Abstract
Motivation: Molecular interaction information, such as protein–protein interactions and protein–small molecule interactions, is indispensable for understanding the mechanism of biological processes and discovering treatments for diseases. Many databases have been built by manual annotation of literature to organize such information into structured form. However, most databases focus on only one type of interactions, which are often not well annotated and integrated with related functional information.
Results: In this study, we integrate molecular interaction information from literature by automatic information extraction and from manually annotated databases. We further integrate the relationships between protein/gene and other bio-entity terms including gene ontology terms, pathways, species and diseases to build an integrated molecular interaction database (IMID). Interactions can be selected by their associated probabilities. IMID allows complex and versatile queries for context-specific molecular interactions, which are not available currently in other molecular interaction databases.
Availability: The database is located at www.integrativebiology.org.
Contact: jinfeng@stat.fsu.edu
1 INTRODUCTION
Molecular interactions, such as protein–protein interaction (PPI) and protein–small molecule interaction, play important roles in almost all biological processes. Information on molecular interactions is indispensable for our understanding of the mechanisms of biological processes and for the development of drugs. Due to the importance of such information, manual annotation has been used to extract it from scientific literature and deposit it into various databases (Alfarano et al., 2005; Aranda et al., 2010; Chatr-aryamontri et al., 2007; Chaurasia et al., 2007; Chen et al., 2009; Han et al., 2004; Jayapandian et al., 2007; Kamburov et al., 2009; Keshava Prasad et al., 2009; Kuhn et al., 2008; Mishra et al., 2006; Pagel et al., 2005; Patil et al., 2011; Prieto and De Las Rivas, 2006; Salwinski et al., 2004; Stark et al., 2006, 2011; von Mering et al., 2005; Xenarios et al., 2002). In recent years, computational methods have also been developed to extract molecular interaction information from the literature…(Baumgartner et al., 2007; Bui et al., 2011; Chowdhary et al., 2009; Giles and Wren, 2008; Huang et al., 2008; Jensen et al., 2006; Krallinger et al., 2008; Wren et al., 2004).
The context of molecular interactions such as the functions of the interactions [related pathways or gene ontology (GO) terms] and the physical nature of the interactions (regulate, inhibit or phosphorylate, etc.) can be very useful for biologists searching for those interactions. However, current databases do not explicitly document such information. In studying molecular interactions, a researcher may wish to find all protein interactions related to a particular biological process/pathway/disease/species, or a combination of these terms (such as a pathway and a species). In addition, the researcher may want to know those interactions related to a particular type of chemical reaction such as phosphorylation. Allowing researchers to perform the above complex searches requires not only integrating the heterogeneous data from different sources, but also organizing them in a highly structured form that enables complicated queries. Availability of such tool will greatly help biologists in their daily research.
In this study, we integrate molecular interaction information from both manually annotated databases and literature (Bell et al., 2011a), and build a relational database, integrated molecular interaction database (IMID, www.integrativebiology.org). Users of IMID can perform complex and versatile queries for context-specific molecular interactions. We expect IMID to be a useful tool for researchers in biology and biomedical sciences.
2 METHODS
The collection of data has been described in detail in a recent study (Bell et al., 2011a). The information in IMID comes from two types of sources: manually curated databases and automatically extracted from literature using a Bayesian network (BN) method (Chowdhary et al., 2009). The information extraction method not only extracts the pairs of interacting molecules, but also the interaction word describing the interactions. The types of information in IMID are as follows: (i) PPIs including physical, regulatory and genetic interactions; (ii) protein–small molecule interactions; and (iii) associations of interactions with other bio-entities such as pathway, species, diseases or GO terms. Information on individual proteins, such as official names, synonyms and species, is obtained from UniProt database (Apweiler et al., 2004). The synonyms of the same proteins are mapped to the same proteins/genes. Interactions from different species are made distinct if possible. An interaction is considered to be related to a particular bio-entity if at least one of the interacting partners is manually annotated to be associated with the bio-entity or the bio-entity term co-occurs with the interaction in a sentence. The association information between proteins and pathways was obtained from Reactome (Vastrik et al., 2007) and Pathway interaction database (Schaefer et al., 2009). The association information between GO terms and proteins was obtained from GO database (Ashburner et al., 2000) and GOA database (Barrell et al., 2009). Both types of associations are manually annotated. When more than one constraint is used in searching PPIs, only the PPIs that are related to all the constraints will be returned. In addition to the above information, probabilities of the interactions are also given. The BN method assigns probabilities to all automatically extracted interactions. Overall, our database has several unique features compared with other databases:
The interactions in the database are linked to their biological context represented by various biological terms. A relational database is built so that users can select a subset of interactions related to one or a combination of the terms.
Users can also select interactions by several other filters such as the type of molecules involved in the interactions (protein–protein or protein–small molecule) and the type of interactions (regulations, genetic interactions or physical interactions). They can also use the interaction words (phosphorylate, methylate, inhibit, etc.) to select a subset of interactions they are interested in.
The database stores both PPIs and protein–small molecule interactions.
The database stores both manually annotated and automatically extracted interaction information. The two types of information can be easily separated using a probability filter.
3 USAGE
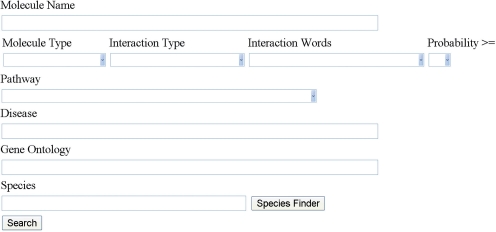
The query interface of IMID (Fig. 1) allows users to perform two types of searches.
Fig. 1.
The user interface of IMID.
(1) Query using any protein/gene or small molecule names in Molecule Name text box. We allow only one protein, gene or small molecule name in this box. Other constraints can be applied using the dropdown and/or text box below the Molecule Name box. Only standard names can be used for search. Using standard ontology systems allows more accurate definition of PPIs and more efficient search performance.
Molecule Type dropdown box allows users to select protein–protein, protein–small molecule or both types of interactions.
Interaction Type dropdown box contains four types of interactions: genetic interaction/association, interaction, physical interaction and regulation. Interactions are grouped to the above types according to the interaction words that are extracted together with the interactions in the text or based on the annotation given in other databases. The type interaction is rather generic, which represents interactions of unknown nature.
Interaction Word dropdown box contains the list of words we used in automatic extraction of molecular interactions. Selecting any word will limit interactions described by that word and its related forms (i.e. -ed, -ing, -s, etc.). Some interaction words carry directionality information, which can be quite useful for network studies (Giles and Wren, 2008). Although we have developed a method for the inference of directionality information (Bell et al., 2011b), the method has not been applied to the large-scale extraction of protein interaction information we performed earlier. As a result, directionality information is not given in the output table. We plan to include this information in future update of the database.
Probability dropdown box discretizes probability into intervals of size 0.1. Manually annotated interactions have probability of 1. Selecting 1 will filter out all automatically extracted interactions.
In addition to the above, Pathway, Disease, GO and Species can also be used individually or in combination to select the subset of PPIs a researcher is interested in.
(2) Without inputting any names at Molecule Name text box, users can search interactions related to other conditions selected. For example, selecting insulin pathway in the Pathway dropdown box and human in Species text box will return all the interactions related to human insulin pathway.
The output interactions are given in a table below the search box containing several types of information including a PubMed ID and links to the PubMed abstracts where the interactions were obtained from. The interactions displayed in the table can be downloaded by the users. For larger download sizes, users are encouraged to contact us for assistance. The interactions shown in the table are automatically plotted as a network for users to visualize using Cytoscape (Smoot et al., 2011). For performance and user experience considerations, only interactions shown in the current page of the table are plotted.
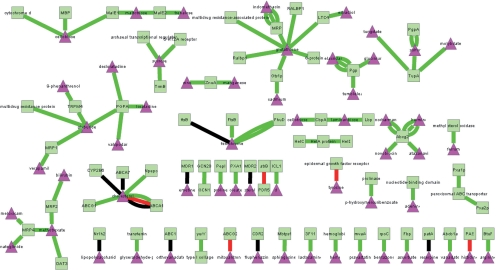
Figure 2 shows the networks for ABC transporter pathway obtained by searching ABC transporter as the pathway name, homo sapiens as species name and a probability cut off of 0.8. Both PPI and protein–small molecule interaction are shown. Squares represent proteins and triangles represent small molecules.
Fig. 2.
A network of ABC transporter pathway.
Funding: National Institutes of Health Grant (R01-HG02518-02 to J.S.L.) in part.
Conflict of Interest: none declared.
REFERENCES
- Alfarano C., et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–D424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R., et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda B., et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell D., et al. The GOA database in 2009–an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:D396–D403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner W.A., Jr, et al. Manual curation is not sufficient for annotation of genomic databases. Bioinformatics. 2007;23:i41–i48. doi: 10.1093/bioinformatics/btm229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell L., et al. Integrated bio-entity network: a system for biological knowledge discovery. PLoS One. 2011a;6:e21474. doi: 10.1371/journal.pone.0021474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell L., et al. Mixture of logistic models and an ensemble approach for extracting protein-protein interactions. ACM Conference on Bioinformatics, Computational Biology and Biomedicine. 2011b:371–375. [Google Scholar]
- Bui Q.C., et al. A hybrid approach to extract protein-protein interactions. Bioinformatics. 2011;27:259–265. doi: 10.1093/bioinformatics/btq620. [DOI] [PubMed] [Google Scholar]
- Chatr-aryamontri A., et al. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaurasia G., et al. UniHI: an entry gate to the human protein interactome. Nucleic Acids Res. 2007;35:D590–D594. doi: 10.1093/nar/gkl817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.Y., et al. HAPPI: an online database of comprehensive human annotated and predicted protein interactions. BMC Genomics. 2009;10(Suppl. 1):S16. doi: 10.1186/1471-2164-10-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary R., et al. Bayesian inference of protein-protein interactions from biological literature. Bioinformatics. 2009;25:1536–1542. doi: 10.1093/bioinformatics/btp245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles C.B., Wren J.D. Large-scale directional relationship extraction and resolution. BMC Bioinformatics. 2008;9(Suppl. 9):S11. doi: 10.1186/1471-2105-9-S9-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K., et al. HPID: the Human Protein Interaction Database. Bioinformatics. 2004;20:2466–2470. doi: 10.1093/bioinformatics/bth253. [DOI] [PubMed] [Google Scholar]
- Huang M., et al. Mining physical protein-protein interactions from the literature. Genome Biol. 2008;9(Suppl. 2):S12. doi: 10.1186/gb-2008-9-s2-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayapandian M., et al. Michigan Molecular Interactions (MiMI): putting the jigsaw puzzle together. Nucleic Acids Res. 2007;35:D566–D571. doi: 10.1093/nar/gkl859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen L.J., et al. Literature mining for the biologist: from information retrieval to biological discovery. Nat. Rev. Genet. 2006;7:119–129. doi: 10.1038/nrg1768. [DOI] [PubMed] [Google Scholar]
- Kamburov A., et al. ConsensusPathDB–a database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37:D623–D628. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshava Prasad T.S., et al. Human Protein Reference Database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krallinger M., et al. Overview of the protein-protein interaction annotation extraction task of BioCreative II. Genome Biol. 2008;9(Suppl. 2):S4. doi: 10.1186/gb-2008-9-s2-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M., et al. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2008;36:D684–D688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra G.R., et al. Human protein reference database–2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel P., et al. The MIPS mammalian protein-protein interaction database. Bioinformatics. 2005;21:832–834. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- Patil A., et al. HitPredict: a database of quality assessed protein-protein interactions in nine species. Nucleic Acids Res. 2011;39:D744–D749. doi: 10.1093/nar/gkq897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto C., De Las Rivas J. APID: Agile Protein Interaction Data Analyzer. Nucleic Acids Res. 2006;34:W298–W302. doi: 10.1093/nar/gkl128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salwinski L., et al. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer C.F., et al. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot M.E., et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C., et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C., et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39:D698–D704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastrik I., et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C., et al. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren J.D., et al. Knowledge discovery by automated identification and ranking of implicit relationships. Bioinformatics. 2004;20:389–398. doi: 10.1093/bioinformatics/btg421. [DOI] [PubMed] [Google Scholar]
- Xenarios I., et al. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]