SUMMARY
Retinoblastoma is an aggressive childhood cancer of the developing retina that is initiated by the biallelic loss of the RB1 gene. To identify the mutations that cooperate with RB1 loss, we performed whole-genome sequencing of retinoblastomas. The overall mutational rate was very low; RB1 was the only known cancer gene mutated. We then evaluated RB1’s role in genome stability and considered nongenetic mechanisms of cancer pathway deregulation. Here we show that the retinoblastoma genome is stable, but multiple cancer pathways can be epigenetically deregulated. For example, the proto-oncogene SYK is upregulated in retinoblastoma and is required for tumor cell survival. Targeting SYK with a small-molecule inhibitor induced retinoblastoma tumor cell death in vitro and in vivo. Thus, RB1 inactivation may allow preneoplastic cells to acquire multiple hallmarks of cancer through epigenetic mechanisms, resulting directly or indirectly from RB1 loss. These data provide novel targets for chemotherapeutic interventions of retinoblastoma.
Retinoblastoma is a rare childhood cancer of the retina that can develop in a sporadic or a heritable form and is fatal if untreated. When the RB1 gene was cloned, it was found to undergo biallelic inactivation in virtually all retinoblastoma tumors1. Since then, hundreds of genetic lesions have been identified in human cancer. These genetic lesions can be grouped based on the signaling pathways they affect that have direct or indirect mechanistic links to many of the common cellular properties or “hallmarks” of cancer. Thus, the rate of cancer progression is related to the kinetics of acquisition of multiple genetic lesions and/or epigenetic changes that ultimately lead to activation of growth-signaling pathways, evasion of cell death and senescence, acquisition of limitless replicative potential, sustained angiogenesis, and local tissue invasion and metastasis2.
RB1 inactivation confers limitless replicative potential to retinoblasts and it is rate limiting for retinoblastoma tumorigenesis3. However, the mechanisms that enable retinoblastoma cells to acquire the additional hallmarks of cancer remain unknown. Evidence from molecular, cellular, and cytogenetic studies suggest that the RB1 protein is required for maintaining chromosomal stability4,5, and its loss leads to chromosome instability (CIN) in cells maintained in culture. These data raise the possibility that RB1 inactivation may underlie the rapid acquisition of cooperating mutations in key cancer pathways through CIN. Alternatively, epigenetic changes may play a more dominant role in cooperating with the loss of RB1 retinoblastoma tumorigenesis. RB1 has been implicated in regulating most major epigenetic processes, including miRNA regulation, DNA methylation, histone modification, and ATP-dependent chromatin reorganization6–10. Thus, inactivation of RB1 in retinoblasts may lead to the rapid epigenetic deregulation of cancer genes that contribute to the essential cellular properties of retinoblastoma.
In this study, we characterized the genetic and epigenetic landscapes of retinoblastoma and explore the role of RB1 in regulating genomic stability. Whole-genome sequencing (WGS) of 4 retinoblastomas and their paired germline DNA samples showed no genetic lesions in known tumor suppressor genes or oncogenes, other than RB1. More importantly, an orthotopic xenograft derived from 1 of the primary tumors showed no evidence of clonal variation or new coding-region mutations. This finding suggests that retinoblastoma’s genome is more stable than previously believed.
Unlike the genetic landscape of retinoblastoma, the epigenetic profile shows profound changes compared to that observed in normal retinoblasts. One of the most striking results was the induction of the expression of the proto-oncogene SPLEEN TYROSINE KINASE (SYK) in human retinoblastoma. SYK is required for tumor cell survival, and inhibition of SYK with a small-molecule inhibitor caused the degradation of MCL1 and caspase-mediated cell death in retinoblastoma cells in culture and in vivo. These findings highlight how comprehensive genetic and epigenetic analyses of tumors can be integrated and lead to the discovery of promising new therapeutic approaches and shed light on the mechanisms underlying the rapid progression of retinoblastoma following RB1 inactivation.
Retinoblastoma Whole-Genome Sequencing
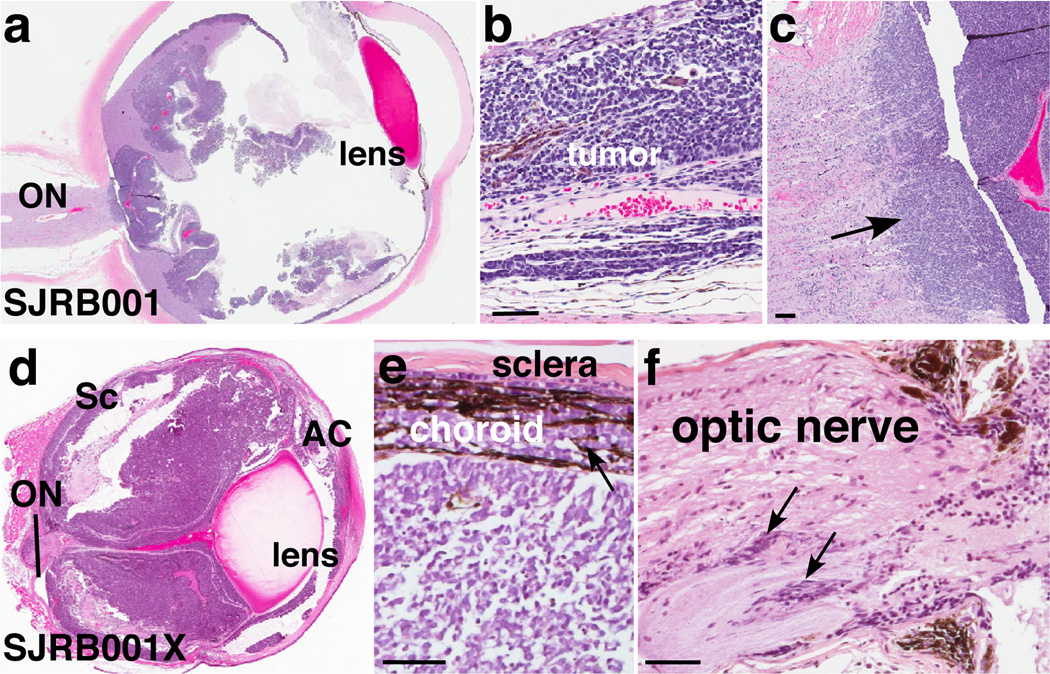
We performed WGS analysis on 4 primary human retinoblastoma samples (Section S1 and Table S1) and from matched normal tissue. Local tumor cell invasion but not metastasis was evident in each patient (Fig. 1a–c and Fig. S1). We generated an orthotopic xenograft, SJRB001X, of the primary tumor SJRB001 by inoculating primary tumor cells into the vitreous of the eyes of immunocompromised mice (Section S2). SJRB001X exhibited molecular, genetic and histopathologic features similar to SJRB001 (Fig. 1d–f, Figs. S2–4, Tables S2–4 and Section S3).
Figure 1. Characterization of retinoblastomas samples.
a–c, Representative retinoblastoma tumor section(SJRB001) stained with hematoxylin and eosin (H&E) showing choroidal and optic nerve invasion (arrow). d–f, H&E-stained section of the SJRB001X orthotopic xenograft with choroidal (e) and optic nerve (f) invasion (arrows). Abbreviations: AC, anterior chamber; ON, optic nerve; Sc, sclera. Scale bars: 25 µm.
Using a paired-end sequencing approach, we generated 1,040.9 Gb of sequence data for the samples described; 956.8 Gb (92%) were successfully mapped to the NCBI 36.1 reference genome (Section S4, Table S5). The average genome coverage was 28.9×, and the average exon coverage was 23.8× with 98.4% of SNPs detected across all 9 genomes showing concordance with their corresponding SNP array genotype calls at the same genomic positions (Table S5). To provide additional sequence coverage, we performed transcriptome sequencing of SJRB001-4 (Section S5, Table S6).
We identified 668 validated somatic sequence mutations and 40 structural variations (SVs) across the 4 cases (Table 1). These included 23 tier-1 mutations in genes, 35 tier-2 mutations in evolutionarily conserved regions of the genome (Section S6), 309 tier-3 mutations in nonrepetitive regions of the genome that are not part of tiers 1 and 2; and 301 tier-4 mutations in repetitive sequences in the genome (Table 1 and Table S7). The average number of sequence mutations was 167 per case (range, 56–258), with only 3.25 mutations per case (range, 0–5) resulting in amino acid changes (Table 1). The estimated mean mutation rate was 6.7×10−7 per base (range 1.03×10−7–2.17×10−8), which is 15-fold less than that in adult tumors analyzed by WGS, except for AML11. The predominant changes were C>A and G>T transversions (Fig. S5), which is consistent with the possibility that some of the transversions result from production of 8-Oxoguanine during oxidative stress. SJRB002 had no somatic sequence variations that resulted in amino acid changes; the only SVs were the loss of heterozygosity (LOH) at the RB1 locus and a gain of chromosome 6p. This suggests that very few genetic lesions are required for retinoblastoma progression after RB1 inactivation.
Table 1.
Summary of Somatic Mutations and Structural Alterations in Retinoblastoma
| 1Sample | 2Tier 1 |
3Non-Silent Tier 1 |
Genes | 5Tier 2 | 6Tier 3 | 7Tier 4 | Total | Mutation Rate |
7Structural Variations |
|---|---|---|---|---|---|---|---|---|---|
| SJRB001 D-G |
7 | 4 |
RB1, CCNC,TMEM195, RHBG |
16 | 117 | 85 | 225 | 1.03×10−7 | 4 |
|
2SJRB001 X-D |
0 | 0 | n.a. | 8 | 68 | 9 | 85 | 5.87×10−8 | 4 |
| SJRB002 D-G |
1 | 0 | n.a. | 1 | 25 | 29 | 56 | 2.17×10−8 | 0 |
| SJRB003 D-G |
7 | 4 |
RB1, HNMT, LHX8, STOML2 |
5 | 67 | 50 | 129 | 5.79×10−8 | 24 |
| SJRB004 D-G |
8 | 5 |
RB1, CD300LG, SDK1, TXK, DMWD |
13 | 100 | 137 | 258 | 8.63×10−8 | 12 |
D refers to diagnostic tumor, G refers to germline (blood DNA) and X refers to xenograft sample.
Tier 1 mutations are found in genes and include exons, 5’ and 3’ UTRs and splice sites. Introns are not included.
Non-silent Tier 1 mutations change amino acids in genes.
All of the somatic mutations in SJRB001 D-G were identified in SJRB001X. This row highlights the new mutations acquired in the xenograft compared to the primary tumor.
Tier 2 mutations are found in regions of the genome that are conserved between humans and mice.
Tier 3 mutations are found in regions of the genome that are not evolutionarily conserved.
Tier 4 mutations are in repetitive regions of the genome.
Structural variations include focal amplifications and deletions, LOH, interchromosomal and intrachromosomal translocations.
Background mutation rate was calculated based on the ratio of Tier 3 mutations to the number of Tier 3 bases covered at least 10x in tumor and germline for each pair.
RB1 gene inactivation in retinoblastoma
Both RB1 alleles were inactivated in each sample (Figs. S6, S7). SJRB002 and SJRB003 had mutations in RB1 combined with copy number–neutral LOH, and SJRB001 and SJRB004 had somatic sequence mutations combined with RB1-promoter hypermethylation (Figs. S6–8). Deep-sequence analysis of the germline sample from SJRB002 revealed that about 10% of reads contained the R445 nonsense mutation, suggesting the presence of germline chimerism for the RB1 mutation (Fig. S6). Combining the WGS data with SNP array data of an additional 42 samples, we found that tumors from patients with lower regional nucleotide diversity were much less likely to undergo LOH at the RB1 locus (Tables S8, S9, Section S7). These data show for the first time a significant association (p=8×10−8, Fisher’s exact test) between a germline genetic variation and mechanism of biallelic RB1 inactivation in retinoblastoma.
Recurrent lesions in retinoblastoma
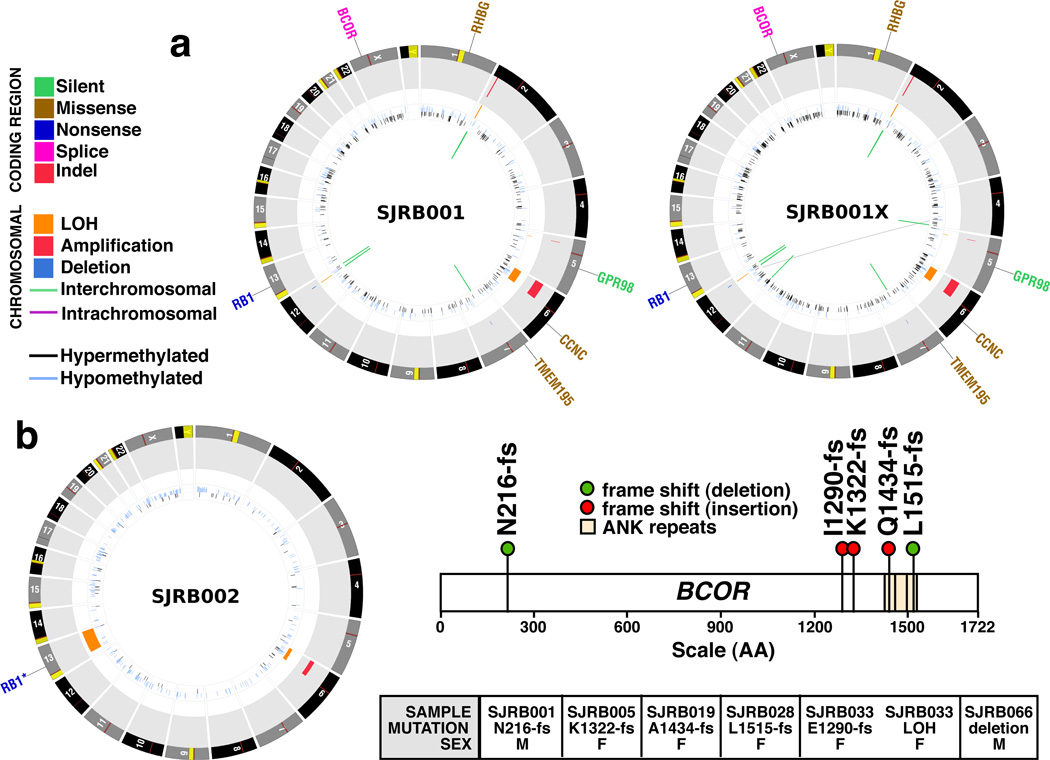
To determine whether any of the 11 genes with somatic mutations that caused amino acid changes or a frameshift in the coding region (Table 1, Fig. 2a,b, Figs. S9, S10, Section S8, Table S10), were recurrently mutated in retinoblastoma, we sequenced all exons from the 11 genes in our recurrent screening cohort of 42 retinoblastomas (Section S4). Only BCOR was recurrently mutated in of retinoblastoma (6/46, 13%). Five of the samples had BCOR mutations that resulted in premature truncation of the encoded protein, and 1 sample had a focal gene deletion (Fig. 2c, Table S11, Fig. S11).
Figure 2. Genomic profiles of SJRB001-2 and SJRB001X.
a,b, CIRCOS plots of genetic alterations in 2 retinoblastomas and the matched orthotopic xenograft. Loss of heterozygosity (orange), amplifications (red), and deletions (blue) are shown. Interchromosomal translocations (green lines) and intrachromosomal translocations (purple lines) are indicated. Sequence mutations in Refseq genes included silent single nucleotide variants (SNVs, green), missense SNVs (brown), nonsense SNVs (dark blue), splice-site mutations (pink), and insertion/deletion mutations (indels, red). c) BCOR mutations identified in the recurrency cohort.
We also used the WGS data to identify somatic SVs including whole-chromosome gains and losses, focal deletions (DEL), insertions (INS), inversions (INV), intrachromosomal rearrangements (ITX), interchromosomal rearrangements (CTX), and regions of LOH (Fig. 2a–c, Section S9, Table S12, Fig. S12). The average number of SVs was 10 per case (range, 0–24) (Table 1). SJRB001 had 4 SVs (2 DEL and 2 INS) including a gain of a region of chromosome 2 spanning MYCN (Table S12) and the only chromosomal lesion in SJRB002 was a gain of 6p, which occurs in about 40% of human retinoblastomas12 (Fig. 2b). Only a few genomic regions were affected by the SVs in SJRB003 and SJRB004 (Figs. S10, S13, S14, Table S12).
Orthotopic retinoblastoma xenograft
The genomic landscape of the orthotopic xenograft was remarkably similar to that of the primary tumor, despite continuous growth and multiple passages in vivo over 9 months (Fig. 2, Table 1). All of the single-nucleotide variants (SNVs) and SVs detected in SJRB001 were retained in SJRB001X. Only 67 new SNVs and 4 SVs were identified in the xenograft, and none targeted annotated genes (Fig. 2, Table 1). Moreover, each mutation was identified at a subclonal level (range, 20%–30%) and the mutant allele frequency for the Tier1-4 lesions in SJRB001 was retained in SJRB001X (Supplementary Fig. 15). This result was surprising because several studies in mice and cell cultures have linked RB1 inactivation to defects in chromosome segregation that result in aneuploidy 4,5,13–15 and CIN4. We measured the distance between sister chromatids, between kinetochores, and the proportion of lagging chromatids in 2 RB1-deficient human retinoblastoma orthotopic xenografts (SJRB001X and SJRB002X)16. Consistent with results from RB1-deficient RPE cells4, the distances between sister chromatids and kinetochores were increased, and there was evidence of lagging chromosomes (Figs. S16, S17). However, less variation in ploidy was observed during spectral karyotype (SKY) analysis of SJRB001X and SJRB002X that was more consistent with the ploidy of wild-type cells (Fig. 3a,b, Table S13). Moreover, CNVs were much lower in our cohort of 46 retinoblastomas than in tumors with known genome instability such as ovarian cancer (Fig. 3c). Together, the cytogenetic data and WGS data suggest that the genome is stable and newly acquired lesions do not provide a selective growth advantage and are thus likely passenger mutations.
Figure 3. Analysis of aneuploidy and CIN in retinoblastoma.
a, Chromosomal missegregation of SJRB001X cells after at least 21 rounds of cell division is plotted in red. b, Representative SKY image of SJRB001X after the third passage in mice. c, Alterations in the 46 Rb cases (Rb) compared to 153 high-grade serous ovarian cancer (Ov) from TGCA. The median chromosomal lesions for retinoblastoma (Rb) was 1.5% and 27.7% for ovarian cancer (Ov).
Identifying deregulated cancer pathways
There are many examples over the past several decades of epigenomic changes such as DNA methylation contributing to tumorigenesis17–19. Indeed, a recent study demonstrated changes in DNA methylation in Wilm’s tumors20 which, tend to have stable genomes like retinoblastomas. To explore whether epigenetic deregulation of genes or pathways promotes tumorigenesis in retinoblastoma, we carried out an integrative analysis of chromatin immunoprecipitation (ChIP) data, DNA-methylation data and gene expression data using order statistics. The SJRB001X sample was used for ChIP assay (Figs. S18–20), and primary tumor and xenograft samples were used for both DNA-methylation and gene expression assays. In all three analyses, experimental results in RB tumors were compared to those from human fetal retinae. A total of 104 genes, including 15 known cancer genes (Fig. 4a and Tables S14, S15), were found to have significant difference between RB tumors and human fetal retina, indicating that several key cancer genes were epigenetically deregulated.
Figure 4. SYK Is Expressed in Retinoblastoma and Is Required for Survival.
a, Whole-genome view of the gene ranks based on integrating ChIP-on-chip, methylation, and gene expression results. Y-axis is –log(p), where p is a p-value of Q-statistic corrected for multiple testing. Significantly (FDR ≤10%) downregulated (green) or upregulated (red) genes are shown. b, c, ChIP validation of histone markers of the SYK promoter including quantification by quantitative PCR (qPCR) with TaqMan probes. Each bar is the mean and standard deviation of triplicate samples. d, SYK gene expression measured by qPCR in fetal week 20 retina (fetal), primary retinoblastoma (tumor), orthotopic xenografts (SJRB001X and SJRB002X) and cell lines. Each bar is the mean and standard deviation of duplicate samples normalized to GPI1 expression. e, Immunoblot of SYK (green) and actin (red) in orthotopic xenografts, human fetal retina, and representative cell lines; black and white representation of the SYK immunoblot is in the lower panel. f, H&E (purple) and anti-SYK (brown) immunohistochemistry of retinoblastoma tissue. g, Immunoprecipitation analysis of SYK and pSYK Y525/526 from Weri1 retinoblastoma cells. h, Viability was measured in triplicate cultures 72 hours after infection of retinoblastoma cells with a lentivirus vector expressing either a control lentivirus or an shRNA against SYK. Scale bars: 10 µm.
SYK is a novel therapeutic target
SYK is the 5th most significant gene identified by the integrative analysis and the only up-regulated kinase gene (Table S15 and Fig. 4a). SYK is expressed throughout the hematopoietic system, regulates immunomodulatory signaling, and has been implicated in several hematologic malignancies21–24. Small-molecule inhibitors of SYK have been developed to treat autoimmune disorders25, and 2 of those agents, BAY61-3606 and R406, have shown efficacy in preclinical leukemia studies26–28.
ChIP-on-chip analysis showed increased activating histone modifications (H3K4me3 and K3K9/14Ac) at SYK’s promoter, and the repressive histone marker (H3k9me3) was unchanged. RNA polymerase II binding to the SYK promoter was also increased (Fig. 4b, Tables S14, S15). These ChIP-on-chip results were validated in independent samples by real-time RT-PCR analysis (Fig. 4c), and we confirmed the increase in SYK gene expression (Fig. 4d). SYK protein levels were higher in human retinoblastoma orthotopic xenografts and cell lines than in human fetal retinae (Fig. 4e). To determine whether SYK is expressed in primary human retinoblastomas, we performed immunohistochemistry on a retinoblastoma tissue microarray (TMA) or whole eye sections. In total, 100% (82/82) showed very strong expression (3+) of SYK in all tumor cells; SYK was not expressed in normal retina (Fig. 4f). SYK’s kinase activity is regulated through autophosphorylation at the Tyr525/526 residues within its catalytic domain. These sites were phosphorylated in retinoblastoma cells, and this phosphorylation was reversed by BAY 61-3606 or R406 exposure (Fig. 4g and data not shown).
To determine whether SYK expression is required for retinoblastoma growth, survival, or both, we generated an shRNA to SYK and cloned it into the lentiviral vector Lenti-SYK-9. Lenti-SYK-9 efficiently knocked-down SYK in retinoblastoma cell lines (Fig. S21) and dramatically increased apoptosis in retinoblastoma cells (Fig. 4h, Fig. S21). Similar results were obtained in vivo using SJRB001X (data not shown). We used an empty lentiviral vector and a lentiviral vector encoding an SYK shRNA that less effectively reduced SYK expression (Lenti-SYK-6) as controls. Cell lines that do not express SYK (BJ, 293T, and uninduced Jurkat cells) were used as controls and the Lenti-SYK-9 lentivirus had no effect on the growth or apoptosis of the control cells.
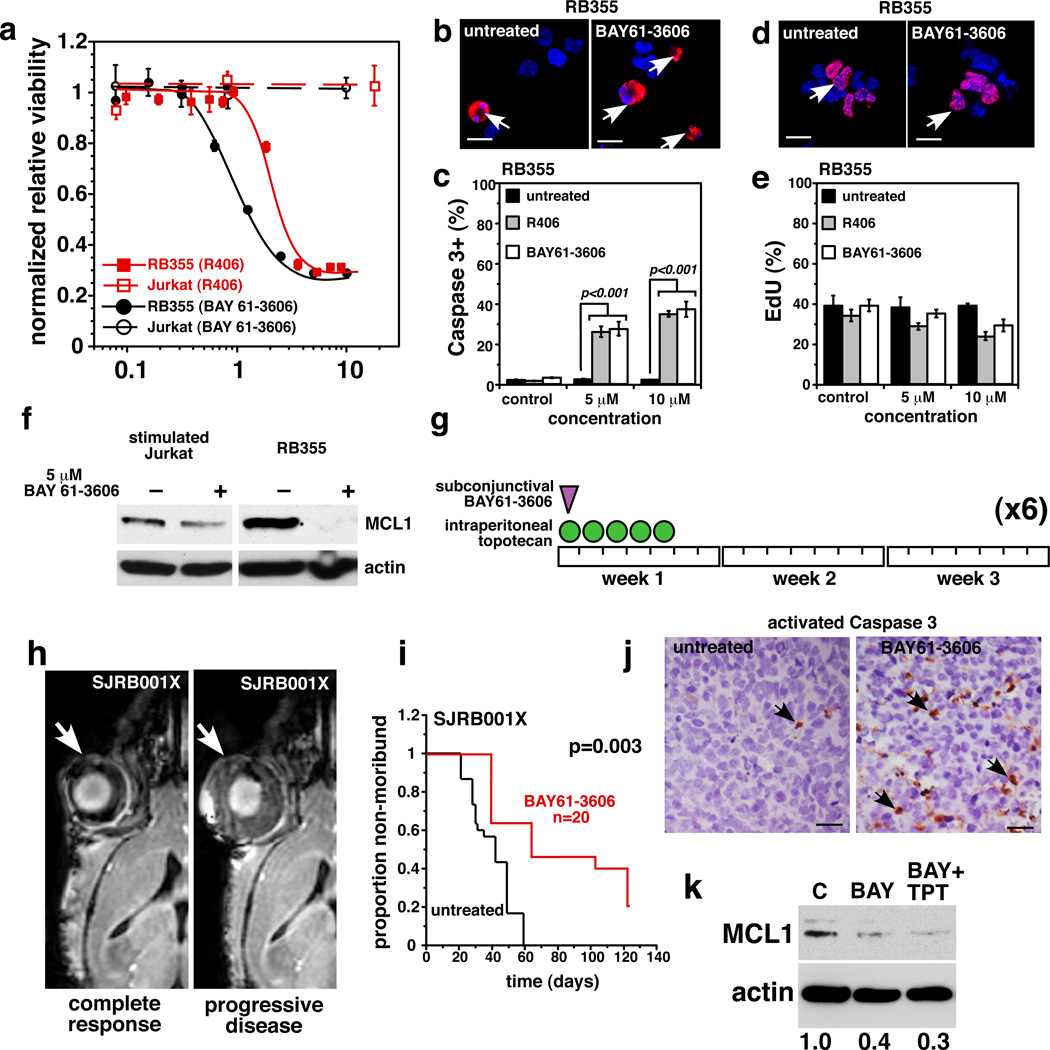
We exposed retinoblastoma cell lines that express high levels of SYK (Weri1 and RB355) to various concentrations of the SYK inhibitors BAY 61-3606 or R406 for 72 hours and then measured cell viability. Jurkat (uninduced) and 293T cells were used as negative controls. Weri1 and RB355 cells were sensitive to both SYK inhibitors, but the Jurkat and 293T cells were unaffected (Fig. 5a). Transmission electron microscopy of retinoblastoma cells treated with the SYK inhibitors showed morphologic features consistent with cell death and mitochondrial defects (Fig. S22); this was confirmed by scoring the proportion of activated caspase-3+ cells (Fig. 5b, c, Fig. S22). Jurkat cells showed no increase in activated caspase-3+ cells after treatment with 10 µM R406 or BAY 61-3606 (data not shown).
Figure 5. Retinoblastoma Cells are Sensitive to SYK Inhibitors.
a, Dose response for SYK inhibitors R406 (red) and BAY 61-3606 (black) in RB355 retinoblastoma cells and a negative control (Jurkat). Each data point is the mean and standard deviation of triplicate samples. b–e Immunofluorescence of activated caspase 3 or EdU(red) before and after treatment of RB355 cells with R406 or BAY 61–3606. A total of 250 cells were scored in duplicate for each sample and each treatment condition to derive the mean and standard deviation. Nuclei were stained with DAPI (blue). f, Treatment of stimulated Jurkat or RB355 cells with 5 µM BAY 61–3606 for 24 hours reduced MCL1 expression. g, Schematic of the treatment schedule for mice with SJRB001X tumors. h, Representative MR images of a mouse whose tumor responded after 4 courses of treatment with BAY 61–3606 (left) and another whose disease progressed during treatment (right). i, Survival curves show that BAY 61–3606+TPT treatment improved outcome. j, Immunostaining for activated caspase 3 (arrows) in untreated or BAY 61–3606–treated eyes. k, untreated or BAY 61-3606–treated eyes. k, Immunoblot showing reduced MCL1 protein after BAY 61–3606 or BAY 61–3606+TPT treatment. Scale bars b, d: 5 µm; j: 10 µm.
The proportions of cells from each line that incorporated EdU were similar, suggesting that retinoblastoma’s cell cycle is not affected by SYK inhibition (Fig. 5d, e, Fig. S22). The effects of targeting SYK were partially rescued by the pan-caspase inhibitor Q-VD-OPH (Fig. S22). Less MitoTracker Red accumulated in retinoblastoma cells exposed to the SYK inhibitors (Fig. S22), yet treatment of Jurkat cells with either inhibitor had no effect on MitoTracker Red accumulation (data not shown). Inhibiting SYK in B-CLL cells reduced their autophosphorylation of SYK and destabilized MCL126,29, the only antiapoptotic member of the BCL-2 family that is upregulated in retinoblastoma (data not shown). Both inhibitors reduced autophosphorylation of SYK on Y525/526 (Fig. 4g) and reduced MCL1 expression coincident with apoptosis (Fig. 5f).
We tested the efficacy of BAY61-3606 in vivo on our SJRB001X model30. The chemotherapy regimen consisted of a single subconjunctival dose of BAY 61–3606 on Day 1 and daily doses of topotecan (TPT) on Days 1–5 until either 6 courses (21 days per course) were administered, or the tumor progressed and surgical enucleation was required (Fig. 5g,h). BAY 61-3606+TPT significantly improved outcome (p=0.003) (Fig. 5i), and its efficacy was correlated with an increase in activated caspase-3+ cells in the treated eyes (Fig. 5j). Previous studies using this model have shown that TPT combined with subconjunctival carboplatin had no effect on tumor response or outcome 30, so the improvement seen here can be attributed to targeting SYK. MCL1 expression was also reduced in the treated eyes, which is consistent with increased apoptosis and targeting SYK in vivo (Fig. 5k). Together, these results suggest that SYK is a promising new target for treating retinoblastoma.
DISCUSSION
It has been suggested that biallelic loss of RB1 directly causes genomic instability, which in turn contributes to the rapid development of overt retinoblastoma. Our data failed to support this hypothesis. The mutational rate and number of SVs per case that we assessed were among the lowest reported in human cancer. Moreover, the only non-silent mutation in SJRB002 was in RB1, and no SVs were detected. The minimal increase in passenger mutations in SJRB001X cells, despite prolonged passage, was also consistent with a relatively stable genome. These results support those from previous studies showing that retinoblastoma karyotypes are stable in vitro and in vivo31,32. Our data suggest that genomic instability is neither a hallmark of retinoblastoma nor sufficient to explain how retinoblastomas progress so rapidly.
We propose that epigenetic mechanisms contribute to retinoblastoma tumorigenesis. We identified several known oncogenes and tumor-suppressor genes with histone modifications and altered DNA methylation that correlated with changes in gene expression. Our key discovery was that SYK is important in retinoblastoma. Retinal progenitor cells and retinal neurons express little to no SYK, and SYK has no known function in the developing visual system. Moreover, no recurrent genetic lesions in SYK are associated with retinoblastoma to suggest that this gene drives tumorigenesis. Only by integrating epigenetic and gene expression analyses, did we identify SYK as an important oncogene in retinoblastoma. This is important not only for expanding our understanding of the biology of retinoblastoma but also for advancing immediate therapeutic options that were not previously considered such as BAY 61-3606 or R406. This study highlights the value of integrating WGS analyses of the genetic and epigenetic features of tumor genomes toward finding a cure for cancers such as retinoblastoma.
METHODS SUMMARY
Full details of sample acquisition, molecular and biochemical procedures, informatics and whole genome sequencing and animal and drug studies are provided in the Supplementary Information. The SJCRH IRB approved experiments involving human subjects and informed consent was obtained from all subjects. For animal studies, all experiments were performed in accordance with federal guidelines and regulations. The SJCRH IACUC approved all animal experiments. The dbGaP accession assigned to this study is phs000352.v1.p1. Lentiviral vectors (GIPZ with Lenti-SYK-9 #V3LHS-366147 and Lenti-SYK-6 #V3LHS-366143) encoding shRNAs to SYK were purchased from OpenBiosystems, Inc.
Supplementary Material
ACKNOWLEDGEMENTS
The whole genome sequencing was supported as part of the St. Jude Children’s Research Hospital – Washington University Pediatric Cancer Genome Project. We thank Jill Lahti, James Dalton and Marc Valentine for help with FISH analysis, Linda Holmfeldt, Jiakun Zhang and Michael Barbato for help with sample preparation, and Virginia Valentine for SKY analysis. We thank Ibrahim Qaddoumi, Carlos Rodriguez-Galindo and Barrett Haik for support of the SJCRH retinoblastoma clinical research and Wei Lei, Daniel McGoldrick, Daniel Alford, Stephen Espy, John Obenauer and Kimberly Johnson for assistance with data acquisition, handling and analysis. We thank Angie McArthur and Cherise Guess for editing the manuscript, Jamshid Temirov for help with sister chromatid analysis, Justin Thurman for help with histology, Fred Krafcik for help with cell culture, Jianrong Wu and Catherine Billups for statistical analysis, Jongrye Jeon for help with lentiviral preparations, and Cori Bradley for assistance with preclinical testing. This work was supported, in part, by Cancer Center Support (CA21765) from the NCI; grants to M.A.D from the NIH (EY014867 and EY018599), the American Cancer Society, and Research to Prevent Blindness Foundation; and the American Lebanese Syrian Associated Charities (ALSAC). M.A.D. is a Howard Hughes Medical Institute Early Career Scientist. This work was also supported by an American Cancer Society Fellowship to A.L.M., the MGH Cancer Center Saltonstall Foundation Scholarship to N.J.D., and funding from AstraZeneca and NIH grants GM81607 and CA64402 to N.J.D.
Footnotes
AUTHOR CONTRIBUTIONS
M.A.D., C.A.B., J.M., J.F-O., R.B. and A.L.M. designed the experiments. L.D. J.Z. led data analysis. M.A.D., L.D., J.Z., X.C., A.U., G.W., J.W., M.R., J.M., S.P., S.M. P.G., G.N., D.Z., C.L., R.S.F., L.L.F., X.H., D.J.D. and K.O. performed data analysis and bioinformatics support. M.A.D.,J.Z., A.U., C.A.B., J.M., J.F-O., R.B. and A.L.M. prepared figures and tables. C.A.B, J.M., F.F-O., R.B. and A.L.M. performed laboratory experiments. J.F-O. created the xenograft. M.W. provided samples. J.E., S.S., C.M. provided assistance with sample processing and handling. A.B. and D.E. provided pathology support. M.A.D., C.N., E.R.M., R.K.W. and J.R.D. supervised the project. M.A.D. wrote the manuscript. J.Z. C.A.B., J.M., J.F-O., L.D., M.W., E.R.M., L.D., S.S., C.M. and J.R.D. critically read and commented on the manuscript.
REFERENCES
- 1.Friend SH, et al. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643–646. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Knudson A. Mutation and Cancer:statistical study of retinoblastoma. PNAS. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manning AL, Longworth MS, Dyson NJ. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes & development. 2010;24:1364–1376. doi: 10.1101/gad.1917310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hernando E, et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- 6.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu J, Ruhf ML, Perrimon N, Leder P. A genome-wide RNA interference screen identifies putative chromatin regulators essential for E2F repression. Proc Natl Acad Sci U S A. 2007;104:9381–9386. doi: 10.1073/pnas.0610279104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benetti R, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:998. doi: 10.1038/nsmb0908-998b. [DOI] [PubMed] [Google Scholar]
- 9.Wen H, Andrejka L, Ashton J, Karess R, Lipsick JS. Epigenetic regulation of gene expression by Drosophila Myb and E2F2-RBF via the Myb-MuvB/dREAM complex. Genes Dev. 2008;22:601–614. doi: 10.1101/gad.1626308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bourgo RJ, et al. SWI/SNF deficiency results in aberrant chromatin organization, mitotic failure, and diminished proliferative capacity. Mol Biol Cell. 2009;20:3192–3199. doi: 10.1091/mbc.E08-12-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ley TJ, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corson TW, Gallie BL. One hit, two hits, three hits, more? Genomic changes in the development of retinoblastoma. Genes Chromosomes Cancer. 2007;46:617–634. doi: 10.1002/gcc.20457. [DOI] [PubMed] [Google Scholar]
- 13.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iovino F, Lentini L, Amato A, Di Leonardo A. RB acute loss induces centrosome amplification and aneuploidy in murine primary fibroblasts. Molecular cancer. 2006;5:38. doi: 10.1186/1476-4598-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amato A, Lentini L, Schillaci T, Iovino F, Di Leonardo A. RNAi mediated acute depletion of retinoblastoma protein (pRb) promotes aneuploidy in human primary cells via micronuclei formation. BMC cell biology. 2009;10:79. doi: 10.1186/1471-2121-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mcevoy J, et al. Coexpression of Normally Incompatible Developmental Pahtways in Retinoblastoma Genesis. Cancer Cell. 2011 doi: 10.1016/j.ccr.2011.07.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feinberg AP, Tycko B. The history of cancer epigenetics. Nature reviews. Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 18.Jones PA, Laird PW. Cancer epigenetics comes of age. Nature genetics. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 19.Laird PW. Cancer epigenetics. Human molecular genetics 14 Spec No 1, R65-76. 2005 doi: 10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- 20.Hansen KD, et al. Increased methylation variation in epigenetic domains across cancer types. Nature genetics. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hahn CK, et al. Proteomic and genetic approaches identify Syk as an AML target. Cancer Cell. 2009;16:281–294. doi: 10.1016/j.ccr.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L, et al. SYK-dependent tonic B-cell receptor signaling is a rational treatment target in diffuse large B-cell lymphoma. Blood. 2008;111:2230–2237. doi: 10.1182/blood-2007-07-100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feldman AL, et al. Overexpression of Syk tyrosine kinase in peripheral T-cell lymphomas. Leukemia. 2008;22:1139–1143. doi: 10.1038/leu.2008.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young RM, et al. Mouse models of non-Hodgkin lymphoma reveal Syk as an important therapeutic target. Blood. 2009;113:2508–2516. doi: 10.1182/blood-2008-05-158618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinblatt ME, et al. An oral spleen tyrosine kinase (Syk) inhibitor for rheumatoid arthritis. The New England journal of medicine. 2010;363:1303–1312. doi: 10.1056/NEJMoa1000500. [DOI] [PubMed] [Google Scholar]
- 26.Baudot AD, et al. The tyrosine kinase Syk regulates the survival of chronic lymphocytic leukemia B cells through PKCdelta and proteasome-dependent regulation of Mcl-1 expression. Oncogene. 2009;28:3261–3273. doi: 10.1038/onc.2009.179. [DOI] [PubMed] [Google Scholar]
- 27.Suljagic M, et al. The Syk inhibitor fostamatinib disodium (R788) inhibits tumor growth in the Emu- TCL1 transgenic mouse model of CLL by blocking antigen-dependent B-cell receptor signaling. Blood. 116:4894–4905. doi: 10.1182/blood-2010-03-275180. [DOI] [PubMed] [Google Scholar]
- 28.Buchner M, et al. Spleen tyrosine kinase inhibition prevents chemokine- and integrin-mediated stromal protective effects in chronic lymphocytic leukemia. Blood. 115:4497–4506. doi: 10.1182/blood-2009-07-233692. [DOI] [PubMed] [Google Scholar]
- 29.Gobessi S, et al. Inhibition of constitutive and BCR-induced Syk activation downregulates Mcl-1 and induces apoptosis in chronic lymphocytic leukemia B cells. Leukemia. 2009;23:686–697. doi: 10.1038/leu.2008.346. [DOI] [PubMed] [Google Scholar]
- 30.Brennan RC, et al. Targeting the p53 pathway in retinoblastoma with subconjunctival Nutlin-3a. Cancer research. 2011;71:4205–4213. doi: 10.1158/0008-5472.CAN-11-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan Y, et al. Engraftment and growth of patient-derived retinoblastoma tumour in severe combined immunodeficiency mice. Eur J Cancer. 2000;36:221–228. doi: 10.1016/s0959-8049(99)00277-4. [DOI] [PubMed] [Google Scholar]
- 32.Squire J, Gallie BL, Phillips RA. A detailed analysis of chromosomal changes in heritable and non-heritable retinoblastoma. Hum Genet. 1985;70:291–301. doi: 10.1007/BF00295364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.