Abstract
Comparative analysis was applied to two cDNA/ESTs libraries (C1 and C2) from Chaetomium cupreum. A total of 5538 ESTs were sequenced and assembled into 2162 unigenes including 585 contigs and 1577 singletons. BlastX analysis enabled the identification of 1211 unigenes with similarities to sequences in the public databases. MFS monosaccharide transporter was found as the gene expressed at the highest level in library C2, but no expression in C1. The majority of unigenes were library specific. Comparative analysis of the ESTs further revealed the difference of C. cupreum in gene expression and metabolic pathways between libraries. Two different sequences similar to the 48-KDa endochitinase and 46-KDa endochitinase were identified in libraries C1 and C2, respectively.
1. Introduction
One of the reasons for environmental disorder is that modern agriculture is an ecologically unbalanced system which has been destroyed by chemical fungicides. Biocontrol is highly interesting alternative method of chemical plant disease control. C. cupreum Ames is an ascomycete fungus with considerable biocontrol potential to plant fungal pathogens, especially several notorious examples belonging to the genera of Pythium, Rhizoctonia, and Pyricularia [1]. In Thailand and China, its biological products have been applied in agricultural disease management [1, 2]; nevertheless, the genetic basis of the defense mechanisms of C. cupreum is not well understood thus inhibiting its application.
ESTs analysis has been proven to be an efficient and valuable tool in obtaining coding gene information, understanding the pathways involved in a given physiological or environmental stimulus [3, 4]. To date, several ESTs studies have been carried out on fungal biocontrol agents, especially to species of Trichoderma. For example, analysis of 8,710 ESTs of T. harzianum CECT 2413 from eight cDNA libraries including those simulating mycoparasitism [5] and of ESTs from four different Trichoderma strains grown under conditions related to biocontrol [6].
In the present study, we performed comparative analysis of two cDNA/ESTs libraries from C. cupreum. An obvious difference in gene expression and metabolic pathways were detected between libraries. This research contributes to elucidating further the mycoparasitisic molecular mechanisms involved in C. cupreum and will help to develop novel strategies in fungal disease management.
2. Materials and Methods
2.1. Fungal Strains and Culture Conditions
The C. cupreum isolate was kindly provided by King Mongkut's Institute of Technology of Ladkrabang, Thailand. Rhizoctonia solani was stored in our laboratory.
C1 was constructed from mycelia of C. cupreum grown on potato dextrose agar (PDA) culture. Library C2 was constructed with mycelia of Rhizoctonia solani cell wall (RsCW) as the carbon source, this was designed to resemble the phases of interaction with R. solani. For C1 library construction, mycelia of C. cupreum were cultured on potato dextrose (PD) medium for 60 h at 27°C and 150 rpm. The biomass was harvested and stored at −80°C until use. For C2 library construction, mycelia were initially grown in PD medium with shaking at 27°C and 150 rpm for 24 h then transferred to SM medium [2.8 g (NH4)2SO4 l−1; 0.6 Urea g l−1; 4 g KH2PO4 l−1; 0.6 g CaCl2 ·2H2O l−1; 0.2 g MgSO4 l−1; 0.01 g FeSO4 ·7H2O l−1; 0.0028 g ZnSO4 ·H2O l−1; 0.0032 g CoCl2 ·6H2O l−1; 5 g RsCW l−1] and incubated for a further 36 h at 27°C and 150 rpm.
2.2. Construction of the cDNA Libraries and DNA Sequencing
Total RNA was extracted using Trizol reagent from mycelia of C. cupreum. Polyadenylated RNA was purified using an Oligotex mRNA Kit (Qiagen). The course of cDNA library construction followed the procedures of Zhang as described before [7]. Unidirectional cDNA libraries were constructed using the pBluescript II plasmid system. Fragments of cDNA clones were sequenced using a T3 primer from the 5′ end with MegaBase1000 DNA sequencer.
2.3. Data Processing and Bioinformatics Analysis
Vector sequences, sequences shorter than 100 bp and containing more than 5% ambiguous bases, were discarded using the Crossmatch program. High-quality sequences were assembled using Phrap (http://www.phrap.org/) and accuracy of contigs was confirmed with Consed [8]. All unigenes were compared against public nonredundant (nr) protein databases using a BlastX search. According to KEGG (Kyoto Encyclopedia of Genes and Genomes) [9], unigenes were assigned to different metabolic pathways with the same criterion as described by Zhang [7]. All high-quality ESTs were submitted to the GenBank database under accession numbers DV544375-DV548659.
3. Results
3.1. ESTs Clustering and Function Assignment
A total of 5,538 cDNA clones with an insert size of more than 700 bp were selected for sequencing, resulting in 4285 ESTs (3066 from C1 and 1219 from C2) after removing sequences representing ribosomal, vector, and low-quality sequences. Minimum, average, and maximum lengths of ESTs were 102, 518, and 795 bp, respectively, with a large fraction falling between 500 and 700 bp (2110 from C1 and 795 from C2) in both libraries.
Using the Phrap and Consed programs, ESTs from both libraries were arranged into 585 contigs and 1577 singletons, giving a total of 2162 unigenes. Each unigene was subjected to analysis against homologous sequences in public protein databases using the BlastX algorithm. Approximately 1211 (56%) of the unigenes were assigned a function with an E-value of 10−5 or lower. The remaining 951 clones had no high homology to genes of known function. A total of 1138 (52.6%) and 691 (32%) unigenes were unique and only expressed in C1 or C2, respectively.
3.2. Exploration of Highly Expressed ESTs
Contigs containing 4 or more ESTs from each library are listed in Table 1. Of the 26 clusters, more than one third were (10/26) expressed only in the C1 library, 2 only in the C2 library, and half (14/29) in both but at a different level.
Table 1.
The difference of high-redundancy genes expression between cDNA libraries from C. cupreum.
| Contig no. | Annotation | C1 | C2 | ||
|---|---|---|---|---|---|
| ESTs | % | ESTs | % | ||
| Contig525 | Glyceraldehyde-3-phosphate dehydrogenase | 109 | 7.41 | 14 | 1.69 |
| Contig531 | Coproporphyrinogen oxidase | 72 | 4.89 | 5 | 0.6 |
| Contig30 | Predicted protein | 56 | 3.81 | 0 | 0 |
| Contig433 | C-4 sterol methyl oxidase | 54 | 3.67 | 0 | 0 |
| Contig313 | Predicted protein | 36 | 2.45 | 0 | 0 |
| Contig2 | Pyruvate decarboxylase | 27 | 1.84 | 0 | 0 |
| Contig39 | Predicted protein | 23 | 1.56 | 0 | 0 |
| Contig507 | Xylulose-5-phosphate phosphoketolase | 21 | 1.43 | 3 | 0.36 |
| Contig 152 | Predicted protein | 20 | 1.36 | 16 | 1.93 |
| Contig523 | EF1-alpha translation elongation factor | 19 | 1.29 | 12 | 1.45 |
| Contig42 | Thiazole biosynthetic enzyme | 19 | 1.29 | 2 | 0.24 |
| Contig520 | Actin | 17 | 1.16 | 4 | 0.48 |
| Contig495 | Glutamine synthetase | 17 | 1.16 | 1 | 0.12 |
| Contig517 | Aspartic protease | 16 | 1.09 | 3 | 0.36 |
| Contig317 | Ammonium transporter | 16 | 1.09 | 0 | 0 |
| Contig419 | ATP synthase protein | 16 | 1.09 | 0 | 0 |
| Contig4 | Alcohol dehydrogenase | 15 | 1.02 | 4 | 0.48 |
| Contig474 | β-1,3-exoglucanase | 14 | 0.95 | 2 | 0.24 |
| Contig494 | ATP citrate lyase | 14 | 0.95 | 1 | 0.12 |
| Contig412 | Histone H2B | 13 | 0.88 | 0 | 0 |
| Contig298 | Pyruvate kinase | 13 | 0.88 | 0 | 0 |
| Contig310 | Aspartate aminotransferase | 11 | 0.75 | 1 | 0.12 |
| Contig408 | Heat shock protein 30 | 11 | 0.75 | 0 | 0 |
| Contig489 | ADP-ATP translocase | 11 | 0.75 | 8 | 0.97 |
| Contig165 | MFS monosaccharide transporter | 0 | 0 | 20 | 2.42 |
| Contig162 | Phosphoenolpyruvate carboxykinase | 0 | 0 | 12 | 1.45 |
Relative values (%): ESTs numbers/all ESTs numbers in C1 or C2 library.
Glyceraldehyde-3-phosphate dehydrogenase was the most highly expressed transcript (109 ESTs) in the C1 library, occurring four times more than in the C2 library. The most highly represented transcripts in the C2 library coded for a putatively major facilitator superfamily (MFS) monosaccharide transporter; no such expression was observed in library C1. The expression of coproporphyrinogen oxidase, thiazole biosynthetic enzyme, glutamine synthetase, ATP citrate lyase, and aspartate aminotransferase were higher in the C1 library. It should be noted that many hits similar to predicted or unknown function proteins were detected in libraries. They are ideal candidates for future study.
3.3. Metabolic Pathways Analysis
Pathways analysis of KEGG was carried out using genes homologous to known functional sequences. A total of 65 and 61 different metabolic pathways were found in the C1 and C2 libraries, respectively. These results show evident difference in metabolic pathways between the libraries (Table 2).
Table 2.
The metabolic pathways difference between cDNA libraries from C. cupreum.
| Map no. | Pathways | ESTs in C1 | ESTs in C2 |
|---|---|---|---|
| 00010 | Glycolysis/gluconeogenesis | 188 | 31 |
| 00020 | Citrate cycle | 47 | 9 |
| 00030 | Pentose phosphate cycle | 35 | 6 |
| 00040 | Pentose and glucuronate interconversions | 6 | 4 |
| 00051 | Fructose and mannose metabolism | 4 | 10 |
| 00052 | Galactose metabolism | 7 | 7 |
| 00053 | Ascorbate and aldarate metabolism | 5 | 5 |
| 00061 | Fatty acid biosynthesis (path 1) | 3 | 1 |
| 00062 | Fatty acid biosynthesis (path 2) | 0 | 1 |
| 00071 | Fatty acid metabolism | 24 | 12 |
| 00100 | Sterol, vitamin K, vitamin E, carotenoids biosynthesis | 1 | 0 |
| 00120 | Bile acid biosynthesis | 19 | 12 |
| 00130 | Ubiquinone biosynthesis | 14 | 3 |
| 00150 | Androgen and estrogen metabolism | 4 | 12 |
| 00190 | Electron transport and oxidative phosphorylation | 34 | 6 |
| 00220 | Urea cycle and metabolism of amino groups | 10 | 0 |
| 00230 | Purine metabolism | 41 | 1 |
| 00240 | Pyrimidine metabolism | 6 | 1 |
| 00251 | Glutamate metabolism | 24 | 4 |
| 00252 | Alanine and aspartate metabolism | 14 | 1 |
| 00260 | Glycine, serine, and threonine metabolism | 2 | 8 |
| 00271 | Methionine metabolism | 12 | 5 |
| 00272 | Cysteine metabolism | 6 | 1 |
| 00280 | Valine, leucine, and isoleucine degradation | 6 | 6 |
| 00290 | Valine, leucine, and isoleucine biosynthesis | 12 | 1 |
| 00300 | Lysine biosynthesis | 4 | 1 |
| 00310 | Lysine degradation | 6 | 5 |
| 00330 | Arginine and proline metabolism | 15 | 6 |
| 00340 | Lysine degradation | 4 | 4 |
| 00350 | Tyrosine metabolism | 25 | 9 |
| 00360 | Phenylalanine metabolism | 9 | 6 |
| 00361 | Gamma-hexachlorocyclohexane degradation | 5 | 6 |
| 00380 | Tryptophan metabolism | 22 | 14 |
| 00400 | Phenylalanine, tyrosine, and tryptophan biosynthesis | 13 | 13 |
| 00410 | B-alanine metabolism | 5 | 5 |
| 00440 | Aminophosphonate metabolism | 4 | 1 |
| 00450 | Selenoamino acid metabolism | 14 | 5 |
| 00472 | D-arginine and ornithine metabolism | 2 | 15 |
| 00480 | Glutathione metabolism | 8 | 2 |
| 00500 | Starch and sucrose metabolism | 16 | 11 |
| 00520 | Nucleotide sugars metabolism | 7 | 4 |
| 00530 | Aminosugars metabolism | 7 | 5 |
| 00550 | Peptide protein biosynthesis | 17 | 0 |
| 00561 | Glycerolipid metabolism | 5 | 9 |
| 00562 | Inositol phosphate metabolism | 4 | 2 |
| 00600 | Sphingoglycolipid metabolism | 4 | 3 |
| 00620 | Pyruvate metabolism | 35 | 10 |
| 00626 | Nitrobenzene degradation | 1 | 1 |
| 00630 | Glyoxylate and dicarboxylate metabolism | 9 | 4 |
| 00640 | Propanoate metabolism | 9 | 6 |
| 00643 | Styrene degradation | 0 | 1 |
| 00650 | Butanoate metabolism | 13 | 6 |
| 00670 | One-carbon pool by folate | 6 | 3 |
| 00680 | Methane metabolism | 35 | 3 |
| 00740 | Riboflavin metabolism | 5 | 5 |
| 00750 | Vitamine B6 metabolism | 0 | 1 |
| 00760 | Nicotinate and nicotinamide metabolism | 4 | 3 |
| 00770 | Pantothenate and CoA biosynthesis | 9 | 1 |
| 00790 | Folate biosynthesis | 4 | 2 |
| 00860 | Porphyrin and chlorophyll metabolism | 180 | 6 |
| 00920 | Sulfur metabolism: reduction and fixation | 2 | 0 |
| 00950 | Alkaloid biosynthesis I | 7 | 0 |
| 00970 | Aminoacyl-tRNA biosynthesis | 11 | 2 |
| 03020 | RNA polymerase | 3 | 1 |
| 03030 | DNA polymerase | 4 | 0 |
| 03130 | Cytochrome C oxidase | 1 | 0 |
| 03140 | Cytochrome C reductase | 10 | 4 |
| 03150 | Succinate dehydrogenase | 3 | 1 |
Glycolysis/gluconeogenesis was the most represented pathway within each library. The second and third most enriched functional pathways in the C1 library were porphyrin and chlorophyll metabolism, which involved 180 ESTs (17.1%), and the citrate cycle, which involved 47 ESTs (4.5%). In the C2 library, the respective pathways were peptideprotein biosynthesis and d-arginine and ornithine metabolism.
It should be noted that the types of genes involved in the same metabolic pathways were greatly different between libraries. For instance, in the glycolysis/gluconeogenesis pathway, the enzymes in the C1 library were enolase, glyceraldehyde 3-phosphate dehydrogenase, fructose 1,6-biphosphate aldolase, and pyruvate decarboxylase, which were assigned to glycolysis; however, in the C2 library, they were fructose-1,6-bisphosphatase, pyruvate carboxylase, and phosphoenolpyruvate carboxykinase, which were assigned to gluconeogenesis. Because glucose is a very important source of nutrition, we speculate that the upregulated genes related to gluconeogenesis observed in the C2 library may be necessary for mycoparasitism, that is, maintenance of fast cell growth rate in response to the competition with the plant fungal pathogen.
3.4. Genes Induced by the Mycoparasitic Process
Differences were observed in gene groups associated with degradation of the cell wall, proteolysis, and toxins production. Seven contigs were presented in both libraries, four were specific to library C1, and eight to library C2 (Table 3), the latter appearing to be induced by the mycoparasitic process directly. Two sequences similar to the 48-KDa endochitinase (GenBank accession nos. DV546055, DV544732, and DV544989) of Aspergillus nidulans and 46-KDa endochitinase (DV547883 and DV547485) of Hypocrea virens were identified in libraries C1 and C2, respectively. Four ESTs from library C1 (DV546459, DV546294, DV544423, and DV546484) and 1 (DV548260) from library C2 shared similarity with serine proteases (Figure 1). One-gene homologue of MAP kinase A (DV548513) was identified in library C2 only (Figure 2).
Table 3.
The different expression of genes related to biocontrol between cDNA libraries from C. cupreum.
| Annotation | C1 | C2 |
|---|---|---|
| ESTs | ESTs | |
| 48 KDa endochitinase | 3 | 0 |
| 46 KDa endochitinase | 0 | 2 |
| β-N-acetylglucosaminidase | 0 | 1 |
| β-1,3-exoglucanase | 22 | 2 |
| β-1,3-endoglucanase | 1 | 0 |
| β-glucosidase D | 1 | 1 |
| β-glucosidase 5 | 6 | 0 |
| β-glucosidase 6 | 1 | 0 |
| Serine protease | 4 | 1 |
| Aspartic proteinase | 20 | 5 |
| Immune antigen 1 | 0 | 1 |
| C-8 sterol isomerase | 2 | 2 |
| Sterol C-22 desaturase | 1 | 1 |
| Sterol-C5-desaturase | 2 | 2 |
| β-endo-1,4-xylanase | 0 | 3 |
| Peptide transporter 2 | 0 | 1 |
| MAP kinase A | 0 | 1 |
| ABC transporters | 0 | 1 |
Figure 1.

DV546294_shared 90% identity with serine proteases of Acremonium sp.
Figure 2.

DV548513 shared 98% identity with MAP kinase of Gibberella intermedia.
4. Discussion
It has been demonstrated that the cell wall of the fungal pathogen can simulate some aspects of the mycoparasitic interactions between biocontrol fungi and its targets [10].
Only a limited amount of overlap (333 unigenes) was observed in both libraries. A total of 1138 and 691 unigenes were unique and only expressed in C1 or C2, respectively. The lack of significant overlap between the individual libraries also suggests a high level of flexibility at the level of gene expression under the examined conditions, some of which may reflect particular requirements for phases of mycoparasitism.
The analysis of the frequency of specific ESTs that form individual contigs can give information about the expression levels of particular genes under different experimental conditions [11]. The most abundant transcripts in library C2 but no expression in C1 were MFS monosaccharide transporters. MFS transporters transport uni-, sym-, and antiporters of sugars, peptides, drugs, and organic and inorganic ions with 12 or 14 transmembrane spanners [12]. In the present study, the high proportion of ESTs expressing a homology to MFS monosaccharide transporters implies that they may be responsible for transport of monosaccharides derived from the degradation of RsCW. This was not consistent with the results of a study of T. harzianum CECT 2413 [13], in which abundant expression of peptide transporter 2 (PTR2) was found in a cDNA library of T. harzianum CECT 2413 when interacted directly with Botrytis cinerea. However, only one EST similar to PTR2 (DV547977) was detected in library C2. We speculate that this may have been caused by the different cultivation times of the two fungi.
Comparison analysis illustrated variations in the proportions of different pathways. Metabolic pathways of ubiquinone biosynthesis; electron transport and oxidative phosphorylation; purine metabolism; pyrimidine metabolism; alanine and aspartate metabolism; valine, leucine, and isoleucine biosynthesis; porphyrin and chlorophyll metabolism were proportionately more represented in library C1. In contrast, pentose and glucuronate interconversions; fructose and mannose metabolism; galactose metabolism; androgen and estrogen metabolism; glycine, serine, and threonine metabolism; valine, leucine, and isoleucine degradation; arginine and proline metabolism; histidine metabolism; tryptophan metabolism; d-arginine and ornithine metabolism; glycerolipid metabolism were overrepresented in library C2. Metabolic pathways of sterol, vitamin K, vitamin E, carotenoids biosynthesis; sulfur metabolism: reduction and fixation; DNA polymerase; cytochrome C oxidase were only observed in library C1, while those of fatty acid biosynthesis (path 2), styrene degradation, and Vitamine B6 metabolism were only observed in library C2.
The results showed that genes related to mycoparasitism were differentially expressed. Two different sequences similar to the 48-KDa endochitinase and 46-KDa endochitinase were identified in libraries C1 and C2, respectively. Since library C1 was obtained from cultivation on PDA medium, the 48 KDa endochitinase homolog might play a role in the dissolution and formation of the cell wall of C. cupreum. Similarly, because library C2 was constructed under conditions associated with mycoparasitism, the 46 KDa endochitinase homolog is expected to be involved in cell wall degradation of the fungal pathogen during the mycoparasitic process.
The conditions used for construction of library C2 were aimed at in vitro simulation of the mycoparasitic process, which is triggered by the recognition of the structural character of the pathogenic fungal cell wall. As a result, the genes involved in signal transduction pathways of mycoparasitism were acquired. Examples include homologue of gene encoding an ABC transporter (ATP-binding cassette transporter, DV548480, Figure 3) and MAP-kinase A (Tmk1 of T. atroviride). Four ESTs have sequence homology to an ABC transporter, it was also observed previously in other fungal pathogens (Gibberella pulicaris and Sclerotinia sclerotiorum) Mehrabi et al. [14] as potential pathogenicity factors responsible for tolerance to phytoalexins or a pathogenicity factor for the host Fleissner et al. [15] and Li et al. [16]. Studies on signal transduction pathways from Trichoderma strains revealed the involvement of MAP-kinases in the mycoparasitic interaction, including production of hydrolytic enzymes such as chitinases and secretion of antibiotic substances [17].
Figure 3.
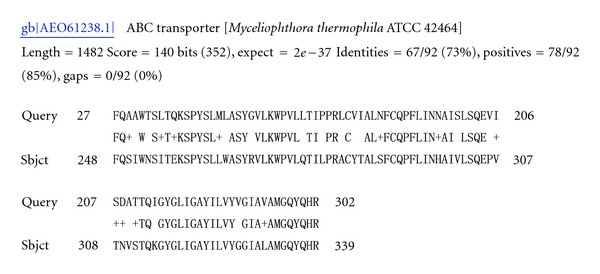
DV548480 shared 73% identity with ABC transporter of Myceliophthora thermophila ATCC 42464.
In this study, we sequenced and analyzed two independent cDNA libraries, providing the first comparative analysis of the transcriptome of C. cupreum under different conditions. The findings provide an entry point for understanding further the molecular mechanisms of this fungus and will also help to advance our efforts in developing novel strategies for biocontrol of fungal diseases.
References
- 1.Soytong K. Application of a New Broad Spectrum Biological Fungicide for Environmental Plant Protection, Biological Control and Biotechnology. HeiLongJiang, China: HeiLongJiang Science and Technology Press; 2003. [Google Scholar]
- 2.Yang Q. Evaluation of Chaetomium for Biological Control of Fusarium Wilt of Tomato in P. R. China, Biological Control and Biotechnology. HeiLongJiang, China: HeiLongJiang Science and Technology Press; 2003. [Google Scholar]
- 3.Adams MD, Kelley JM, Gocayne JD, et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252(5013):1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- 4.Bouck A, Vision T. The molecular ecologist’s guide to expressed sequence tags. Molecular Ecology. 2007;16(5):907–924. doi: 10.1111/j.1365-294X.2006.03195.x. [DOI] [PubMed] [Google Scholar]
- 5.Vizcaíno JA, González FJ, Suárez MB, et al. Generation, annotation and analysis of ESTs from Trichoderma harzianum CECT 2413. BMC Genomics. 2006;7:p. 193. doi: 10.1186/1471-2164-7-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vizcaíno JA, Redondo J, Suárez MB, et al. Generation, annotation, and analysis of ESTs from four different Trichoderma strains grown under conditions related to biocontrol. Applied Microbiology and Biotechnology. 2007;75(4):853–862. doi: 10.1007/s00253-007-0885-0. [DOI] [PubMed] [Google Scholar]
- 7.Zhang HY, Yang Q. Expressed sequence tags-based identification of genes in the biocontrol agent Chaetomium cupreum . Applied Microbiology and Biotechnology. 2007;74(3):650–658. doi: 10.1007/s00253-006-0701-2. [DOI] [PubMed] [Google Scholar]
- 8.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Research. 1998;8(3):195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 9.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanz L, Montero M, Redondo J, Llobell A, Monte E. Expression of an α-1,3-glucanase during mycoparasitic interaction of Trichoderma asperellum . FEBS Journal. 2005;272(2):493–499. doi: 10.1111/j.1742-4658.2004.04491.x. [DOI] [PubMed] [Google Scholar]
- 11.Ebbole DJ, Jin Y, Thon M, et al. Gene discovery and gene expression in the rice blast fungus, Magnaporthe grisea: analysis of expressed sequence tags. Molecular Plant-Microbe Interactions. 2004;17(12):1337–1347. doi: 10.1094/MPMI.2004.17.12.1337. [DOI] [PubMed] [Google Scholar]
- 12.Pao SS, Paulsen IT, Saier MH. Major facilitator superfamily. Microbiology and Molecular Biology Reviews. 1998;62(1):1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vizcaíno JA, Cardoza RE, Hauser M, et al. ThPTR2, a di/tri-peptide transporter gene from Trichoderma harzianum . Fungal Genetics and Biology. 2006;43(4):234–246. doi: 10.1016/j.fgb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Mehrabi R, van der Lee T, Waalwijk C, Kema GHJ. MgSlt2, a cellular integrity MAP kinase gene of the fungal wheat pathogen Mycosphaerella graminicola, is dispensable for penetration but essential for invasive growth. Molecular Plant-Microbe Interactions. 2006;19(4):389–398. doi: 10.1094/MPMI-19-0389. [DOI] [PubMed] [Google Scholar]
- 15.Fleissner A, Sopalla C, Weltring KM. An ATP-binding cassette multidrug-resistance transporter is necessary for tolerance of Gibberella pulicaris to phytoalexins and virulence on potato tubers. Molecular Plant-Microbe Interactions. 2002;19(2):102–108. doi: 10.1094/MPMI.2002.15.2.102. [DOI] [PubMed] [Google Scholar]
- 16.Li R, Roger R, Lone B, et al. Interaction of Sclerotinia sclerotiorum with a resistant Brassica napus cultivar: expressed sequence tag analysis identifies genes associated with fungal pathogenesis. Fungal Genetics and Biology. 2004;41(8):735–753. doi: 10.1016/j.fgb.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Reithner B, Schuhmacher R, Stoppacher N, Pucher M, Brunner K, Zeilinger S. Signaling via the Trichoderma atroviride mitogen-activated protein kinase Tmk1 differentially affects mycoparasitism and plant protection. Fungal Genetics and Biology. 2007;44(11):1123–1133. doi: 10.1016/j.fgb.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


