Abstract
Codeine is bioactivated to morphine, a strong opioid agonist, by the hepatic cytochrome P450 2D6 (CYP2D6); hence, the efficacy and safety of codeine as an analgesic are governed by CYP2D6 polymorphisms. Codeine has little therapeutic effect in patients who are CYP2D6 poor metabolizers, whereas the risk of morphine toxicity is higher in ultrarapid metabolizers. The purpose of this guideline (periodically updated at http://www.pharmgkb.org) is to provide information relating to the interpretation of CYP2D6 genotype test results to guide the dosing of codeine.
FOCUSED LITERATURE REVIEW
A systematic literature review was conducted that focused on CYP2D6 and its relevance in codeine use (see Supplementary Data online). This guideline was developed based on interpretation of the literature by the authors and by experts in the field.
GENE: CYP2D6
Background
More than 80 CYP2D6 alleles have been defined by the Cytochrome P450 Nomenclature Committee at http://www.cypalleles.ki.se. Clinical phenotype data are available for many common alleles (see Supplementary Tables S1–S5 online); however, many alleles are not typically tested for in clinical trials. Their clinical phenotype is predicted on the basis of the expected functional impact of their defining genetic variation or is extrapolated from results of in vitro functional studies using different substrates.
Interpretation of genetic test results
Most clinical laboratories report CYP2D6 genotype using the star (⋆) allele nomenclature and may provide interpretation of the patient’s predicted metabolizer phenotype. Single-nucleotide polymorphisms (SNPs) and other sequence variations, including insertions and deletions, are determined by genetic laboratory tests. The reference SNP number (rs number) for a given SNP defines the specific genomic nucleotide alteration. Each star (⋆) allele (or haplotype) is defined by the presence of a specific combination of SNPs and/or other sequence alterations within the CYP2D6 gene locus. The key alleles are shown in Supplementary Table S1 online and the key allele-defining SNPs and their respective impacts on CYP2D6 enzyme function are provided in Supplementary Table S2 online. Genetic results are reported as a diplotype, which includes one maternal and one paternal allele (e.g., CYP2D6⋆1/⋆4; Supplementary Table S3 online). In some cases, patients have more than two copies of the CYP2D6 gene. Those alleles are denoted by an “xN” following the allele designation (e.g., CYP2D6⋆2xN; duplication) (see Supplementary Data online for details). Additional details about allele nomenclature and definitions can be found at http://www.cypalleles.ki.se/cyp2d6.htm, and information regarding the effects of allelic variation on CYP2D6 substrates can be found at http://www.pharmgkb.org/search/annotatedGene/cyp2d6/haplotype.jsp. CYP2D6 allele frequencies differ substantially between racial and ethnic groups. Supplementary Table S1 online summarizes the most important allele frequencies for the major races and Hispanics.
The combination of alleles is used to determine a patient’s diplotype. CYP2D6 alleles are characterized as wild-type (normal function), reduced-function, or nonfunctional based on the expected activity level of the enzyme for which they encode. Each functional group is assigned an activity value ranging from 0 to 1.0 (e.g., 0 for nonfunctional, 0.5 for reduced function, and 1.0 for fully functional).1 Supplementary Table S4 online describes the activity score values assigned to selected alleles. If multiple copies of the CYP2D6 gene are detected, the activity score is multiplied by the number of copies of each allele present. The total CYP2D6 activity score is the sum of the values assigned to each allele, which typically ranges from 0 to 3.0 but may exceed 3.0 in rare cases.1,2
The CYP2D6 activity score relates to the phenotype classification system as follows (Table 1): patients with an activity score of 0 are poor metabolizers, those with a score of 0.5 are considered intermediate metabolizers, and those with a score of 1.0, 1.5 or 2.0 represent a range of extensive metabolizers. Patients with a score >2.0 are classified as ultrarapid metabolizers. The extensive metabolizer phenotype represents normal (wild-type) enzyme activity.1–3 The incidence of poor and ultrarapid metabolizers varies greatly (0–10% and 0–29%, respectively) among various populations. Selected CYP2D6 activity scores are provided in Supplementary Table S3 online, and predicted phenotypes of common diplotypes are summarized in Supplementary Table S5 online.
Table 1.
Assignment of likely codeine metabolism phenotypes based on CYP2D6 diplotypes
| Likely phenotypea | Activity score | Genotypes | Examples of diplotypes |
|---|---|---|---|
| Ultrarapid metabolizer (∼1–2% of patients) | >2.0 | An individual carrying more than two copies of functional alleles | *1/*1xN, *1/*2xN |
| Extensive metabolizer (∼77–92% of patients) | 1.0–2.0b | An individual carrying two alleles encoding full or reduced function or one full function allele together with either one nonfunctional or one reduced-function allele | *1/*1, *1/*2, *2/*2, *1/*41,*1/*4,*2/*5, *10/*10 |
| Intermediate metabolizer (∼2–11% of patients) | 0.5b | An individual carrying one reduced and one nonfunctional allele | *4/*10, *5/*41 |
| Poor metabolizer (∼5–10% of patients) | 0 | An individual carrying no functional alleles | *4/*4, *4/*5, *5/*5, *4/*6 |
The frequency estimates are based on data from Caucasians and may differ substantially for other ethnicities. See Supplementary Data online for estimates of phenotype frequencies among different ethnic/geographic groups.
Note that some investigators define patients with an activity score of 0.5 and 1.0 as intermediate metabolizers and define patients with an activity score of 1.5 and 2.0 as extensive metabolizers. Classifying patients with an activity score of 1.0 as extensive metabolizers in this guideline is based on data specific for formation of morphine from codeine in these patients.13
It is noted that subjects with genotypes giving rise to an activity score of 1.0, caused by a diplotype containing one functional and one nonfunctional allele or containing two reduced-function alleles, are classified by some investigators as “intermediate metabolizers.” Regardless of the term used to describe such individuals (extensive metabolizers or intermediate metabolizers), they have lower CYP2D6 activity than subjects with two fully functional alleles (activity score of 2.0) and higher activity as compared with subjects with one reduced and one nonfunctional allele (with an activity score of 0.5). Given that there is no gold standard for phenotype classification, genotypes that result in CYP2D6 activity scores of 1.0, 1.5, and 2.0 are grouped together as extensive metabolizers in this guideline, which is based on data specific for codeine metabolism (Supplementary Table S6 online).
It should be noted that reference laboratories providing clinical CYP2D6 genotyping may use varying methods to assign phenotypes. Therefore it is advisable to note a patient’s CYP2D6 diplotype and to calculate an activity score before making therapeutic decisions about codeine therapy.
AVAILABLE GENETIC TEST OPTIONS
Several academic and commercial clinical laboratories offer genetic testing for CYP2D6. Specific information (on several of these laboratories, on the assays, and links to the product labels) can be found in the supplementary information and at http://www.pharmgkb.org/resources/forScientificUsers/pharmacogenomic_tests.jsp.
Incidental findings
Currently, there are no diseases or conditions known to be linked to variations in the CYP2D6 gene independent of drug metabolism and response. Although there are some isolated reports of CYP2D6 genetic effects on phenotypes other than in relation to drug response,4,5 these findings have not been consistently reproduced in other studies.
Other considerations
Modification of the predicted phenotype by drug interactions
The CYP2D6 metabolizer phenotype may undergo alteration in a patient who is taking drugs that inhibit CYP2D6 activity. A list of CYP2D6 inhibitors can be found at http://medicine.iupui.edu/clinpharm/ddis/table.aspx. For this purpose, drugs are classified as strong, moderate, or weak inhibitors, based on the US Food and Drug Administration (FDA) guidance on drug interaction studies (http://www.fda.gov/Drugs/DevelopmentApprovalProcess/DevelopmentResources/DrugInteractionsLabeling/ucm081177.htm#classInhibit). Borges et al. demonstrated that incorporating the impact of drug interactions by CYP2D6 inhibitors improves the ability of the activity score to predict the extent of tamoxifen metabolism by CYP2D6.2 For patients on strong inhibitors, the CYP2D6 activity score is adjusted to 0 and the predicted phenotype is that of a poor metabolizer. For patients who are on weak or moderate CYP2D6 inhibitors, the activity score is multiplied by 0.5 and then converted to the predicted phenotype.2
CYP2D6 does not appear to be induced by concurrent medications; however, there are limited data showing that CYP2D6 enzyme activity increases during pregnancy. Wadelius et al. demonstrated that CYP2D6 activity increases during pregnancy by showing that the value of the dextromethorphan/dextrorphan metabolic ratio was reduced by 53%; Heikkinen et al. demonstrated that the norfluoxetine/fluoxetine metabolic ratio increased 2.4-fold.6,7 The apparent oral clearance of metoprolol was shown to increase four- to fivefold during pregnancy.8 Although mean CYP2D6 activity appears to increase during pregnancy, the large interindividual variability in the increase, and the limited number of subjects studied, make it difficult to make any recommendations regarding adjustment of the activity scores of functional alleles during pregnancy. The CYP2D6 activity scores of nonfunctional alleles are not affected by pregnancy.
CODEINE
Background
Codeine is an opioid analgesic indicated for the relief of mild to moderately severe pain. The analgesic properties of codeine stem from its conversion to morphine and morphine-6-glucuronide because codeine’s affinity for μ-opioid receptors is 200-fold weaker than that of morphine.9,10 Both codeine and morphine also have antitussive effects.
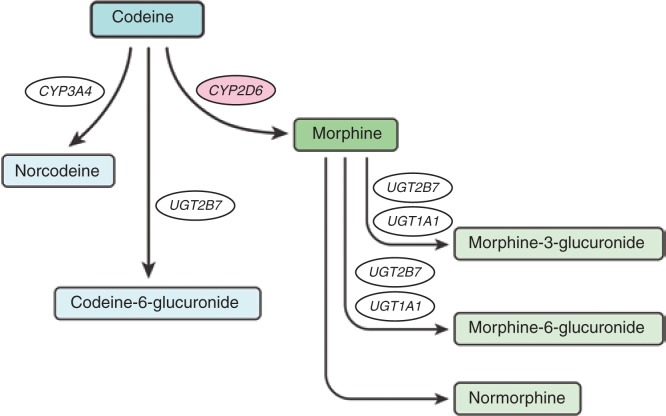
O-Demethylation of codeine into morphine by CYP2D6 represents a minor pathway in extensive metabolizers, accounting for only 5–10% of codeine clearance in such individuals; however, this pathway is essential for its opioid activity (Figure 1). The percentage of codeine converted into morphine can be affected by drug interactions and can be much higher in ultrarapid metabolizers.11 Morphine is further glucuronidated to morphine-3-glucuronide and morphine-6-glucuronide. Morphine-6-glucuronide is known to have analgesic activity in humans, whereas morphine-3-glucuronide is generally not considered to possess analgesic properties. Approximately 80% of an administered dose of codeine is converted to inactive metabolites by glucuronidation to codeine-6-glucuronide via UDPglucuronosyltransferase (UGT) 2B7, and by N-demethylation to norcodeine via CYP3A4. The analgesic activity of codeine-6-glucuronide in humans is unknown, whereas norcodeine is thought to have no analgesic properties.
Figure 1.
Codeine metabolism pathway.
Codeine analgesia is closely related to CYP2D6 pharmacogenetics. The association between CYP2D6 metabolizer phenotype and the formation of morphine from codeine is well defined. Pharmacokinetic and pharmacodynamic studies show a decrease in morphine levels and a decrease in analgesia in poor metabolizers receiving codeine as compared with extensive metabolizers.12,13 Mikus et al. reported a decreased incidence of gastrointestinal side effects (i.e., constipation) in poor metabolizers as compared with extensive metabolizers;14 a later study by the same group of investigators found that central side effects (e.g., sedation, nausea, and dry mouth) did not differ between poor metabolizers and extensive metabolizers.12 Further studies are needed to fully investigate the impact of the CYP2D6 poor metabolizer phenotype on the adverse-effect profile of codeine.
On the other hand, increased conversion to morphine in CYP2D6 ultrarapid metabolizers can result in toxic systemic concentrations of morphine11 even at low codeine doses. The most common adverse reactions to codeine include drowsiness, lightheadedness, dizziness, sedation, shortness of breath, nausea, vomiting, and sweating. Serious adverse reactions include respiratory depression and, to a lesser degree, circulatory depression, respiratory arrest, shock, and cardiac arrest. Pharmacokinetic studies show increased conversion of codeine to morphine in ultrarapid metabolizers as compared with extensive metabolizers. 15 Case reports detail the occurrence of severe or lifethreatening side effects following standard doses of codeine in ultrarapid metabolizers.11,16,17
Despite these data, codeine continues to be widely used, and most patients receive codeine without prior CYP2D6 genotyping. This guideline recommends using alternative analgesics in patients who are CYP2D6 poor metabolizers or ultrarapid metabolizers. It is important to recognize that, in addition to codeine, several other opioids are metabolized, at least in part, by CYP2D6. The opioids tramadol, hydrocodone, and oxycodone are O-demethylated by CYP2D6 to O-desmethyltramadol, hydromorphone, and oxymorphone, respectively.
Tramadol in its available racemic form is extensively metabolized via several pathways, including CYP2D6-mediated oxidation to O-desmethyltramadol, which has a 200-fold greater affinity for μ-opioid receptors than the parent drug does.10,18 Consequently, (+)-O-desmethyltramadol is principally responsible for opioid receptor-mediated analgesia, whereas (+)- and (−)-tramadol contribute to analgesia by inhibiting reuptake of the neurotransmitters serotonin and noradrenaline. As compared with CYP2D6 extensive metabolizers, poor metabolizers have been shown to have much lower median values of area under the concentration–time curves for the active metabolite after a dose of tramadol.19,20 In addition, several prospective clinical trials have shown that, as compared with CYP2D6 extensive metabolizers, poor metabolizers more often fail to exhibit analgesia in response to tramadol.18,19,21 Judging from this evidence, it is likely that tramadol has reduced clinical efficacy in CYP2D6 poor metabolizers.
Pharmacokinetic studies showed higher peak plasma concentrations of (+)-O-desmethyltramadol after a dose of tramadol, and also greater analgesia, stronger miosis, and higher incidence of nausea in ultrarapid metabolizers as compared with extensive metabolizers.22 There is one case report of respiratory depression in a CYP2D6 ultrarapid metabolizer with renal impairment after postsurgical tramadol treatment.23 On the basis of these data, the use of analgesics other than tramadol may be preferable in CYP2D6 poor metabolizers and also in ultrarapid metabolizers.
Hydrocodone is biotransformed by CYP2D6 into hydromorphone, which has a 10- to 33-fold greater affinity for μ-opioid receptors as compared with the parent drug. Relative to extensive metabolizers, poor metabolizers have been shown to have lower peak concentrations of hydromorphone after a dose of hydrocodone24; however, CYP2D6 metabolizer status does not appear to affect response to hydrocodone.24,25 To date, there is no information on the pharmacokinetics of hydrocodone in CYP2D6 ultrarapid metabolizers. Therefore, there is insufficient evidence as to whether poor metabolizers can be expected to have decreased analgesia when treated with hydrocodone, and whether ultrarapid metabolizers have an increased risk of toxicity with normal doses of hydrocodone.
Approximately 11% of an oxycodone dose is O-demethylated by CYP2D6 to the minor metabolite oxymorphone, which has a 40-fold higher affinity and eightfold higher potency for μ-opioid receptors as compared with the parent drug.10,26 As compared with extensive metabolizers, poor metabolizers have been shown to have lower peak concentrations of oxymorphone after a dose of oxycodone.27,28 However, prospective clinical studies have produced conflicting data pertaining to the associations between CYP2D6 metabolizer phenotype and the analgesic effect and toxicity of oxycodone. In two studies in healthy volunteers, differential analgesia response to experimental pain stimuli was observed between extensive and poor metabolizers and also between ultrarapid and extensive/poor metabolizers.29,30 However, clinical studies in postoperative patients and in patients with cancer failed to demonstrate a significant difference in analgesia or side effects in response to oxycodone across CYP2D6 phenotypes.27,28 Physiologic alterations (e.g., miosis) after dosing with oxycodone correlate best with exposure to the parent compound.31 It is therefore premature to recommend routine therapy adjustment for oxycodone on the basis of CYP2D6 genotype.
The differing levels of evidence for the association of CYP2D6 phenotype with hydrocodone and oxycodone analgesia as compared with codeine and tramadol may be because of the difference in the relative roles of the parent drug and circulating metabolites in analgesia among these CYP2D6 substrates.31
To avoid treatment complications, opioids that are not metabolized by CYP2D6, including morphine, oxymorphone, buprenorphine, fentanyl, methadone, and hydromorphone,32 along with nonopioid analgesics, may be considered as alternatives for use in CYP2D6 poor metabolizers and in ultrarapid metabolizers, depending on the type and chronicity of the pain being treated.
Other genes affecting codeine
Glucuronidation of codeine and of morphine is mediated by the polymorphic UGT2B7 enzyme.33 Although the production of morphine-6-glucuronide is almost exclusively catalyzed by UGT2B7, several isoforms of the UGT1A subfamily are also involved in the formation of morphine-3-glucuronide. Conflicting evidence exists regarding the impact of the UGT2B7⋆2 variant on the glucuronidation of codeine.34 Polymorphisms in the MDR1 transporter (ABCB1) gene also appear to have a modest association with opioid dose requirements.35 The response to codeine may also be influenced by polymorphisms in drug response genes including, but not limited to, the opioid receptor μ1 gene OPRM1; however, the importance of this gene to clinical outcome is not yet fully appreciated.35
LINKING GENETIC VARIABILITY TO VARIABILITY IN DRUG-RELATED PHENOTYPES
There is substantial evidence linking CYP2D6 genotype to variability in codeine efficacy and toxicity (see Supplementary Table S6 online). Decreased codeine analgesia has been observed in poor metabolizers, whereas severe or life-threatening toxicity after normal doses of codeine has been documented in ultrarapid metabolizers. This body of evidence, rather than randomized clinical trials involving pharmacogenetic testing, provides the basis of the therapeutic recommendations in Table 2.
Table 2.
Codeine therapy recommendations based on CYP2D6 phenotype
| Phenotype | Implications for codeine metabolism | Recommendations for codeine therapy | Classification of recommendation for codeine therapya |
|---|---|---|---|
| Ultrarapid metabolizer | Increased formation of morphine following codeine administration, leading to higher risk of toxicity | Avoid codeine use due to potential for toxicity. Consider alternative analgesics such as morphine or a nonopioid. Consider avoiding tramadol.b | Strong |
| Extensive metabolizer | Normal morphine formation | 15–60 mg every 4 h as needed for pain (label recommendation) | Strong |
| Intermediate metabolizer | Reduced morphine formation | Begin with 15–60 mg every 4 h as needed for pain. If no response, consider alternative analgesics such as morphine or a nonopioid. Monitor tramadol use for response. | Moderate |
| Poor metabolizer | Greatly reduced morphine formation following codeine administration, leading to insufficient pain relief | Avoid codeine use due to lack of efficacy. Consider alternative analgesics such as morphine or a nonopioid. Consider avoiding tramadol.b | Strong |
Rating scheme is described in Supplementary Data online.
Although detailed recommendations for using CYP2D6 phenotype in tramadol therapy are beyond the scope of this guideline, there is strong evidence for decreased efficacy of tramadol in poor metabolizers and a single case report of toxicity in an ultrarapid metabolizer with renal impairment following tramadol postsurgery. Use of other analgesics in CYP2D6 poor metabolizers and ultrarapid metabolizers may therefore be preferable.18,19,21,23
CYP2D6 GENETIC TEST INTERPRETATION AND SUGGESTED CLINICAL ACTION
Table 2 summarizes the therapeutic recommendations for codeine based on CYP2D6 phenotype. A standard starting dose of codeine, as recommended in the product label, is warranted in patients with an extensive metabolizer phenotype (i.e., a CYP2D6 activity score of 1.0 to 2.0). Likewise, a standard starting dose of codeine is warranted in patients with an intermediate metabolizer phenotype (i.e., a CYP2D6 activity score of 0.5); these patients should be monitored closely for less than optimal response and should be offered an alternative analgesic if required. If the CYP2D6 substrate tramadol is selected as alternative therapy in intermediate metabolizers, close monitoring should be carried out because of the possibility of low response.
If clinical genotyping identifies a patient as a CYP2D6 poor metabolizer (i.e., a CYP2D6 activity score of 0), current evidence suggests that the use of codeine be avoided because of the possibility of lack of effect, and that an alternative analgesic should be used. That is, it may be preferable to use an analgesic other than the CYP2D6 substrate tramadol in poor metabolizers. There is insufficient evidence in the literature to recommend a higher dose of codeine in poor metabolizers, especially given that adverse effects do not differ between poor metabolizers and extensive metabolizers.12
In a patient identified as a CYP2D6 ultrarapid metabolizer (i.e., a CYP2D6 activity score of >2.0), the choice of an alternative analgesic should be made to avoid the risk of severe toxicity associated with a “normal” dose of codeine. That is, it may be preferable to use an analgesic other than the CYP2D6 substrate tramadol in ultrarapid metabolizers.
OTHER CONSIDERATIONS: BREASTFED INFANTS
Codeine and its metabolites, including morphine, are secreted in human breast milk, but the amount is typically low and dose dependent. However, breastfeeding women with an ultrarapid metabolizer phenotype may achieve high serum levels of morphine on standard codeine therapy.36 This may lead to high concentrations of morphine in breast milk and dangerously high serum morphine levels in their breastfed infants.37 Notably, there was a reported case of fatal opioid poisoning in a breastfed neonate as a result of the codeine treatment taken by the mother, an ultrarapid metabolizer.38 Case reports such as this prompted the FDA to change the codeine product label to include information on the increased risk of morphine overdose in breastfed infants whose mothers are taking codeine and are ultrarapid metabolizers. Additional information about codeine use in breastfeeding mothers can be found elsewhere.39 Codeine is not recommended in children less than 2 years of age and presumably would carry similar dangers in other neonates and young children who are themselves ultrarapid metabolizers.17
POTENTIAL BENEFITS AND RISKS FOR THE PATIENT
The potential benefit of CYP2D6 genotype testing is that patients with genotypes that are associated with a higher risk of ineffective analgesia or of an adverse event may be identified, and alternative analgesics may be administered to these patients. CYP2D6 genotyping is reliable when performed in qualified laboratories. However, as with any laboratory test, a possible area of risk is an error in genotyping, which could have long-term adverse health implications for the patient.
CAVEATS: APPROPRIATE USE AND/OR POTENTIAL MISUSE OF GENETIC TESTS
One of the challenges with using clinical pharmacogenetic testing for codeine dosing is the need for results at the time an analgesic regimen is chosen. The ideal situation is to preemptively genotype patients who may need pain control in the future. For instance, preemptive CYP2D6 genotyping has been implemented in patients treated for acute lymphoblastic leukemia, who often require pain control medication during their therapy.40 Like all diagnostic tests, the CYP2D6 genotype test is one of multiple pieces of information that clinicians should consider in guiding their therapeutic choice for each patient.
Disclaimer
Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines reflect expert consensus based on clinical evidence and peer-reviewed literature available at the time they are written and are intended only to assist clinicians in decision making and to identify questions for further research. New evidence may have emerged since the time a guideline was submitted for publication. Guidelines are limited in scope and are not applicable to interventions or diseases not specifically identified. Guidelines do not account for all individual variations among patients and cannot be considered inclusive of all proper methods of care or exclusive of other treatments. It remains the responsibility of the health-care provider to determine the best course of treatment for a patient. Adherence to any guideline is voluntary, with the ultimate determination regarding its application to be made solely by the clinician and the patient. CPIC assumes no responsibility for any injury to persons or damage to persons or property arising out of or related to any use of CPIC’s guidelines, or for any errors or omissions.
Supplemental Material
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/cpt
Acknowledgments
We acknowledge the critical input of S. Cross, H.J. Guchelaar, J. Hoffman, T. Manolio, P. Mummaneni, M. Relling, and S. Scott and members of the Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network, funded by the National Institutes of Health (NIH). This work was supported by NIH U01 GM061373, R01 GM088076, K24DA00417, PharmGKB (R24-GM61374), ALSAC, and the Agency for Healthcare Research and Quality R01 HS19818-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Agency for Healthcare Research and Quality.
Footnotes
CONFLICT OF INTEREST
The authors declared no conflict of interest.
References
- 1.Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther. 2008;83:234–242. doi: 10.1038/sj.clpt.6100406. [DOI] [PubMed] [Google Scholar]
- 2.Borges S, et al. Composite functional genetic and comedication CYP2D6 activity score in predicting tamoxifen drug exposure among breast cancer patients. J Clin Pharmacol. 2010;50:450–458. doi: 10.1177/0091270009359182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaedigk A, Gotschall RR, Forbes NS, Simon SD, Kearns GL, Leeder JS. Optimization of cytochrome P4502D6 (CYP2D6) phenotype assignment using a genotyping algorithm based on allele frequency data. Pharmacogenetics. 1999;9:669–682. doi: 10.1097/01213011-199912000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Kirchheiner J, Lang U, Stamm T, Sander T, Gallinat J. Association of CYP2D6 genotypes and personality traits in healthy individuals. J Clin Psychopharmacol. 2006;26:440–442. doi: 10.1097/01.jcp.0000229484.52955.22. [DOI] [PubMed] [Google Scholar]
- 5.Zackrisson AL, Lindblom B, Ahlner J. High frequency of occurrence of CYP2D6 gene duplication/multiduplication indicating ultrarapid metabolism among suicide cases. Clin Pharmacol Ther. 2010;88:354–359. doi: 10.1038/clpt.2009.216. [DOI] [PubMed] [Google Scholar]
- 6.Wadelius M, Darj E, Frenne G, Rane A. Induction of CYP2D6 in pregnancy. Clin Pharmacol Ther. 1997;62:400–407. doi: 10.1016/S0009-9236(97)90118-1. [DOI] [PubMed] [Google Scholar]
- 7.Heikkinen T, Ekblad U, Palo P, Laine K. Pharmacokinetics of fluoxetine and norfluoxetine in pregnancy and lactation. Clin Pharmacol Ther. 2003;73:330–337. doi: 10.1016/s0009-9236(02)17634-x. [DOI] [PubMed] [Google Scholar]
- 8.Högstedt S, Lindberg B, Peng DR, Regårdh CG, Rane A. Pregnancyinduced increase in metoprolol metabolism. Clin Pharmacol Ther. 1985;37:688–692. doi: 10.1038/clpt.1985.114. [DOI] [PubMed] [Google Scholar]
- 9.Mignat C, Wille U, Ziegler A. Affinity profiles of morphine, codeine, dihydrocodeine and their glucuronides at opioid receptor subtypes. Life Sci. 1995;56:793–799. doi: 10.1016/0024-3205(95)00010-4. [DOI] [PubMed] [Google Scholar]
- 10.Volpe DA, et al. Uniform assessment and ranking of opioid Mu receptor binding constants for selected opioid drugs. Regul Toxicol Pharmacol. 2011;59:385–390. doi: 10.1016/j.yrtph.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Gasche Y, et al. Codeine intoxication associated with ultrarapid CYP2D6 metabolism. N Engl J Med. 2004;351:2827–2831. doi: 10.1056/NEJMoa041888. [DOI] [PubMed] [Google Scholar]
- 12.Eckhardt K, Li S, Ammon S, Schänzle G, Mikus G, Eichelbaum M. Same incidence of adverse drug events after codeine administration irrespective of the genetically determined differences in morphine formation. Pain. 1998;76:27–33. doi: 10.1016/s0304-3959(98)00021-9. [DOI] [PubMed] [Google Scholar]
- 13.Lötsch J, Rohrbacher M, Schmidt H, Doehring A, Brockmöller J, Geisslinger G. Can extremely low or high morphine formation from codeine be predicted prior to therapy initiation? Pain. 2009;144:119–124. doi: 10.1016/j.pain.2009.03.023. [DOI] [PubMed] [Google Scholar]
- 14.Mikus G, et al. Effect of codeine on gastrointestinal motility in relation to CYP2D6 phenotype. Clin Pharmacol Ther. 1997;61:459–466. doi: 10.1016/S0009-9236(97)90196-X. [DOI] [PubMed] [Google Scholar]
- 15.Kirchheiner J, et al. Pharmacokinetics of codeine and its metabolite morphine in ultra-rapid metabolizers due to CYP2D6 duplication. Pharmacogenomics J. 2007;7:257–265. doi: 10.1038/sj.tpj.6500406. [DOI] [PubMed] [Google Scholar]
- 16.Dalén P, Frengell C, Dahl ML, Sjöqvist F. Quick onset of severe abdominal pain after codeine in an ultrarapid metabolizer of debrisoquine. Ther Drug Monit. 1997;19:543–544. doi: 10.1097/00007691-199710000-00011. [DOI] [PubMed] [Google Scholar]
- 17.Ciszkowski C, Madadi P, Phillips MS, Lauwers AE, Koren G. Codeine, ultrarapid-metabolism genotype, and postoperative death. N Engl J Med. 2009;361:827–828. doi: 10.1056/NEJMc0904266. [DOI] [PubMed] [Google Scholar]
- 18.Poulsen L, Arendt-Nielsen L, Brøsen K, Sindrup SH. The hypoalgesic effect of tramadol in relation to CYP2D6. Clin Pharmacol Ther. 1996;60:636–644. doi: 10.1016/S0009-9236(96)90211-8. [DOI] [PubMed] [Google Scholar]
- 19.Stamer UM, Musshoff F, Kobilay M, Madea B, Hoeft A, Stuber F. Concentrations of tramadol and O-desmethyltramadol enantiomers in different CYP2D6 genotypes. Clin Pharmacol Ther. 2007;82:41–47. doi: 10.1038/sj.clpt.6100152. [DOI] [PubMed] [Google Scholar]
- 20.Borlak J, Hermann R, Erb K, Thum T. A rapid and simple CYP2D6 genotyping assay–case study with the analgetic tramadol. Metab Clin Exp. 2003;52:1439–1443. doi: 10.1016/s0026-0495(03)00256-7. [DOI] [PubMed] [Google Scholar]
- 21.Stamer UM, et al. Impact of CYP2D6 genotype on postoperative tramadol analgesia. Pain. 2003;105:231–238. doi: 10.1016/s0304-3959(03)00212-4. [DOI] [PubMed] [Google Scholar]
- 22.Kirchheiner J, Keulen JT, Bauer S, Roots I, Brockmöller J. Effects of the CYP2D6 gene duplication on the pharmacokinetics and pharmacodynamics of tramadol. J Clin Psychopharmacol. 2008;28:78–83. doi: 10.1097/JCP.0b013e318160f827. [DOI] [PubMed] [Google Scholar]
- 23.Stamer UM, Stüber F, Muders T, Musshoff F. Respiratory depression with tramadol in a patient with renal impairment and CYP2D6 gene duplication. Anesth Analg. 2008;107:926–929. doi: 10.1213/ane.0b013e31817b796e. [DOI] [PubMed] [Google Scholar]
- 24.Otton SV, Schadel M, Cheung SW, Kaplan HL, Busto UE, Sellers EM. CYP2D6 phenotype determines the metabolic conversion of hydrocodone to hydromorphone. Clin Pharmacol Ther. 1993;54:463–472. doi: 10.1038/clpt.1993.177. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan HL, et al. Inhibition of cytochrome P450 2D6 metabolism of hydrocodone to hydromorphone does not importantly affect abuse liability. J Pharmacol Exp Ther. 1997;281:103–108. [PubMed] [Google Scholar]
- 26.Davis MP, Varga J, Dickerson D, Walsh D, LeGrand SB, Lagman R. Normal-release and controlled-release oxycodone: pharmacokinetics, pharmacodynamics, and controversy. Support Care Cancer. 2003;11:84–92. doi: 10.1007/s00520-002-0385-9. [DOI] [PubMed] [Google Scholar]
- 27.Zwisler ST, Enggaard TP, Mikkelsen S, Brosen K, Sindrup SH. Impact of the CYP2D6 genotype on post-operative intravenous oxycodone analgesia. Acta Anaesthesiol Scand. 2010;54:232–240. doi: 10.1111/j.1399-6576.2009.02104.x. [DOI] [PubMed] [Google Scholar]
- 28.Andreassen TN, et al. Do CYP2D6 genotypes reflect oxycodone requirements for cancer patients treated for cancer pain? A cross-sectional multicentre study. Eur J Clin Pharmacol. 2011 doi: 10.1007/s00228-011-1093-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zwisler ST, et al. The hypoalgesic effect of oxycodone in human experimental pain models in relation to the CYP2D6 oxidation polymorphism. Basic Clin Pharmacol Toxicol. 2009;104:335–344. doi: 10.1111/j.1742-7843.2009.00378.x. [DOI] [PubMed] [Google Scholar]
- 30.Samer CF, et al. Genetic polymorphisms and drug interactions modulating CYP2D6 and CYP3A activities have a major effect on oxycodone analgesic efficacy and safety. Br J Pharmacol. 2010;160:919–930. doi: 10.1111/j.1476-5381.2010.00709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lalovic B, Kharasch E, Hoffer C, Risler L, Liu-Chen LY, Shen DD. Pharmacokinetics and pharmacodynamics of oral oxycodone in healthy human subjects: role of circulating active metabolites. Clin Pharmacol Ther. 2006;79:461–479. doi: 10.1016/j.clpt.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 32.Rollason V, Samer C, Piguet V, Dayer P, Desmeules J. Pharmacogenetics of analgesics: toward the individualization of prescription. Pharmacogenomics. 2008;9:905–933. doi: 10.2217/14622416.9.7.905. [DOI] [PubMed] [Google Scholar]
- 33.Coffman BL, King CD, Rios GR, Tephly TR. The glucuronidation of opioids, other xenobiotics, and androgens by human UGT2B7Y(268) and UGT2B7H(268) Drug Metab Dispos. 1998;26:73–77. [PubMed] [Google Scholar]
- 34.Court MH, et al. Evaluation of 3’-azido-3’-deoxythymidine, morphine, and codeine as probe substrates for UDP-glucuronosyltransferase 2B7 (UGT2B7) in human liver microsomes: specificity and influence of the UGT2B7*2 polymorphism. Drug Metab Dispos. 2003;31:1125–1133. doi: 10.1124/dmd.31.9.1125. [DOI] [PubMed] [Google Scholar]
- 35.Lötsch J, et al. Cross-sectional analysis of the influence of currently known pharmacogenetic modulators on opioid therapy in outpatient pain centers. Pharmacogenet Genomics. 2009;19:429–436. doi: 10.1097/fpc.0b013e32832b89da. [DOI] [PubMed] [Google Scholar]
- 36.Willmann S, Edginton AN, Coboeken K, Ahr G, Lippert J. Risk to the breast-fed neonate from codeine treatment to the mother: a quantitative mechanistic modeling study. Clin Pharmacol Ther. 2009;86:634–643. doi: 10.1038/clpt.2009.151. [DOI] [PubMed] [Google Scholar]
- 37.Madadi P, et al. Pharmacogenetics of neonatal opioid toxicity following maternal use of codeine during breastfeeding: a case-control study. Clin Pharmacol Ther. 2009;85:31–35. doi: 10.1038/clpt.2008.157. [DOI] [PubMed] [Google Scholar]
- 38.Koren G, Cairns J, Chitayat D, Gaedigk A, Leeder SJ. Pharmacogenetics of morphine poisoning in a breastfed neonate of a codeine-prescribed mother. Lancet. 2006;368:704. doi: 10.1016/S0140-6736(06)69255-6. [DOI] [PubMed] [Google Scholar]
- 39.Madadi P, et al. Guidelines for maternal codeine use during breastfeeding. Can Fam Physician. 2009;55:1077–1078. [PMC free article] [PubMed] [Google Scholar]
- 40.Crews KR, et al. Development and implementation of a pharmacistmanaged clinical pharmacogenetics service. Am J Health Syst Pharm. 2011;68:143–150. doi: 10.2146/ajhp100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.