Abstract
Motivation: Pathway diagrams from PubMed and World Wide Web (WWW) contain valuable highly curated information difficult to reach without tools specifically designed and customized for the biological semantics and high-content density of the images. There is currently no search engine or tool that can analyze pathway images, extract their pathway components (molecules, genes, proteins, organelles, cells, organs, etc.) and indicate their relationships.
Results: Here, we describe a resource of pathway diagrams retrieved from article and web-page images through optical character recognition, in conjunction with data mining and data integration methods. The recognized pathways are integrated into the BiologicalNetworks research environment linking them to a wealth of data available in the BiologicalNetworks' knowledgebase, which integrates data from >100 public data sources and the biomedical literature. Multiple search and analytical tools are available that allow the recognized cellular pathways, molecular networks and cell/tissue/organ diagrams to be studied in the context of integrated knowledge, experimental data and the literature.
Availability: BiologicalNetworks software and the pathway repository are freely available at www.biologicalnetworks.org.
Contact: baitaluk@sdsc.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Biological pathways provide intuitive views of the myriad of interactions involved in biological processes. A typical signaling pathway, for example, can represent receptor-binding events, protein complexes, phosphorylation reactions, translocations and transcriptional regulation, usually depicted with a set of symbols, with lines and arrows. Automated detection of genes and proteins and inference of signaling network architecture and dynamics from general (Peng et al., 2008; Rodriguez-Esteban et al., 2009; Shatkay et al., 2006) as well as domain specific (Brass et al., 2008; Conrad et al., 2004; Kaplow et al., 2009; Nir et al., 2010; Neumann et al., 2006; Peng et al., 2008) imaging data, has been of increasing interest during last years. These sophisticated methods were implemented only for a specific domain and do not provide public services for extracted pathways analysis or a software for a broader use. Thus in spite of the importance of collecting and maintaining pathway information, with the attendant biological annotations and experimental data, pathways construction, curation and integration with public databases and experimental data remain a tedious and time-consuming task (Cerami et al., 2011; Kanehisha et al., 2010; Kelder et al., 2009; Schaefer et al., 2009).
A small number of systems have considered the issue of figure retrieval: Sub-cellular Location Image Finder (Murphy et al., 2004) that retrieves fluorescence micrograph images from a single-journal corpus; BioText (Hearst et al., 2007) and Yale Image Finder (YIF) (Xu et al., 2008) that performs text queries of figures legends. None of these methods analyzes information embedded in the image itself, nor do they compare with and supplement knowledge available in public databases and PudMed.
Here we present the system that addresses the problems of extraction, comparison, searching and managing biological knowledge embedded in pathway imaging data. The system provides a repository of freely available pathways recognized from imaging data. The repository allows biologists to explore, search over pathway components in open access publications and World Wide Web (WWW) and construct user's pathways and interaction networks. These pathways can be visualized and dynamically analyzed in concert with data integrated from other public databases and literature.
2 SYSTEM DESCRIPTION
2.1 Methods
The process of image recognition, data extraction and integration consists of several steps (Supplementary Fig. S1). First, objects and relations are extracted from the image, together with their coordinates. This is done using mathematical morphology and binary analysis routines of ImageJ (http://rsbweb.nih.gov/ij/). The image is transferred to binary gray-scale 32-bit RGB mode by applying a threshold adjustment with a Huang Filter. Special mathematical morphology ‘Opening’ after ‘Closing’ operations are applied to reduce the number of domains for recognition by removing areas that are too small. Finally, an ‘analyze particles’ procedure applied to the last image retrieves all possible candidates for nodes as objects represented by points of an enclosing polygon.
Text recognition is done separately with preliminary image processing (Kou et al., 2007; Li et al., 2008). Cleaning non-textual elements using properties that characterize horizontal text objects, such as alignment, height–width ratio, character separation and connectedness, is performed. ‘Cognitive OpenOCR (Cuneiform)’ (http://en.openocr.org) software is used for batch image text recognition and ‘AutoIt v3’ (http://www.autoitscript.com) for automated batch operations. The scanning procedure is executed both horizontally and vertically to extract horizontal and vertical text.
The next step involves the extracted text, objects and relations being sent to the IntegromeDB (Baitaluk et al., 2010) (back-end database) data integration pipeline to check the consistency of the data (Supplementary Fig. S2 and Table S1). We check that all recognized objects are genes/proteins, processes, cell types, diseases or any other object type constituting BioNets ontology (Baitaluk et al., 2010; Kozhenkov et al., 2011) in our database. Recognized relations are compared with the existing (literature or pubic databases) interactions, reactions and relations integrated in the IntegromeDB from >100 of public databases. Only those recognized relations that are supported by at least one type of evidence from our integrated database are included in the final pathway.
We scanned a collection of >150 journals, 50 000 articles and ~25 000 figures available in PubMed Central and WWW through the Google Image service API by querying terms from Pathway Ontology (e.g. ‘Nicotine pathway’), Gene Ontology Biological Process (e.g. ‘molecular synthesis’), ‘$GeneName pathway’ (e.g. ‘NF-B pathway’), etc. After filtering out images not containing objects/relations, top 1012 pathway/network diagrams (richest in number of literature-supported relations) are stored on a remote server and the Lucene open-source search engine (http://lucene.apache.org) is used to index, retrieve and rank the image text descriptions (using the default statistical ranking). In case of publication, the image description is the image legend, whereas in the case of a web page, the specifically designed algorithm retrieves the most appropriate description from the web page text surrounding the image. Image publication date and source journal are stored as separate fields that can also be used to sort the results.
2.2 Quality evaluation
Out of the top 1012 pathways, 600 were randomly chosen for expert evaluation (see also Section 2 in Supplementary Material). We asked six experts (each evaluating 100 different pathways/images) to tell which pair of pathway image is close or equivalent from the point of view of a biological user. The recognized pathways were considered close/equivalent to its source image if the difference between them was <10% of the total number of objects and relations in the pathway. In the experiments in which the experts made 600 similarity assertions on different random pathways, the quality of recognition was estimated as ~87%, i.e. 521/600 recognized pathways were qualified as equivalent to its image by the experts.
For additional quality evaluation and community assessment, every pathway in the repository has a ‘Like/Don’t Like' option button (Fig. 2I) and a Report Form for the user's opinion and to leave comments on the quality of the pathway. This feedback will be used in refining our data mining and integration methods.
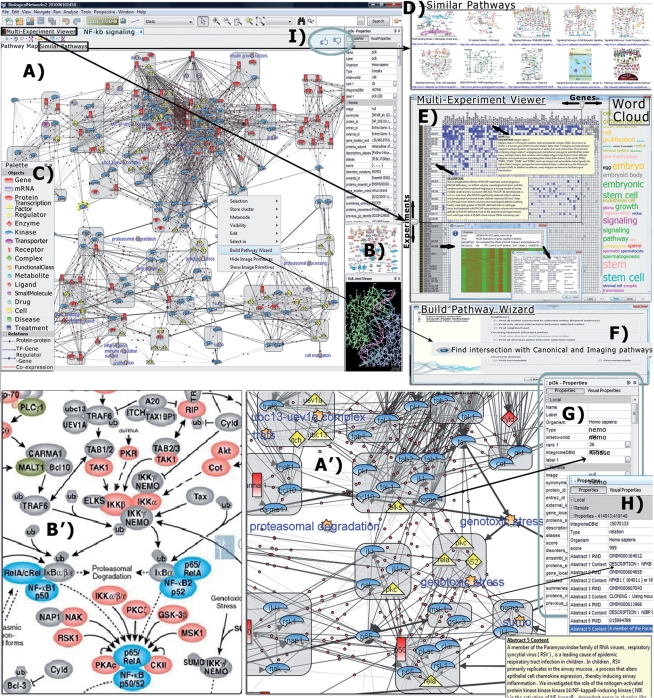
Fig. 2.
Data associated with a Pathway in BiologicalNetworks system. (A and A′) NF-kB signaling pathway extracted from image (B and B′). (C) Pathway Palette explaining pathway elements. (D) Pathways similar (in gene/protein members) to opened pathway are shown on the ‘Similar Pathways’ tabbed panel. (E) Multi-experiment viewer represents microarray experiments that are most relevant (by number of differentially expressed pathway's gene members) searched over a wide variety of cell and tissue types. (F) Build Pathway Wizard (BPW) assists the user in finding regulatory paths, protein–protein, genetic and functional links, between selected objects, searches for common targets or regulators for the group of molecules, finds connection to canonical and other imaging pathways. (G) Node properties, (H) Link properties, including Abstracts from PudMed publications from which the link was extracted. (I) ‘Like/Don’t Like' buttons for pathway quality evaluation and users' comments.
2.3 Data access
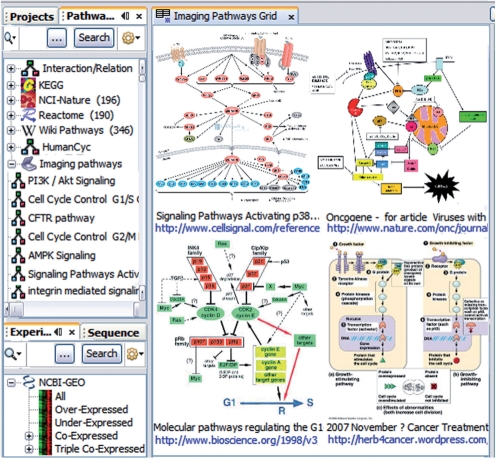
The constantly growing ‘Imaging Pathways’ repository currently contains 1012 pathways. To access ‘Imaging Pathways’ repository, run BiologicalNetworks integrated environment and open Pathway Data Panel on the left (Fig. 1). The pathways recognized from imaging data are under ‘Imaging Pathways’ node in the pathways data tree. An example of NF-kB signaling pathway (Fig. 2) and related data are also accessible upon launching the program from the Welcome Screen (click on the ‘Imaging Pathways’ demo project). Additionally, BiologicalNetworks' back-end database IntegromeDB integrates Reactome, KEGG, BioCarta, NCI-Nature pathways, WikiPathways and HumanCyc thus making the BiologicalNetworks the richest compendium of integrated pathways.
Fig. 1.
Pathways extracted from imaging data for the ‘p53’ query.
2.4 Functionality
‘Imaging Pathways’ can be explored in conjunction with public knowledge and experimental data (Fig. 2) collected in the BiologicalNetworks' back-end database, IntegromeDB, for a variety of species from >100 data sources (Baitaluk et al., 2010). BiologicalNetworks allows the building of integrative models starting from a given pathway (Fig. 2A, A', B and B') exploring gene/protein (Fig. 2G) and interaction/relation (Fig. 2H) properties, interaction networks (Fig. 2D and F), microarrays and other experimental data (e.g. transcriptomic, metabolomic, proteomic) (Fig. 2E), functional sequences, drugs, tissues, developmental stages and other types of data (Kozhnekov et al., 2010).
Users can start with a simple search over the pathway repository from the Quick Search bar located above the Pathway Data Panel (Fig. 1, left) for pathways, gene/protein descriptions and other metadata. For example, searching ‘p53’ in the current pathway collection produces only 14 results when run over figure caption text (as most of the current search engines do), but returns 68 results if analyzing information embedded in the image itself (Fig. 1). The search results can be viewed either in a tree list view or in a grid view (Fig. 1), which is especially useful for seeing commonalities among topics, such as all the pathways that include a given gene, or seeing all pathways/images of embryo development of some species. Text description as well as source image together with image caption, references to PubMed articles and links to source web pages are provided for every pathway (Fig. 1). The ‘Similar Pathways’ Window/Tab (Fig. 2D) shows pathways having the highest number of common objects (gene, proteins, etc.) with the opened pathway. Advanced functionality on pathways exploration is described in the Supplementary File and Video Tutorial (Demo #5, ‘Pathway Collection’) on the BiologicalNetworks main page.
3 DISCUSSION AND FUTURE WORK
The compendium of pathways is constantly growing, and thus we will be adding more pathways into compendium. We estimate that the total size of pathway imaging data available in the PubMed and WWW is at least ~100 times bigger than what we have currently processed. We will be adding additional data types such as miRNA to our list of recognized object types and will integrate pathways repository with a gene regulatory modules discovery tool available in BiologicalNetworks to enable exploration and discovery of regulatory regions and regulatory modules using canonical pathway enrichment tool. We will also maintain accuracy by high-level curation of our pathways. We plan to extend image and accompanying metadata search. Our text and object recognition methods will be refined to include other types of pathways (e.g. biochemical reactions, high-level cellular processes) and derive the relation direction and type. We will improve the database's check/verify procedures by clustering and comparison of similar or relative pathways to find doubtful components.
Additional improvements of the system will include: (i) further examination of the images for the potentially novel interactions; (ii) using NLP technology to decipher abbreviations used in the image by reference to the figure legend or an external synonyms table and usage of the figure legend in case when objects are labeled by numbers; (iii) using NLP technology to decipher color-coded information (see Section 8 in Supplementary Material).
Funding: This work was supported by the National Institute of Health grant (R01GM084881 to M.B.).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Baitaluk M., Ponomarenko J. Semantic integration of data on transcriptional regulation. Bioinformatics. 2010;26:1651–1661. doi: 10.1093/bioinformatics/btq231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass A.L., et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Cerami E.G., et al. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:685–690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad C., et al. Automatic identification of subcellular phenotypes on human cell arrays. Genome Res. 2004;14:1130–1136. doi: 10.1101/gr.2383804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearst M.A., et al. BioText search engine: beyond abstract search. Bioinformatics. 2007;23:2196–2197. doi: 10.1093/bioinformatics/btm301. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., et al. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:355–360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplow I.M., et al. RNAiCut: automated detection of significant genes from functional genomic screens. Nat. Methods. 2009;6:476–477. doi: 10.1038/nmeth0709-476. [DOI] [PubMed] [Google Scholar]
- Kelder T., et al. Mining biological pathways using WikiPathways Web Services. PLoS One. 2009;4:e6447. doi: 10.1371/journal.pone.0006447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou Z., et al. A stacked graphical model for associating sub-images with sub-captions. Pac. Symp. Biocomput. 2007;2007:257–268. [PMC free article] [PubMed] [Google Scholar]
- Kozhenkov S., et al. BiologicalNetworks 2.0-integrative view of genome biology data. BMC Bioinformatics. 2010;11:610. doi: 10.1186/1471-2105-11-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozhenkov S., et al. BiologicalNetworks - tools enabling the integration of multi-scale data for the host-pathogen studies. BMC Syst. Biol. 2011;5:7. doi: 10.1186/1752-0509-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., et al. A figure image processing system. Graphics recognition, recent advances and new opportunities. Lect. Notes Comput. Sci. 2004;5046:191–201. [Google Scholar]
- Murphy R.F., et al. Proceedings of the IASTED International Conference on Knowledge Sharing and Collaborative Engineering (KSCE-2004). US Virgin Islands: CTA Press, St Thomas; 2004. Extracting and structuring sub-cellular location information from on-line journal articles: the sub-cellular location image finder; pp. 109–114. [Google Scholar]
- Neumann B., et al. High-throughput RNAi screening by time-lapse imaging of live human cells. Nat. Methods. 2006;3:385–390. doi: 10.1038/nmeth876. [DOI] [PubMed] [Google Scholar]
- Nir O., et al. Inference of RhoGAP/GTPase regulation using single-cell morphological data from a combinatorial RNAi screen. Genome Res. 2010;20:372–380. doi: 10.1101/gr.100248.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H. Bioimage informatics: a new area of engineering biology. Bioinformatics. 2008;24:1827–1836. doi: 10.1093/bioinformatics/btn346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Esteban R., Iossifov I. Figure mining for biomedical research. Bioinformatics. 2009;25:2082–2084. doi: 10.1093/bioinformatics/btp318. [DOI] [PubMed] [Google Scholar]
- Schaefer C.F., et al. The Pathway Interaction Database. Nucleic Acids Res. 2009;37:674–679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatkay H., et al. Integrating image data into biomedical text categorization. Bioinformatics. 2006;22:446–453. doi: 10.1093/bioinformatics/btl235. [DOI] [PubMed] [Google Scholar]
- Xu S., et al. Yale Image Finder (YIF): a new search engine for retrieving biomedical images. Bioinformatics. 2008;24:1968–1970. doi: 10.1093/bioinformatics/btn340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.