Abstract
Northern peatlands are a large repository of atmospheric carbon due to an imbalance between primary production by plants and microbial decomposition. The James Bay Lowlands (JBL) of northern Ontario are a large peatland-complex but remain relatively unstudied. Climate change models predict the region will experience warmer and drier conditions, potentially altering plant community composition, and shifting the region from a long-term carbon sink to a source. We collected a peat core from two geographically separated (ca. 200 km) ombrotrophic peatlands (Victor and Kinoje Bogs) and one minerotrophic peatland (Victor Fen) located near Victor Bog within the JBL. We characterized (i) archaeal, bacterial, and fungal community structure with terminal restriction fragment length polymorphism of ribosomal DNA, (ii) estimated microbial activity using community level physiological profiling and extracellular enzymes activities, and (iii) the aeration and temperature dependence of carbon mineralization at three depths (0–10, 50–60, and 100–110 cm) from each site. Similar dominant microbial taxa were observed at all three peatlands despite differences in nutrient content and substrate quality. In contrast, we observed differences in basal respiration, enzyme activity, and the magnitude of substrate utilization, which were all generally higher at Victor Fen and similar between the two bogs. However, there was no preferential mineralization of carbon substrates between the bogs and fens. Microbial community composition did not correlate with measures of microbial activity but pH was a strong predictor of activity across all sites and depths. Increased peat temperature and aeration stimulated CO2 production but this did not correlate with a change in enzyme activities. Potential microbial activity in the JBL appears to be influenced by the quality of the peat substrate and the presence of microbial inhibitors, which suggests the existing peat substrate will have a large influence on future JBL carbon dynamics.
Keywords: peatlands, T-RFLP, microbial community, microbial activity, carbon dioxide, James Bay Lowlands, CLPP
Introduction
Peatlands are important to the global carbon cycle as they are large repositories of atmospheric carbon, with estimates ranging between 270 and 455 Pg (1015) carbon, despite only covering 3% of the Earth’s surface (Gorham, 1991; Turunen et al., 2002). This is due to a relative imbalance between rates of net primary production by plants and microbial decomposition (Clymo, 1984), resulting from persistently water saturated environments and consequent anoxia (Moore and Basiliko, 2006). Such conditions also favor microbial CH4 production, thus peatlands are also significant sources of atmospheric CH4 (Moore et al., 1998; Laine et al., 2007; Roulet et al., 2007), which has a global warming potential 23 times higher than CO2 under the IPCC’s 100-year timeframe (Forster et al., 2007).
The James Bay Lowlands (JBL) are a large peatland-complex that form the southeast part of the Hudson Bay Lowlands, Canada, the second largest peatland area in the world (325,000 km2) after the Western Siberian Lowlands (Riley, 1982; Gorham, 1991). Few studies have been conducted on carbon biogeochemistry in the JBL (Roulet, 2000; Yu et al., 2010), which represents a massive atmospheric carbon store characterized by a mosaic of peatland types that were formed under varying environmental conditions and may differ in their sensitivity to environmental change (Martini, 2006). The JBL therefore represents both an important frontier in peatland ecology and a significant factor for the atmospheric carbon balance.
Concern has been raised over the stability of the JBL carbon store as climate models predict an increase in mean annual air temperature of 3–4°C by 2020 and 5–10°C by 2050, with only minor increases in precipitation of 10–20% in this region by the end of the twenty-first century (Hengeveld, 2000; Gagnon and Gough, 2005). Warmer temperatures are expected to increase evapotranspiration resulting in lower water table levels (Gorham, 1991; Roulet et al., 1992). Short-term water table level fluctuations may increase decomposition as the presence of bimolecular oxygen (O2) activates phenol oxidases responsible for the degradation of phenolic compounds (Sinsabaugh, 2010). This change can release hydrolase enzymes, responsible for labile carbon degradation, from inhibition and has been referred to as the “enzymic latch theory” (Freeman et al., 2001, 2004). Continued lowering of the water table height may lead to a complete shift in plant community composition (Weltzin et al., 2003; Laiho, 2006; Robroek et al., 2007), potentially altering litter decomposability (Dorrepaal et al., 2005), heterotrophic respiration (Moore and Basiliko, 2006), and increased CH4 production (Hines et al., 2008).
Many studies have demonstrated that temperature and hydrology-related redox state directly control carbon mineralization in peatlands (e.g., Moore and Dalva, 1993; Updegraff et al., 1998; Keller et al., 2004). However, microorganisms ultimately control peat decomposition and evidence indicates that the soil microbial community composition is important to ecosystem processes (Balser and Firestone, 2005; Reed and Martiny, 2007); but its effects on carbon mineralization are unresolved (Waldrop and Firestone, 2006; Bardgett et al., 2008; McGuire and Treseder, 2010). Soil pH has been identified as one of the most influential factors controlling microbial diversity and community composition across soils types (Fierer and Jackson, 2006; Allison and Treseder, 2008; Rousk et al., 2010). However, peatland microbial diversity has not been extensively categorized (Kraigher et al., 2006) but differences in microbial community structure have been identified between peatland types (i.e., bogs and fens; Jaatinen et al., 2007; Ausec et al., 2009; Peltoniemi et al., 2009) as well as among depths down the peat profile (Morales et al., 2006; Jaatinen et al., 2007), potentially due to changes in pH.
Differences in microbial community composition among peatlands should be expected as vegetation varies from poorly decomposable mosses and shrubs in ombrotrophic bogs that only receive nutrients from dry and wet deposition (Aerts et al., 1999) to more easily decomposable sedges, herbs, and shrubs in minerotrophic fens that receive nutrients from groundwater (Bragazza et al., 2007). Although ecosystem models assume that microbial communities are functionally redundant (Reed and Martiny, 2007) differences in microbial activity have been observed among peatland habitats and depths (Fisk et al., 2003). Moreover, plant litter decomposition rates have been shown to vary depending on both the structure of the microbial community and the past resource inputs a community has experienced (Strickland et al., 2009). Given the contrasting botanical composition between bog and fen peatlands, microbial communities may preferentially utilize their “native” organic compounds to a greater extent than microbes that have been exposed to peat with a different chemical composition.
In this study we used multiple approaches to characterize depth-dependent microbial community structure and function in two bogs and a fen within the JBL. This included molecular fingerprinting techniques, two independent estimates of microbial activity and in vitro assays of microbial responses to changes in aeration and temperature. We hypothesized that (i) pH is the dominant control of microbial community composition and activity among sites and depths, with more diverse communities observed at higher pH, (ii) substrate utilization patterns of carbon compounds will differ between the bogs and the fen, with the fen preferentially mineralizing labile compounds and bogs mineralizing more chemically complex compounds, (iii) measures of microbial activity and community composition correlate with carbon mineralization, and (iv) higher peat temperature and aeration results in greater rates of microbial activity, with the largest response occurring in surface peat.
Materials and Methods
Sample collection and preparation
The climate of the JBL is described as being humid microthermal-subarctic (Dcf), according to the Köppen Climate Classification System (Christopherson and Byrne, 2009). Annual total precipitation ranges from 610 to 660 mm and mean temperatures range from −29 to −15°C in January and 12 to 18°C in July between 50° and 52°N (Martini, 2006). The region is essentially roadless, thus it is difficult to access and required the use of helicopters.
Three sampling locations were chosen based on their nutrient status and distance from one another. Kinoje (51°35′N 82°12′W) and Victor (52°42′N 85°49′W) Bogs are separated by 206 km and have similar characteristics. Both are ombrotrophic, characterized by Sphagnum mosses and lichen (>50%), ericaceous shrubs (primarily members of the genera Vaccinium and Kalmia; >50% cover), and stunted black spruce (Picea mariana; ∼10% cover). Victor Fen is located near Victor Bog (∼1.1 km), but is minerotrophic and dominated by Carex spp. (>50%), with patches of Sphagnum and brown mosses (Drepanocladus, Scorpidium, Aulacomnium; <50% cover), ericaceous shrubs (<25% cover), and larch (Larix laricina; ∼10%). Due to logistical constraints, we were able to collect one peat core (Russian pear corer) per site in the summer of 2009 to a depth of 2 m. All cores were collected from hollows and immediately frozen at −18°C at a nearby research station prior to transport back to Toronto.
Subsamples from each core were taken at 0–10 cm (Surface, above the water table), 50–60 cm (Middle, in a zone of fluctuating water table), and 100–110 cm (Deep, always below the water table). Surface samples from both bog cores were weakly decomposed and classified as H3 using the von Post system of humification (Rydin and Jeglum, 2006), while the fen surface was classed as H4. All middle and deep samples were highly decomposed and categorized as H8 and H9, respectively. To preserve the in situ microbial community from being altered by thaw–refreeze cycles, each sub-section was cut vertically in half while still frozen. The microbial community composition and activity was assessed on one of the sections while the second vertical section was subjected to a temperature and aeration manipulation experiment (below). When required for analysis the peat was thawed at 21°C and gently homogenized to reduce the variation within the sub-sample. Peat moisture content was determined gravimetrically by drying 1 g of wet peat at 105°C for 24 h and peat pH was measured in reverse osmosis purified water after 1 h (soil-to-water ratio of 1:4).
Profiling the microbial community
Archaeal, bacterial, and fungal community structure within the peat samples was profiled using terminal restriction fragment length polymorphism analysis (T-RFLP). DNA was extracted from each peat sample using the MoBio PowerSoil DNA Isolation Kit (MoBio Laboratories Inc., Carlsbad, CA, USA) according to the manufacturer’s instructions. To minimize DNA extraction bias (e.g., Feinstein et al., 2009), three successive extractions were pooled per peat sample. Archaeal rDNA genes were amplified from total DNA using an unlabeled forward primer Ar109 (5′-ACK GCT CAG TAA CAC GT-3′) and the fluorescently labeled reverse primer Ar912r (5′-[6FAM]CTC CCC CGC CAA TTC CTT TA-3′; Lueders and Friedrich, 2000). Bacterial rDNA genes were amplified using a fluorescently labeled forward primer Eu27f (5′ [6FAM]AGA GTT TGA TCM TGG CTC AG-3′) and an unlabeled reverse primer Eu1492r (5′-ACG GYT ACC TTG TTA CGA CTT-3′; Bräuer et al., 2006). Fungal rDNA genes were amplified with an unlabeled forward primer Fu-817f (5′-TTA GCA TGG AAT AAT RRA ATA GGA-3′) and a fluorescently labeled reverse primer Fu-1536r (5′-[6FAM]ATT AGC AAT GCY CTA TCC CCA-3′; Edel-Hermann et al., 2004). The reaction mixture (50 μl) consisted of 20 ng soil DNA, 36.5 μl H2O, 5 μl 10× buffer, 4 μl MgCl2, 1.25 μl dNTPs, 2 μl primers (forward and reverse), and 0.25 μl Taq (Sigma-Aldrich, USA). DNA was amplified by PCR using a Primus 96 plus thermal cycler (MWG Biotech, Germany) with the following reaction conditions: 5 min 94°C, followed by 30 cycles of 94°C for 1 min, annealing temperature 1 min, 72°C for 2 min and a final extension at 72°C for 10 min. The annealing temperature for the bacterial primers was 50 and 52°C for archaeal and fungal primers. For each sample, the PCR product of three reactions was pooled to minimize amplification bias.
The PCR products were purified in order to remove residual primers using a GenElute PCR clean-up kit (Sigma-Aldrich, USA) according to the manufacturer’s instructions and quantified using a Nanodrop spectrophotometer (Thermo Scientific, USA). Archaeal T-RFLP analysis followed the method outlined by Lueders and Friedrich (2000). Aliquots of purified archaeal amplicons (50 ng) were digested using TaqI (3 U) and 1 μl of appropriate buffer supplied by the manufacturer (New England BioLabs, USA) and 1 μl of bovine serum albumin were combined to a total volume of 10 μl and digested at 65°C for 2 h. Bacterial T-RFLP analysis followed the method outlined by Lukow et al. (2000). Aliquots of purified bacterial amplicons (50 ng) were digested with the restriction endonuclease MspI (10 U) and 1 μl of appropriate buffer supplied by the manufacturer to a total volume of 10 μl and digested at 37°C for 3 h. Fungal T-RFLP analysis followed the method outlined by Edel-Hermann et al. (2004). Aliquots of purified fungal amplicons (50 ng) were double digested with AluI and MboI (5 U) and 1 μl of the appropriate buffer supplied by the manufacturer to a total volume of 10 μl and digested at 37°C for 3 h. Terminal restriction fragments were measured on an ABI3730 DNA Analyzer (Applied Biosystems, Foster City, CA, USA) with GeneScan1000-ROX™ fragment size standards (Applied Biosystems) at the University of Guelph Lab Services (ON, Canada). Terminal restriction fragment (T-RFs) lengths between 50 and 1200 bp were determined relative to the standards. T-RFs differing by less that 2 bp were considered to be the same fragment and their data combined and only T-RFs that represented ≥ 1% of the total sample were included for statistical analysis. The abundance (A) of each T-RF was calculated as:
where ni is the peak height of one T-RF, N is the sum of all peak heights in that sample, and multiplied by 10,000 so the data could be analyzed using traditional ecological methods. Peak heights were used in preference to peak area due to overlapping peaks (Blackwood et al., 2003) and this method is more accurate at establishing relative gene abundances (Lueders and Friedrich, 2003).
Microbial biomass carbon (MB-C) and nitrogen (MB-N) was determined using slight modifications to the chloroform (CHCl3) fumigation extraction method by Vance et al. (1987). Three replicate 10 g (wet weight) samples of peat from the sub-sections were each split into two 5 g samples One sample was placed inside a vacuum desiccator and fumigated with ethanol-free CHCl3 in the dark for 24 h. Due to the high moisture content of the peat soil, 0.5 ml of CHCl3 was added directly to the surface to aid cell lysis (Ocio and Brookes, 1990) and a beaker of 1 N NaOH was placed inside the desiccator to remove any excess CO2. CHCl3 vapor and residue was removed through repeated evacuation. All peat samples were sealed in containers with 40 ml of 0.5 M K2SO4, shaken for 1 h at 200 rpm on an oscillating shaker, and filtered through 0.45 μm glass fiber filters. The dissolved organic carbon (DOC) and N (DON) concentration of the peat soil extracts was measured with a TOC/TN analyzer (IL550, Lachat Instruments, USA). MB-C and MB-N was calculated by subtracting the non-fumigated extractable DOC/DON from the fumigated extractable DOC/DON after correcting for differences in initial peat moisture content.
Estimating the activity of the microbial community
Microbial metabolic potential within each JBL sample was estimated by measuring the mineralization rates of four natural and six synthetic substrates of varying chemical complexity and molecular weight. Briefly, synthetic substrates were selected based on their prevalence in natural plant tissues and ranged from simple, low molecular weight compounds to large chemically complex substances. These substrates included: alkali lignin and methylcellulose, potentially representative of major, complex, high carbon concentration, structural compounds in many plants and p-coumaric acid (a lignin derivative from non-woody plant tissues), sodium benzoate (an aromatic organic acid and potential lignin derivative), glucose (a cellulose derivative), and a commercially available mixture of amino acids (protein derivatives, Sigma-Aldrich catalog # R7131). The pH of the synthetic solutions was adjusted to that of reverse osmosis water to avoid a pH effect. Dissolved organic matter extracts from four plant types (two sedge and two Sphagnum) were made by autoclaving chopped plant material in reverse osmosis water and filtered through a 0.45-μm glass fiber filer. Each substrate was adjusted to a standard concentration of 1 mg carbon ml−1.
Substrate induced respiration (SIR) assays were conducted by adding 10 ml of substrate (10 mg carbon) to 5 g of peat (wet mass) in 30 ml serum vials each capped with a rubber stopper and crimped. Reverse osmosis water acted as the control (basal microbial activity). Vials were vigorously shaken by hand for 30 s to ensure mixing and incubated under oxic conditions for 48 h and anoxic conditions for 72 h. Anoxic conditions were created through repeated evacuation with a vacuum pump and back-flushing with nitrogen (N2) gas and brought to atmospheric pressure. Gas samples were taken using a 1-ml syringe following mixing of the gas in the vial and CO2 concentrations were measured using an infrared gas analyzer (Qubit Systems, Kingston, ON, Canada). CO2 production was monitored at five times periods (0, 4, 16, 24, 48 h) for the oxic assay and six time periods (additional measurement at 72 h) for the anoxic assay and was calculated and expressed as total mass of CO2 produced (using the ideal gas law) per gram of dry peat. Preliminary analysis revealed that CO2 production rates increased exponentially after 16 (oxic) and 48 (anoxic) h. Thus to represent the initial responses of in situ microbial community to substrate addition, comparisons of CO2 production were made after 16 and 48 h for oxic and anoxic conditions, respectively. SIR data are expressed as the ratios of CO2 production divided by basal (control) respiration, allowing inter-site comparisons.
Potential extracellular enzyme activities (EEA) were determined following the method outlined by Saiya-Cork et al. (2002). Briefly, the activity of 1,4-β-glucosidase, cellobiohydrolase, 1,4,-β-N-acetyl-glucosaminidase, phosphatase, and sulfatase were determined fluorometrically using 200 μM methylumbelliferone (MUB)-linked substrates. Peroxidase and phenol oxidase activities were determined with a colorimetric assay using 25 mM l-3,4,-dihydroxy-phenylalanine (l-DOPA) as a substrate; 10 μl of 0.3% H2O2 was added to the peroxidase assay. A soil slurry was created by combining 0.5 g of peat (wet mass) to 125 ml of acetate buffer (50 mM, pH 5) and homogenized for 60 s with a hand blender. A preliminary study showed that estimated enzyme activity was not influenced by the mass of peat used (max. 2.5 g). For each enzyme assay, 200 μl of soil slurry and 50 μl of specific substrate were loaded onto a 96-well microplate with eight analytical replicates. Each plate contained a positive and negative control. All assays were incubated at 21°C for 2 h, except for phosphatase and 1,4,-β-N-acetyl-glucosaminidase assays which were incubated for 0.5 h. To stop the reaction in the fluorometric assays 10 μl of 0.5 N NaOH was added at the end of the incubation period. Fluorescence (excitation energy 365 nm, emission 460 nm) and absorbance (460 nm) was measured using a multi-plate spectrophotometer (Synergy HT, Biotek, USA).
Temperature and aeration control of microbial activity
To test how future environmental change might influence microbial activity, peat microcosms for each sub-sample were established by adding 5 g of homogenized wet peat to 20 ml serum vials. Each vial was capped with a rubber stopper and crimped. Vials were randomly allocated to one of four treatments with two temperatures and either oxic or anoxic conditions: 4°C oxic, 4°C anoxic, 14°C oxic, and 14°C anoxic. Each treatment had three replicates. Anoxic conditions were established by repeated evacuation and back-flushing with N2 gas and brought to atmospheric pressure. Gas samples were taken at 90, 144, and 270 h and transferred to 10 ml pre-evacuated glass 10 ml crimped glass vials with butyl rubber stoppers (Geo-Microbial Technologies Inc., Ochelata, OK, USA), where they were later measured for CO2 and CH4 concentration using a SRI 8610-0040 gas chromatograph with a FID and methanizer (SRI Instruments, Torrance, CA, USA). As the enzyme assays required destructive sampling (0.5 g was removed), one replicate per treatment was analyzed for each enzyme activity per time period. The mass of the remaining peat was recorded and the treatment conditions re-established. The fractional rate of increase in microbial activity in the 14°C over 4°C temperature (the Q10) was calculated for each site and depth.
Statistics
Enzyme activity and SIR ratios were analyzed with non-metric multidimensional scaling (NMDS) ordination using Bray–Curtis distance, while T-RF proportional abundance data (square-root transformed) was analyzed with Sørensen’s distance. All the NMDS ordinations were run using a random starting configuration, a maximum of 250 iterations and an instability criterion of 0.00001; two dimensions were used for all plots and Monte Carlo tests (1,000 randomized runs) was used to determine significance. The Shannon–Wiener index (Hill et al., 2003) was used to estimate T-RF diversity. Analysis of variance (ANOVA) with Tukey’s post hoc test was used to determine differences in basal respiration. Classification and regression tree analysis (CART) was conducted to partition the relative effects of peat sample and the experimental variables on CO2 and CH4 production in the microcosm experiment. Relationships between CO2 and CH4 concentration were determined using Pearson’s correlation analysis. Exploratory analysis of the measured variables was analyzed in two steps, as there are more variables than peat samples, following a similar method to Hudson et al. (2003). Firstly, variables that correlated with basal CO2 respiration, pH, enzyme activity, and SIR data to a probability level of < 0.1 were identified. Then, backward stepwise linear multiple regression was used to identify the most important variables. The residuals met the parametric assumptions of the model. All statistics were performed following tests for normality and heteroscedasticity. Data were natural log transformed when parametric assumptions were not met and pH data were log10 transformed prior to analysis. All statistical analyses were performed using R v. 2.13. (R Development Core Team, 2011) with the package vegan for NMDS ordination (Oksanen et al., 2011) and mvpart for CART analysis (De’ath, 2011).
Results
Peat pH
Victor Fen surface peat had the highest pH of 5.8 and there was no difference in pH between middle and deep samples (pH 5.0). Kinoje and Victor bogs were more acidic and had the same pH value of 3.4 at surface and similar pH values of 3.2 and 3.4 were observed in the middle peat samples and 3.8 and 3.5 in the deep peat, respectively.
Microbial community characterization
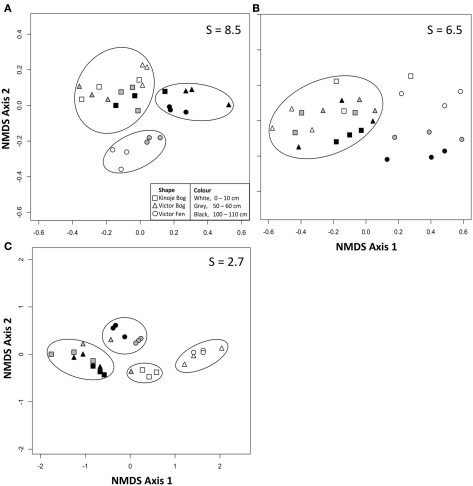
NMDS ordination showed that the bacterial T-RFs among the peat samples appeared to form a single cluster (Figure 1A), as many abundant T-RFs were found among the peat samples (Table A1 in Appendix). However, bacterial richness was greatest in Kinoje and Victor bogs and many of the same taxa were present in the two peatlands although more unique T-RFs were found at Victor Bog primarily at 0–10 cm and 50–60 cm (Table A1 in Appendix). NMDS ordination of the archaeal T-RFs showed clustering among the peatland types (Figure 1B), however many of the same T-RFs were identified at each site and clustering appears to be driven by a few rare T-RFs (Table A1 in Appendix). There was not enough dissimilarity among the fungal T-RFs to separate by peat sample with NMDS ordination.
Figure 1.
Two dimensional non-metric multidimensional scaling (NMDS) plots of (A) bacterial terminal restriction fragments (T-RFs) and (B) archaeal T-RFs identified at three depths from peat cores collected from peatlands within the James Bay Lowlands. S indicates the stress value as a percentage.
Kinoje Bog MB-C at 0–10 cm was approximately six times greater than at both Victor Bog and Fen (Table 1). MB-C generally decreased with depth except at Victor Bog 50–60 cm, which had a biomass 1.7 times greater than the surface. The carbon:nitrogen ratio typically decreased from the surface to deep samples, but the ratio was similar among all depth samples at Victor Bog.
Table 1.
Microbial biomass C and N within peat soil at three depths from cores collected from three James Bay peatlands (n = 1).
| Peatland | Sample depth (cm) | MB-C (μg g−1) | MB-N (μg g−1) | C:N |
|---|---|---|---|---|
| Kinoje Bog | 0–10 | 1463 | 75.5 | 19.7 |
| 50–60 | 211 | 14.9 | 14.7 | |
| 100–110 | 61 | 6.5 | 10.7 | |
| Victor Bog | 0–10 | 254 | 9.9 | 26.8 |
| 50–60 | 431 | 22.6 | 19.5 | |
| 100–110 | 92 | 4.6 | 22.8 | |
| Victor Fen | 0–10 | 210 | 17.7 | 11.8 |
| 50–60 | 9 | 5.9 | 13.5 | |
| 100–110 | 80 | 14.8 | 5.5 |
Microbial activity
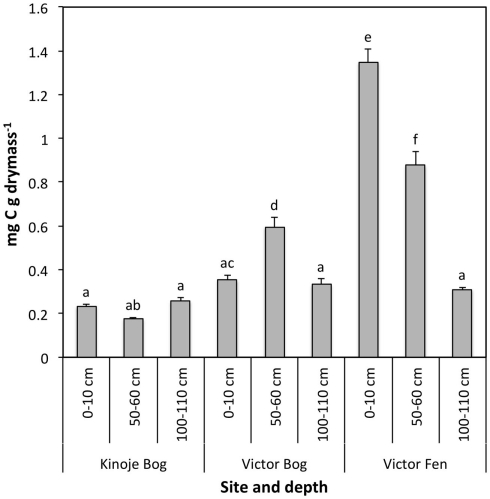
Aerobic basal CO2 respiration was significantly higher in the Victor Fen surface and middle depth peat samples compared with the deep fen peat and peat from all depths at both Kinoje and Victor Bogs (F4,18 = 84.0, p < 0.001; Figure 2). There was little difference in CO2 production among the bog peat samples, except for Victor Bog middle depth where CO2 production was approximately two times greater than surface and deep peat. Patterns of anaerobic basal respiration were similar to aerobic basal respiration and were strongly correlated (data not shown).
Figure 2.
Basal respiration (mean ± 1 SE; n = 3) in an oxic environment after 24 h in peat collected at three depths across three James Bay peatlands. Different letters indicate significance difference at p < 0.05.
The overall magnitude of substrate utilization in the oxic SIR assay generally increased with depth as indicated by the mean ratio, except for the Victor Bog middle depth sample (Table 2). Deep peat had the highest SIR ratios on average and the NMDS ordination of the oxic SIR ratios revealed that most of the deep peat samples clustered together (Figure 3A). Kinoje Bog and Victor Bog had similar responses to substrate addition in both magnitude and relative ability to mineralize different substrates, while Victor Fen formed a separate cluster on the NMDS ordination, particularly as the surface and middle samples had higher average ratios than the two bogs. In contrast, substrate utilization in the anoxic SIR assay was greatest in the surface samples, except at Victor Bog where the middle peat sample was highest on average. NMDS ordination of the anoxic SIR ratios showed little clustering among the peat depth samples within the sites, but Kinoje and Victor Bog appear to have a similar response. There was little clustering among the Victor Fen samples (Figure 3B).
Table 2.
Substrate induced respiration ratios (substrate/basal respiration; mean ± 1 SE) under oxic (16 h) and anoxic conditions (48 h) of peat soil at three depths from peat cores collected from the James Bay Lowlands.
| Study site | Depth (cm) | Synthetic substrates |
|||||
|---|---|---|---|---|---|---|---|
| Alkali lignin | Amino acids | Glucose | Methyl cellulose | p-coumaric acid | Sodium benzoate | ||
| OXIC | |||||||
| Kinoje Bog | 0–10 | 2.08 (0.24) | 1.08 (0.10) | 0.89 (0.06) | 1.25 (0.12) | 0.92 (0.09) | 0.66 (0.05) |
| 50–60 | 2.03 (0.08) | 2.06 (0.13) | 0.99 (0.08) | 1.27 (0.09) | 0.94 (0.04) | 0.86 (0.06) | |
| 100–110 | 1.98 (0.25) | 1.49 (0.17) | 1.04 (0.12) | 0.94 (0.03) | 0.84 (0.09) | 0.99 (0.07) | |
| Victor Bog | 0–10 | 1.98 (0.20) | 1.67 (0.12) | 1.66 (0.02) | 1.13 (0.07) | 0.78 (0.01) | 0.93 (0.11) |
| 50–60 | 1.24 (0.12) | 1.28 (0.10) | 0.72 (0.05) | 0.78 (0.07) | 0.66 (0.06) | 0.62 (0.04) | |
| 100–110 | 3.48 (0.04) | 2.37 (0.41) | 1.69 (0.17) | 1.74 (0.14) | 1.48 (0.13) | 0.97 (0.03) | |
| Victor Fen | 0–10 | 1.62 (0.17) | 2.28 (0.13) | 3.27 (0.25) | 0.88 (0.04) | 1.47 (0.13) | 2.06 (0.08) |
| 50–60 | 1.96 (0.08) | 2.16 (0.14) | 2.47 (0.06) | 1.09 (0.12) | 1.23 (0.14) | 0.78 (0.09) | |
| 100–110 | 1.85 (0.06) | 2.71 (0.15) | 1.49 (0.12) | 1.02 (0.08) | 0.96 (0.05) | 0.81 (0.06) | |
| ANOXIC | |||||||
| Kinoje Bog | 0–10 | 2.04 (0.74) | 2.65 (0.24) | 2.48 (0.92) | 1.99 (0.46) | 1.40 (0.19) | 0.65 (0.09) |
| 50–60 | 1.30 (0.33) | 1.36 (0.22) | 1.44 (0.34) | 1.06 (0.06) | 0.89 (0.20) | 0.40 (0.13) | |
| 100–110 | 1.97 (0.26) | 1.26 (0.30) | 1.30 (0.08) | 1.03 (0.11) | 0.95 (0.08) | 0.51 (0.09) | |
| Victor Bog | 0–10 | 0.91 (0.24) | 0.93 (0.05) | 1.26 (0.27) | 1.08 (0.04) | 0.52 (0.04) | 0.47 (0.05) |
| 50–60 | 2.13 (0.45) | 1.82 (0.08) | 1.71 (0.16) | 1.39 (0.22) | 1.11 (0.21) | 0.89 (0.04) | |
| 100–110 | 1.50 (0.30) | 1.26 (0.44) | 1.09 (0.25) | 1.10 (0.41) | 0.86 (0.18) | 0.53 (0.15) | |
| Victor Fen | 0–10 | 2.58 (0.61) | 3.02 (0.50) | 5.24 (1.52) | 1.02 (0.12) | 1.51 (0.05) | 2.81 (0.49) |
| 50–60 | 1.57 (0.18) | 2.96 (0.35) | 2.91 (1.03) | 0.94 (0.24) | 0.25 (0.06) | 0.76 (0.08) | |
| 100–110 | 1.17 (0.04) | 1.96 (0.59) | 1.52 (0.05) | 0.85 (0.11) | 0.81 (0.15) | 0.47 (0.15) | |
| Study site | Depth (cm) |
Organic substrates |
|||||
| Rich fen sedge | Int. fen sedge | Int. fen Sphagnum | Poor fen Sphagnum | Mean ratio | |||
| OXIC | |||||||
| Kinoje Bog | 0–10 | 2.97 (0.65) | 1.75 (0.03) | 3.72 (0.94) | 4.65 (0.48) | 2.00 | |
| 50–60 | 3.80 (0.48) | 2.36 (0.25) | 4.36 (0.27) | 4.37 (0.59) | 2.30 | ||
| 100–110 | 6.01 (0.58) | 2.26 (0.38) | 4.36 (0.52) | 4.92 (0.51) | 2.48 | ||
| Victor Bog | 0–10 | 3.12 (0.30) | 2.26 (0.29) | 5.46 (0.13) | 6.68 (0.53) | 2.57 | |
| 50–60 | 3.11 (0.40) | 1.78 (0.22) | 2.83 (0.13) | 4.53 (0.32) | 1.75 | ||
| 100–110 | 9.53 (0.55) | 4.85 (1.33) | 6.55 (1.23) | 6.60 (0.64) | 3.93 | ||
| Victor Fen | 0–10 | 3.63 (0.39) | 3.28 (0.38) | 2.51 (0.15) | 3.42 (0.33) | 2.44 | |
| 50–60 | 4.88 (0.30) | 5.72 (0.54) | 3.09 (0.29) | 4.48 (0.42) | 2.79 | ||
| 100–110 | 7.25 (0.88) | 4.85 (0.42) | 4.58 (0.02) | 6.24 (0.25) | 3.18 | ||
| ANOXIC | |||||||
| Kinoje Bog | 0–10 | 2.06 (0.50) | 1.89 (0.63) | 2.69 (0.08) | 3.91 (1.06) | 2.18 | |
| 50–60 | 1.65 (0.30) | 1.50 (0.42) | 2.49 (0.31) | 2.16 (0.61) | 1.43 | ||
| 100–110 | 2.26 (0.49) | 3.08 (0.26) | 2.16 (0.57) | 2.62 (0.27) | 1.71 | ||
| Victor Bog | 0–10 | 1.31 (0.21) | 1.15 (0.04) | 1.82 (0.42) | 1.43 (0.53) | 1.09 | |
| 50–60 | 2.22 (0.36) | 1.91 (0.19) | 3.01 (0.42) | 2.94 (0.78) | 1.91 | ||
| 100–110 | 1.99 (0.46) | 2.27 (0.44) | 2.75 (0.59) | 2.80 (0.50) | 1.62 | ||
| Victor Fen | 0–10 | 4.3 (0.94) | 5.79 (1.31) | 3.55 (0.58) | 4.99 (0.30) | 3.48 | |
| 50–60 | 5.25 (1.56) | 6.32 (0.65) | 4.15 (0.32) | 6.56 (1.93) | 3.17 | ||
| 100–110 | 5.35 (0.82) | 5.95 (0.80) | 4.55 (0.91) | 6.36 (1.31) | 2.90 | ||
Int. represents Intermediate
Figure 3.
Two dimensional non-metric multidimensional scaling (NMDS) plots of (A) oxic (after 16 h) and (B) anoxic (after 48 h) substrate induced respiration responses of peat microbial communities and (C) initial extracellular enzyme activity (EEA) from three peat cores collected from the James Bay Lowlands. S indicates the stress value as a percentage and the circles grouping data are guides only.
Enzyme activity generally decreased with depth, but the Kinoje Bog middle depth sample had higher B-xylosidase, cellobiohydrolase, sulfatase, and peroxidase activities (Table 3). NMDS ordination of the initial EEAs showed that Victor Bog and Fen surface samples had similar activities (Figure 3C). While Kinoje Bog surface peat had much lower enzyme activities (between 2.5 and 6 times; Table 3) the samples did cluster close to the Victor peatland surface samples. Similarly, Kinoje and Victor bog middle and deep samples clustered together, whereas Victor Fen middle and deep peat formed their own group as enzyme activities were much higher (range 0.5–12.7 times).
Table 3.
Activity of enzymes (mean ± 1 SE nmol h−1 g−1) within peat soil collected at three depths (n = 3) from peat cores collected in the James Bay Lowlands.
| Sample location | Sample depth (cm) | N-acetyl-beta-d-glucosaminide | Phosphatase | β-glucosidase | Sulfatase | B-xylosidase | Cello-biohydrolase | Per-oxidase | Phenol oxidase |
|---|---|---|---|---|---|---|---|---|---|
| Kinoje Bog | 0–10 | 360.6 (67.5) | 8261.1 (1058.4) | 1096.8 (101.5) | 4.8 (0.9) | 36.6 (0.6) | 88.1 (5.8) | 1.9 (0.7) | 0.3 (0.1) |
| 50–60 | 16.1 (9.6) | 735.9 (291.5) | 339.2 (80) | 23.6 (3.4) | 78.2 (12) | 21.7 (5.5) | 5.6 (0.8) | 2.1 (0.8) | |
| 100–110 | 48.7 (9.3) | 1845 (244.3) | 338.5 (48) | 0 (0) | 45.5 (14.3) | 21.8 (1.7) | 2.4 (1.3) | 0 (0) | |
| Victor Bog | 0–10 | 900.2 (333) | 45151.8 (16612.7) | 2611.9 (500.3) | 32.3 (20.3) | 196.3 (94.4) | 248.5 (34.2) | 9.6 (1.3) | 1.6 (0.9) |
| 50–60 | 139.3 (76) | 2164.9 (1264) | 1047.2 (313.6) | 20.6 (15.7) | 119.1 (5.1) | 64.6 (36.7) | 2.1 (1.1) | 3.4 (2.4) | |
| 100–110 | 41.2 (3.5) | 1126.9 (334.7) | 387.2 (81.3) | 0 (0) | 89.4 (14.3) | 4.7 (0.5) | 4.2 (2.3) | 1 (1) | |
| Victor Fen | 0–10 | 1667.6 (112.5) | 39231.2 (4371.8) | 3913.3 (200.2) | 145.2 (52.2) | 362.5 (52.6) | 415.5 (16.6) | 29.4 (6.5) | 3.5 (1.9) |
| 50–60 | 1777.5 (266.8) | 3227.4 (60.1) | 2686.2 (150.8) | 85.3 (15) | 145 (5.7) | 344.3 (24) | 17.3 (2.6) | 2 (0.7) | |
| 100–110 | 647.8 (69.9) | 1072.6 (280.5) | 2199.1 (217.8) | 0 (0) | 79.6 (2.1) | 275 (22.6) | 17.2 (3.9) | 1.1 (0.6) |
Influence of temperature and aeration on microbial activities
Temperature only affected potential CO2 production rates in the surface peat samples at Victor Bog and Victor Fen (Figure 4), where rates were highest in the oxic treatment at 14°C and lowest in the anoxic treatment at 4°C. Although potential CO2 production rates in an oxic environment were greatest at Victor Fen, the largest effect of temperature was observed at Victor Bog in both aeration conditions. Mean Q10 values at Victor Bog were 1.57 ± 0.05 and 2.34 ± 0.07, and 1.46 ± 0.03 and 1.52 ± 0.16 at Victor Fen in an oxic and anoxic environment, respectively. In contrast, similar potential CO2 production rates were observed in the middle and deep samples at both Victor Bog and Victor Fen. Interestingly, temperature and aeration states did not have an affect on potential CO2 production rates at Kinoje Bog, where higher rates were found in deeper peat. Overall, anoxic potential CO2 production rates followed a similar pattern to oxic production rates, and there was a strong correlation between the two (t52 = 15.5, p < 0.001, r2 = 0.82).
Figure 4.
Classification and regression tree (CART) analysis to describe the effect of temperature (4 and 14°C) and aeration (anoxic and oxic) on average CO2 production rates (mg CO2 g drymass−1 day−1) after 270 h (n = 3) in a microcosm experiment using peat soil collected at three depths (0–10, 50–60, and 100–110 cm) from peatlands in the James Bay Lowlands, Canada. At each split the following information is given: the variable that splits the observation into “daughter” nodes, the classification criterion (e.g., peatland, depth, aeration, or temperature), the average potential CO2 production rate and the number of observations in each group (ni).
Similarly, temperature had no effect on potential CH4 production rates in any peat sample and there was no difference in potential CH4 production rates among the middle and deep peat samples at all three peatlands (Figure 5). Compared to middle and deep peat samples the potential CH4 production rates in surface peat at Victor Fen were 2 times greater and 3.7 times greater at Victor Bog, but were approximately 50% lower at Kinoje Bog. Moreover, anoxic CH4 production rates correlated with oxic (t52 = 5.2, p < 0.001, r2 = 0.34) and anoxic (t52 = 4.7, p < 0.001, r2 = 0.29) CO2 production rates.
Figure 5.
Classification and regression tree (CART) analysis to describe the effect of temperature and aeration (anoxic and oxic) on potential CH4 production rate (μg CH4 g drymass−1 day−1) after 270 h (n = 3) in a microcosm experiment using peat soil collected at three depths (0–10, 50–60, and 100–110 cm) from peatlands in the James Bay Lowlands, Canada. See Figure 4 for CART explanation.
Measured EEA in the microcosm experiment at 90, 180, and 270 h did not correlate with CO2 production in either the oxic or anoxic environments, except for a weak correlation between oxic CO2 production and sulfatase (t106 = 4.3, p < 0.001, r2 = 0.15), but this was primarily driven by the absence of enzyme activity in deep peat.
Interactions among measured variables
Although basal CO2 respiration correlated well for several variables with r values ranging from 0.6 to 0.85 (Table 4), only pH was included in the final regression model (basal CO2 mg g−1 = 0.07 + 0.48(pH); F1,7 = 8.5, p < 0.05, adjusted r2 = 0.49). Similarly, pH could also explain anoxic SIR values (Anoxic NMDS SIR = 1.41–0.35(pH); F1,7 = 32.3, p < 0.001, adjusted r2 = 0.77), but not oxic SIR values. Despite pH correlating well with several enzyme activities, it was not significant in the final regression model. Interestingly, bacterial and fungal diversity did not correlate with pH, but good correlations were observed for measures of archaeal diversity and pH. Overall enzyme activity (NMDS values) correlated with bacterial diversity, but the regression was not significant, but there was no relationship with archaeal or fungal diversity.
Table 4.
Pearson’s product moment correlations between measures of microbial activity/pH and potential explanatory variables.
| Dependent variable | Independent variable | Test statistic | r | Consistent with hypothesis |
|---|---|---|---|---|
| Basal CO2 mg | pH | t7 = 2.9, p < 0.05 | 0.74 | Yes |
| Archaea Shannon | t7 = 2.0, p < 0.1 | 0.6 | Yes | |
| NMDS enzyme | t7 = 2.2, p < 0.1 | 0.65 | Yes | |
| β-1-4-glucosidase | t7 = 3.9, p < 0.05 | 0.83 | Yes | |
| Cellobiohydrolase | t7 = 3.4, p < 0.05 | 0.79 | Yes | |
| β-N-acetyl-glucosaminidase | t7 = 4.3, p < 0.05 | 0.85 | Yes | |
| Peroxidiase | t7 = 3.1, p < 0.05 | 0.76 | Yes | |
| Phenol oxidase | t7 = 2.5, p < 0.05 | 0.69 | Yes | |
| Sulfatase | t7 = 5.3, p < 0.001 | 0.9 | Yes | |
| B-xylosidase | t7 = 4.1, p < 0.01 | 0.84 | Yes | |
| NMDS initial EEA | NMDS bacteria | t7 = 2.2, p < 0.01 | −0.64 | Yes |
| NMDS oxic SIR | MB-C | t7 = 2.4, p < 0.05 | −0.68 | Yes |
| MB-N | t7 = 2.2, p < 0.01 | −0.65 | Yes | |
| NMDS archaea | t7 = 2.2, p < 0.01 | −0.63 | Yes | |
| NMDS anoxic SIR | pH | t7 = 5.7, p < 0.001 | −0.91 | Yes |
| β-1-4-glucosidase | t7 = 1.9, p < 0.1 | −0.59 | Yes | |
| Cellobiohydrolase | t7 = 2.5, p < 0.05 | −0.68 | Yes | |
| β-N-acetyl-glucosaminidase | t7 = 2.5, p < 0.05 | −0.67 | Yes | |
| Peroxidase | t7 = 2.9, p < 0.05 | −0.73 | Yes | |
| Sulfatase | t7 = 1.9, p < 0.1 | −0.58 | Yes | |
| pH | Archaea Shannon | t7 = 2.5, p < 0.05 | 0.68 | Yes |
| NMDS Archaea | t7 = 2.2, p < 0.01 | −0.63 | Yes | |
| β-1-4-glucosidase | t7 = 3.4, p < 0.05 | 0.79 | Yes | |
| Cellobiohydrolase | t7 = 4.2, p < 0.01 | 0.85 | Yes | |
| β-N-acetyl-glucosaminidase | t7 = 3.6, p < 0.01 | 0.8 | Yes | |
| Peroxidase | t7 = 6.5, p < 0.001 | 0.93 | Yes | |
| Sulfatase | t7 = 2.9, p < 0.05 | 0.73 | Yes | |
| Xylase | t7 = 2.1, p < 0.01 | 0.63 | Yes |
NMDS represents the first axis from the respective NMDS plot.
Discussion
Microbial community characterization
The same dominant microbial taxa identified using T-RFLP were found among all three peatlands despite differences in geographic location, nutrient status, and plant community composition. This contradicts our initial hypothesis and previous studies where differences in microbial community composition have been identified among peatlands with different vegetation characteristics (Ausec et al., 2009; Peltoniemi et al., 2009; Andersen et al., 2010). However, this finding is consistent with previous peatland studies comparing bacterial communities in two similar fens in Slovenia (Kraigher et al., 2006) and archaea and bacteria in Sphagnum dominated bogs across the northeastern USA (Basiliko et al., 2003; Morales et al., 2006). In contrast to previous studies (Bååth and Anderson, 2003; Fierer and Jackson, 2006; Rousk et al., 2010), where higher bacterial diversity is associated with more neutral pH, a greater number of taxa were actually detected in the more acidic bogs rather than the fen. Moreover, there was no correlation between pH and the microbial community structure or diversity indices. However, the range of peat pH values in the JBL was relatively small, which may explain why diversity was not explained by pH. Furthermore, pH also appeared not to influence microbial community composition in other peatland studies (Kraigher et al., 2006; Ausec et al., 2009), suggesting that the relationship between pH and microbial diversity in general might be less important in peatland ecosystems.
Richness was also similar among the depths with few taxa confined to a single depth profile, in contrast to Morales et al. (2006) who found a greater number of bacterial T-RFs in surface peat (0–15 cm) than in deep peat (1 m). Although this may have been due to the relatively low detected richness level relative to Morales et al. (2006), it suggests that at least the dominant members of the microbial community are always present, despite presumed differences in oxygen content with depth.
Little is known about the role of fungi in peatlands, but diverse communities have been found beneath the Sphagnum mat and in close association with ericaceous shrubs (Thormann, 2006; Artz et al., 2007) and different fungal communities have been linked to the vegetation and nutrient status of peatlands (Artz et al., 2007; Trinder et al., 2008). Fungi are generally thought to predominate in peatland lawns and hummocks (Jaatinen et al., 2007) as they cannot utilize alternative electron acceptors in anoxic environments and successfully compete with bacteria for carbon substrates (Killham and Prosser, 2007). Thus it was interesting that most fungal taxa were identified at all three depths among the three JBL peatlands. Fungi are typically most competitive under an oxic environment, however they can carry out fermentation. So their abundance at depth may in part be due to a lack of alternative inorganic electron acceptors (e.g., sulfate) that might otherwise have facilitated anaerobic bacterial respiration in these deep anoxic peat samples. This might also imply that facultative fungi supply fermentation products that ultimately fuel methanogenesis (Conrad, 2007).
Consistent with prior studies, microbial biomass decreased with depth probably due to the presence of more labile organic matter and higher redox states typically found near the surface of peat profiles (Blodau and Moore, 2002; Blodau et al., 2004; Basiliko et al., 2007). Although microbial biomass across sites was generally quite small compared to other bogs and fens (Moore and Basiliko, 2006), it was similar to that in mined peatlands in more southern regions of Canada that have relatively low nutrient and carbon availability (Basiliko et al., 2007); which might indicate that substrate or nutrient availability for heterotrophic microbial communities is low even in the fen site. Decreasing MB-C:MB-N ratios with depth has been observed elsewhere (e.g., Basiliko et al., 2007) and is probably due to decreasing fungal:bacterial ratios with depth and the fact that bacteria typically have higher anabolic N requirements (Killham and Prosser, 2007). Bacteria often predominate in deeper more anoxic peat layers as they can utilize alternative electron acceptors (Killham and Prosser, 2007). Microbial biomass did not correlate with potential CO2 production rates in contrast to previous studies (Blodau et al., 2004; Basiliko et al., 2007). However, this may simply reflect the low microbial biomass and low amount of labile carbon in our sites.
Microbial activities
Despite the similarity of the microbial communities among the three peatlands, there were differences in basal respiration, initial enzyme activities and substrate utilization patterns. Greater microbial activity was observed in the fen compared to the bogs, possibly due to the higher in situ nutrient concentrations and greater carbon substrate availability typically found in fens (Moore and Basiliko, 2006; Knorr and Blodau, 2009). Overall the average oxic SIR ratio response was very similar among deep peat samples in all three peatlands and greater than those in both the surface and middle peat samples. Deeper peat is older and typically has lower levels of labile carbon (Glatzel et al., 2003; Artz et al., 2006). Thus, the microbial community at these depths was probably substrate limited relative to the surface peat. However, this trend disappeared in the anoxic SIR assay, as there was little difference in the response among depths between Kinoje and Victor bogs. In contrast, Victor Fen had greater average SIR values in the surface peat sample, likely as a result of higher redox substrate concentrations (Knorr and Blodau, 2009; Webster and McLaughlin, 2010).
This apparent contradiction between microbial activity and community composition may be explained by differences in peat organic matter quality. Plant litter from bogs and fens have different nutrient concentrations (Bragazza et al., 2007) and therefore microorganisms in these contrasting habitats will have different nutrient requirements. Recently, Straková et al. (2011) identified litter type as the main factor affecting microbial EEA in bog peat soil. Similar to our study the authors reported higher enzyme activities in peat composed of vascular plants, typical in fens, rather than Sphagnum dominated peat and also failed to identify a strong relationship between microbial community composition and EEA. Moreover, the differences in SIR activities, identified by the NMDS ordination, among sites was primarily due to the magnitude of the response, but the relative substrate utilization patterns (i.e., among the 10 substrates) were actually similar among the three peatlands despite our hypothesis that the two bogs and fen would preferentially mineralize different substrates, probably because the microbial communities were similar. The subdued response to substrate addition in the bogs may be due to inhibitory effects of Sphagnum leachates on microbial activity (Verhoeven and Toth, 1995; Hättenschwiler and Vitousek, 2000; Hájek et al., 2010). Furthermore, vegetation type has been shown to influence microbial activity more than water table drawdown (Trinder et al., 2008; Straková et al., 2011) and small changes in the plant community composition can have profound affects on microbial processes, such as methanogenesis (Hines et al., 2008).
The relatively high N-acetyl-beta-d-glucosaminidase activity throughout the peat profile suggests a high metabolic demand for N released during chitin turnover (Sinsabaugh et al., 1993) and corresponds with presence and identification of fungal taxa at all peat depths. This contradicts our previous finding that carbon:nitrogen ratio decreases with depth and might represent bacterial dominance of decomposition due to higher N demand. However, it is not certain whether fungi or bacteria produced these enzymes or what the specific activity of the fungi is in deep peat. Interestingly, a recent meta-analysis found that shifts in bacterial to fungal dominance across environmental gradients often do not correlate with changes in element cycling and may be due to niche overlap between fungal and bacterial communities (Strickland and Rousk, 2010). In an antibiotic inhibition assay, Winsborough and Basiliko (2010) showed that bacteria dominated peatland microbial activity; while fungal activity did increase in acidic dry bogs it was still lower than bacterial activity. Although we do not have direct measurements of fungal biomass, these results suggest that our understanding of the relationship between fungi and ecosystem function in peat soil is incomplete.
We did not find a relationship between microbial community composition and activities in contrast to our hypothesis. Although this could be due to the issues of the taxa-level resolution using T-RFLP small-subunit rDNA, this finding highlights the importance of carbon substrate as a proximate control of microbial community dynamics because similar communities can perform different functions given different resources and conditions.
Environmental change
As predicted and consistent with previous studies (Moore and Dalva, 1993; Updegraff et al., 1998; Keller et al., 2004), an increase in aeration of the peat soil stimulated CO2 production rates, but only in Victor Bog and Fen, and temperature only affected rates in the surface peat samples. This suggests that oxygen availability is more important than temperature in increasing mineralization rates, possibly brought about by an increase in enzyme activity in response to a release from inhibition through the “enzymic latch theory” (Freeman et al., 2001, 2004). However, CO2 production did not correlate with enzyme activity probably because the enzymes measured in this study may only be a subset of those actually involved in peat decomposition (Horwath, 2007). Potential CO2 production rates among the depths were lower in anoxic conditions but there was a strong correlation between oxic and anoxic CO2 production consistent with previous investigations (Moore and Dalva, 1997; Yavitt et al., 1997; Glatzel et al., 2004). These findings suggest that similar peat properties control the activities of both aerobic and anaerobic microbial metabolism (Glatzel et al., 2004; Basiliko et al., 2007). Similarly, CH4 production rates were low throughout the peat profile but still within the range of previously reported values (Moore and Dalva, 1993; Glatzel et al., 2004; Basiliko et al., 2007).
Conclusion
Microbial community composition was very similar among peatlands and at depths within the JBL despite differences in geographic location and nutrient status. In contrast, microbial activity appears to be determined by the quality of the peat substrate and the presence of potential microbial inhibitors. As climate change is expected to cause a shift in JBL plant community composition, this study suggests that the microbial community will respond quickly to changes in plant litter and root exudate quality but the existing peat substrate will probably have a large influence on future microbial activity. For example, a shift from Sphagnum to sedge-dominated peatlands may not necessarily result in the expected increase in carbon mineralization due to the antimicrobial properties of the Sphagnum-peat. Interestingly, we identified fungal taxa in deep peat but it is unclear what anaerobic processes are occurring and how these organisms influence carbon cycling. Thus, more in depth profiling of the JBL microbial community and identification of the potential constraints on microbial activity is required in order to better predict future peatland carbon dynamics.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors gratefully acknowledge Benoit Hamel, Mark Crofts, and Adam Kinnunen for collection of peat samples, Varun Gupta and Charlotte Hewins for assistance with laboratory analyses and the two reviewers for comments and suggestions to improve the manuscript. Funding was provided by Ontario Ministry for Natural Resources, The National Science Foundation Enzymes in the Environment Research Coordination Network, The Corning Institute for Education and Research, and The Reinberger Foundation.
Appendix
Table A1.
Operational taxonomic units (OTU) of archaea, bacteria, and fungi identified at three depths in peatlands across the James Bay Lowlands, Canada using terminal restriction fragment length polymorphism analysis (T-RFLP) of rDNA.
| Site | T-RF length | Proportion within each depth |
||
|---|---|---|---|---|
| 0–10 cm | 50–60 cm | 100–110 cm | ||
| ARCHAEA | ||||
| Kinoje bog | 51 | – | – | 0.73 |
| 178 | 1.00 | 0.25 | – | |
| 384 | – | – | 0.02 | |
| 483 | – | 0.75 | 0.25 | |
| Victor bog | 51 | 0.03 | – | – |
| 178 | 0.03 | 0.03 | – | |
| 193 | 0.07 | 0.07 | – | |
| 384 | 0.43 | 0.45 | 0.60 | |
| 483 | 0.41 | 0.42 | 0.40 | |
| 729 | 0.03 | 0.03 | – | |
| Victor fen | 51 | – | 0.07 | – |
| 67 | – | – | 0.04 | |
| 75 | – | 0.05 | 0.11 | |
| 83 | 0.11 | 0.10 | 0.42 | |
| 249 | – | 0.04 | – | |
| 277 | 0.27 | 0.26 | 0.11 | |
| 384 | 0.28 | 0.20 | 0.14 | |
| 483 | 0.34 | 0.23 | 0.13 | |
| 729 | – | 0.04 | 0.05 | |
| BACTERIA | ||||
| Kinoje bog | 59 | – | – | 0.02 |
| 61 | 0.07 | – | – | |
| 66 | 0.88 | 0.15 | 0.04 | |
| 82 | – | 0.08 | 0.02 | |
| 89 | – | 0.41 | 0.01 | |
| 136 | – | – | 0.02 | |
| 140 | 0.02 | 0.13 | – | |
| 144 | 0.03 | 0.03 | 0.02 | |
| 160 | – | 0.02 | – | |
| 165 | – | – | 0.12 | |
| 260 | – | 0.10 | 0.04 | |
| 429 | – | 0.02 | – | |
| 447 | – | 0.03 | – | |
| 476 | – | – | 0.17 | |
| 481 | – | – | 0.26 | |
| 484 | – | – | 0.24 | |
| 491 | – | – | 0.03 | |
| 509 | – | 0.02 | – | |
| Victor bog | 54 | 0.06 | – | – |
| 59 | 0.04 | – | – | |
| 61 | 0.01 | 0.07 | 0.04 | |
| 66 | 0.37 | 0.13 | 0.08 | |
| 89 | 0.03 | 0.08 | 0.10 | |
| 110 | 0.02 | – | – | |
| 123 | 0.02 | – | – | |
| 133 | – | 0.03 | – | |
| 136 | 0.09 | 0.16 | 0.15 | |
| 140 | 0.07 | – | – | |
| 144 | 0.08 | 0.14 | 0.20 | |
| 147 | 0.01 | – | – | |
| 160 | 0.01 | – | – | |
| 165 | 0.01 | 0.07 | 0.08 | |
| 260 | 0.04 | 0.06 | – | |
| 280 | – | 0.03 | – | |
| 284 | 0.04 | – | – | |
| 301 | 0.02 | – | – | |
| 429 | 0.03 | – | – | |
| 432 | 0.01 | – | – | |
| 447 | – | 0.03 | – | |
| 450 | 0.01 | – | – | |
| 476 | – | 0.15 | 0.14 | |
| 484 | – | – | 0.22 | |
| 496 | 0.02 | – | – | |
| 519 | – | 0.04 | – | |
| Victor fen | 66 | 0.30 | 0.60 | 0.30 |
| 79 | – | – | 0.07 | |
| 89 | – | 0.04 | – | |
| 97 | – | 0.04 | – | |
| 144 | – | – | 0.19 | |
| 176 | 0.04 | – | – | |
| 484 | 0.21 | 0.08 | 0.30 | |
| 496 | – | – | 0.13 | |
| 513 | 0.21 | 0.12 | – | |
| 519 | 0.10 | 0.06 | – | |
| 521 | 0.15 | 0.06 | – | |
| 193 | 0.34 | – | 0.05 | |
| FUNGI | ||||
| Kinoje bog | 150 | 0.32 | 0.83 | 0.85 |
| 178 | – | 0.03 | – | |
| 193 | 0.34 | – | 0.05 | |
| 294 | – | 0.08 | – | |
| 306 | 0.15 | 0.06 | 0.10 | |
| 328 | 0.20 | – | – | |
| Victor bog | 150 | 0.64 | 0.82 | 0.18 |
| 153 | – | – | 0.82 | |
| 193 | 0.29 | 0.04 | – | |
| 294 | – | 0.06 | – | |
| 306 | 0.07 | 0.03 | – | |
| 799 | – | 0.05 | – | |
| Victor fen | 150 | 0.09 | 0.61 | 0.24 |
| 178 | – | – | 0.06 | |
| 193 | 0.72 | 0.17 | 0.16 | |
| 306 | 0.19 | 0.22 | 0.53 | |
Each OTU is represented by a terminal restriction fragment (T-RF) expressed as a proportion of the total sample (e.g., Kinoje Bog 0–10 cm).
References
- Aerts A. R., Verhoeven J. T. A., Whigham D. F. (1999). Plant-mediated controls on nutrient cycling in temperate fens and bogs. Ecology 80, 2170–2181 10.1890/0012-9658(1999)080[2170:PMCONC]2.0.CO;2 [DOI] [Google Scholar]
- Allison S. D., Treseder K. K. (2008). Warming and drying suppress microbial activity and carbon cycling in boreal forest soils. Glob. Chang. Biol. 14, 2898–2909 10.1111/j.1365-2486.2008.01549.x [DOI] [Google Scholar]
- Andersen R., Grasset L., Thormann M., Rochefort L., Francez A.-J. (2010). Changes in microbial community structure and function following Sphagnum peatland restoration. Soil Biol. Biochem. 42, 291–301 10.1016/j.soilbio.2009.11.006 [DOI] [Google Scholar]
- Artz R. R. E., Anderson I. C., Chapman S. J., Hagn A., Schloter M., Potts J. M., Campbell C. D. (2007). Changes in fungal community composition in response to vegetational succession during the natural regeneration of cutover peatlands. Microb. Ecol. 54, 508–522 10.1007/s00248-007-9220-7 [DOI] [PubMed] [Google Scholar]
- Artz R. R. E., Chapman S., Campbell C. (2006). Substrate utilisation profiles of microbial communities in peat are depth dependent and correlate with whole soil FTIR profiles. Soil Biol. Biochem. 38, 2958–2962 10.1016/j.soilbio.2006.04.017 [DOI] [Google Scholar]
- Ausec L., Kraigher B., Mandic-Mulec I. (2009). Differences in the activity and bacterial community structure of drained grassland and forest peat soils. Soil Biol. Biochem. 41, 1874–1881 10.1016/j.soilbio.2009.06.010 [DOI] [Google Scholar]
- Bååth E., Anderson T.-H. (2003). Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques. Soil Biol. Biochem. 35, 955–963 10.1016/S0038-0717(03)00154-8 [DOI] [Google Scholar]
- Balser T. C., Firestone M. K. (2005). Linking microbial community composition and soil processes in a California annual grassland and mixed-conifer forest. Biogeochemistry 73, 395–415 10.1007/s10533-004-0372-y [DOI] [Google Scholar]
- Bardgett R. D., Freeman C., Ostle N. J. (2008). Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2, 805–814 10.1038/ismej.2008.58 [DOI] [PubMed] [Google Scholar]
- Basiliko N., Blodau C., Roehm C., Bengtson P., Moore T. R. (2007). Regulation of decomposition and methane dynamics across natural, commercially mined, and restored northern peatlands. Ecosystems 10, 1148–1165 10.1007/s10021-007-9083-2 [DOI] [Google Scholar]
- Basiliko N., Yavitt J. B., Dees P. M., Merkel S. M. (2003). Methane biogeochemistry and methanogen communities in two northern peatland ecosystems, New York state. Geomicrobiol. J. 20, 563–577 10.1080/713851165 [DOI] [Google Scholar]
- Blackwood C. B., Marsh T., Kim S.-H., Paul E. A. (2003). Terminal restriction fragment length polymorphism data analysis for quantitative comparison of microbial communities. Appl. Environ. Microbiol. 69, 926–932 10.1128/AEM.69.2.926-932.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blodau C., Basiliko N., Moore T. R. (2004). Carbon turnover in peatland mesocosms exposed to different water table levels. Biogeochemistry 67, 331–351 10.1023/B:BIOG.0000015788.30164.e2 [DOI] [Google Scholar]
- Blodau C., Moore T. R. (2002). Macroporosity affects water movement and pore water sampling in peat soils. Soil Sci. 167, 98–109 10.1097/00010694-200202000-00002 [DOI] [Google Scholar]
- Bragazza L., Siffi C., Iacumin P., Gerdol R. (2007). Mass loss and nutrient release during litter decay in peatland: the role of microbial adaptability to litter chemistry. Soil Biol. Biochem. 39, 257–267 10.1016/j.soilbio.2006.07.014 [DOI] [Google Scholar]
- Bräuer S. L., Cadillo-Quiroz H., Yashiro E., Yavitt J. B., Zinder S. H. (2006). Isolation of a novel acidiphilic methanogen from an acidic peat bog. Nature 442, 192–194 10.1038/nature04810 [DOI] [PubMed] [Google Scholar]
- Christopherson R., Byrne M.-L. (2009). Geosystems, An Introduction to Physical Geography. 2nd Canadian Edn Toronto: Pearson Education [Google Scholar]
- Clymo R. S. (1984). The limits to peat bog growth. Philos. Trans. R. Soc. Lond. B Biol. Sci. 303, 605–654 10.1098/rstb.1984.0002 [DOI] [Google Scholar]
- Conrad R. (2007). Microbial ecology of methanogens and methanotrophs. Adv. Agron. 96, 1–63 10.1016/S0065-2113(07)96005-8 [DOI] [Google Scholar]
- De’ath G. (2011). mvpart: Multivariate partitioning. R package version 1.4-0. Available at: http://CRAN.R-project.org/package=mvpart
- Dorrepaal E., Cornelissen J. H. C., Aerts R., Wallén B., Logtestijn R. S. P. Van. (2005). Are growth forms consistent predictors of leaf litter quality and decomposability across peatlands along a latitudinal gradient? J. Ecol. 93, 817–828 10.1111/j.1365-2745.2005.01024.x [DOI] [Google Scholar]
- Edel-Hermann V., Dreumont C., Pérez-Piqueres A., Steinberg C. (2004). Terminal restriction fragment length polymorphism analysis of ribosomal RNA genes to assess changes in fungal community structure in soils. FEMS Microbiol. Ecol. 47, 397–404 10.1016/S0168-6496(04)00002-9 [DOI] [PubMed] [Google Scholar]
- Feinstein L. M., Sul W. J., Blackwood C. B. (2009). Assessment of bias associated with incomplete extraction of microbial DNA from soil. Appl. Environ. Microbiol. 75, 5428–5433 10.1128/AEM.00120-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N., Jackson R. B. (2006). The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 103, 626–631 10.1073/pnas.0507535103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk M. C., Ruether K. F., Yavitt J. B. (2003). Microbial activity and functional composition among northern peatland ecosystems. Soil Biol. Biochem. 35, 591–602 10.1016/S0038-0717(03)00053-1 [DOI] [Google Scholar]
- Forster P., Ramaswamy V., Artaxo P., Berntsen T., Betts R., Fahey D. W., Haywood J., Lean J., Lowe D. C., Myhre G., Nganga J., Prinn R., Raga G., Schulz M., Van Dorland R., IPCC (2007). “Changes in atmospheric constituents and in radiative forcing,” in Climate Change 2007: The Physical Science Basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change, eds Solomon S., Qin D., Manning M., Chen Z., Marquis M., Averyt K. B., Tignor M., Miller H. L. (Cambridge: Cambridge University Press; ), 210–215 [Google Scholar]
- Freeman C., Ostle N., Fenner N., Kang H. (2004). A regulatory role for phenol oxidase during decomposition in peatlands. Soil Biol. Biochem. 36, 1663–1667 10.1016/j.soilbio.2004.07.012 [DOI] [Google Scholar]
- Freeman C., Ostle N., Kang H. (2001). An enzymic “latch” on a global carbon store. Nature 409, 149. 10.1038/35051650 [DOI] [PubMed] [Google Scholar]
- Gagnon A. S., Gough W. A. (2005). Climate change scenarios for the Hudson Bay region: an intermodel comparison. Clim. Change 69, 269–297 10.1007/s10584-005-1815-8 [DOI] [Google Scholar]
- Glatzel S., Basiliko N., Moore T. R. (2004). Carbon dioxide and methane production potentials of peat from natural, harvested and restored sites, Eastern Québec, Canada. Wetlands 24, 261–267 10.1672/0277-5212(2004)024[0261:CDAMPP]2.0.CO;2 [DOI] [Google Scholar]
- Glatzel S., Kalbitz K., Dalva M., Moore T. R. (2003). Dissolved organic matter properties and their relationship to carbon dioxide efflux from restored peat bogs. Geoderma 113, 397–411 10.1016/S0016-7061(02)00372-5 [DOI] [Google Scholar]
- Gorham E. (1991). Northern peatlands: role in the carbon cycle and probable responses to climatic warming. Ecol. Appl. 1, 182. 10.2307/1941811 [DOI] [PubMed] [Google Scholar]
- Hájek T., Ballance S., Limpens J., Zijlstra M., Verhoeven J. T. A. (2010). Cell-wall polysaccharides play an important role in decay resistance of Sphagnum and actively depressed decomposition in vitro. Biogeochemistry 103, 45–57 10.1007/s10533-010-9444-3 [DOI] [Google Scholar]
- Hättenschwiler S., Vitousek P. (2000). The role of polyphenols in terrestrial ecosystem nutrient cycling. Trends Ecol. Evol. (Amst.) 15, 238–243 10.1016/S0169-5347(00)01861-9 [DOI] [PubMed] [Google Scholar]
- Hengeveld H. G. (2000). Projections for Canada’s Climate Future: a Discussion of Recent Simulations with the Canadian Global Climate Model. Downsview, ON: Meteorological Service Of Canada [Google Scholar]
- Hill T. C. J., Walsh K. A., Harris J. A., Moffett B. F. (2003). Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43, 1–11 10.1111/j.1574-6941.2003.tb01040.x [DOI] [PubMed] [Google Scholar]
- Hines M. E., Duddleston K. N., Rooney-Varga J. N., Fields D., Chanton J. P. (2008). Uncoupling of acetate degradation from methane formation in Alaskan wetlands: connections to vegetation distribution. Global Biogeochem. Cycles 22, GB2017. 10.1029/2006GB002903 [DOI] [Google Scholar]
- Horwath W. H. (2007). “Carbon cycling and formation of soil organic matter,” in Soil Microbiology, Ecology, and Biochemistry, 3rd Edn, ed. Paul E. A. (Oxford: Academic Press; ), 119–144 [Google Scholar]
- Hudson J. J., Dillon P. J., Somers K. M. (2003). Long-term patterns in dissolved organic carbon in boreal lakes: the role of incident radiation, precipitation, air temperature, southern oscillation and acid deposition. Hydrol. Earth Syst. Sci. 7, 390–398 10.5194/hess-7-390-2003 [DOI] [Google Scholar]
- Jaatinen K., Fritze H., Laine J., Laiho R. (2007). Effects of short- and long-term water-level drawdown on the populations and activity of aerobic decomposers in a boreal peatland. Glob. Chang. Biol. 13, 491–510 10.1111/j.1365-2486.2006.01312.x [DOI] [Google Scholar]
- Keller J. K., White J. R., Bridgham S. D., Pastor J. (2004). Climate change effects on carbon and nitrogen mineralization in peatlands through changes in soil quality. Glob. Chang. Biol. 10, 1053–1064 10.1111/j.1529-8817.2003.00770.x [DOI] [Google Scholar]
- Killham K., Prosser J. I. (2007). “The prokaryotes” in Soil Microbiology, Ecology, and Biochemistry, 3rd Edn, ed. Paul E. A. (Oxford: Academic Press; ), 119–144 [Google Scholar]
- Knorr K.-H., Blodau C. (2009). Impact of experimental drought and rewetting on redox transformations and methanogenesis in mesocosms of a northern fen soil. Soil Biol. Biochem. 41, 1187–1198 10.1016/j.soilbio.2009.02.030 [DOI] [Google Scholar]
- Kraigher B., Stres B., Hacin J., Ausec L., Mahne I., Vanelsas J., Mandicmulec I. (2006). Microbial activity and community structure in two drained fen soils in the Ljubljana Marsh. Soil Biol. Biochem. 38, 2762–2771 10.1016/j.soilbio.2006.04.031 [DOI] [Google Scholar]
- Laiho R. (2006). Decomposition in peatlands: reconciling seemingly contrasting results on the impacts of lowered water levels. Soil Biol. Biochem. 38, 2011–2024 10.1016/j.soilbio.2006.02.017 [DOI] [Google Scholar]
- Laine A., Wilson D., Kiely G., Byrne K. A. (2007). Methane flux dynamics in an Irish lowland blanket bog. Plant Soil 299, 181–193 10.1007/s11104-007-9374-6 [DOI] [Google Scholar]
- Lueders T., Friedrich M. (2000). Archaeal population dynamics during sequential reduction processes in rice field soil. Appl. Environ. Microbiol. 66, 2732–2742 10.1128/AEM.66.7.2732-2742.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lueders T., Friedrich M. W. (2003). Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by using defined template mixtures of methanogenic pure cultures and soil DNA extracts. Appl. Environ. Microbiol. 69, 320–326 10.1128/AEM.69.1.320-326.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukow T., Dunfield P., Liesack W. (2000). Use of the T-RFLP technique to assess spatial and temporal changes in the bacterial community structure within an agricultural soil planted with transgenic and non-transgenic potato plants. FEMS Microbiol. Ecol. 32, 241–247 10.1111/j.1574-6941.2000.tb00717.x [DOI] [PubMed] [Google Scholar]
- Martini I. P. (2006) “The cold-climate peatlands of the Hudson Bay Lowland, Canada: brief overview of recent work” in Peatlands: Evolution and Records of Environmental and Climate Changes, eds Martini I. P., Cortizas A. M., Chesworth W. (Amsterdam: Elsevier; ), 53–84 [Google Scholar]
- McGuire K. L., Treseder K. K. (2010). Microbial communities and their relevance for ecosystem models: decomposition as a case study. Soil Biol. Biochem. 42, 529–535 10.1016/j.soilbio.2009.11.016 [DOI] [Google Scholar]
- Moore T. R., Basiliko N. (2006) “Decomposition in boreal peatlands” in Ecological Studies, Vol. 188. Boreal Peatland Ecosystems, eds Wieder R. K., Vitt D. H. (Berlin: Springer-Verlag; ), 125–143 [Google Scholar]
- Moore T. R., Dalva M. (1993). The influence of temperature and water table position on carbon dioxide and methane emissions from laboratory columns of peatland soils. J. Soil Sci. 44, 651–664 10.1111/j.1365-2389.1993.tb02330.x [DOI] [Google Scholar]
- Moore T. R., Dalva M. (1997). Methane and carbon dioxide exchange potentials of peat soils in aerobic and anaerobic laboratory incubations. Soil Biol. Biochem. 29, 1157–1164 10.1016/S0038-0717(97)00037-0 [DOI] [Google Scholar]
- Moore T. R., Roulet N. T., Waddington J. M. (1998). Uncertainty in predicting the effect of climatechange on the carbon cycling of Canadian peatlands. Clim. Change 40, 229–245 10.1023/A:1005408719297 [DOI] [Google Scholar]
- Morales S. E., Mouser P. J., Ward N., Hudman S. P., Gotelli N. J., Ross D. S., Lewis T. A. (2006). Comparison of bacterial communities in New England Sphagnum bogs using terminal restriction fragment length polymorphism (T-RFLP). Microb. Ecol. 52, 34–44 10.1007/s00248-005-0264-2 [DOI] [PubMed] [Google Scholar]
- Ocio J., Brookes P. C. (1990). Soil microbial biomass measurements in sieved and unsieved soil. Soil Biol. Biochem. 22, 999–1000 10.1016/0038-0717(90)90016-S [DOI] [Google Scholar]
- Oksanen J., Blanchet F. G., Kindt R., Legendre P., O’Hara R. B., Simpson G. L., Solymos P., Stevens M. H. H., Wagner H. (2011). Vegan: Community Ecology Package. R package version 1.17-9. Available at: http://CRAN.R-project.org/package=vegan
- Peltoniemi K., Fritze H., Laiho R. (2009). Response of fungal and actinobacterial communities to water-level drawdown in boreal peatland sites. Soil Biol. Biochem. 41, 1902–1914 10.1016/j.soilbio.2009.06.018 [DOI] [Google Scholar]
- R Development Core Team (2011). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available at: http://www.R-project.org/
- Reed H. E., Martiny J. B. H. (2007). Testing the functional significance of microbial composition in natural communities. FEMS Microbiol. Ecol. 62, 161–170 10.1111/j.1574-6941.2007.00386.x [DOI] [PubMed] [Google Scholar]
- Riley J. L. (1982). Hudson Bay Lowland flouristic inventory, wetlands catalogue and conservation strategy. Nat. Can. 109, 543–555 [Google Scholar]
- Robroek B. J. M., Limpens J., Breeuwer A., Schouten M. G. C. (2007). Effects of water level and temperature on performance of four Sphagnum mosses. Plant Ecol. 190, 97–107 10.1007/s11258-006-9193-5 [DOI] [Google Scholar]
- Roulet N. T. (2000). Peatlands, carbon storage, greenhouse gases, and the Kyoto protocol: prospecs and significance for Canada. Wetlands 20, 605–615 10.1672/0277-5212(2000)020[0605:PCSGGA]2.0.CO;2 [DOI] [Google Scholar]
- Roulet N. T., Lafleur P. M., Richard P. J. H., Moore T. R., Humphreys E. R., Bubier J. (2007). Contemporary carbon balance and late Holocene carbon accumulation in a northern peatland. Glob. Chang. Biol. 13, 397–411 10.1111/j.1365-2486.2006.01292.x [DOI] [Google Scholar]
- Roulet N. T., Moore T., Bubier J., Lafleur P. M. (1992). Northern fens: methane flux and climate change. Tellus 44B, 100–105 [Google Scholar]
- Rousk J., Bååth E., Brookes P. C., Lauber C. L., Lozupone C., Caporaso J. G., Knight R., Fierer N. (2010). Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 1340–1351 10.1038/ismej.2010.58 [DOI] [PubMed] [Google Scholar]
- Rydin H., Jeglum J. K. (2006). The Biology of Peatlands. Oxford: Oxford University Press [Google Scholar]
- Saiya-Cork K. R., Sinsabaugh R. L., Zak D. R. (2002). The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Biol. Biochem. 34, 1309–1315 10.1016/S0038-0717(02)00074-3 [DOI] [Google Scholar]
- Sinsabaugh R. L. (2010). Phenol oxidase, peroxidase and organic matter dynamics of soil. Soil Biol. Biochem. 42, 391–404 10.1016/j.soilbio.2009.10.014 [DOI] [Google Scholar]
- Sinsabaugh R. L., Antibus R. K., Linkins A. E., McClaugherty C. A., Rayburn L., Repert D., Weiland T. (1993). Wood decomposition: nitrogen and phosphorus dynamics in relation to extracellular enzyme activity. Ecology 74, 1586–1593 10.2307/1940086 [DOI] [Google Scholar]
- Straková P., Niemi R. M., Freeman C., Peltoniemi K., Toberman H., Heiskanen I., Fritze H., Laiho R. (2011). Litter type affects the activity of aerobic decomposers in a boreal peatland more than site nutrient and water table regimes. Biogeosciences 8, 2741–2755 10.5194/bg-8-2741-2011 [DOI] [Google Scholar]
- Strickland M. S., Osburn E., Lauber C., Fierer N., Bradford M. A. (2009). Litter quality is in the eye of the beholder: initial decomposition rates as a function of inoculum characteristics. Funct. Ecol. 23, 627–636 10.1111/j.1365-2435.2008.01515.x [DOI] [Google Scholar]
- Strickland M. S., Rousk J. (2010). Considering fungal:bacterial dominance in soils – methods, controls, and ecosystem implications. Soil Biol. Biochem. 42, 1385–1395 10.1016/j.soilbio.2010.05.007 [DOI] [Google Scholar]
- Thormann M. (2006). Diversity and function of fungi in peatlands: a carbon cycling perspective. Can. J. Soil Sci. 86, 281–293 10.4141/S05-082 [DOI] [Google Scholar]
- Trinder C. J., Johnson D., Artz R. R. E. (2008). Interactions among fungal community structure, litter decomposition and depth of water table in a cutover peatland. FEMS Microbiol. Ecol. 64, 433–448 10.1111/j.1574-6941.2008.00487.x [DOI] [PubMed] [Google Scholar]
- Turunen J., Tomppo E., Tolonen K., Reinikainen A. (2002). Estimating carbon accumulation rates of undrained mires in Finland – application to boreal and subarctic regions. Holocene 12, 69–80 10.1191/0959683602hl522rp [DOI] [Google Scholar]
- Updegraff K., Bridgham S. D., Pastor J., Weishampel P. (1998). Hysteresis in the temperature response of carbon dioxide and methane production in peat soils. Biogeochemistry 43, 253–272 10.1023/A:1006097808262 [DOI] [Google Scholar]
- Vance E., Brookes P., Jenkinson D. (1987). An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 19, 703–707 10.1016/0038-0717(87)90052-6 [DOI] [Google Scholar]
- Verhoeven J. T. A., Toth E. (1995). Decomposition of Carex and Sphagnum litter in fens: effect of litter quality and inhibition by living tissue homogenates. Soil Biol. Biochem. 27, 271–275 10.1016/0038-0717(94)00183-2 [DOI] [Google Scholar]
- Waldrop M. P., Firestone M. K. (2006). Response of microbial community composition and function to soil climate change. Microb. Ecol. 52, 716–724 10.1007/s00248-006-9100-6 [DOI] [PubMed] [Google Scholar]
- Webster K. L., McLaughlin J. W. (2010). Importance of the water table in controlling dissolved carbon along a fen nutrient gradient. Soil Sci. Soc. Am. J. 74, 2254. 10.2136/sssaj2009.0111 [DOI] [Google Scholar]
- Weltzin J. F., Bridham S. C., Pastor J., Chen J., Harth C. (2003). Potential effects of warming and drying on peatland plant community composition. Glob. Chang. Biol. 9, 141–151 10.1046/j.1365-2486.2003.00571.x [DOI] [Google Scholar]
- Winsborough C., Basiliko N. (2010). Fungal and bacterial activity in northern peatlands. Geomicrobiol. J. 27, 315–320 10.1080/01490450903424432 [DOI] [Google Scholar]
- Yavitt J. B., Williams C. J., Wieder R. K. (1997). Production of methane and carbon dioxide in peatland ecosystems across North America: effects of temperature, aeration, and organic chemistry of peat. Geomicrobiol. J. 14, 299–316 10.1080/01490459709378054 [DOI] [Google Scholar]
- Yu Z., Loisel J., Brosseau D. P., Beilman D. W., Hunt S. J. (2010). Global peatland dynamics since the last glacial maximum. Geophys. Res. Lett. 37, 1–5 10.1029/2010GL043584 [DOI] [Google Scholar]