Abstract
Transport proteins function in the translocation of ions, solutes and macromolecules across cellular and organellar membranes. These integral membrane proteins fall into >600 families as tabulated in the Transporter Classification Database (www.tcdb.org). Recent studies, some of which are reported here, define distant phylogenetic relationships between families with the creation of superfamilies. Several of these are analyzed using a novel set of programs designed to allow reliable prediction of phylogenetic trees when sequence divergence is too great to allow the use of multiple alignments. These new programs, called SuperfamilyTree1 and 2 (SFT1 and 2), allow display of protein and family relationships, respectively, based on thousands of comparative BLAST scores rather than multiple alignments. Superfamilies analyzed include: (1) Aerolysins, (2) RTX Toxins, (3) Defensins, (4) Ion Transporters, (5) Bile/Arsenite/Riboflavin Transporters, (6) Cation: Proton Antiporters, and (7) the Glucose/Fructose/Lactose superfamily within the prokaryotic phosphoenol pyruvate-dependent Phosphotransferase System. In addition to defining the phylogenetic relationships of the proteins and families within these seven superfamilies, evidence is provided showing that the SFT programs outperform programs that are based on multiple alignments whenever sequence divergence of superfamily members is extensive. The SFT programs should be applicable to virtually any superfamily of proteins or nucleic acids.
Key Words: Transport proteins, Membranes, Protein superfamilies, Phylogenetic analyses, Novel programs, Comparisons
Introduction
Phylogenetic programs can help us visualize the relationships of transport proteins belonging to the same superfamily (sharing a common evolutionary ancestor). There are several methods currently available that measure the evolutionary divergence amongst related proteins using different algorithms and techniques. Prior to the development of the SuperfamilyTree (SFT) programs, these methods relied on multiple alignments to properly determine the relationships within a superfamily. However, when proteins within a superfamily are too distantly related to one another, they will not align properly, and as a result, a reliable and accurate phylogenetic tree will not be generated [Ogdenw and Rosenberg, 2006]. The SFT programs use thousands of BLAST bit scores instead of multiple alignments, thus avoiding the pitfalls often encountered when determining phylogeny for distantly related proteins [Yen et al., 2009, 2010].
In this paper, we compare the SFT1 and 2 programs with various programs used to derive phylogenetic trees based on multiple alignments. The superfamilies analyzed include three pore-forming toxin superfamilies [(1) Aerolysin, (2) RTX toxin and (3) Defensin superfamilies], three secondary carrier superfamilies [(4) Ion Transporters, IT, (5) Bile/Arsenite/Riboflavin Transporters, BART, and (6) Cation: Proton Antiporters, CPA, superfamilies] and a single group translocating superfamily, the (7) Phosphotransferase System (PTS-GFL) superfamily (see Transporter Classification Database, TCDB; www.tcdb.org). We show that whenever members of a superfamily are sufficiently close in sequence, trees derived by methods based on multiple alignments or BLAST bit scores give excellent agreement. However, when sequences are too divergent to allow construction of reliable multiple alignments, the SFT programs outperform all others. The work reported here provides a more detailed comparative analysis of the applicability of these programs to phylogenetic tree construction. It also reveals, for the first time, the phylogenetic relationships of the proteins and families that comprise these seven superfamilies. The results (a) substantiated predetermined family assignments in TCDB, (b) allowed us to correct a few errors in these assignments and (c) verified the utility of these programs as applied to a wide variety of proteins and nucleic acid homologues. Their use in both functional prediction and consideration of the biological/historical relationships of the members of each superfamily are discussed.
Methods
Using the TCDB database [Saier et al., 2006, 2009], we can generate a temporary database file containing proteins that define all members within our superfamily of interest. This database file is used to define the criteria for superfamily definition and how that superfamily would later be broken down into families or subfamilies. The division of proteins into superfamilies, families, and subfamilies was conducted according to assignments in TCDB.
Multiple Alignment Methods
In previous publications, we compared multiple programs designed to establish homology and derive multiple alignment-based phylogenetic trees [Matias et al., 2010; Wang et al., 2009; Young et al., 1999]. Only some of these programs were used in the present comparative analyses.
For neighbor-joining [Gascuel and Steel, 2006; Saitou and Nei, 1987], parsimony [Felsenstein, 1996; Kolaczkowski and Thornton, 2004], and Bayesian methods [Huelsenbeck et al., 2008], the resulting database was used to create a multiple alignment. In the neighbor-joining method, the multiple alignment was used to generate a neighbor-join phylogenetic tree using the ClustalX program [Thompson et al., 1997]. The resulting file was then viewed as a radial phylogenetic tree using the Tree View (TV) program [Zhai et al., 2002].
In the parsimony method, the multiple alignment was used to create a PHYLIP file [Felsenstein, 2004]. Then the program ProtPars (http://evolution.genetics.washington.edu/phylip/doc/protpars.html) was used to generate 100 parsimony trees, which were then consolidated in to a single consensus tree. This tree was then viewed as a radial tree using the TV program.
In the Bayesian method, the multiple alignment was used to create a NEXUS file [Maddison et al., 1997]. Then the program, MrBayes [Huelsenbeck and Ronquist, 2001], was used to generate a Bayesian tree. The resulting tree was then viewed as a radial phylogenetic tree using the TV program.
SFT Methods
The SFT programs were designed to provide reliable phylogenetic data for distantly related homologues, particularly when sequence divergence is too great to allow construction of accurate multiple alignments. Family assignments in TCDB were based on proof of homology using rigorous statistical approaches as described in Saier et al. [2009]. The SFT programs provide confirmation of these assignments and go further to define the relationships of the proteins (SFT1) and family or subfamilies (SFT2) within superfamilies. This had never been possible before.
For the SFT methods, temporary databases, generated from TCDB, were used for rapid sequence similarity searches. Using these databases, we used PSI-BLAST [Altschul et al., 1990, 1997] to search the NCBI protein database and matched up potential members for each family. BLAST hits were then classified and sorted into respective families and subfamilies. This approach differs from that described previously in Yen et al. [2009, 2010] by streamlining the procedures of operation automating data entry and stabilizing the operating sequence.
The resulting database files were then used to generate a phylogenetic tree using the SFT1 program by generating comparative BLAST bit score matrices of the superfamily through 100 repeat shuffles. The programs, Fitch and Consense [Fitch and Margoliash, 1967] (http://evolution.genetics.washington.edu/phylip/doc/protpars.html), utilized the matrix information to generate 100 phylogenetic trees and consolidate those trees into a single consensus tree. The resulting SFT1 tree shows the relative phylogenetic positions of all members of the families within the superfamily. The information from the SFT1 program is then used to combine sequences in each of the constituent families into a single file. The same programs and methods are applied to the newly formed database files to generate an SFT2 tree. These trees are viewed as radial phylogenetic trees using the TV program [Zhai et al., 2002]. The procedures used are described in a step-by-step fashion on our online Wiki (web address: http://132.239.144.24/?p=78). These programs can be downloaded from TCDB's Biotools section and installed following directions found on the SFT Wiki webpage.
Results
The Aerolysin Superfamily
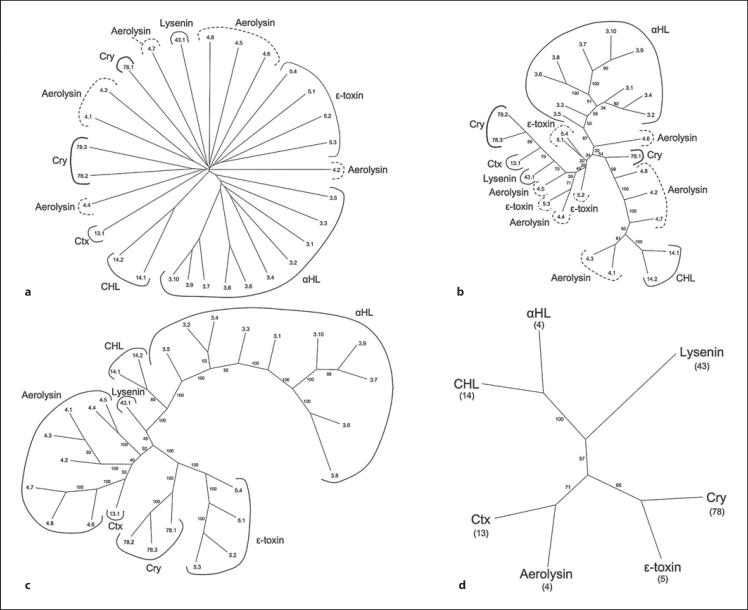
The Aerolysin superfamily [Iacovache et al., 2008] consists of seven families in TCDB, several of which were not known to be related prior to these studies. We have established (unpubl. results) that six of these families are related using standard criteria as described in earlier papers [Chang et al., 2004; Matias et al., 2010; Povolotsky et al., 2010; Saier, 1994; Saier et al., 2009; Wang et al., 2009]. The six families are the α-hemolysins (αHL, TC# 1.C.3), aerolysins (TC# 1.C.4), ∊-toxins (TC# 1.C.5), cytotoxins (Ctx, TC# 1.C.13), cytohemolysins (CHL, TC# 1.C.14), and crystal proteins (Cry, 1.C.78). Additionally, we have evidence that the Lysenin family (TC# 1.C.43) is distantly related (unpubl. results).
Similar to Yen et al. [2010], we constructed phylogenetic trees using four programs as shown in figure 1. Figure 1a shows the tree based on a ClustalX program-generated multiple alignment using the TV program (neighbor-joining). Figure 1b shows a tree, also using a ClustalX-generated multiple alignment and TV, but based on the ProtPars program (Phylip package, parsimony). Figure 1c shows a tree based on the SFT1 program, and figure 1d shows a consensus tree based on the SFT2 program. The results show that the SFT1 and SFT2 programs are superior to classical programs based on multiple alignments when sequence-divergent proteins are compared as described below.
Fig. 1.
Phylogenetic (Fitch) trees for the Aerolysin superfamily using the proteins in TCDB as of February 2011. Four different methods of tree construction were used: ClustalX-based neighbor-joining (a), ProtPars-based parsimony (b), the BLAST-derived SFT1 program results showing all Aerolysin superfamily members (c), and the SFT2-based tree showing all Aerolysin superfamily families (d). a–c Numbers indicate the protein TC#s (last two digits of the complete TC#). d Family abbreviations are presented with TC family numbers in parenthesis. b–d Small numbers adjacent to the branches represent the ‘bootstrap’ values, indicating the reliability of the branching order. See TCDB for protein identification.
As shown in the ClustalX-based neighbor-joining tree (fig. 1a), many of the family members segregate into the expected families according to assignments in TCDB. However, the large aerolysin family and the smaller Cry family within this superfamily are not cohesive. The four major branches of the aerolysin family all stem from different points near the center of the tree, and the 3 members of the Cry family form two different clusters. No obvious relationships between these clusters were apparent.
In the parsimony tree (fig. 1b), we also observed that members of the aerolysin family are distributed on four different branches, but the protein cluster memberships differ significantly from those in figure 1a. Additionally, the four members of the ∊-toxin family are separated from each other, present on three different branches, and these are separated from each other by Cry family members. The results clearly suggest that these two programs were not capable of detecting the correct phylogenetic relationships.
These results should be contrasted with those shown in figure 1c and d, obtained using the SFT1 and SFT2 programs, respectively. In figure 1c, each family forms a distinct cluster. Thus, all aerolysin family members are present on a single branch (lower left-hand side) and the ∊-toxin family forms a single coherent cluster (lower right-hand side). The single Ctx family member included in this study appears to be the most closely related to the aerolysins as also indicated in the SFT2 tree (see below). Members of other large families (e.g. the αHL family) also cluster together. While this is sometimes true of the figure 1a and b trees, the clustering pattern of the members are not always the same. Remaining families are not large enough to make similar comparisons.
Figure 1d shows the relationships of the different families to each other. The bacterial αHL and CHL families cluster together on this tree, in agreement with the patterns shown in figure 1a and c but not b. Additionally, the bacterial Ctx family and the ubiquitous aerolysin family cluster together as expected from figure 1c, but this relationship cannot be deduced from figure 1a or b. Finally, the Cry and ∊-toxin families form a cluster in figure 1c and d, but not in figure 1a or b. These results clearly demonstrate the superiority of the SFT programs over classical multiple alignment-based programs.
The RTX Toxin Superfamily
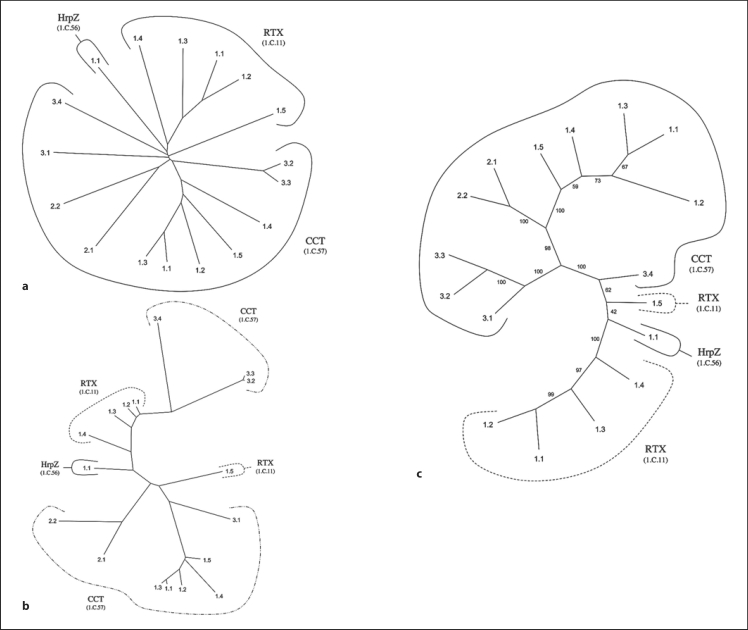
The RTX toxin superfamily consists of three currently recognized families, all derived from bacteria. These families include: RTX (TC# 1.C.11), HrpZ (TC# 1.C.56), and CCT (TC# 1.C.57). These three families include pore-forming exotoxins possessing multiple domains capable of insertion into the membranes of target species [Davies et al., 2001, 2002; Genisyuerek et al., 2011; Lee et al., 2001; Reineke et al., 2007]. We established in figures S1A–C and figures S2A–C (web address: http://www.biology.ucsd.edu/∼msaier/supmat/SFT/) that these three families were related using standard criteria as described in earlier papers [Chang et al., 2004; Matias et al., 2010; Povolotsky et al., 2010; Saier, 1994; Saier et al., 2009; Wang et al., 2009].
Figure 2 shows phylogenetic trees for TC members of this superfamily determined using three programs, (a) ClustalX (Neighbor-Joining), (b) MrBayes (Bayesian), and (c) SFT1. In all three trees, some degree of family intermixing was observed, but depending on the program, the nature of this intermixing differed. For example, in figure 2, the single HrpZ family member (TC# 1.C.56.1.1) clusters loosely with the RTX family (TC# 1.C.11). In figure 2a and c, CCT family members cluster together separately from the other proteins, but in figure 2b, CCT members can be found on two distinct branches.
Fig. 2.
Phylogenetic (Fitch) trees for the RTX toxin superfamily using the proteins in TCDB as of February 2011. Three different methods of tree construction were used: ClustalX-based neighbor-joining (a), MrBayes-based Bayesian (b), and the BLAST-derived SFT1 program showing all RTX toxin superfamily members (c). In all three figures, numbers indicate the protein TC#s. Family TC#s are indicated within parentheses under the family abbreviation. c Small numbers adjacent to the branches represent the ‘bootstrap’ values, indicating the reliability of the branching order. See TCDB for protein identification.
We compared two proteins, gi547678 from the HrpZ family and gi73853298 from the RTX toxin family. After identifying the hydrophobic transmembrane regions, we compared residues 18–370 of the RTX toxin homologue with residues 1–347 of the HrpZ homologue. This comparison yielded a GAP score of 12.3 standard deviations with 34.2% identity and 25.8% similarity (fig. S2, see web address above). These scores are sufficient to establish homology between the two families. This high degree of similarity explains the observed intermixing between members of RTX and HrpZ in all three phylogenetic trees.
In summary, the results obtained with the three programs differed from each other, where figure 2a and c gave more similar phylogenetic relationships and greater coherence among members of the RTX toxin and CCT families. Thus, it can be concluded that when proteins are aligned properly, ClustalX and Bayesian trees show reasonable agreements with the SFT1 tree.
The Defensin Superfamily
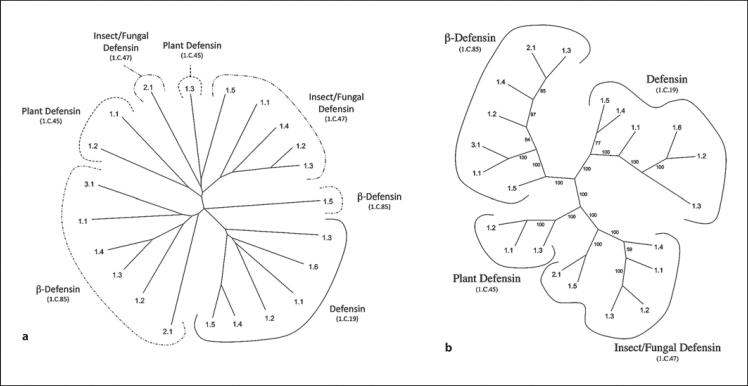
The Defensin superfamily consists of four recognized families of peptide toxins. These small toxins form oligomeric pores of variable sizes. Zhu et al. [2005] have described this superfamily, noting that in addition to the pore-forming peptide toxins, some are sweet tasting proteins and others are animal toxins that instead of forming pores, exert their toxic effects by targeting ion channels. This superfamily is sometimes referred to as the cysteine-stabilized αβ-superfamily because members exhibit a single α-helix with an αβ-cysteine motif and two C-terminal β-strands [Zhu et al., 2005].
The pore-forming defensin families are: (1) the mammalian defensins (TC# 1.C.19), (2) the plant defensins (TC# 1.C.45), (3) the fungal/insect defensins (TC# 1.C.47) and (4) the β-defensins (TC# 1.C.85). This last mentioned family includes three subfamilies: mammalian pore-forming toxins, snake venom myotoxins that modify voltage-sensitive Na+ channels and exhibit analgesic effects, and bird gallinacins that serve as antimicrobial agents (see TCDB; www.tcdb.org).
Two phylogenetic trees for the defensin superfamily, generated using (1) a ClustalX-based multiple alignment with the TV program versus (2) the SFT1 program with TV, are shown in figure 3a and b, respectively. The proteins in TCDB are included. In addition to these two trees, two more phylogenetic trees were generated using ClustalX-based multiple alignments and MrBayes with TV (Bayesian) as well as the SFT2 program using TV (see figures S3A and S3B, respectively, at web address given above).
Fig. 3.
Phylogenetic (Fitch) trees for the Defensin superfamily using the proteins in TCDB as of February 2011. Two different methods of tree construction were used: ClustalX-based neighbor-joining (a) and the BLAST-derived SFT1 (b). Both trees show all Defensin superfamily members. Numbers indicate the protein TC#s, while family TC#s are indicated within parentheses under the family name. b Small numbers adjacent to the branches represent the ‘bootstrap’ values, indicating the relative reliability of the branching order.
Interestingly, when the ClustalX and MrBayes trees were examined, we discovered that both trees separated one member of the β-defensin family (TC# 1.C.85.1.5) from all the other members of this family. In the insect/fungal defensin family (TC# 1.C.47), members were also separated, but in different ways. In the Bayesian tree, proteins 1.1 through 1.4 cluster together, but 1.5 and 2.1 cluster separately. In the ClustalX tree, the same is true, but the two latter proteins cluster loosely with one of the plant defensins (TC# 1.C.45.1.3). Also in this tree, the plant defensins and the insect/fungal defensins cluster loosely together. In the Bayesian tree, these proteins also cluster loosely with the mammalian defensins. In both trees, only the mammalian defensins form a coherent group. This can be contrasted with the results obtained with the SFT1 program (fig. 3b). In this tree, each of the four defensin families cluster separately and coherently. The plant defensins cluster loosely with the insect/fungal defensins, while the β-defensins, derived from various animals including mammals, cluster loosely with the mammalian defensins. It is interesting to note that while the ClustalX and Bayesian trees did not detect family relationships, the proteins for which the family relationships were not correctly depicted are the most distant members of that family in the SFT1 tree. Once again, these facts reveal the greater power of the SFT programs to detect correct distant relationships.
The IT Superfamily
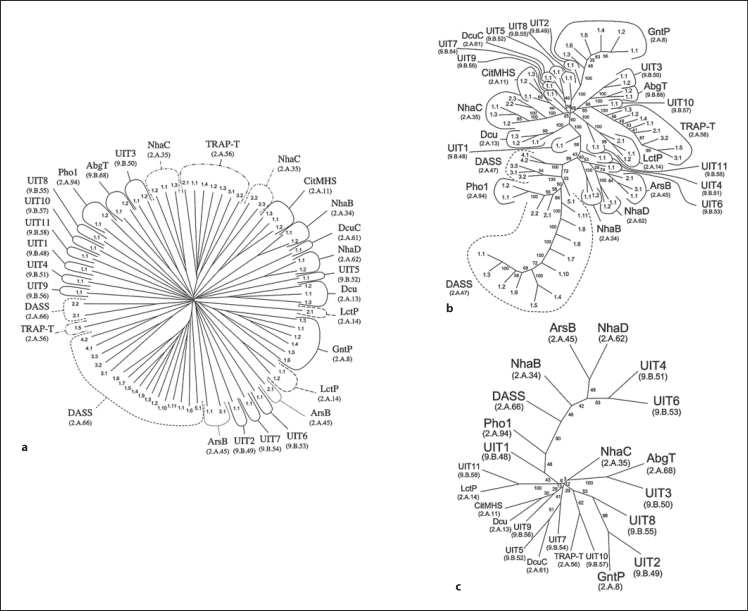
The IT superfamily has been described by Prakash et al. [2003]. This superfamily consists of 13 families of known function plus 11 more families of unknown function. All functionally characterized members of this superfamily transport ionic species; not one has been shown to transport a neutral (non-charged) molecule [Prakash et al., 2003].
Phylogenetic trees obtained for this superfamily are depicted in figure 4, where these three trees show the results of the ClustalX, SFT1 and SFT2 programs, respectively. The ClustalX and SFT1 programs show the positions of all the proteins within the 24 families of the IT superfamily in TCDB, while figure 4c shows the relationships of these families to each other. Interestingly, the IT superfamily includes members that can function as either secondary or primary active transporters, e.g. the DASS family (TC# 2.A.47) includes members that function exclusively as secondary carriers, while the ArsB family (TC# 2.A.45) can function by either primary or secondary active transport depending upon the availability of an ArsA ATPase [Bhattacharjee, 2000; Castillo and Saier, 2010; Rosen et al., 1995]. In addition, members of the TRAP-T family, while functioning exclusively as secondary active transporters, require the presence of an auxiliary 4 TMS membrane protein as well as an extracytoplasmic solute-binding receptor [Kelly and Thomas, 2001; Mulligan et al., 2007; Rabus et al., 1999]. Thus, the IT superfamily includes members that function by different mechanisms.
Fig. 4.
Phylogenetic (Fitch) trees for the IT superfamily using the proteins in TCDB. Three different methods of tree construction were used: ClustalX-based neighbor-joining (a), the BLAST-derived SFT1 approach showing all IT superfamily members (b), and the SFT2 approach showing all IT superfamily families (c). a, b Numbers indicate the protein TC#s (last two digits of the complete TC#). c Family abbreviations are presented with TC family numbers in parentheses. b, c Small numbers adjacent to the branches represent the ‘bootstrap’ values, indicating the reliability of the branching order. See TCDB for protein identification.
In figure 4a, it is interesting to observe the clustering patterns of proteins according to their family memberships within the ClustalX tree. It can be seen, for example, that certain families, like members of the DASS family within the IT superfamily, occur on two branches of the phylogenetic tree. Members of the LctP family occur on two different branches; members of the NhaC family occur on two branches; ArsB family members occur on two branches, and TRAP-T family members occur on two distant branches. All other members cluster into their appropriate families as designated in TCDB.
In comparison, it can be seen in the SFT1 tree (fig. 4b) that nearly all currently recognized proteins within a single family cluster tightly together. The DASS family members are found in two nearby clusters, separated only by members of the Pho1 family. This intermixing of family members suggests that certain members of the DASS family are closely related to members of the Pho1 family.
The configuration of figure 4c, which presents the relative positions of the 24 families, based on the SFT2 program, provides a guide to potential functions of several of the uncharacterized Unknown IT (UIT) families. For example, UIT11 clusters together with Lactate and Glycolate porters of the LctP family, suggesting that members of the UIT11 family may also transport short-chain monocarboxylates. The CitMHS and Dcu families transport di- and tri-carboxylic acids, and therefore it is not surprising that they cluster together. Since UIT9 is included in the same branch of the tree, one can predict that members of this family also transport di- and tri-carboxylates. This suggestion is further supported by the fact that the next closest functionally characterized family within the IT superfamily, DcuC, also transports dicarboxylates. Since UIT5 and UIT7 are found on the same branch as DcuC, one can suggest that all of these families function in di- and tri-carboxylic acid transport.
Continuing with this logic, the UIT10 family clusters with the TRAP-T family. However, TRAP-T family members are known to transport a wide variety of organic substances, including dicarboxylates, sugar acids, taurine, keto-monocarboxylates, ectoine, glutamate, and chlorobenzoate. The common feature of all these substances is that they contain a single carboxyl group. Therefore, it can be postulated that UIT10 also transports monocarboxylates. Closest to the branch bearing the TRAP-T family is the GntP family. All members of the GntP family transport monocarboxylates, most of them being sugar acids, but a few transport D-serine and D-glycerate. For this reason, we can postulate that UIT2 and UIT8 similarly transport monocarboxylates, since these three families cluster together. Finally, UIT3 clusters together with the AbgT family which transports p-aminobenzoyl-glutamate, another organic anion. It seems that most functionally characterized superfamily members transport anionic substances with the exceptions only of the NhaB, NhaC and NhaD families, which primarily catalyze transport of monovalent cations. It is worthy of note, that all functionally characterized families in the lower half of the tree except NhaC transport organic anions, suggesting that similar substrates are recognized by transporters within most of the UIT families within the IT superfamily.
Functional Predictions of UIT Family Members Based on Genomic Context
UIT1 was examined using the SEED database [Overbeek et al., 2005]. Searches revealed that this family includes a protein that is encoded within the same operon as two other proteins, malonyl-CoA synthetase and malonyl-CoA decarboxylase in Rhizobium leguminosarm bv. trifolii [An and Kim, 1998]. It therefore may transport malonate or acetate. The UIT1 family has therefore been redesignated as TC# 2.A.101 rather than TC# 9.B.48, its previously assigned TC number.
The BART Superfamily
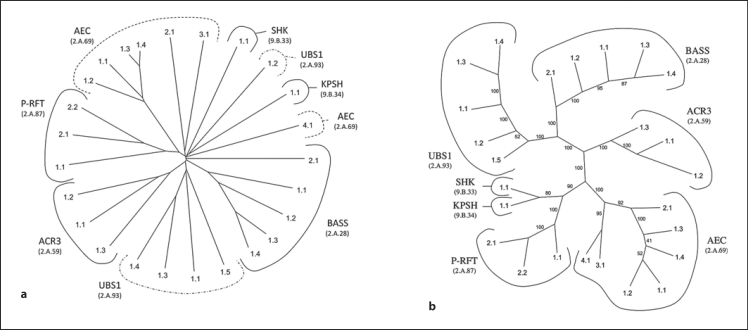
The BART superfamily has been described by Mansour et al. [2007]. Phylogenetic trees were constructed for the proteins within this superfamily included in TCDB using 4 different programs: ClustalX (Neighbor-Joining), MrBayes (Bayesian), SFT1 and SFT2. ClustalX proved to be better than MrBayes as the latter failed in several instances to group together members of certain families. Figure 5a and b shows the ClustalX and SFT1 trees, respectively. The other two trees can be viewed in figures S4A and S4B on our website. The BASS family generally clusters adjacent to the UBS1 family. In the UBS1 family, SFT1 clusters all members together, but one of these proteins (TC# 2.A.93.1.2) failed to cluster with the other members on the ClustalX tree. This protein proved to be relatively distant from the other members of this family (fig. 5b). The fact that SFT1 was able to cluster all of these proteins together reveals its superiority in detecting distant relationships. It should be noted that both the Bayesian and ClustalX trees, both of which are based on multiple alignments, made the same mistakes. While the Bayesian tree made the most such mistakes, the SFT1 program apparently made none.
Fig. 5.
Phylogenetic trees for the BART superfamily using the proteins in TCDB. a ClustalX. b SFT1. The conventions of presentation are the same as for figure 3. For family and protein identification, see TCDB.
In figure 5b, the SHK (TC# 9.B.33) and KPSH (TC# 9.B.34) families cluster together. This fact agrees with the observation that members of both families consist of N-terminal 5 TMS BART superfamily domains followed by catalytic domains [Mansour et al., 2007]. However, while SFT1 identified this relationship, ClustalX did not. In the latter tree, these families branch from each other near the base of the tree, suggesting a lack of resolution. P-RFT clusters loosely with the SHK and KPSH proteins in the SFT trees in agreement with the fact that all three families have the basic 5 TMS element and lack a duplication of this unit. Once again, the ClustalX program failed to detect this relationship.
The AEC family (TC# 2.A.69) shows similar relationships. Thus, in the SFT1 tree, all of these proteins cluster loosely together in agreement with family assignments. However, in the ClustalX tree, proteins of the AEC family are localized to 3 distinct branches. The proteins of subfamily 1 are found together; the proteins of subfamilies 2 and 3 cluster loosely together, and subfamily 4 is on a distinct branch arising from the base of the tree. It appears that SFT1 is superior in detecting these distant relationships. Finally, the ACR3 family (TC# 2.A.59) members cluster together on both trees.
The SFT2 program, in which the positions of proteins within each of the families are integrated so that only family relationships are depicted, reveals clustering patterns in agreement with those observed in figure 5b (see figure S4B on our website). Thus, SHK and KPSH cluster tightly together with P-RFT branching more distantly at the top of the tree. Further, BASS and UBS1 cluster tightly together, with ACR3 and AEC branching more distantly. It is interesting to note that all of the families at the top of the tree have the basic 5 TMS unit while all families at the bottom of the tree have 10 TMSs with just one exception. This protein, in the UBS1 family (TC# 2.A.93.1.2), has 6 TMSs and is the most distant member of this family. It is possible that it should be assigned to a distinct family.
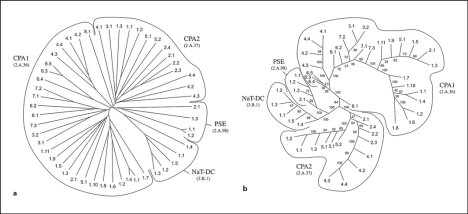
The CPA Superfamily
The CPA superfamily currently consists of 4 families: CPA1 (TC# 2.A.36), CPA2 (TC# 2.A.37), PSE (TC# 2.A.94), and NaT-DC (TC# 3.B.1). While the first three families include secondary active transporters, the last one (NaT-DC) utilizes organo-acid decarboxylation to drive Na+ efflux [Dimroth et al., 2001]. The trees generated by ClustalX and SFT1 are shown in figure 6. In addition to these two trees, an additional tree was generated using the SFT2 program (see figure S5 on our website). In all three trees, the PSE and NaT-DC families clustered fairly closely together while the CPA1 and CPA2 families clustered more loosely together but more distantly from the PSE and NaT-DC families.
Fig. 6.
Phylogenetic trees for the CPA superfamily using the proteins in TCDB. a ClustalX. b SFT1. The conventions of presentation are the same as for figure 3. For family and protein identification, see TCDB.
Starting with the smallest family, the PSE family, we see that the two trees exhibit complete agreement with respect to the branching positions of the proteins. Similarly, for the NaT-DC family, we find almost identical branching patterns except that the positions of proteins 1.1 and 1.5 are switched. On both trees, all members of the CPA1 family segregate from members of the CPA2 family.
The largest family included within the CPA superfamily is the CPA1 family. This family consists of 8 subfamilies. The largest of these subfamilies is subfamily 1. In both ClustalX and SFT1 trees, subfamily 1 member cluster together and distantly from all other members of the CPA1 family, but proteins 2.1 and 5.1 are included within this subfamily. These two proteins are the only members of their respective subfamilies included within TCDB. TC Blast searches reveal that both 2.1 and 5.1 are more closely related to members of subfamily 1 than they are to any of the other subfamilies, and thus, they can be viewed as distant members of subfamily 1. With respect to subfamilies 3 and 4, both are coherent, with members grouping together on a single branch in both the ClustalX and SFT1 trees. Moreover, subfamilies 3 and 4 are more closely related to subfamilies 6 and 7 in both trees. Only in the SFT1 tree does subfamily 3 associate with family 7 while family 4 associates with family 6. In the ClustalX tree, members of subfamilies 6 and 7 are intermixed.
The second largest family in the CPA superfamily is the CPA2 family. Once again, substantial agreement is observed between the two trees shown in figure 6. Thus, subfamily 1 comprises a clear cluster most closely related to the proteins in subfamilies 3 and 5. Subfamily 2 is also coherent, although subfamily 4, coherent in the SFT1 tree, is segregated in the ClustalX tree. This presumably reflects the greater distance of protein 4.1 from proteins 4.2, 4.3 and 4.4 as observed in both trees. Thus, in this case, we see excellent agreement when comparing the two trees with only a few exceptions.
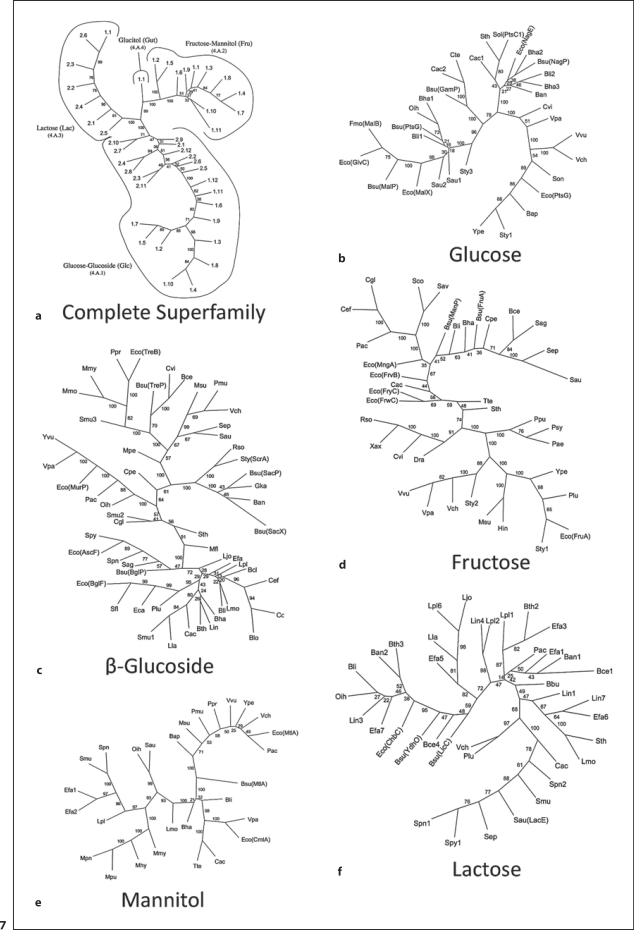
The PTS-GFL Superfamily
The permeases of the bacterial phosphotransferase system (PTS) have been extensively characterized [Barabote and Saier, 2006; Lengeler and Jahreis, 2009]. This functional superfamily consists of seven recognized families, four of which include members that are homologous and therefore comprise a phylogenetic superfamily [Nguyen et al., 2006]. The three remaining families include the mannose family, where all constituents of these enzyme complexes are evolutionarily distinct from all other PTS families, the Ascorbate/Galactitol superfamily, and the non-transporting Dihydroxyactetone family [Barabote and Saier, 2006; Saier et al., 2005].
The four families within the first of these PTS superfamilies include the glucose-glucoside (Glc; TC# 4.A.1) family, the fructose-mannitol (Fru; TC# 4.A.2) family, the Lactose-N,N′-diacetylchitobiose-β-glucoside (Lac; TC# 4.A.3) family, and the glucitol (Gut; TC# 4.A.4) family [Hvorup et al., 2003; Nguyen et al., 2006; Saier et al., 2005]. While the complete superfamily tree is presented in figure 7a, those for the individual families and subfamilies are shown in figure 7b–f, where the first two of these families are divided into two subfamilies. All TC entries included within these four families were included in the phylogenetic tree generated by SFT1 (fig. 7a). It can be seen that all proteins within the glucose-glucoside family cluster together on a single branch. All of the monosaccharide permeases in this family (TC# 4.A.1.1) occur at the bottom of this branch, while all glycoside permeases (TC# 4.A.1.2) cluster together on the upper portion of this branch. The lactose (TC# 4.A.3.1) and diacetylchitobiose (TC# 4.A.3.2) families cluster together on the upper left-hand side of the tree. The third branch (upper right-hand side) includes the glucitol family (Gut; TC# 4.A.4) and the fructose-mannitol family (Fru; TC# 4.A.2). This observation suggests that glucitol permeases, the only PTS permeases that are split into two polypeptide chains, may be most closely related to the fructose permeases. This is of substantial interest in view of our early suggestion that the fructose permeases were primordial [Saier et al., 1985].
Fig. 7.
Phylogenetic (Fitch) trees for the PTS-GFL superfamily using the proteins examined previously by Nguyen et al. 2006]. Only the BLAST-based SFT1 method of tree construction was used. a Numbers indicate the protein TC#s, while family TC#s are indicated within parentheses under the family designation. Small numbers adjacent to the branches represent the ‘bootstrap’ values, indicating the reliability of the branching order. At the ends of each branch, the protein abbreviation is presented as in table 1 of Nguyen et al. [2006].
Phylogenetic trees for large numbers of PTS permeases were generated using the SFT1 program. The proteins included within this study were the same ones included in the previous study by Nguyen et al. [2006] where the ClustalX and TV programs were used. The trees reported by Nguyen et al. [2006] are shown in figure 1A–F of that paper, and they correspond to the trees shown in figure 7 in this paper, generated with the SFT1 program. The correspondence between these two methods of phylogenetic analysis is substantial although a few minor differences can be observed, particularly for those proteins close to the center of the tree.
While proteins frequently cluster according to the phylogenetic groupings of their source organisms, exceptions can be observed. These exceptions may represent cases of horizontal gene transfer. Due to their clustering with proteins from one bacterial phylum, the source organism from which these genes were transferred can be predicted.
Escherichia coli contains several paralogs of PTS permeases that fall within the fructose family. These include FrvB, FrwC, FryC, and MngA [Nguyen et al., 2006]. These proteins are distantly related in both trees, and they therefore cluster loosely together. This observation suggests that they arose by gene duplication events early during the evolution of the proteobacteria. Examination of the remaining trees shown in figure 7 in this paper and figure 1A–F in Nguyen et al. [2006] confirms the close correspondence between positions of proteins within the trees generated by these two programs. Since the former trees are based on BLAST scores while the latter are based on multiple alignments, we feel that the results reinforce the conclusion of the reliability of both programs when sequences are not so diverse as to prevent construction of a reliable multiple alignment.
Discussion
In this paper, we have conducted phylogenetic analysis of some of the largest superfamilies of integral membrane transport proteins. In doing so, we have intentionally analyzed superfamilies with tremendous sequence divergence between members. Whenever this sequence divergence was too great to allow construction of reliable multiple alignments, the SFT1 and 2 programs proved superior to all tested alternative programs.
Since SFT1 and 2 use larger protein databases to help define each protein family/subfamily, when compared to other phylogenetic programs and methods, the SFT1 and 2 programs are usually more accurate and reliable in determining the correct phylogenetic relationships within a superfamily. The usage of protein databases coupled with the novel usage of BLAST bit score comparison matrices allow the SFT programs to properly determine the phylogeny of superfamilies containing more evolutionarily divergent members [Yen et al., 2009, 2010]. Using other phylogenetic programs and methods, more distantly related proteins are usually either grouped together due to long-branch clustering or excluded from clusters and given their own distinctive isolated branches [Felsenstein, 1978; Kolaczkowski and Thornton, 2009; Ogdenw and Rosenberg, 2006; Siddall and Whiting, 1999]. The minor drawback to using BLAST bit score comparisons for determining phylogeny is that the ‘bootstrap’ values become less indicative of the reliability and accuracy of observed clustering patterns for very closely related proteins. However, BLAST bit scores allow the SFT programs to generate trees within a fraction of the time it would take to create phylogenetic trees of comparable reliability using other phylogenetic methods [Felsenstein, 2004; Liu et al., 2010].
In conclusion, the SFT programs serve as an invaluable tool for the prediction of phylogenetic relationships amongst proteins sharing a common ancestor. Using novel techniques, the SFT programs can quickly generate accurate and reliable phylogenetic trees for superfamilies of transport proteins. The SFT programs and methods have a wide range of applications for the study of any kind of homologous protein or nucleic acid sequences, and are useful for predicting and evaluating the evolutionary, functional, mechanistic, and structural relationships of proteins to each other and of nucleic acids or nucleic acid-based entities to each other.
Acknowledgements
The authors wish to thank Carl H. Welliver for assistance with manuscript preparation. The work reported in this paper was supported by NIH grant: 2 R01 GM077402.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JH, Kim YS. A gene cluster encoding malonyl-CoA decarboxylase (MatA), malonyl-CoA synthetase (MatB) and a putative dicarboxylate carrier protein (MatC) in Rhizobium trifolii–cloning, sequencing, and expression of the enzymes in Escherichia coli. Eur J Biochem. 1998;257:395–402. doi: 10.1046/j.1432-1327.1998.2570395.x. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee H, Zhou T, Li J, Gatti DL, Walmsley AR, Rosen BP. Structure-function relationships in an anion-translocating ATPase. Biochem Soc Trans. 2000;28:520–526. [PubMed] [Google Scholar]
- Castillo R, Saier MH. Functional promiscuity of homologues of the bacterial ArsA ATPases. Int J Microbiol. 2010;2010:187–373. doi: 10.1155/2010/187373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AB, Lin R, Keith Studley W, Tran CV, Saier MH., Jr Phylogeny as a guide to structure and function of membrane transport proteins. Mol Membr Biol. 2004;21:171–181. doi: 10.1080/09687680410001720830. [DOI] [PubMed] [Google Scholar]
- Davies RL, Campbell S, Whittam TS. Mosaic structure and molecular evolution of the leukotoxin operon (lktCABD) in Mannheimia (Pasteurella) haemolytica, Mannheimia glucosida, and Pasteurella trehalosi. J Bacteriol. 2002;184:266–277. doi: 10.1128/JB.184.1.266-277.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies RL, Whittam TS, Selander RK. Sequence diversity and molecular evolution of the leukotoxin (lktA) gene in bovine and ovine strains of Mannheimia (Pasteurella) haemolytica. J Bacteriol. 2001;183:1394–1404. doi: 10.1128/JB.183.4.1394-1404.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimroth P, Jockel P, Schmid M. Coupling mechanism of the oxaloacetate decarboxylase Na+ pump. Biochim Biophys Acta. 2001;1505:1–14. doi: 10.1016/s0005-2728(00)00272-3. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Cases in which parsimony and compatibility methods will be positively misleading. Syst Zool. 1978;27:401–410. [Google Scholar]
- Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Inferring Phylogenies. Sunderland: Sinauer Associates; 2004. [Google Scholar]
- Fitch WM, Margoliash E. Construction of phylogenetic trees. Science. 1967;155:279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- Gascuel O, Steel M. Neighbor-joining revealed. Mol Biol Evol. 2006;23:1997–2000. doi: 10.1093/molbev/msl072. [DOI] [PubMed] [Google Scholar]
- Genisyuerek S, Papatheodorou P, Guttenberg G, Schubert R, Benz R, Aktories K. Structural determinants for membrane insertion, pore formation and translocation of Clostridium difficile toxin B. Mol Microbiol. 2011;79:1643–1654. doi: 10.1111/j.1365-2958.2011.07549.x. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Joyce P, Lakner C, Ronquist F. Bayesian analysis of amino acid substitution models. Philos Trans R Soc Lond B Biol Sci. 2008;363:3941–3953. doi: 10.1098/rstb.2008.0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Hvorup R, Chang AB, Saier MH., Jr Bioinformatic analyses of the bacterial L-ascorbate phosphotransferase system permease family. J Mol Microbiol Biotechnol. 2003;6:191–205. doi: 10.1159/000077250. [DOI] [PubMed] [Google Scholar]
- Iacovache I, van der Goot FG, Pernot L. Pore formation: an ancient yet complex form of attack. Biochim Biophys Acta. 2008;1778:1611–1623. doi: 10.1016/j.bbamem.2008.01.026. [DOI] [PubMed] [Google Scholar]
- Kelly DJ, Thomas GH. The tripartite ATP-independent periplasmic (TRAP) transporters of bacteria and archaea. FEMS Microbiol Rev. 2001;25:405–424. doi: 10.1111/j.1574-6976.2001.tb00584.x. [DOI] [PubMed] [Google Scholar]
- Kolaczkowski B, Thornton JW. Performance of maximum parsimony and likelihood phylogenetics when evolution is heterogeneous. Nature. 2004;431:980–984. doi: 10.1038/nature02917. [DOI] [PubMed] [Google Scholar]
- Kolaczkowski B, Thornton JW. Long-branch attraction bias and inconsistency in Bayesian phylogenetics. PLoS One. 2009;4:e7891. doi: 10.1371/journal.pone.0007891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Klusener B, Tsiamis G, Stevens C, Neyt C, Tampakaki AP, Panopoulos NJ, Nöller J, Weiler EW, Cornelis GR, Mansfield JW, Nürnberger T. HrpZ(Psph) from the plant pathogen Pseudomonas syringae pv. phaseolicola binds to lipid bilayers and forms an ion-conducting pore in vitro. Proc Natl Acad Sci USA. 2001;98:289–294. doi: 10.1073/pnas.011265298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler JW, Jahreis K. Bacterial PEP-dependent carbohydrate: phosphotransferase systems couple sensing and global control mechanisms. Contrib Microbiol. 2009;16:65–87. doi: 10.1159/000219373. [DOI] [PubMed] [Google Scholar]
- Liu K, Linder CR, Warnow T. Multiple sequence alignment: a major challenge to large-scale phylogenetics. PLoS Curr. 2010;2:RRN1198. doi: 10.1371/currents.RRN1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison DR, Swofford DL, Maddison WP. NEXUS: an extensible file format for systematic information. Syst Biol. 1997;46:590–621. doi: 10.1093/sysbio/46.4.590. [DOI] [PubMed] [Google Scholar]
- Mansour NM, Sawhney M, Tamang DG, Vogl C, Saier MH., Jr The bile/arsenite/riboflavin transporter (BART) superfamily. FEBS J. 2007;274:612–629. doi: 10.1111/j.1742-4658.2006.05627.x. [DOI] [PubMed] [Google Scholar]
- Matias MG, Gomolplitinant KM, Tamang DG, Saier MH., Jr Animal Ca2+ release-activated Ca2+ (CRAC) channels appear to be homologous to and derived from the ubiquitous cation diffusion facilitators. BMC Res Notes. 2010;3:158. doi: 10.1186/1756-0500-3-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan C, Kelly DJ, Thomas GH. Tripartite ATP-independent periplasmic transporters: application of a relational database for genome-wide analysis of transporter gene frequency and organization. J Mol Microbiol Biotechnol. 2007;12:218–226. doi: 10.1159/000099643. [DOI] [PubMed] [Google Scholar]
- Nguyen TX, Yen MR, Barabote RD, Saier MH., Jr Topological predictions for integral membrane permeases of the phosphoenolpyruvate:sugar phosphotransferase system. J Mol Microbiol Biotechnol. 2006;11:345–360. doi: 10.1159/000095636. [DOI] [PubMed] [Google Scholar]
- Ogdenw TH, Rosenberg MS. Multiple sequence alignment accuracy and phylogenetic inference. Syst Biol. 2006;55:314–328. doi: 10.1080/10635150500541730. [DOI] [PubMed] [Google Scholar]
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Rückert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povolotsky TL, Orlova E, Tamang DG, Saier MH. Defense against cannibalism: the SdpI family of bacterial immunity/signal transduction proteins. J Membr Biol. 2010;235:145–162. doi: 10.1007/s00232-010-9260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Cooper G, Singhi S, Saier MH., Jr The ion transporter superfamily. Biochim Biophys Acta. 2003;1618:79–92. doi: 10.1016/j.bbamem.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Rabus R, Jack DL, Kelly DJ, Saier MH., Jr TRAP transporters: an ancient family of extracytoplasmic solute-receptor-dependent secondary active transporters. Microbiology. 1999;145:3431–3445. doi: 10.1099/00221287-145-12-3431. [DOI] [PubMed] [Google Scholar]
- Reineke J, Tenzer S, Rupnik M, Koschinski A, Hasselmayer O, Schrattenholz A, Schild H, von Eichel-Streiber C. Autocatalytic cleavage of Clostridium difficile toxin B. Nature. 2007;446:415–419. doi: 10.1038/nature05622. [DOI] [PubMed] [Google Scholar]
- Rosen BP, Bhattacharjee H, Shi W. Mechanisms of metalloregulation of an anion-translocating ATPase. J Bioenerg Biomembr. 1995;27:85–91. doi: 10.1007/BF02110335. [DOI] [PubMed] [Google Scholar]
- Saier MH., Jr Computer-aided analyses of transport protein sequences: gleaning evidence concerning function, structure, biogenesis, and evolution. Microbiol Rev. 1994;58:71–93. doi: 10.1128/mr.58.1.71-93.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Jr, Grenier FC, Lee CA, Waygood EB. Evidence for the evolutionary relatedness of the proteins of the bacterial phosphoenolpyruvate:sugar phosphotransferase system. J Cell Biochem. 1985;27:43–56. doi: 10.1002/jcb.240270106. [DOI] [PubMed] [Google Scholar]
- Saier MH, Hvorup RN, Barabote RD. Evolution of the bacterial phosphotransferase system: from carriers and enzymes to group translocators. Biochem Soc Trans. 2005;33:220–224. doi: 10.1042/BST0330220. [DOI] [PubMed] [Google Scholar]
- Saier MH, Jr, Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–D186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Jr, Yen MR, Noto K, Tamang DG, Elkan C. The Transporter Classification Database: recent advances. Nucleic Acids Res. 2009;37:D274–D278. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Siddall ME, Whiting MF. Long-branch abstractions. Cladistics. 1999;15:9–24. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Dukarevich M, Sun EI, Yen MR, Saier MH., Jr Membrane porters of ATP-binding cassette transport systems are polyphyletic. J Membr Biol. 2009;231:1–10. doi: 10.1007/s00232-009-9200-6. [DOI] [PubMed] [Google Scholar]
- Yen MR, Chen JS, Marquez JL, Sun EI, Saier MH. Multidrug resistance: phylogenetic characterization of superfamilies of secondary carriers that include drug exporters. Methods Mol Biol. 2010;637:47–64. doi: 10.1007/978-1-60761-700-6_3. [DOI] [PubMed] [Google Scholar]
- Yen MR, Choi J, Saier MH., Jr Bioinformatic analyses of transmembrane transport: novel software for deducing protein phylogeny, topology, and evolution. J Mol Microbiol Biotechnol. 2009;17:163–176. doi: 10.1159/000239667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young GB, Jack DL, Smith DW, Saier MH., Jr The amino acid/auxin:proton symport permease family. Biochim Biophys Acta. 1999;1415:306–322. doi: 10.1016/s0005-2736(98)00196-5. [DOI] [PubMed] [Google Scholar]
- Zhai Y, Tchieu J, Saier MH., Jr A web-based Tree View (TV) program for the visualization of phylogenetic trees. J Mol Microbiol Biotechnol. 2002;4:69–70. [PubMed] [Google Scholar]
- Zhu S, Gao B, Tytgat J. Phylogenetic distribution, functional epitopes and evolution of the CSalphabeta superfamily. Cell Mol Life Sci. 2005;62:2257–2269. doi: 10.1007/s00018-005-5200-6. [DOI] [PMC free article] [PubMed] [Google Scholar]