMP trafficking and PI3Kδ expression linked?
In considering the mechanisms by which MT1-MMP regulates PI3Kδ expression, our interest focused initially on the ability of the protease to hydrolyze an expanding number of potential substrates that can be linked to the inflammatory response (e.g., chemokines, growth factors, cytokines, and complement components) (Barbolina and Stack 2008; Butler et al. 2008). In an unpredicted manner, however, defects in PI3Kδ/Akt/GSK3 activity, Mi-2/NuRD expression, and proinflammatory gene regulation observed in MT1-MMP-null macrophages were all rescued by re-expressing MT1-MMP in a catalytically inactive form. Recent studies have suggested that MT1-MMP can regulate cell function via protease-independent pathways that impact various signal transduction cascades as well as energy metabolism, but in each of these systems, the MT1-MMP cytosolic tail played a required role in mediating these effects (D'Alessio et al. 2008; Mori et al. 2009; Sakamoto and Seiki 2009, 2010; Gonzalo et al. 2010; Proulx-Bonneau et al. 2011)—a structural requirement distinct from that observed in our study, where neither the MT1-MMP cytosolic tail nor transmembrane domains are required for restoring macrophage immune responses. To our knowledge, the notion that proteolytically inactive MT1-MMP controls cell function at the level of gene regulation has not been considered previously, nor has MT1-MMP deficiency been heretofore associated with enhanced functional responses.
Confronted with these results and a lack of precedent regarding mechanisms by which catalytically inactive MT1-MMP might affect gene expression, we noted that an expanding list of transmembrane receptors and growth factor and adhesion molecules, ranging from EGF and IGF receptors to CD44 and heparin-binding EGF, traffic to the nuclear compartment, where they activate transcriptional programs (Liao and Carpenter 2007; Hieda et al. 2008; Dunham-Ems et al. 2009; Lee et al. 2009; Sehat et al. 2010; Wang et al. 2010a). Unlike MT1-MMP, all other membrane-anchored molecules identified to date gain access to the nuclear compartment via trafficking signals encoded within either their cytosolic tail or transmembrane domains (Sehat et al. 2010; Wang et al. 2010a). The nuclear trafficking routes used by MT1-MMP, where neither of these domains are required, remain undefined, and further studies will be needed to determine whether MT1-MMP associates with other partners that serve as cotransporters (e.g., CD44 or pericentrin) (Golubkov et al. 2005; Sillibourne et al. 2007; Barbolina and Stack 2008; Delaval and Doxsey 2010). Regardless of the particular path taken, our finding that a cytosol-directed MT1-MMP mutant can also access the nuclear compartment indicates that cell surface localization per se is not a required step in this process. Interestingly, these findings complement a series of reports documenting the nuclear localization of several secreted MMP family members, including a single report describing nuclear trafficking of MT1-MMP in hepatocellular carcinomas (Kwan et al. 2004; Limb et al. 2005; Si-Tayeb et al. 2006; Eguchi et al. 2008; Yang et al. 2010). To date, however, the functional significance of nuclear MT1-MMP in cancer cells has not been explored, while nuclear-associated functions of secreted MMPs have primarily focused on the induction of apoptosis-related pathways that require the catalytically active form of the enzymes (Kwan et al. 2004; Limb et al. 2005; Si-Tayeb et al. 2006; Eguchi et al. 2008; Yang et al. 2010). Furthermore, none of the nuclear function-associated activities assigned to the secreted MMPs have been confirmed in widely available knockout mouse models.
In considering potential mechanisms by which nuclear MT1-MP might control PI3Kδ-dependent immunomodulatory pathways in macrophages, our attention was drawn to recent studies suggesting that the secreted MMP stromelysin-1 can localize to the nuclear compartment, where it displays transcription factor-like activity by controlling CTGF expression (Eguchi et al. 2008). While the organization of the PI3Kδ promoter is complex, we found that intranuclear MT1-MMP likewise associates with, and activates, a highly conserved transcription factor-binding cluster within exon −1 of mouse PI3KCD, the most proximal (i.e., 11 kb upstream of the start site) of the five untranslated exons identified up to 81 kb upstream of the translational start codon in exon 1 (Kok et al. 2009). The identification of full-length MT1-MMP in the nucleoplasm is similar to that described for other nuclear trafficking transmembrane proteins where the lipophilic membrane-spanning domain has been released intact from the plasma membrane (Liao and Carpenter 2007; Low et al. 2010; Sehat et al. 2010; Wang et al. 2010a,b). The extrusion of a transmembrane protein into the nucleoplasm is an area of active research for all nuclear trafficking transmembrane proteins, with recent studies supporting a translocon Sec61-dependent process (Liao and Carpenter 2007; Wang et al. 2010b). Further efforts are required to determine the processes responsible for MT1-MMP membrane translocation as well as the identity of the MT1-MMP domains involved in direct or indirect PI3KCD-binding interactions that likely require accessory DNA-binding partners similar to those described recently for nucleus-directed transmembrane receptors (e.g., Huo et al. 2010). Independent of the final mechanisms, it is doubtful that MT1-MMP exerts its myriad effects on macrophage function by acting solely as a transactivator of PI3Kδ expression. Nevertheless, it seems similarly unlikely that the ability of MT1-MMP to traffic to the nuclear compartment, bind to PI3Kδ promoter elements, and drive expression of a minimal PI3Kδ promoter construct all reflect events unrelated to its immunoregulatory activity. Finally, it is noteworthy that the catalytic subunit of PI3Kδ has also been identified as one of a cohort of MT1-MMP-regulated target genes in cancer cells (Rozanov et al. 2008). While the mechanism of action and functional impact of MT1-MMP-dependent PI3Kδ activation in cancer cells remain to be examined, the ability of the proteinase to regulate PI3Kδ expression in normal as well as neoplastic cells suggests that the enzyme exerts more complex effects on cell function than entertained previously.
MT1-MMP: a reciprocating regulator of ECM remodeling and immune function
Given the impact of MT1-MMP on macrophage immune function, the question arises as to the pathophysiologically relevant processes that regulate MT1-MMP expression and trafficking in vivo. Under basal conditions, macrophage MT1-MMP expression is maintained at low levels (Shankavaram et al. 2001), thus allowing for maximal expression of a wide range of gene products associated with the immune response. As inflammatory signals wane and macrophages transition from a host defensive stance to one dedicated to ECM remodeling and resolution, increased levels of MT1-MMP would not only support extracellular proteolytic events critical for tissue remodeling, but also trigger the PI3Kδ-dependent down-regulation of proinflammatory gene responses in tandem with the up-regulation of anti-inflammatory cytokines. Whether MT1-MMP trafficking itself is under constitutive or regulated expression is currently unknown. Indeed, many of the other membrane-anchored molecules that are known to traffic to the nucleus do so under the control of constitutive processes that remain unexplored (Wang et al. 2010a). Nonetheless, our data indicate that MT1-MMP is poised to serve as a novel and heretofore unrecognized epigenetic regulatory element that controls an array of inflammatory responses by modulating PI3Kδ signaling as well as downstream chromatin remodeling complexes. Given these findings, previously documented defects in MT1-MMP−/− macrophages, lymphocytes, dendritic cells, and hematopoietic stem cell function (Sithu et al. 2007; Schneider et al. 2008; Vagima et al. 2009; Xiong et al. 2009; Gawden-Bone et al. 2010) may well require re-evaluation within the context of previously unappreciated changes in MT1-MMP-dependent gene regulation.
Materials and methods
Isolation and culture of mouse primary cells
Bone marrow macrophages were prepared as described using cells isolated from 2- to 4-wk-old male wild-type (Mmp14+/+) or MT1-MMP-null (Mmp14−/−) Swiss Black mice (Holmbeck et al. 1999; Sakamoto and Seiki 2009). Briefly, bone marrow cells were incubated for 24 h in α-MEM supplemented with 10% heat-inactivated fetal bovine serum (HI-FBS), 1% penicillin-streptomycin solution (Invitrogen), and 10 ng/mL M-CSF (R&D Systems). Nonadherent cells were collected and incubated in medium with M-CSF for 7 d in nontissue culture-treated plastic dishes (Hikita et al. 2006). Mouse dermal fibroblasts were isolated as described (Sabeh et al. 2004).
Fluorescent matrix degradation assays
Fluorescently labeled films of gelatin were generated with AlexaFluor derivatives (Molecular Probes) using the manufacturer's protocol as described (Sabeh et al. 2004; Li et al. 2008). Chamber coverglass slips (Corning) were pretreated with 50 μg/mL poly-L-lysine and 0.5% gluteraldehyde and coated with labeled matrix, followed by quenching with sodium borohydride. Bone marrow-derived macrophages (1 × 105) were plated on fluorescently labeled matrix films and cultured in heat-inactivated FBS-supplemented medium in the absence or presence of 10 ng/mL TNF-α (R&D systems). At the end of the culture period, samples were fixed and stained with antibodies directed against F4/80 (Abcam) or cortactin (Abcam).
Migration assays
In vitro chemotaxis assays were performed using Transwell dishes (5-μm pore size; Corning) (Kellermann et al. 1999). Macrophages (1 × 106 in 0.1 mL) were placed in the upper well of Transwell inserts with the filter surfaces left untreated or coated with gelatin or fibronectin (Sigma). The lower well contained culture medium (0.6 mL) supplemented with MCP-1 (R&D Systems). Cells that had migrated to the lower surface of the filters were harvested by incubation in EDTA/PBS and counted.
3D invasion assay
3D collagen gels were formed in 3-μm-pore Transwell inserts at a final concentration of 2.2 mg/mL type I collagen as described (Sabeh et al. 2009a). Wild-type or MT1-MMP−/− macrophages (2 × 105) in medium supplemented with 10% HI-FBS were cultured atop collagen gels with 100 nM MCP-1 added to the lower compartment of the Transwell chambers with or without 10 μM GM6001. The number of invading cells per high-powered field (hpf) was quantified by phase-contrast microscopy (Sabeh et al. 2009a).
Bone marrow transplantation and the isolation of peritoneal macrophages
Five-week-old wild-type mice were irradiated (1200 rads) and transplanted with bone marrow from littermate Mmp14−/− or Mmp14+/+ mice as described (Xiong et al. 2009). Bone marrow cell suspensions were prepared from the femurs of Mmp14−/− and littermate Mmp14+/+ controls, and 5 × 106 cells were infused into recipient mice via tail vein injection. Six weeks following transplantation, mice reconstituted with Mmp14+/+ or Mmp14−/− bone marrow cells were injected intraperitoneally with 1 mL of 3% Brewer thioglycollate medium. Four days later, peritoneal exudate cells were collected and enriched for macrophages by surface adherence (Xiong et al. 2009).
Fluorescence-activated cell sorter analysis (FACS)
FACS was performed as described (Serbina and Pamer 2006). Briefly, mouse peritoneal exudate cells were suspended in FACS buffer (2% BSA, 0.1% sodium azide in PBS) and incubated with Fc block (eBioscience), FITC-conjugated Mac3 (M3/84), and APC-conjugated Ly6C (HK1.4) antibodies or the corresponding isotype controls (eBioscience) for 15 min at 25°C prior to analysis by FACS Canto II (BD Bioscience).
Lentiviral gene transfer
293T cells were transfected with pLentilox IRES EGFP plasmids expressing various MT1-MMP mutant cDNAs and packaging vectors (Addgene) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Lentiviral supernatant was collected after 36 h, and subconfluent monolayers of macrophages were incubated in the supernatant with 8 μg/mL polybrene (Millipore) for 12 h at 37°C. Transgene expression was confirmed 4 d postinfection when all studies were performed.
Transcriptional profiling
Total RNA was isolated from MT1-MMP-null mouse macrophages following infection with either an HA-tagged MT1-MMP or EGFP lentiviral expression vector. Following a 96-h culture period, total mRNA was isolated, labeled, and hybridized to mouse 430 2.0 cDNA microarrays (Affymetrix). Three replicate sets of each sample were analyzed at the University of Michigan Microarray Core. Differentially expressed probe sets were determined using a minimum fold change of 1.5 and a maximum P-value of 0.005. GO analysis was performed to identify biological processes regulated by MT1-MMP. GO coefficients were calculated as −log(P-value) (Chun et al. 2004; Rowe et al. 2009).
qPCR
RNA was isolated from bone marrow-derived macrophages or thioglycollate-elicited peritoneal macrophages harvested from bone marrow-transplanted mice using TRIzol extraction (Invitrogen). In selected experiments, bone marrow macrophages were transduced with lentiviral control vector, full-length MT1-MMP, or various MT1-MMP mutant constructs. Four days posttransfection, cells were preincubated with the PI3K inhibitor LY294002 (20 μM; EMD), the PI3Kδ specific inhibitor IC87114 (10 μM; Selleck), or the GSK3 inhibitor SB216763 (10 μM; Sigma-Aldrich) for 1 h prior to stimulation with 1 μg/mL LPS (Sigma-Aldrich) for 2 h (Martin et al. 2005). qPCR was performed in triplicate samples using SYBR Green PCR master mix (Applied Biosystems) according to the manufacturer's instructions. For each transcript examined, mRNA levels were normalized to Gapd mRNA levels. The primers used were constructed as described in the Supplemental Material. Semiquantitative RT–PCR was performed with primers for mouse MMP-2, MMP-8, MMP-13, MT1-MMP, MT2-MMP, and MT3-MMP as described previously (Sabeh et al. 2009a). All other primers were constructed as described in the Supplemental Material.
Western blot analysis
Western blots were performed as described with antibodies directed against MT1-MMP (Epitomics), Mi-2β (AbCam), HDAC2 (Santa Cruz Biotechnology), total histone 3 (Millipore), p110δ (Santa Cruz Biotechnology), pAKT Ser 473 (Cell Signaling), total Akt (Cell Signaling), pGSK3β(Ser 9) (Cell Signaling), total GSK3β (Cell Signaling), or tubulin (Santa Cruz Biotechnology) (Li et al. 2008). Bound primary antibodies were detected with horseradish peroxidase-conjugated species-specific secondary antibodies (Santa Cruz Biotechnology) using the Super Signal Pico system (Pierce).
Cytokine assays
Wild-type or MT1-MMP-null macrophages were stimulated with LPS (1 μg/mL) for 12 h, and the culture supernatants were collected. For in vivo experiments, 4-wk-old wild-type and MT1-MMP heterozygote littermates received a single 3 mg/kg dose of LPS via intraperitoneal injection with serum prepared from blood collected by cardiac puncture 6 h postinjection. Murine IL-6, IL-10, and TNFα levels were measured in mouse serum or culture supernatants using a Bio-plex bead-based cytokine assay (Bio-Rad) (Ito et al. 2009). IL-12b levels were measured by sandwich ELISA using polyclonal rabbit anti-IL-12 as described (Chensue et al. 1995).
Restriction enzyme accessibility assay
Wild-type or MT1-MMP-null macrophages were cultured with or without 1 μg/mL LPS for 4 h. In selected experiments, cells were preincubated with DMSO, the PI3K inhibitor LY294002 (20 μM), or the GSK3 inhibitor SB216763 (10 μM) for 1 h prior to LPS stimulation. Cells were then harvested in ice-cold 2 mM EDTA and washed in PBS, and nuclei were isolated by incubating the cell pellet in NP-40 lysis buffer (10 mM Tris at pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40, 0.15 mM spermine, 0.5 mM spermidine) for 5 min at 4°C followed by centrifugation at 1000 rpm (Weinmann et al. 2001). Pelleted nuclei were digested with SpeI (100 U; New England Biolabs) for 15 min at 37°C, followed by genomic DNA isolation using the Qiagen DNeasy kit (Qiagen). Purified DNA (10 μg) was digested overnight with KpnI (100 U; New England Biolabs) and SphI (100 U; New England Biolabs) followed by QIAquick PCR purification (Qiagen) (Ramirez-Carrozzi et al. 2006). Samples were resolved on a 0.8% agarose gel and transferred to a nylon membrane (Hybond-XL, Amersham Pharmacia) for Southern blot analysis. Hybridization was performed using an IL-12b (+64 to +437)-specific probe, which was amplified from mouse genomic DNA by PCR (primers: forward, 5′-CTCTCCTCTTCCCTGTCGCTAACTC-3′; reverse, 5′-CCTTTCTATCAAATACACATCTGTCC-3′) (Zhou et al. 2010). Purified PCR products were labeled with 32P using the RadPrime DNA labeling system (Invitrogen), and blots were analyzed by Phosphor Screen (Molecular Dynamics). Percent DNA cleavage by SpeI was calculated as the ratio of the intensity of the 1287/1612-base-pair (bp) cleaved band to the 1287/1612-bp band plus the 2205-bp uncleaved promoter fragment using Quantity One software (Bio-Rad) (Zhou et al. 2010).
ChIP assay
To assess interactions between Mi2-β, HDAC-2, AcH4, or H3K27Me3 and the IL-12b promoter, wild-type or MT1-MMP-null macrophages were cultured with or without LPS for 4 h and fixed for 10 min with 1% formaldehyde, and a ChIP assay was performed as described (Gilchrist et al. 2006). In selected experiments, macrophages were pretreated with LY294002 (20 μM) or SB216763 (10 μM) for 1 h prior to stimulation. Cross-linking was terminated by the addition of 125 mM glycine (pH 2.5) for 10 min. Nuclei were extracted in lysis buffer (50 mM Tris at pH 8.0, 2 mM EDTA, 0.1% NP-40, 10% glycerol supplemented with a protease inhibitor cocktail) for 15 min on ice and resuspended in SDS lysis buffer (50 mM Tris at pH 8.1, 1% SDS, 10 mM EDTA with the protease inhibitor cocktail) after pelleting at 1000g. Chromatin was sheared by sonication (four 30-sec pulses at setting 6; VirTis Virsonic 100 sonicator), centrifuged to pellet debris, and then diluted 1:10 in RIPA buffer (50 mM Tris at pH 8.0, 0.5% NP-40, 0.2 M NaCl, 0.5 mM EDTA with a protease inhibitor cocktail). After preclearing with salmon sperm–saturated protein A for 15 min, immunoprecipitations were carried out overnight at 4°C with the following primary antibodies: anti-Mi2-β, anti-HDAC2, anti-acetyl H4 (Millipore), anti-H3K27Me3 (Millipore), or control IgG (Santa Cruz Biotechnology). After washing three times in a high-salt buffer (20 mM Tris at pH 8.0, 0.1% SDS, 1% NP-40, 2 mM EDTA, 500 mM NaCl) and three times in a low-salt buffer (1× Tris/EDTA, TE), immune complexes were extracted in 1× TE containing 1% SDS, and protein–DNA cross-links were dissociated by heating overnight at 65°C. Following proteinase K digestion, DNA was recovered using a Qiagen PCR purification kit (Qiagen). To assess binding interactions between MT1-MMP and the PI3Kδ promoter, ChIP was performed as described above, but with wild-type or MT1-MMP-null macrophages lentivirally transduced with HA-tagged MT1-MMP or a control vector 4 d prior to assay. DNA–protein complexes were immunoprecipitated with an anti-HA antibody (Santa Cruz Biotechnology).
PCR amplification was performed in a final volume of 25 μL containing 2 μL of extracted DNA. The number of cycles used for each primer set was 25–32, depending on the linear range. The conditions for PCR amplification were as follows: denaturing for 45 sec at 95°C, annealing for 1 min, and extension for 2 min at 72°C. Sequences for IL12b promoter-specific primers were constructed as described previously (Gilchrist et al. 2006). Primers specific for the p110δ promoter were constructed based on previously described murine PI3KCD promoter sequences (Supplemental Table 1; Kok et al. 2009). qPCR amplification was performed on genomic DNA obtained from each sample before immunoprecipitation as “Input.”
Luciferase assay
A PI3KCD exon −1 promoter region DNA fragment (−500 to 100) was subcloned into pGL4.23 (Promega) upstream of the firefly luciferase reporter gene using specific primers (forward, 5′-CAGCTGACCTTCTCATCTCTAA-3′; reverse, 5′-AAGGACCACCACCAA-3′). Wild-type or MT1-MMP-null macrophages (1 × 106) were cotransfected with 1 μg of pGL4.23 firefly luciferase reporter construct or empty vector, 0.3 μg of Renilla luciferase reporter control (Promega), and 1 μg of a MT1-MMP pcDNA3.1 expression vector or empty vector by electroporation using a Mouse Macrophage Nucleofector kit (Lonza). Cells were harvested 24 h post-transfection, and the luminescence was measured according to the manufacturer's protocol.
Construction of expression plasmids and lentivirus production
HA-tagged full-length MT1-MMP, a catalytically inactive, full-length form of MT1-MMP that harbors an E240 → A substitution in the catalytic domain (E/A 240), a cytosolic tail-truncated mutant (ΔCT; ΔArg 563–Val 582), a hemopexin domain-deleted mutant (ΔPEX; ΔCys 318–Gly 535), and a His-tagged transmembrane domain-deleted mutant (ΔTM; ΔGly 535–Val 582) have been described previously (Hotary et al. 2003; Li et al. 2008). The signal peptide of the ΔTM construct (Met 1–Phe 33) was deleted to generate ΔSigΔTM. An HA-tagged MT1-MMP chimeric protein that combines the extracellular domain of MT1-MMP (Met 1–Gly 535) with the MT6-MMP GPI anchor (Asp 531–Arg 562) (MT1ΔTM/MT6-MMP GPI) was assembled as described (Li et al. 2008). The control vector carried an EGFP cDNA alone, and all cDNAs were subcloned into the pLenti lox IRES EGFP vector.
Microscopy
Confocal imaging was performed on a laser scanning fluorescent microscope (model FV500, Olympus) equipped with acquisition software (Fluo View; Olympus) using a 60× water immersion objective at 25°C. Equal photomultiplier tube intensity and gain settings were used in acquiring images. For double- or triple-color imaging, selective excitations at 466 nm, 549 nm, and 633 nm were used. For the visualization of intranuclear MT1-MMP localization, wild-type or MT1-MMP-null macrophages lentivirally transduced with various tagged MT1-MMP mutants were fixed and stained with either HA-antibody (Santa Cruz Biotechnology) or His-antibody (Santa Cruz Biotechnology) together with DAPI, and Z-slices were taken with confocal microscopy. 3D reconstructed images were made with Imaris software (Bitplane Scientific).
Subcellular fractionation
Wild-type macrophages transduced with HA-tagged MT1-MMP were subjected to high-salt nuclear fractionation as described (Sehat et al. 2010). Briefly, cells were harvested in buffer A (10 mM HEPES at pH 7.5, 10 mM KCl, 2 mM MgCl2, 1 mM EDTA, 0.01% NP-40, protease inhibitor cocktail) and disrupted by 20 passes through a 21-gauge needle. After centrifugation at 500g for 5 min, the supernatant was designated as the cytosolic fraction, and the pellet was washed twice with buffer A and extracted with 50 μL of buffer A supplemented with 500 mM NaCl and 25% glycerol on an end-over-end rotator for 30 min at 4°C. After centrifugation at 12,000g for 5 min, the supernatant and pellet were designated as the soluble and nuclear pellet fractions, respectively. Subcellular fractions were analyzed by SDS-PAGE under reducing conditions, followed by probing with anti-HA antibody.
Statistical analysis
Statistical analyses were performed using a Student's t-test.
Acknowledgments
This work was supported by NIH grants R01 CA116516, CA071699, CA088308 (to S.J.W.), and R01 HL62400 (to B.T.B.); the Breast Cancer Research Foundation (to S.J.W.); and NIH P60 DK020572 for the MDRTC Cell and Molecular Biology Core at the University of Michigan.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.178749.111.
References
- Barbolina MV, Stack MS 2008. Membrane type 1-matrix metalloproteinase: Substrate diversity in pericellular proteolysis. Semin Cell Dev Biol 19: 24–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurel E, Michalek SM, Jope RS 2010. Innate and adaptive immune responses regulated by glycogen synthase kinase-3 (GSK3). Trends Immunol 31: 24–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler GS, Dean RA, Tam EM, Overall CM 2008. Pharmacoproteomics of a metalloproteinase hydroxamate inhibitor in breast cancer cells: Dynamics of membrane type 1 matrix metalloproteinase-mediated membrane protein shedding. Mol Cell Biol 28: 4896–4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Kozarekar P, Pavlaki M, Chiarelli C, Bahou WF, Zucker S 2004. Distinct roles for the catalytic and hemopexin domains of membrane type 1-matrix metalloproteinase in substrate degradation and cell migration. J Biol Chem 279: 14129–14139 [DOI] [PubMed] [Google Scholar]
- Chensue SW, Ruth JH, Warmington K, Lincoln P, Kunkel SL 1995. In vivo regulation of macrophage IL-12 production during type 1 and type 2 cytokine-mediated granuloma formation. J Immunol 155: 3546–3551 [PubMed] [Google Scholar]
- Chun TH, Sabeh F, Ota I, Murphy H, McDonagh KT, Holmbeck K, Birkedal-Hansen H, Allen ED, Weiss SJ 2004. MT1-MMP-dependent neovessel formation within the confines of the three-dimensional extracellular matrix. J Cell Biol 167: 757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TH, Hotary KB, Sabeh F, Saltiel AR, Allen ED, Weiss SJ 2006. A pericellular collagenase directs the 3-dimensional development of white adipose tissue. Cell 125: 577–591 [DOI] [PubMed] [Google Scholar]
- Chun TH, Inoue M, Morisaki H, Yamanaka I, Miyamoto Y, Okamura T, Sato-Kusubata K, Weiss SJ 2010. Genetic link between obesity and MMP14-dependent adipogenic collagen turnover. Diabetes 59: 2484–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougoule C, Le Cabec V, Poincloux R, Al Saati T, Mege JL, Tabouret G, Lowell CA, Laviolette-Malirat N, Maridonneau-Parini I 2010. Three-dimensional migration of macrophages requires Hck for podosome organization and extracellular matrix proteolysis. Blood 115: 1444–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alessio S, Ferrari G, Cinnante K, Scheerer W, Galloway AC, Roses DF, Rozanov DV, Remacle AG, Oh ES, Shiryaev SA, et al. 2008. Tissue inhibitor of metalloproteinases-2 binding to membrane-type 1 matrix metalloproteinase induces MAPK activation and cell growth by a non-proteolytic mechanism. J Biol Chem 283: 87–99 [DOI] [PubMed] [Google Scholar]
- Delaval B, Doxsey SJ 2010. Pericentrin in cellular function and disease. J Cell Biol 188: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham-Ems SM, Lee YW, Stachowiak EK, Pudavar H, Claus P, Prasad PN, Stachowiak MK 2009. Fibroblast growth factor receptor-1 (FGFR1) nuclear dynamics reveal a novel mechanism in transcription control. Mol Biol Cell 20: 2401–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi T, Kubota S, Kawata K, Mukudai Y, Uehara J, Ohgawara T, Ibaragi S, Sasaki A, Kuboki T, Takigawa M 2008. Novel transcription-factor-like function of human matrix metalloproteinase 3 regulating the CTGF/CCN2 gene. Mol Cell Biol 28: 2391–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippov S, Koenig GC, Chun TH, Hotary KB, Ota I, Bugge TH, Roberts JD, Fay WP, Birkedal-Hansen H, Holmbeck K, et al. 2005. MT1-matrix metalloproteinase directs arterial wall invasion and neointima formation by vascular smooth muscle cells. J Exp Med 202: 663–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster KS, McCrary WJ, Ross JS, Wright CF 2006. Members of the hSWI/SNF chromatin remodeling complex associate with and are phosphorylated by protein kinase B/Akt. Oncogene 25: 4605–4612 [DOI] [PubMed] [Google Scholar]
- Fukao T, Koyasu S 2003. PI3K and negative regulation of TLR signaling. Trends Immunol 24: 358–363 [DOI] [PubMed] [Google Scholar]
- Gawden-Bone C, Zhou Z, King E, Prescott A, Watts C, Lucocq J 2010. Dendritic cell podosomes are protrusive and invade the extracellular matrix using metalloproteinase MMP-14. J Cell Sci 123: 1427–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann F, Jung S, Littman DR 2003. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity 19: 71–82 [DOI] [PubMed] [Google Scholar]
- Ghosn EE, Cassado AA, Govoni GR, Fukuhara T, Yang Y, Monack DM, Bortoluci KR, Almeida SR, Herzenberg LA 2010. Two physically, functionally, and developmentally distinct peritoneal macrophage subsets. Proc Natl Acad Sci 107: 2568–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, Roach JC, Kennedy K, Hai T, Bolouri H, Aderem A 2006. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature 441: 173–178 [DOI] [PubMed] [Google Scholar]
- Golubkov VS, Boyd S, Savinov AY, Chekanov AV, Osterman AL, Remacle A, Rozanov DV, Doxsey SJ, Strongin AY 2005. Membrane type-1 matrix metalloproteinase (MT1-MMP) exhibits an important intracellular cleavage function and causes chromosome instability. J Biol Chem 280: 25079–25086 [DOI] [PubMed] [Google Scholar]
- Gonzalo P, Guadamillas MC, Hernandez-Riquer MV, Pollan A, Grande-Garcia A, Bartolome RA, Vasanji A, Ambrogio C, Chiarle R, Teixido J, et al. 2010. MT1-MMP is required for myeloid cell fusion via regulation of Rac1 signaling. Dev Cell 18: 77–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M 2010. Immunity, inflammation, and cancer. Cell 140: 883–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha M, Mackman N 2002. The phosphatidylinositol 3-kinase-Akt pathway limits lipopolysaccharide activation of signaling pathways and expression of inflammatory mediators in human monocytic cells. J Biol Chem 277: 32124–32132 [DOI] [PubMed] [Google Scholar]
- Hieda M, Isokane M, Koizumi M, Higashi C, Tachibana T, Shudou M, Taguchi T, Hieda Y, Higashiyama S 2008. Membrane-anchored growth factor, HB-EGF, on the cell surface targeted to the inner nuclear membrane. J Cell Biol 180: 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikita A, Yana I, Wakeyama H, Nakamura M, Kadono Y, Oshima Y, Nakamura K, Seiki M, Tanaka S 2006. Negative regulation of osteoclastogenesis by ectodomain shedding of receptor activator of NF-κB ligand. J Biol Chem 281: 36846–36855 [DOI] [PubMed] [Google Scholar]
- Holmbeck K, Bianco P, Caterina J, Yamada S, Kromer M, Kuznetsov SA, Mankani M, Robey PG, Poole AR, Pidoux I, et al. 1999. MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell 99: 81–92 [DOI] [PubMed] [Google Scholar]
- Hotary KB, Allen ED, Brooks PC, Datta NS, Long MW, Weiss SJ 2003. Membrane type I matrix metalloproteinase usurps tumor growth control imposed by the three-dimensional extracellular matrix. Cell 114: 33–45 [DOI] [PubMed] [Google Scholar]
- Huber AR, Weiss SJ 1989. Disruption of the subendothelial basement membrane during neutrophil diapedesis in an in vitro construct of a blood vessel wall. J Clin Invest 83: 1122–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo L, Wang YN, Xia W, Hsu SC, Lai CC, Li LY, Chang WC, Wang Y, Hsu MC, Yu YL, et al. 2010. RNA helicase A is a DNA-binding partner for EGFR-mediated transcriptional activation in the nucleus. Proc Natl Acad Sci 107: 16125–16130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Schaller M, Hogaboam CM, Standiford TJ, Sandor M, Lukacs NW, Chensue SW, Kunkel SL 2009. TLR9 regulates the mycobacteria-elicited pulmonary granulomatous immune response in mice through DC-derived Notch ligand delta-like 4. J Clin Invest 119: 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermann SA, Hudak S, Oldham ER, Liu YJ, McEvoy LM 1999. The CC chemokine receptor-7 ligands 6Ckine and macrophage inflammatory protein-3β are potent chemoattractants for in vitro- and in vivo-derived dendritic cells. J Immunol 162: 3859–3864 [PubMed] [Google Scholar]
- Kessenbrock K, Plaks V, Werb Z 2010. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell 141: 52–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok K, Nock GE, Verrall EA, Mitchell MP, Hommes DW, Peppelenbosch MP, Vanhaesebroeck B 2009. Regulation of p110δ PI 3-kinase gene expression. PLoS ONE 4: e5145 doi: 10.1371/journal.pone.0005145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan JA, Schulze CJ, Wang W, Leon H, Sariahmetoglu M, Sung M, Sawicka J, Sims DE, Sawicki G, Schulz R 2004. Matrix metalloproteinase-2 (MMP-2) is present in the nucleus of cardiac myocytes and is capable of cleaving poly (ADP-ribose) polymerase (PARP) in vitro. FASEB J 18: 690–692 [DOI] [PubMed] [Google Scholar]
- Lai D, Wan M, Wu J, Preston-Hurlburt P, Kushwaha R, Grundstrom T, Imbalzano AN, Chi T 2009. Induction of TLR4-target genes entails calcium/calmodulin-dependent regulation of chromatin remodeling. Proc Natl Acad Sci 106: 1169–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JL, Wang MJ, Chen JY 2009. Acetylation and activation of STAT3 mediated by nuclear translocation of CD44. J Cell Biol 185: 949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Ota I, Yana I, Sabeh F, Weiss SJ 2008. Molecular dissection of the structural machinery underlying the tissue-invasive activity of membrane type-1 matrix metalloproteinase. Mol Biol Cell 19: 3221–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HJ, Carpenter G 2007. Role of the Sec61 translocon in EGF receptor trafficking to the nucleus and gene expression. Mol Biol Cell 18: 1064–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limb GA, Matter K, Murphy G, Cambrey AD, Bishop PN, Morris GE, Khaw PT 2005. Matrix metalloproteinase-1 associates with intracellular organelles and confers resistance to lamin A/C degradation during apoptosis. Am J Pathol 166: 1555–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, Bourguignon L, Hung MC 2001. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol 3: 802–808 [DOI] [PubMed] [Google Scholar]
- Lo HW, Cao X, Zhu H, Ali-Osman F 2010. Cyclooxygenase-2 is a novel transcriptional target of the nuclear EGFR–STAT3 and EGFRvIII–STAT3 signaling axes. Mol Cancer Res 8: 232–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low PC, Misaki R, Schroder K, Stanley AC, Sweet MJ, Teasdale RD, Vanhaesebroeck B, Meunier FA, Taguchi T, Stow JL 2010. Phosphoinositide 3-kinase δ regulates membrane fission of Golgi carriers for selective cytokine secretion. J Cell Biol 190: 1053–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Li XY, Hu Y, Rowe RG, Weiss SJ 2010. MT1-MMP controls human mesenchymal stem cell trafficking and differentiation. Blood 115: 221–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Rehani K, Jope RS, Michalek SM 2005. Toll-like receptor-mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat Immunol 6: 777–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias-Roman S, Galvez BG, Genis L, Yanez-Mo M, de la Rosa G, Sanchez-Mateos P, Sanchez-Madrid F, Arroyo AG 2005. Membrane type 1-matrix metalloproteinase is involved in migration of human monocytes and is regulated through their interaction with fibronectin or endothelium. Blood 105: 3956–3964 [DOI] [PubMed] [Google Scholar]
- Monick MM, Carter AB, Robeff PK, Flaherty DM, Peterson MW, Hunninghake GW 2001. Lipopolysaccharide activates Akt in human alveolar macrophages resulting in nuclear accumulation and transcriptional activity of β-catenin. J Immunol 166: 4713–4720 [DOI] [PubMed] [Google Scholar]
- Mori H, Gjorevski N, Inman JL, Bissell MJ, Nelson CM 2009. Self-organization of engineered epithelial tubules by differential cellular motility. Proc Natl Acad Sci 106: 14890–14895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser DM, Edwards JP 2008. Exploring the full spectrum of macrophage activation. Nat Rev Immunol 8: 958–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C, Ding A 2010. Nonresolving inflammation. Cell 140: 871–882 [DOI] [PubMed] [Google Scholar]
- Ohtake Y, Tojo H, Seiki M 2006. Multifunctional roles of MT1-MMP in myofiber formation and morphostatic maintenance of skeletal muscle. J Cell Sci 119: 3822–3832 [DOI] [PubMed] [Google Scholar]
- Papakonstanti EA, Zwaenepoel O, Bilancio A, Burns E, Nock GE, Houseman B, Shokat K, Ridley AJ, Vanhaesebroeck B 2008. Distinct roles of class IA PI3K isoforms in primary and immortalised macrophages. J Cell Sci 121: 4124–4133 [DOI] [PubMed] [Google Scholar]
- Proulx-Bonneau S, Guezguez A, Annabi B 2011. A concerted HIF-1α/MT1-MMP signalling axis regulates the expression of the 3BP2 adaptor protein in hypoxic mesenchymal stromal cells. PLoS ONE 6: e21511 doi: 10.1371/journal.pone.0021511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST 2006. Selective and antagonistic functions of SWI/SNF and Mi-2β nucleosome remodeling complexes during an inflammatory response. Genes Dev 20: 282–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehani K, Wang H, Garcia CA, Kinane DF, Martin M 2009. Toll-like receptor-mediated production of IL-1Ra is negatively regulated by GSK3 via the MAPK ERK1/2. J Immunol 182: 547–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommel C, Camps M, Ji H 2007. PI3K δ and PI3K γ: Partners in crime in inflammation in rheumatoid arthritis and beyond? Nat Rev Immunol 7: 191–201 [DOI] [PubMed] [Google Scholar]
- Rowe RG, Weiss SJ 2009. Navigating ECM barriers at the invasive front: The cancer cell–stroma interface. Annu Rev Cell Dev Biol 25: 567–595 [DOI] [PubMed] [Google Scholar]
- Rowe RG, Li XY, Hu Y, Saunders TL, Virtanen I, Garcia de Herreros A, Becker KF, Ingvarsen S, Engelholm LH, Bommer GT, et al. 2009. Mesenchymal cells reactivate Snail1 expression to drive three-dimensional invasion programs. J Cell Biol 184: 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozanov DV, Savinov AY, Williams R, Liu K, Golubkov VS, Krajewski S, Strongin AY 2008. Molecular signature of MT1-MMP: Transactivation of the downstream universal gene network in cancer. Cancer Res 68: 4086–4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeh F, Ota I, Holmbeck K, Birkedal-Hansen H, Soloway P, Balbin M, Lopez-Otin C, Shapiro S, Inada M, Krane S, et al. 2004. Tumor cell traffic through the extracellular matrix is controlled by the membrane-anchored collagenase MT1-MMP. J Cell Biol 167: 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeh F, Li XY, Saunders TL, Rowe RG, Weiss SJ 2009a. Secreted versus membrane-anchored collagenases: Relative roles in fibroblast-dependent collagenolysis and invasion. J Biol Chem 284: 23001–23011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeh F, Shimizu-Hirota R, Weiss SJ 2009b. Protease-dependent versus -independent cancer cell invasion programs: Three-dimensional amoeboid movement revisited. J Cell Biol 185: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadhu C, Masinovsky B, Dick K, Sowell CG, Staunton DE 2003. Essential role of phosphoinositide 3-kinase δ in neutrophil directional movement. J Immunol 170: 2647–2654 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Seiki M 2009. Cytoplasmic tail of MT1-MMP regulates macrophage motility independently from its protease activity. Genes Cells 14: 617–626 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Seiki M 2010. A membrane protease regulates energy production in macrophages by activating hypoxia-inducible factor-1 via a non-proteolytic mechanism. J Biol Chem 285: 29951–29964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider F, Sukhova GK, Aikawa M, Canner J, Gerdes N, Tang SM, Shi GP, Apte SS, Libby P 2008. Matrix-metalloproteinase-14 deficiency in bone-marrow-derived cells promotes collagen accumulation in mouse atherosclerotic plaques. Circulation 117: 931–939 [DOI] [PubMed] [Google Scholar]
- Sehat B, Tofigh A, Lin Y, Trocme E, Liljedahl U, Lagergren J, Larsson O 2010. SUMOylation mediates the nuclear translocation and signaling of the IGF-1 receptor. Sci Signal 3: ra10 doi: 10.1126/scisignal.2000628 [DOI] [PubMed] [Google Scholar]
- Serbina NV, Pamer EG 2006. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat Immunol 7: 311–317 [DOI] [PubMed] [Google Scholar]
- Shankavaram UT, Lai WC, Netzel-Arnett S, Mangan PR, Ardans JA, Caterina N, Stetler-Stevenson WG, Birkedal-Hansen H, Wahl LM 2001. Monocyte membrane type 1-matrix metalloproteinase. Prostaglandin-dependent regulation and role in metalloproteinase-2 activation. J Biol Chem 276: 19027–19032 [DOI] [PubMed] [Google Scholar]
- Sillibourne JE, Delaval B, Redick S, Sinha M, Doxsey SJ 2007. Chromatin remodeling proteins interact with pericentrin to regulate centrosome integrity. Mol Biol Cell 18: 3667–3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K, Monvoisin A, Mazzocco C, Lepreux S, Decossas M, Cubel G, Taras D, Blanc JF, Robinson DR, Rosenbaum J 2006. Matrix metalloproteinase 3 is present in the cell nucleus and is involved in apoptosis. Am J Pathol 169: 1390–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sithu SD, English WR, Olson P, Krubasik D, Baker AH, Murphy G, D'Souza SE 2007. Membrane-type 1-matrix metalloproteinase regulates intracellular adhesion molecule-1 (ICAM-1)-mediated monocyte transmigration. J Biol Chem 282: 25010–25019 [DOI] [PubMed] [Google Scholar]
- Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, Floss T, Goettlicher M, Noppinger PR, Wurst W, et al. 2007. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3β activity. Nat Med 13: 324–331 [DOI] [PubMed] [Google Scholar]
- Vagima Y, Avigdor A, Goichberg P, Shivtiel S, Tesio M, Kalinkovich A, Golan K, Dar A, Kollet O, Petit I, et al. 2009. MT1-MMP and RECK are involved in human CD34+ progenitor cell retention, egress, and mobilization. J Clin Invest 119: 492–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goethem E, Poincloux R, Gauffre F, Maridonneau-Parini I, Le Cabec V 2010. Matrix architecture dictates three-dimensional migration modes of human macrophages: Differential involvement of proteases and podosome-like structures. J Immunol 184: 1049–1061 [DOI] [PubMed] [Google Scholar]
- Vignali M, Hassan AH, Neely KE, Workman JL 2000. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol 20: 1899–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YN, Yamaguchi H, Hsu JM, Hung MC 2010a. Nuclear trafficking of the epidermal growth factor receptor family membrane proteins. Oncogene 29: 3997–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YN, Yamaguchi H, Huo L, Du Y, Lee HJ, Lee HH, Wang H, Hsu JM, Hung MC 2010b. The translocon Sec61β localized in the inner nuclear membrane transports membrane-embedded EGF receptor to the nucleus. J Biol Chem 285: 38720–38729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann AS, Plevy SE, Smale ST 1999. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity 11: 665–675 [DOI] [PubMed] [Google Scholar]
- Weinmann AS, Mitchell DM, Sanjabi S, Bradley MN, Hoffmann A, Liou HC, Smale ST 2001. Nucleosome remodeling at the IL-12 p40 promoter is a TLR-dependent, Rel-independent event. Nat Immunol 2: 51–57 [DOI] [PubMed] [Google Scholar]
- Weiss SJ 1989. Tissue destruction by neutrophils. N Engl J Med 320: 365–376 [DOI] [PubMed] [Google Scholar]
- Xiong W, Knispel R, MacTaggart J, Greiner TC, Weiss SJ, Baxter BT 2009. Membrane-type 1 matrix metalloproteinase regulates macrophage-dependent elastolytic activity and aneurysm formation in vivo. J Biol Chem 284: 1765–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Candelario-Jalil E, Thompson JF, Cuadrado E, Estrada EY, Rosell A, Montaner J, Rosenberg GA 2010. Increased intranuclear matrix metalloproteinase activity in neurons interferes with oxidative DNA repair in focal cerebral ischemia. J Neurochem 112: 134–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Apte SS, Soininen R, Cao R, Baaklini GY, Rauser RW, Wang J, Cao Y, Tryggvason K 2000. Impaired endochondral ossification and angiogenesis in mice deficient in membrane-type matrix metalloproteinase I. Proc Natl Acad Sci 97: 4052–4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Collins CA, Wu P, Brown EJ 2010. Protein tyrosine phosphatase SHP-1 positively regulates TLR-induced IL-12p40 production in macrophages through inhibition of phosphatidylinositol 3-kinase. J Leukoc Biol 87: 845–855 [DOI] [PubMed] [Google Scholar]