Abstract
The xanthophyll carotenoids lutein and zeaxanthin constitute the major carotenoids of the macular pigment in the human retina where they are thought to act in part to prevent light induced oxidative damage associated with age-related macular degeneration (AMD). The highly selective uptake of these pigments is mediated by specific carotenoid-binding proteins (GSTP1 and StARD3) recently identified in our laboratory. Carotenoids are hydrophobic in nature, so we first systematically optimized carotenoid preparations that are nano-dispersed in aqueous buffers, and then we used a new-generation surface plasmon resonance (SPR) protocol called FastStep™, which is significantly faster than conventional SPR assays. We have explored carotenoid-binding interactions of five proteins: human serum albumin (HSA), β-lactoglobulin (LG), steroidogenic acute regulatory domain proteins (StARD1, StARD3) and glutathione S- transferase Pi isoform (GSTP1). HSA and LG showed relatively weak interaction with carotenoids (KD > 1 μM). GSTP1 evidenced high affinity and specificity towards zeaxanthin and meso-zeaxanthin with KD values 0.14 ± 0.02 μM and 0.17 ± 0.02 μM, respectively. StARD3 expressed a relative high specificity towards lutein with a KD value of 0.59 ± 0.03 μM, whereas StARD1 exhibited a relatively low selectivity and affinity (KD > 1 μM) towards the various carotenoids tested.
Keywords: Carotenoids, FastStep™, Macula, Retina, Surface plasmon resonance, Xanthophylls
Introduction
The carotenoids comprise a group of over 600 yellow and orange-red pigments synthesized by plants and micro-organisms that are widely distributed in nature. Structurally, carotenoids can be broadly classified into hydrocarbon polyenes (carotenes) and their oxygenated derivatives (xanthophylls) [1]. Lutein, zeaxanthin, and lutein’s metabolite meso-zeaxanthin are the key xanthophyll carotenoids that provide yellow color to the macula of the primate retina. They are present in a 1:1:1 ratio at a total concentration that is close to 1 mM at the center of the macula (the fovea) [2, 3]. This unusually high concentration of just lutein and the zeaxanthins is unique to the eyes of humans and fellow primates; no other mammals concentrate carotenoids in ocular tissue beyond trace amounts. Epidemiological studies reveal that high carotenoid intake and serum levels decrease the risk of neovascular age-related macular degeneration (NV-AMD) by up to 43 % [4–9]. It has been proposed that the macular pigments play an important role in filtering phototoxic blue light [10]. The macular carotenoids are also effective antioxidants that may help to protect against oxidative stress associated with age-related macular degeneration (AMD) [11, 12]. Direct tissue analyses of macular pigments in autopsy eyes have also demonstrated an inverse correlation between lower carotenoid levels and higher risk of AMD [13].
Physiologically, whenever there is selective uptake of a biological compound into a target tissue, it is likely to be mediated by specific binding proteins [14]. There have been numerous reports on carotenoid-binding proteins among micro-organisms and invertebrates [15]. In humans, carotenoids are mainly derived from the diet and are transported in the blood by non-specific carrier proteins such as serum albumin, lactoglobulin, and various lipoproteins [16]. In the past, our laboratory has identified and characterized several specific, high-affinity lutein- and zeaxanthin-binding proteins in the human macula [17, 18]. We identified StARD3 (steroidogenic acute regulatory domain protein 3) as the lutein-binding protein and glutathione S-transferase Pi isoform (GSTP1) as the zeaxanthin-binding protein in the human macula by co-purification with endogenous carotenoids, immunohistochemical localization in appropriate cellular layers, and confirmation of binding interactions by classical binding studies and through usage of a first-generation surface plasmon resonance (SPR) biosensor platform.
The classical methods of in vitro assessment of protein carotenoid interactions include equilibrium dialysis, ultra-filtration, ultra-centrifugation, HPLC, etc. The sensitivity and reproducibility of these traditional methods are low, especially for ligands with extremely high affinity or low aqueous solubility [19]. SPR-based biosensors have drawn attention in recent years because of their abilities to analyze protein-ligand interactions rapidly and sensitively [20]. Binding events caused by molecular interactions can be monitored in real-time, making SPR a powerful nondestructive and label-free transduction principle for biosensing [21]. These label-free interactions provide direct and detailed feedback about the kinetics and affinity of the complex formation. This facilitates higher throughput screening of a variety of ligands in shorter amounts of time in a reproducible and sensitive manner [21].
Carotenoids are hydrophobic in nature, making it difficult to study these molecules in an aqueous environment [22]. These compounds develop high nonspecific binding on the sensor surface due to its highly charged functional groups, which creates additional challenges for a SPR-based platform. In this study, we optimized carotenoid preparations that are nano-dispersed in an aqueous environment, and we then implemented a new generation SPR platform using the FastStep™ protocol that allows for rapid and complete assessment of binding interactions in a manner of minutes.
Materials and methods
Chemicals
Amine coupling reagents N-hydroxysuccinimide, 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride, and 1 M sodium ethanolamine hydrochloride (pH 8.5) were used according to the recommendations of the manufacturer (GE Healthcare, Wauwatosa, WI). Fatty-acid-free human serum albumin (HSA), β-lactoglobulin (LG) (Sigma-Aldrich, St. Louis, MO), glutathione S-transferase Pi isoform (GSTP1) (Oxford Biomedical Research, Rochester Hills, MI), and the binding domains of StARD1 and StARD3 (expressed in our laboratory) were used as supplied. StARD1 protein binding domain (residues 63–285) was expressed in BL21 (DE3) cells (Invitrogen, Carlsbad, CA) using an expression vector, pTWIN-N-His6-N-62 StAR (a gift from Professor Walter L. Miller, UCSF). StARD3 protein binding domain (residues 216–444) was expressed in BL21 (DE3) cells (Invitrogen, Carlsbad, CA) using an expression vector, pET22b-His-StARTdomain (a gift from Professor James H. Hurley, NIH) [23]. The carotenoids shown in Fig. S-1 (supplementary material) were generous gifts from a variety of manufacturers: (3R, 3′R)-zeaxanthin (ZeaVision, St. Louis, MO), (3R, 3′S-meso)-zeaxanthin (DSM, Basel, Switzerland), (3R, 3′R, 6′R)-lutein (Kemin Health, Des Moines, IA) and (3S, 3′S)-astaxanthin (Cardax Pharmaceutical, Aiea, HI). All carotenoids were crystalline, with >98% isomeric and chemical purity confirmed by HPLC.
Instrumentation
SPR analyses were conducted at 25°C using a fully automated SensiQ Pioneer optical biosensor equipped with COOH5 sensor chips (ICX Nomadics, Oklahoma City, OK) [24]. Unlike many commercially available SPR instruments, SensiQ Pioneer is capable of mixing the buffer and sample streams immediately prior to the entrance of the flow cell (Fig. 1). This unique feature helps to vary the analyte concentration during the association phase to achieve a step-gradient analysis [25]. The instrument is programmed to run either two-fold or three-fold dilution series. The sensor is designed to a Kretschmann’s configuration, whereby monochromatic light is reflected from the sensing surface over a range of incident angles, and the reflectance minimum will arise with respect to the incident angle and is detected by a photodiode array. The sensing surface is a planar glass chip with 50 nm gold film coating. Using this platform, a protein surface and an unmodified reference surface were prepared for simultaneous analyses. Immediately after chip docking, each channel was primed with water. Prior to each interaction analysis, the biosensor detector response was normalized by an automated procedure, and the instrument was primed five times with freshly degassed running buffer. Data were collected at 2 Hz.
Fig. 1.
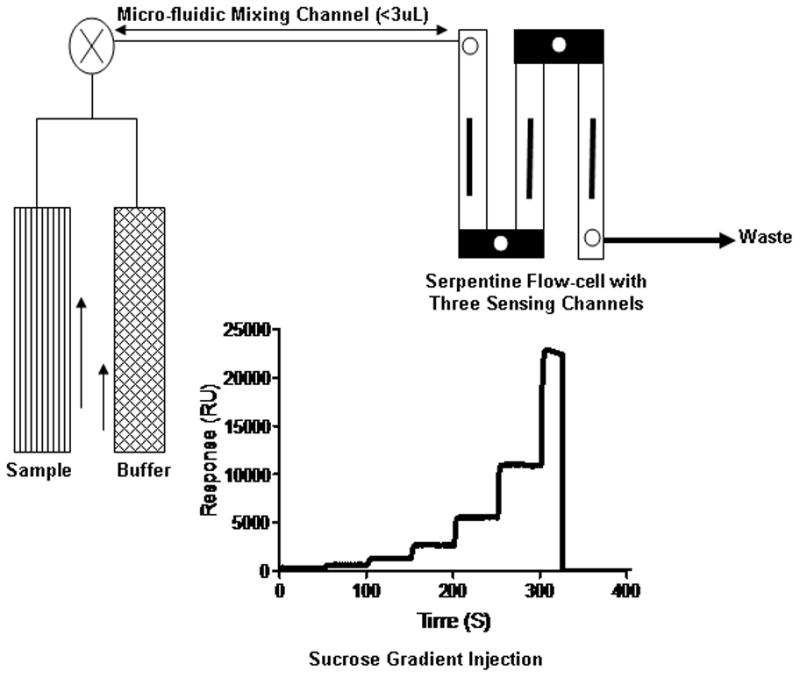
Schematic drawing of SensiQ Pioneer fluidics. The sample and buffer are injected simultaneously into a mixing chamber prior to the injection into the flow cell.
Protein immobilization and analyte interactions using FastStep™ assay
HSA, LG, GSTP1 and StARD domains (10 μg/mL in 10 mM sodium acetate, pH 4.5–5.0) were each immobilized on individual sensor chip surfaces using a standard amine-coupling protocol (flow rate of 10 μl/min) to obtain a density of 10–12 kRU. Each of the five carotenoids was dissolved in sucrose monolaurate (2 mM) (Mitsubishi Chemicals, Tokyo, Japan) to achieve high concentration, and 10 mM PBS (pH 7.4) with 0.01% Triton X-100 and 0.4 mM sucrose monolaurate was used as the running buffer. Typically, the carotenoid concentration series spanned 0.01–10 μM, and the final sucrose monolaurate concentration was 0.4 mM. Five blanks were analyzed at the beginning of the analysis, and the remaining blanks were randomly injected throughout the analysis for double-referencing purposes [26]. A FastStep™ concentration series of sucrose (20 % w/v in running buffer) was injected to create the concentration gradient for analysis. The analyte concentration series was injected as two-fold dilutions in running buffer using FastStep™ gradient injection mode with a flow rate of 200 μl/minute.
Data processing and analysis
SPR response data (sensorgrams) were zeroed at the beginning of each injection and double referenced [26]. For equilibrium analysis, the responses at equilibrium were plotted against the analyte concentration and fit to a 1:1 binding isotherm using the Qdat™ analysis software (Biologic Software, Campbell, Australia) and GraphPad Prism 5.04 (GraphPad Software Inc., La Jolla, USA).
Results and discussion
Preparation of soluble carotenoids in aqueous buffers
Carotenoids tend to stick to the biosensor surface films because of their conjugated double bonds and hydrophobicity. This results in high non-specific binding (NSB) on the sensor surface. Initially, we dissolved crystalline carotenoids in organic solvents such as DMSO, ethanol and THF and diluted them into the assay buffer with a final organic solvent concentration of 3–5%, but carotenoid molecules, because of their hydrophobic nature, can form aggregates in an aqueous solution depending on the polarity of the environment [27]. The observed binding in these cases was most likely due to the non-specific sticking of these precipitates to the hydro-gel matrix and to the sensor surface. The carotenoids lose their fine structure of the absorption spectrum, and the absorption maxima shift > 50 nm [28]. There are several studies reported on the aqueous solubility of carotenoids with the help of various carrier molecules [29–32]. We tried common detergents such as SDS, CHAPS, and Tween-20 to solubilize carotenoids in the running buffer, but these detergents did not reduce NSB on the sensor surface. Next, we used sucrose monolaurate for the nano-dispersion of carotenoids in aqueous buffers. The carotenoid-sucrose monolaurate complex was prepared as previously reported [33]. The final sucrose monolaurate concentration was 0.4 mM.
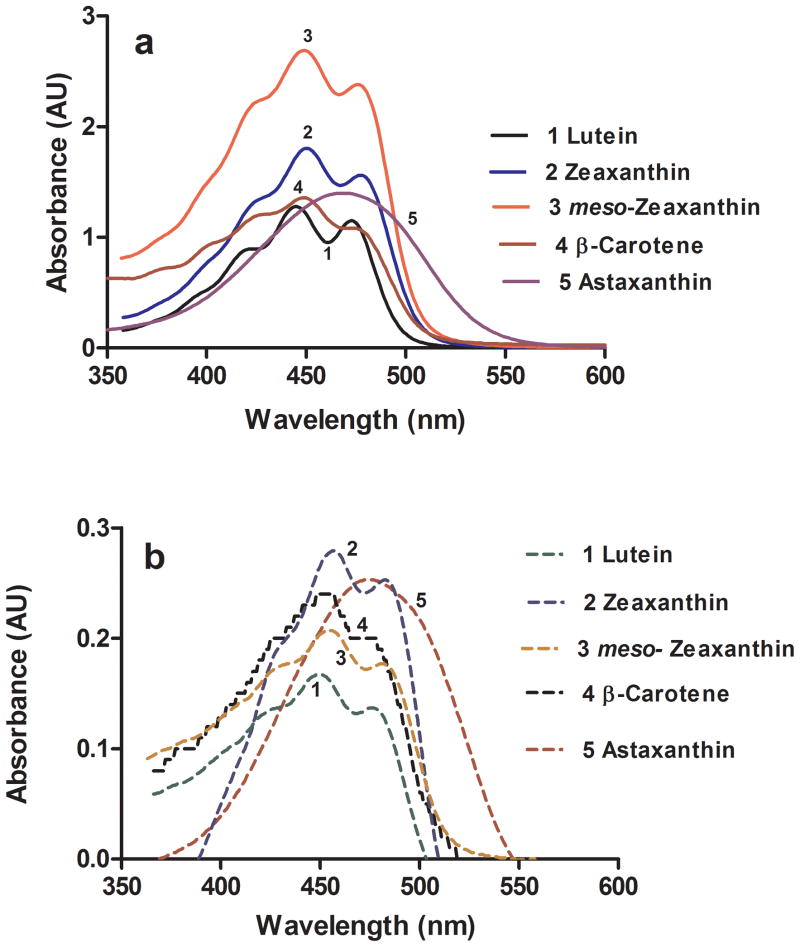
Fig. 2a shows the typical spectral signatures of carotenoids taken in methanol. The spectra also show the presence of some minor amounts of breakdown products as well (except for astaxanthin), which are evidenced by the sub-bands in the short-wavelength region of the spectrum. As shown in Fig. 2b, the carotenoid nano-dispersions in sucrose monolaurate retained their spectral signatures with slight changes in their absorption maxima which could be due to the changes in electronic state of carotenoids in aqueous buffers [32]. The carotenoid-sugar ester nano-dispersions were stable and able to pass through a 0.2 μM filter, and they did not precipitate in aqueous buffers. The running buffer with 0.4 mM sucrose monolaurate was used in the assay to subtract any NSB binding effect due to the sucrose monolaurate in the analyte. The surfaces that were densely polyethylene glycated (COOH5 sensor chips) displayed much lower NSB.
Fig. 2.
UV-visible spectrum of crystalline carotenoids in methanol (a), UV-visible spectrum of carotenoids in sucrose monolaurate-PBS buffer (b).
Standard versus FastStep™ injections
The FastStep™ injection method is designed to test the molecular interaction in a wide concentration range with a single injection. These concentration series are created inside the SensiQ Pioneer instrument in an automated process. It uses high refractive index sucrose (20%) to establish the concentration gradient, and Qdat™ software applies these gradients to the analyte injection during analysis.
We validated the FastStep™ protocol by performing both standard as well as FastStep™ injections for HSA, LG and GSTP1. The proteins were immobilized to a density of ~10,000 RU on a COOH5 chip. The five pure carotenoids, nano-dispersed in 0.01% Trition X-100 and 0.4 mM sucrose monolaurate, flow over the sensing surface in two-fold dilution series in both methods. The standard injection method requires the preparation of analyte dilution manually in contrast to FastStep™, in which we need to prepare only the single top concentration for each analyte. Since the carotenoids have fast association rates with these proteins, they reached equilibrium very quickly during the association phase. The responses at equilibrium were plotted to a simple 1:1 binding isotherm model, as shown in Fig S-2 and S-3 (supplementary material). The equilibrium dissociation constants (KD) for both the standard injection and FastStep™ methods are presented in Table 1. The results show an excellent correlation between these two methods, so the FastStep™ assay was used for the other protein-carotenoid experiments reported in this paper. FastStep™ injection was previously studied for a well characterized stable enzyme carbonic anhydrase (CAII) [25]. The authors compared the interaction of CAII with its sulfonamide-based inhibitors using both standard and FastStep™ injections. They found similar kinetic results using these two modes of injection. To confirm the reliability of the FastStep™ technique, we denatured GSTP1 using guanidine hydrochloride (6 M) and injected zeaxanthin over the surface. As shown in Fig. S-5 (supplementary material), there is no specific binding interaction observed.
Table 1.
Equilibrium dissociation constants (KD) determined by FastStep™ surface plasmon resonance (SPR) at 25°C.
| Binding proteinsa | ||||||||
|---|---|---|---|---|---|---|---|---|
| Carotenoids | HSA (FS) (μM) | HSA (SI) (μM) | LG (FS) (μM) | LG (SI) (μM) | GSTP1 (FS) (μM) | GSTP1 (SI) (μM) | StARD1 (FS) (μM) | StARD3 (FS) (μM) |
| Astaxanthin | 1.56 ± 0.03 | 1.47 ± 0.01 | 1.90 ± 0.05 | 1.92 ± 0.04 | 1.16 ± 0.02 | 1.26 ± 0.02 | 1.96 ± 0.09 | 2.09 ± 0.09 |
| β-Carotene | 1.44 ± 0.07 | 1.53 ± 0.02 | 2.40 ± 0.05 | 2.27 ± 0.03 | 1.09 ± 0.01 | 1.19 ± 0.02 | 1.79 ± 0.08 | 2.30 ± 0.06 |
| Lutein | 1.16 ± 0.04 | 1.11 ± 0.05 | 2.48 ± 0.08 | 1.73 ± 0.02 | 1.30 ± 0.01 | 1.33 ± 0.02 | 1.15 ± 0.06 | 0.59 ± 0.03 |
| meso-Zeaxanthin | 1.30 ± 0.06 | 2.00 ± 0.03 | 2.70 ± 0.02 | 2.72 ± 0.03 | 0.18 ± 0.02 | 0.19 ± 0.03 | 1.36 ± 0.06 | 1.63 ± 0.07 |
| Zeaxanthin | 1.60 ± 0.05 | 1.71 ± 0.01 | 2.30 ± 0.02 | 3.07 ± 0.04 | 0.14 ± 0.02 | 0.15 ± 0.02 | 1.32 ± 0.09 | 1.60 ± 0.01 |
Abbreviations: HSA, human serum albumin; LG, lactoglobulin; GSTP1, glutathione S-transferase pi isoform; StARD1 and StARD3, steroidogenic acute regulatory domain proteins, 1 & 3, respectively. SI, standard injection; FS, Faststep™ injection. The standard error represents the residual of the model fit.
One of the advantages of FastStep™ is that it requires remarkably less time (~4 min) than the conventional method, which usually takes more than 6 hours. It is well documented that proteins on the sensor surface may lose their activity over time [34]. Using FastStep™, we could characterize a large number of samples using the same surface without losing their activity. Since sample dilution in FastStep™ is in-situ, it further reduces the sample preparation time.
Interaction of carotenoids with their binding proteins
We studied several proteins known to interact with carotenoids and compared their binding affinities and specificities. The proteins were immobilized onto a COOH5 chip at a density of ~10,000 RU using the standard amine coupling. The carotenoids were injected over the protein surface in seven-concentration dilution series. Two to three replicates were injected for each carotenoid, and interactions were reproducible. The equilibrium binding constants are tabulated in Table 1. As shown in Fig. 3 to 5, carotenoids bound in a concentration dependent manner, and they reached equilibrium quite rapidly during the association phase.
Fig. 3.
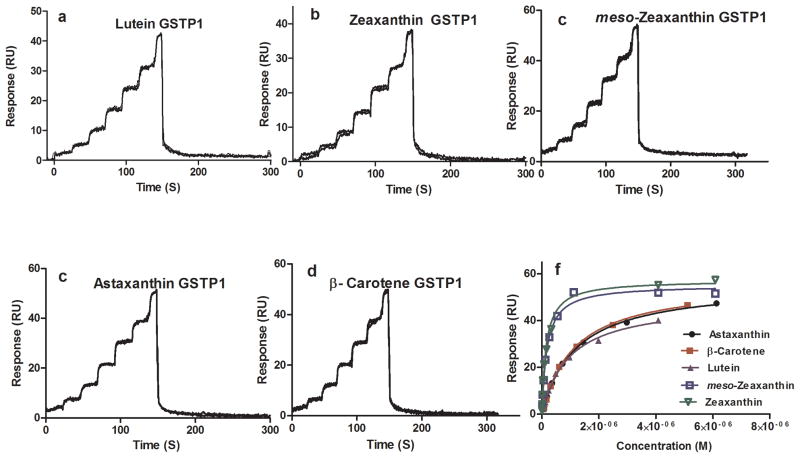
GSTP1 carotenoid FastStep™ SPR assay. Sensorgrams of five different carotenoids (a–e), equilibrium binding isotherm plot (f).
Fig. 5.
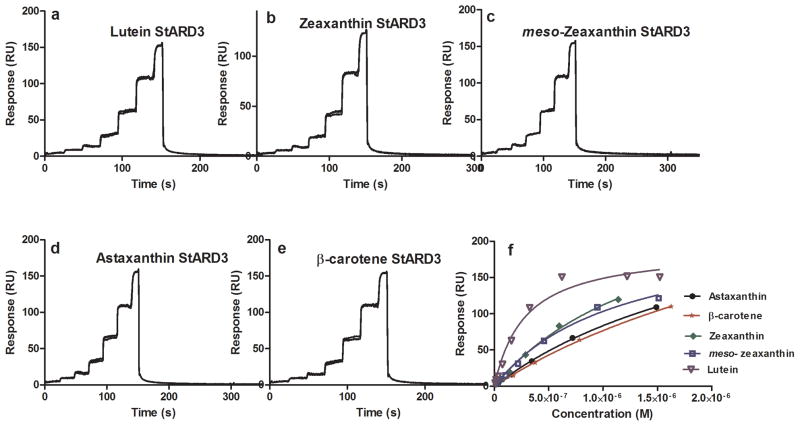
StARD3 carotenoid FastStep™ SPR assay. Sensorgrams of five different carotenoids (a–e), equilibrium binding isotherm plot (f).
HSA and LG are the most abundant proteins present in human serum and milk, respectively. They are known to bind many drugs and hydrophobic compounds reversibly [35]. In this study, HSA and LG showed relatively weak interactions with carotenoids (KD >1 μM, Table 1), consistent with their role as nonspecific carrier proteins for carotenoids and many other hydrophobic ligands. The sensorgrams and binding isotherm plots for these interactions are shown in Fig. S-2 and S-3 (supplementary material).
GSTP1 showed high affinity and specificity for zeaxanthin and meso-zeaxanthin with KD values 0.14 ± 0.02 μM and 0.17 ± 0.02 μM, respectively (Table 1). These results confirm our published findings that GSTP1 is the physiologically relevant binding protein for zeaxanthin in the human macula [18]. Fig. 3 and S-4 (supplementary material) show the sensorgrams and equilibrium binding plots for GSTP1.
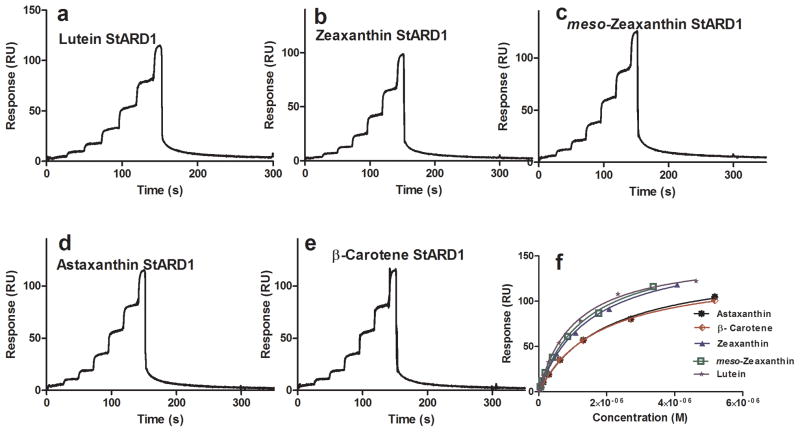
StARD proteins consist of a 15-member family of human proteins that bind and transport hydrophobic ligands [36]. Our attention was initially drawn to this family of proteins when it was reported that the silkworm gut and silk gland lutein-binding protein (CBP) had significant homology to members of the human StARD family, particularly StARD1 and StARD3. In 2009, we reported that an antibody to silkworm carotenoid-binding protein (anti-CBP) had positive immunoreactivity against partially purified lutein-binding protein isolated from the human retina on immunoblots, and it specifically labeled cone inner segments on sections of monkey fovea [37]. We recently identified StARD3 (also known as MLN64) as the lutein-binding protein in the human retina [17]. StARD3 showed a relative high specificity towards lutein with a KD value of 0.59 ± 0.03 μM using FastStep™ (Table 1). All other carotenoids tested had a KD above 1 μM. StARD1 is not present in the retina, but it is abundantly expressed in another carotenoid-rich tissue, the corpus luteum of mammalian ovary [38–41]. In contrast to StARD3, StARD1 exhibited a relatively low selectivity and affinity (KD >1 μM) towards the various carotenoids tested. Fig. 4 and 5 show the equilibrium plots and sensorgrams for StARD1 and StARD3, respectively.
Fig. 4.
StARD1 carotenoid FastStep™ SPR assay. Sensorgrams of five different carotenoids (a–e), equilibrium binding isotherm plot (f).
Our previous studies, confirmed by western blot and immunolocalization experiments, revealed that the macular regions in primate retina are enriched by StARD3 and GSTP1 proteins [17, 18]. These proteins, with their high affinity binding sites are responsible for the abundant concentration and stability of the macular pigments lutein and zeaxanthin. These results further re-inforce our in vivo studies conducted by resonance Raman Imaging for measuring macular pigment carotenoid distribution [42]. These proteins are known to have functions other than carotenoid binding; StARD1 and StARD3 are reported to act as intracellular transporters in cholesterol metabolism [43]. Further characterization of the binding site is required to determine whether or not carotenoids and other ligands are binding in the same region as StARD3. Likewise, GSTP1 is known to be expressed in other tissues such as liver, and it is well documented for its role in the detoxification of xenobiotics [44]. We also reported that the GSTP1-carotenoid complex enhances the antioxidant capacity of the carotenoid ligand [45]. Similar effects of StARD3 are yet to be studied.
Based on affinity and specificity, carotenoid-binding proteins can be classified into two groups. Class 1 carotenoid binding proteins generally have strong affinity (0.1–0.6 μM) for just one or two carotenoids. The Class 1 proteins in this study include GSTP1 (zeaxanthin and meso-zeaxanthin) and StARD3 (lutein). HSA, LG and StARD1 can be classified as Class 2 binders with moderate affinity (KD >1 μM) and relatively low selectivity.
Conclusions
FastStep™-SPR based biosensor technology can be applied to study carotenoid affinities with target proteins reliably and reproducibly. Biosensor-based assays should facilitate further study of the functional roles of carotenoid binding, transport and metabolic proteins in the human retina. The FastStep™ assays are fast, less labor intensive and equally sensitive when compared with conventional SPR assays. The in-situ sample dilutions and gradient analyte injections in FastStep™ make it an excellent tool for the rapid screening of fragment libraries in drug discovery.
Supplementary Material
Highlights.
The interactions of carotenoid-binding proteins using SPR biosensor are reported.
The FastStep™ SPR assay improved analyte throughput with simple assay set up.
The binding kinetics of xanthophyll-binding proteins can be studied using SPR.
Acknowledgments
This work was supported by National Institute of Health Grant EY-11600 and grants from the Macula Vision Research Foundation, the Lowy Foundation, and Research to Prevent Blindness, and the Foundation Fighting Blindness. We would like to acknowledge Professor Walter L. Miller (UCSF) and Professor James H. Hurley (NIH) for providing us StARD1 and StARD3 plasmids, respectively.
Abbreviations
- AMD
age related macular degeneration
- EDTA
ethylenediamine-N,N,N′,N′-tetraacetic acid
- FS
fast step™
- GSTP1
glutathione S-transferase π isoform
- HSA
human serum albumin
- KD
equilibrium dissociation constant
- LG
β-lactoglobulin
- NSB
non-specific binding
- PBS
phosphate buffered saline
- RU
resonance unit
- SI
standard injection
- SPR
surface plasmon resonance
- StARD
steroidogenic acute regulatory domain protein
- Triton X-100
polyethylene glycol p-(1,1,3,3-tetramethylbutyl)-phenyl ether
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sandmann G. Eur J Biochem. 1994;223:7–24. doi: 10.1111/j.1432-1033.1994.tb18961.x. [DOI] [PubMed] [Google Scholar]
- 2.Bone RA, Landrum JT, Hime GW, Cains A, Zamor J. Invest Ophthalmol Vis Sci. 1993;34:2033–2040. [PubMed] [Google Scholar]
- 3.Bone RA, Landrum JT, Tarsis SL. Vision Res. 1985;25:1531–1535. doi: 10.1016/0042-6989(85)90123-3. [DOI] [PubMed] [Google Scholar]
- 4.Moeller SM, Jacques PF, Blumberg JB. J Am Coll Nutr. 2000;19:522S–527S. doi: 10.1080/07315724.2000.10718975. [DOI] [PubMed] [Google Scholar]
- 5.Haddad WM, Souied E, Coscas G, Soubrane G. Bull Soc Belge Ophtalmol. 2006:15–22. [PubMed] [Google Scholar]
- 6.Bernstein PS, Zhao D, Wintch SW, Ermakov IV, McClane RW, Gellermann W. Ophthalmology. 2002;109:1780–1787. doi: 10.1016/s0161-6420(02)01173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bone RA, Landrum JT, Guerra LH, Ruiz CA. J Nutr. 2003;133:992–998. doi: 10.1093/jn/133.4.992. [DOI] [PubMed] [Google Scholar]
- 8.Eye Disease Case-Control Study Group. Arch Ophthalmol. 1993;111:104–109. doi: 10.1001/archopht.1993.01090010108035. [DOI] [PubMed] [Google Scholar]
- 9.Mares-Perlman JA, Brady WE, Klein R, Klein BE, Bowen P, Stacewicz-Sapuntzakis M, Palta M. Arch Ophthalmol. 1995;113:1518–1523. doi: 10.1001/archopht.1995.01100120048007. [DOI] [PubMed] [Google Scholar]
- 10.Landrum JT, Bone RA, Kilburn MD. Adv Pharmacol. 1997;38:537–556. doi: 10.1016/s1054-3589(08)60998-9. [DOI] [PubMed] [Google Scholar]
- 11.Kvansakul J, Rodriguez-Carmona M, Edgar DF, Barker FM, Kopcke W, Schalch W, Barbur JL. Ophthalmic Physiol Opt. 2006;26:362–371. doi: 10.1111/j.1475-1313.2006.00387.x. [DOI] [PubMed] [Google Scholar]
- 12.Dagnelie G, Zorge IS, McDonald TM. Optometry. 2000;71:147–164. [PubMed] [Google Scholar]
- 13.Bone R, Landrum J, Mayne S, Gomez C, Tibor S, Twaroska E. Invest Ophthalmol Vis Sci. 2001;42:235–240. [PubMed] [Google Scholar]
- 14.Li B, Vachali P, Bernstein PS. Photochemical & Photobiological Sciences. 2010;9:1418–1425. doi: 10.1039/c0pp00126k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhosale P, Bernstein PS. Arch Biochem Biophys. 2007;458:121–127. doi: 10.1016/j.abb.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Agamey A, Cantrell A, Land EJ, McGarvey DJ, Truscott TG. Photochem Photobiol Sci. 2004;3:802–811. doi: 10.1039/b315651f. [DOI] [PubMed] [Google Scholar]
- 17.Li B, Vachali P, Frederick JM, Bernstein PS. Biochemistry. 2011;50:2541–2549. doi: 10.1021/bi101906y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhosale P, Larson AJ, Frederick JM, Southwick K, Thulin CD, Bernstein PS. J Biol Chem. 2004;279:49447–49454. doi: 10.1074/jbc.M405334200. [DOI] [PubMed] [Google Scholar]
- 19.Day YS, Myszka DG. J Pharm Sci. 2003;92:333–343. doi: 10.1002/jps.10293. [DOI] [PubMed] [Google Scholar]
- 20.Cooper M. Analytical and Bioanalytical Chemistry. 2003;377:834–842. doi: 10.1007/s00216-003-2111-y. [DOI] [PubMed] [Google Scholar]
- 21.Richard BM, Anna JT. Handbook of Surface Plasmon Resonance. Royal Society of Chemistry; 2008. [Google Scholar]
- 22.Namitha KK, Negi PS. Crit Rev Food Sci Nutr. 2010;50:728–760. doi: 10.1080/10408398.2010.499811. [DOI] [PubMed] [Google Scholar]
- 23.Tsujishita Y, Hurley JH. Nat Struct Biol. 2000;7:408–414. doi: 10.1038/75192. [DOI] [PubMed] [Google Scholar]
- 24.Chinowsky TM, Quinn JG, Bartholomew DU, Kaiser R, Elkind JL. Sensors and Actuators B: Chemical. 2003;91:266–274. [Google Scholar]
- 25.Rich RL, Quinn JG, Morton T, Stepp JD, Myszka DG. Anal Biochem. 2010;407:270–277. doi: 10.1016/j.ab.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myszka DG. J Mol Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 27.Köhn S, Kolbe H, Korger M, Köpsel C, Mayer B, Auweter H, Lüddecke E, Bettermann H, Martin H-D. In: Carotenoids. Britton G, Liaaen-Jensen S, Pfander H, editors. Birkhäuser; Basel: 2008. pp. 53–98. [Google Scholar]
- 28.Takagi S, Nakano S, Kameyama K, Takagi T. Agric Biol Chem. 1987;51:1561–1566. [Google Scholar]
- 29.Borel P, Grolier P, Armand M, Partier A, Lafont H, Lairon D, Azais-Braesco V. J Lipid Res. 1996;37:250–261. [PubMed] [Google Scholar]
- 30.Davarnejad R, Kassim KM, Ahmad Z, Sata SA. Journal of Chemical & Engineering Data. 2009;54:2200–2207. [Google Scholar]
- 31.Takagi S, Takeshi Y, Kunio T, Takagi T. Agric Biol Chem. 1987;51:1567–1572. [Google Scholar]
- 32.Takagi S, Takeda K, Kameyama K, Takagi T. Agric Biol Chem. 1982;46:2035–2040. [Google Scholar]
- 33.Fullmer L, Emmick T. Carotenoid nanodispersions for use in water-based systems and a process for their preparation. 641,774. United States Patent. 2005
- 34.Rich RL, Day YS, Morton TA, Myszka DG. Anal Biochem. 2001;296:197–207. doi: 10.1006/abio.2001.5314. [DOI] [PubMed] [Google Scholar]
- 35.Peters JT. All About Albumin. Academic Press; San Diego: 1995. pp. 76–132. [Google Scholar]
- 36.Alpy F, Tomasetto C. J Cell Sci. 2005;118:2791–2801. doi: 10.1242/jcs.02485. [DOI] [PubMed] [Google Scholar]
- 37.Bhosale P, Li B, Sharifzadeh M, Gellermann W, Frederick JM, Tsuchida K, Bernstein PS. Biochemistry. 2009;48:4798–4807. doi: 10.1021/bi9004478. [DOI] [PubMed] [Google Scholar]
- 38.Devoto L, Kohen P, Gonzalez RR, Castro O, Retamales I, Vega M, Carvallo P, Christenson LK, Strauss JF., 3rd J Clin Endocrinol Metab. 2001;86:5633–5639. doi: 10.1210/jcem.86.11.7982. [DOI] [PubMed] [Google Scholar]
- 39.Kiriakidou M, McAllister JM, Sugawara T, Strauss JF., 3rd J Clin Endocrinol Metab. 1996;81:4122–4128. doi: 10.1210/jcem.81.11.8923870. [DOI] [PubMed] [Google Scholar]
- 40.Sierralta WD, Kohen P, Castro O, Munoz A, Strauss JF, 3rd, Devoto L. Mol Cell Endocrinol. 2005;242:103–110. doi: 10.1016/j.mce.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 41.Czeczuga-Semeniuk E, Wolczynski S. Oncol Rep. 2005;14:1385–1392. [PubMed] [Google Scholar]
- 42.Bernstein PS, Yoshida MD, Katz NB, McClane RW, Gellermann W. Invest Ophthalmol Vis Sci. 1998;39:2003–2011. [PubMed] [Google Scholar]
- 43.Lavigne P, Najmanivich R, LeHoux JG. In: Cholesterol Binding and Cholesterol Transport Proteins. Harris JR, editor. Springer; Netherlands: 2010. pp. 425–437. [Google Scholar]
- 44.Parker LJ, Italiano LC, Morton CJ, Hancock NC, Ascher DB, Aitken JB, Harris HH, Campomanes P, Rothlisberger U, De Luca A, Lo Bello M, Ang WH, Dyson PJ, Parker MW. Chemistry. 2011;17:7806–7816. doi: 10.1002/chem.201100586. [DOI] [PubMed] [Google Scholar]
- 45.Bhosale P, Bernstein PS. Biochim Biophys Acta. 2005;1740:116–121. doi: 10.1016/j.bbadis.2005.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.