Abstract
Loss-of-function mutations in the EDR1 gene of Arabidopsis confer enhanced resistance to Golovinomyces cichoracearum (powdery mildew). Disease resistance mediated by the edr1 mutation is dependent on an intact salicylic acid (SA) signaling pathway, but edr1 mutant plants do not constitutively express the SA-inducible gene PR-1, and are not dwarfed. To identify other components of the EDR1 signaling network, we screened for mutations that enhanced the edr1 mutant phenotype. Here we describe an enhancer of edr1 mutant, eed3, which forms spontaneous lesions in the absence of pathogen infection, constitutively expresses both SA and methyl-jasmonate (JA) inducible defense genes and is dwarfed. Positional cloning of eed3 revealed that the mutation causes a premature stop codon in GLUCAN SYNTHASE-LIKE 5 (GSL5, also known as POWDERY MILDEW RESISTANT 4, PMR4), which encodes a callose synthase required for pathogen-induced callose production. Significantly, gsl5 single mutants do not constitutively express PR-1 or AtERF1 (a JA-inducible gene) and are not dwarfed. Thus, loss of both EDR1 and GSL5 function has a synergistic effect. Our data suggest that EDR1 and GSL5 negatively regulate SA and JA production and/or signaling by independent mechanisms, and that negative regulation of defense signaling by GSL5 may be independent of callose production.
To identify genes that regulate plant defense responses, we previously screened for Arabidopsis thaliana mutants with increased resistance to virulent pathogens. A mutant that displayed enhanced disease resistance (edr1) to Pseudomonas syringae and Golovinomyces cichoracearum (powdery mildew) was found (Frye and Innes, 1998). The edr1-1 mutation causes a premature stop codon in the EDR1 gene, which encodes a protein kinase with similarity to CTR1 (Frye et al., 2001), a negative regulator of ethylene responses (Kieber et al., 1993; Cao et al., 1997). The enhanced resistance of the edr1 mutant is suppressed by mutations that block salicylic acid (SA) perception (npr1/nim1), or reduce SA production (sid2, eds1 and pad4; (Frye et al., 2001; Tang et al., 2005). The transgenic expression of NahG, which lowers endogenous SA levels, also eliminates the edr1-mediated enhanced disease resistance phenotype (Frye et al., 2001). In contrast to the clear requirement for SA signaling in edr1-mediated resistance, neither ethylene nor jasmonic acid appear to be necessary, as mutations in the ETHYLENE INSENSITIVE 2 (EIN2) gene or CORONATINE INSENSITIVE 1 (COI1) gene do not alter edr1-mediated disease resistance (Frye et al., 2001).
SA is a central regulator of plant defense responses (Delaney et al., 1994). Exogenous application of SA induces a heightened state of resistance to numerous pathogens (Vernooij et al., 1994), while reduction of endogenous SA levels makes plants more susceptible to both virulent and avirulent pathogens (Delaney et al., 1994; Nawrath and Metraux, 1999; Wildermuth et al., 2001). SA levels are elevated during pathogen infection and appear to enhance resistance in part by potentiating production of reactive oxygen species (Shirasu et al., 1997), along with inducing a large suite of defense genes (Glazebrook et al., 2003; Tsuda et al., 2008). Understanding how SA levels are regulated, and how SA regulates defense responses is a major goal of the plant-microbe interaction field. We wish to understand the link between EDR1 function and activation of SA-mediated defenses.
Although the EDR1 protein appears to function as a negative regulator of SA-mediated defenses, the edr1 mutant does not contain significantly elevated SA levels and it does not constitutively express the PR-1 gene (Frye et al., 2001), which is often used as an indicator of SA signaling. Instead, the edr1 mutant behaves as if it was primed for defense gene induction, similar to plants treated with beta-aminobutyric acid (BABA) (van Hulten et al., 2006). This primed state leads to a more rapid and more robust defense response upon pathogen infection (Frye and Innes, 1998; Frye et al., 2001; Conrath et al., 2002; Ton et al., 2005). BABA-induced priming is dependent in part on the hormone abscisic acid (ABA), as Arabidopsis mutants defective in ABA synthesis (aba1-5) or ABA signaling (abi4-1) are impaired in BABA-induced resistance to the necrotrophic fungal pathogens Alternaria brassicicola and Plectosphaerella cucumerina (Ton and Mauch-Mani, 2004). The role of ABA in this resistance appears to be linked to production of callose, a pathogen-induced β-1,3-glucan thought to function as a physical and chemical barrier to entry of fungal hyphae. BABA-treated plants show an earlier and more pronounced accumulation of callose upon pathogen infection, but callose accumulation is absent in BABA-treated abi4-1 plants. Furthermore, a mutation in the callose synthase gene GLUCAN SYNTHASE-LIKE GSL5 (also known as POWDERY MILDEW RESISTANT 4; PMR4 but referred to as GSL5 in the remainder of this paper), blocks BABA-induced callose accumulation and resistance to A. brassicicola and P. cucumerina (Ton and Mauch-Mani, 2004).
The role of ABA signaling and callose synthesis in edr1-mediated disease resistance is not yet clear. However, the edr1 mutant displays enhanced sensitivity to ABA in a seedling germination assay (Wawrzynska et al., 2008). In addition, edr1-mediated resistance can be suppressed by a gain-of-function mutation in the KEEP ON GOING (KEG) gene (Wawrzynska et al., 2008), which encodes a RING finger E3-ubiquitin ligase thought to target the transcription factor ABSCISIC ACID-INSENSITIVE5 (ABI5) for proteasome-mediated degradation (Stone et al., 2006). Loss-of-function keg mutant plants accumulate high levels of ABI5 and are hypersensitive to ABA (Stone et al., 2006). The gain-of-function keg4 mutation, which suppresses edr1-mediated resistance, also suppresses the hypersensitivity of edr1 to ABA (Wawrzynska et al., 2008). Other phenotypes of the edr1 mutant are enhanced drought-induced growth inhibition and enhanced ethylene-induced senescence (Frye et al., 2001; Tang et al., 2005), which are consistent with an overall enhanced sensitivity of edr1 plants to ABA (Wawrzynska et al., 2008). There may also be a link to callose synthesis, as counterintuitively, loss of GSL5 function, renders Arabidopsis resistant to powdery mildew infection via a mechanism that highly resembles edr1-mediated resistance (Jacobs et al., 2003; Nishimura et al., 2003). Similar to infection of edr1 plants, infection of pmr4-1 plants with G. cichoracearum induces large lesions that are not observed in wild-type plants, and a more rapid induction of the defense gene PR1 (Vogel and Somerville, 2000). As observed with edr1, pmr4-1-mediated resistance can be suppressed by mutations that reduce SA synthesis or SA perception (Nishimura et al., 2003). Despite these phenotypic similarities, the mechanistic links between callose synthesis, ABA signaling, SA signaling, and EDR1 function remain unknown.
To identify additional components of the EDR1 signaling network, we performed a mutant screen to identify both enhancers and suppressors of the edr1 mutant phenotype (Wawrzynska et al., 2008). Such edr1 suppressors/enhancers may uncover additional components of the SA regulatory system, pathways that interact with the EDR1 pathway in the regulation of defense responses and/or senescence as well as downstream signaling components. Here, we report the identification of an edr1 enhancer mutant that shows severe growth retardation, necrotic lesions, and elevated PR-1 expression even in the absence of powdery mildew infection. Mapping of this enhancer mutation revealed that it was caused by a premature stop codon in the callose synthase gene GSL5, indicating that simultaneous loss of EDR1 and GSL5 has a synergistic effect on the regulation of defense responses.
RESULTS
The eed3 mutation causes retarded growth and necrotic lesion phenotypes that are dependent on the edr1 mutation
We screened a population of Arabidopsis mutants (60,000 M2 seedlings derived from ~3500 ethyl methane-sulfonate mutagenized M1 edr1 plants) to identify mutations that suppressed or enhanced the edr1 disease resistance phenotype (Wawrzynska et al., 2008). We identified nine edr1 enhancers, i.e. plants displaying necrotic lesions prior to powdery mildew infection. The mutant with the severest phenotype was selected for further characterization, and was named eed3 for enhancer of edr1 mutant 3. In comparison to wild-type Col-0 and edr1 plants, edr1eed3 double mutant plants were dwarfed and displayed large necrotic lesions, mainly on the older leaves starting to appear after 4–5 weeks of growth under short day (9 hrs) conditions (Fig. 1). Crosses with WT Col-0 and edr1 plants revealed that the edr1eed3 mutant phenotype required both mutations. We therefore proceeded with isolation of the EED3 gene.
Fig. 1.
Phenotype of the edr1eed3 mutant. A, edr1 and edr1eed3 plants at 5 weeks of growth under short days. B, Close-up of the edr1eed3 mutant. Note the large necroses on the edges of older leaves and small necroses on the younger leaves.
Identification of the EED3 gene
To map the eed3 mutation, we crossed the edr1eed3 mutant to Landsberg erecta (Ler) and then selected F2 plants that displayed a normal edr1 mutant phenotype (i.e. wild-type appearance prior to inoculation with powdery mildew, then development of lesions after inoculation). These plants were confirmed to be homozygous for edr1 using PCR and then allowed to self. The resulting F3 families were scored for segregation of the dwarfed phenotype. 218 F3 plants (derived from selfing 37 F2 parents homozygous for edr1) displaying the dwarf phenotype were selected for DNA isolation and scoring of molecular markers (see Methods).
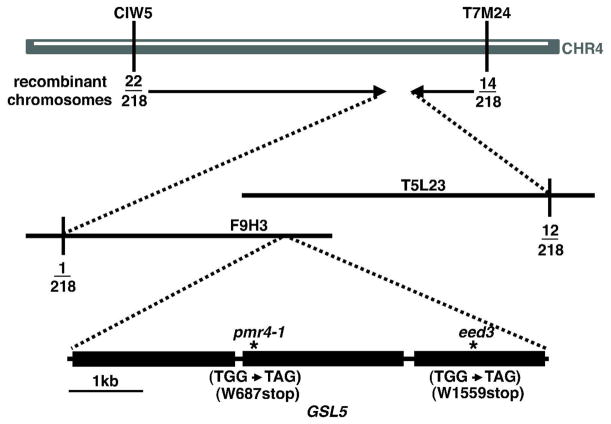
Initially, the eed3 mutation was mapped to a region between the microsatellite markers CIW5 and T7M24 on chromosome 4 (Fig. 2). To further localize the mutated gene, we created PCR markers at intervals between these two markers using small insertions/deletions that are polymorphic between Ler and Col-0 (Jander et al., 2002). Scoring of these additional markers localized the mutation to a 160-kb interval (covering the 3′-end of bacterial artificial chromosome (BAC) F9H3 (GenBank accession AF071527) and the 5′-end of BAC T5L23 (GenBank accession AC005142) defined by one recombinant at the left border and twelve recombinants at the right border (Fig. 2). This region harbors 47 genes (from At4g03390 to At4g03730). Among them we found the GSL5 gene (At4g03550) encoding callose synthase. Loss-of-function mutants of GLS5 are known to have enhanced resistance to powdery mildew (Jacobs et al., 2003; Nishimura et al., 2003). We therefore amplified GSL5 from the edr1eed3 mutant and sequenced it. Sequencing revealed a G to A transition in the 3rd exon, which caused a premature stop codon at amino acid 1559 of this 1780 amino acid protein. Because the premature stop codon is 24 amino acids upstream of an RXTG motif believed to be involved in UDP-glucose binding in this enzyme family (Inoue et al., 1996; Ostergaard et al., 2002), and eliminates the last five transmembrane domains, it is highly unlikely that the truncated protein would have any callose synthase activity.
Fig. 2.
Positional cloning of EED3. Vertical lines indicate the positions of markers that defined the eed3 genetic interval. Fractions indicate the number of recombination events identified between the indicated markers and the EED3 gene out of the total number of chromosomes scored. BAC clones spanning the region to which eed3 was mapped are indicated. Sequencing of candidate genes in this interval revealed a G to A transition in At4g03550 (GSL5) resulting in a premature stop codon at tryptophan 1559. Also shown is the genomic structure of the GSL5 gene with exons indicated by black boxes and positions of the pmr4-1 and eed3 mutations indicated by the * symbol.
Phenotype of edr1 pmr4-1 double mutant
To confirm that loss of GSL5 function enhances the edr1 phenotype, we crossed edr1 plants to the previously described pmr4-1 mutant, which contains a stop codon at amino acid 687, near the beginning of the second exon (Fig. 2) (Vogel and Somerville, 2000; Nishimura et al., 2003). In the F2 generation, we observed dwarfed plants with lesions segregating at a ratio of 4:70, which is not significantly different from the expected ratio of 1:15 (chi-squared = 0.09, P=0.76). Genotyping of three dwarfed plants revealed that all were homozygous for both edr1 and pmr4-1, while genotyping of three sibling plants with wild-type stature showed that none were homozygous for both mutations. We thus conclude that the loss of GSL5 function does indeed dramatically enhance the edr1 phenotype and that pmr4-1 and edr1 mutations have a synergistic effect.
Powdery mildew growth is inhibited in edr1eed3 plants
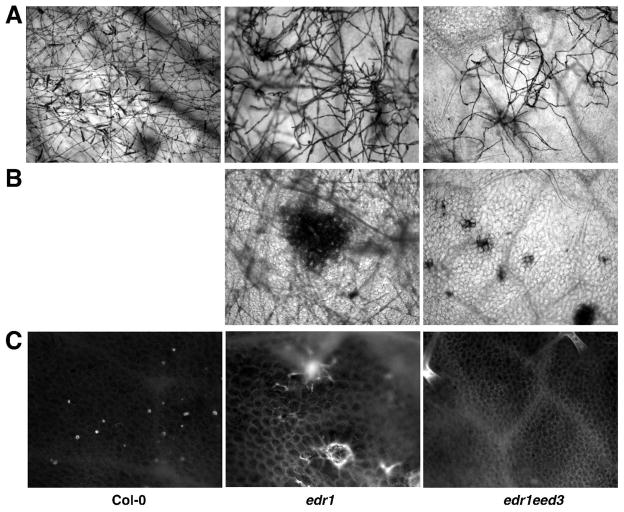
To further characterize edr1eed3 double mutants, we looked at the development of G. cichoracearum and host cell death using trypan blue staining. We also examined callose deposition during powdery mildew infection with aniline blue staining. Spores of G. cichoracearum germinated and produced appressorial germ tubes on WT Col-0, edr1 and edr1eed3 leaves 1 day after inoculation (data not shown). By 3 days after inoculation, G. cichoracearum developed extensive branched hyphae on both Col-0 and edr1 leaves, while fungal growth was significantly inhibited on leaves of edr1eed3 (data not shown). By 5 days, extensive hyphae nearly covered both Col-0 and edr1 leaves, but not edr1eed3 leaves (Fig. 3A). Many conidiophores formed on Col-0 leaves while significantly fewer formed on edr1 leaves, consistent with our previous analyses of the edr1 mutant (Frye and Innes, 1998). These observations demonstrated that the growth of G. cichoracearum was affected at a late stage of the infection process in edr1 mutant, but at an earlier stage in the edr1eed3 mutant. The edr1 mutant displayed large patches of dead mesophyll cells 5 days after infection (Fig. 3B), while edr1eed3 plants displayed large necroses even before infection. This massive mesophyll cell death prior to infection was not observed on WT Col-0 leaves, nor in pmr4-1 mutant leaves (Nishimura et al., 2003; Vogel and Somerville, 2000).
Fig. 3.
Response of Arabidopsis WT Col-0, edr1 and edr1eed3 plants to G. cichoracearum. A, Fungal hyphae growing on the surface of WT Col-0, edr1 and edr1eed3 leaves 5 days after infection, stained with trypan blue. Note extensive hyphae and conidia produced on WT Col-0 leaves 5 days after infection, but very few conidia on edr1 leaves and limited hyphae growth on edr1eed3 leaves. B, Extensive mesophyll cell death in edr1 and edr1eed3 mutant leaves 5 days after infection revealed by trypan blue staining C, Aniline blue staining for callose deposition in mesophyll cell walls of WT Col-0, edr1 and edr1eed3 leaves 5 days after inoculation.
Both WT Col-0 and edr1 plants displayed punctate staining of callose in the cell walls of epidermal cells 5 days after inoculation (Fig. 3C); however, only edr1 showed callose staining in large clumps of mesophyll cells (Fig. 3). This is consistent with our previous results (Frye and Innes, 1998). As expected, no callose deposition was observed in the leaves of edr1eed3 (Fig. 3C).
GSL5 negatively regulates defense gene expression
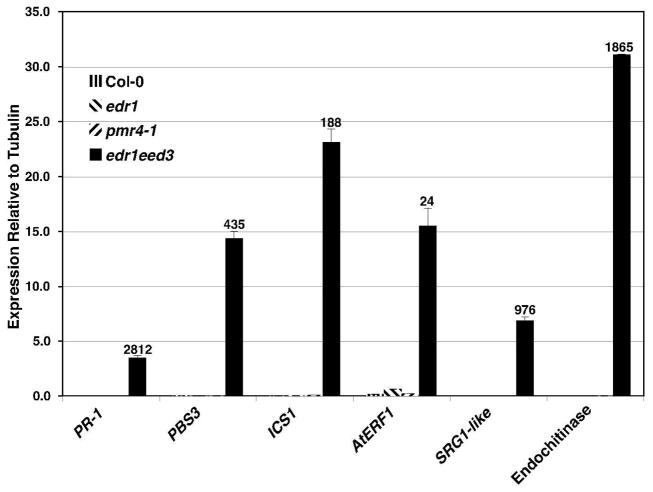
Because the pmr4-1 mutant displays a more rapid induction of SA-inducible genes upon pathogen attack (Vogel and Somerville, 2000), and because mutants with constitutively elevated SA levels typically have retarded growth (Jirage et al., 2001; Mauch et al., 2001; Andreasson et al., 2005; Mateo et al., 2006), we decided to analyze the edr1eed3 double mutant for expression of defense genes to see if these were constitutively elevated. Specifically, we performed quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) analysis on PR-1 (At2g14610, a marker for SA-dependent gene expression; (Bowling et al., 1994)), PBS3 (At5g13320, involved in regulation of SA levels; (Nobuta et al., 2007)), ICS1 (At1g74710, involved in SA biosynthesis; (Wildermuth et al., 2001)), AtERF1 (At4g17500, an APETALA2/ETHYLENE RESPONSE FACTOR transcription factor family member that is strongly inducible by methyl jasmonate (Nemhauser et al., 2006) and which is upregulated in the edr1 mutant; (Wawrzynska et al., 2008)), SRG1-like (At4g10500, a stress-induced ascorbate oxidase upregulated in pmr4-1 after powdery mildew infection (Nishimura et al., 2003)), and an endochitinase (At2g43570, also upregulated in pmr4-1 following powdery mildew infection (Nishimura et al., 2003) and by the SA-responsive transcription factor WRKY70 (Li et al., 2004)). RNA was isolated from rosette leaves of 4.5 week old plants grown in the absence of pathogens under 9 hr days, which was approximately 1 week after lesions began appearing on edr1eed3 plants. All six of these defense genes were dramatically elevated in edr1eed3 plants compared to WT Col-0, edr1, and pmr4-1 plants (Fig. 4). This result suggests that callose or callose synthase acts to suppress both SA and JA-induced defense signaling in edr1 mutant plants and that loss of both PMR4 and EDR1 has a synergistic effect on both the SA and JA signaling pathways.
Fig. 4.
Synergistic effect of edr1 and eed3 mutations on both SA- and JA-inducible defense gene expression. qRT-PCR analysis was performed on the indicated defense genes in WT Col-0, edr1, pmr4-1 and edr1eed3 mutants grown for 4.5 weeks under 9 hr days. Total RNA was isolated from leaves. Values are the average with SD of three technical replicates and are normalized relative to the TUA3 alpha tubulin gene. Numbers above the edr1eed3 bar indicate the fold increase in expression relative to expression in the edr1 or pmr4-1 single mutant (the mutant with the higher expression was used as the denominator). The whole experiment was repeated three times with similar results. AGI numbers for the indicated genes are: PR-1, At2g14610; PBS3, At5g13320; ICS1, At1g74710; AtERF1, At4g17500; SRG1-like, At4g10500; endochitinase, At2g43570.
DISCUSSION
Arabidopsis contains a family of 12 putative callose synthase genes (GSL1 through GSL12) (Enns et al., 2005). Very limited information is available regarding the biological functions of individual GSL family members and the role of callose in plant development. Presumably, each member of the family mediates the synthesis of callose in different tissues and/or under different environmental/developmental conditions (Richmond and Somerville, 2000; Verma and Hong, 2001). GSL5 appears to be the main biosynthetic enzyme responsible for callose deposition in response to biotic, abiotic, and chemical stresses (Nishimura et al., 2003). However, GSL5 transcript can be detected in all plant organs, including root, rosette leaf, stems and flowers, and is apparently partially redundant in function with GSL1 during normal plant development (Enns et al., 2005). Plants heterozygous for a gsl1 mutation and homozygous for a gsl5 mutation are dwarfed and nearly infertile. The infertility is caused in part by non-viability of gsl1gsl5 pollen, which lack a callose wall between the microspores of the pollen tetrad (Enns et al., 2005). Based on our experiments and those of (Nishimura et al., 2003), we conclude that during normal plant development GSL5 also functions as a negative regulator of SA and JA biosynthesis and/or signaling.
A gsl5 single mutant (pmr4-1) grows at the same rate as wild-type and does not outwardly appear to be stressed, although when grown under short-day conditions leaves are epinastic (Vogel and Somerville, 2000). Unlike previously identified classes of Arabidopsis disease-resistant mutants, cpr, lsd, and acd (Bowling et al., 1994; Dietrich et al., 1994), gsl5 plants do not constitutively express PR-1. However, when a gsl5 mutation was combined with the edr1 mutation, we observed a strong induction of the SA biosynthetic gene ICS1 and the SA-inducible gene PR-1 (Fig. 4). We thus conclude that the EDR1 kinase must function in part to negatively regulate SA biosynthesis in the gsl5 mutant background. When infected with powdery mildew, gsl5 mutants develop lesions reminiscent of hypersensitive cell death (Vogel and Somerville, 2000), similar to those observed for the edr1 mutant (Frye and Innes, 1998). Also like the edr1 mutant, blocking the SA signaling pathway in a gsl5 mutant suppresses the lesion phenotype and the enhanced resistance to powdery mildew (Nishimura et al., 2003). This indicates that the enhanced resistance of gsl5 as well as of edr1 is caused by a hyperactive SA response, and not a direct result of the loss of callose in gsl5 mutants, or kinase action in edr1 mutants. Combining gsl5 and edr1 mutations has a synergistic effect on SA signaling, which suggests that GSL5 and EDR1 may negatively regulate SA signaling by separate pathways.
GSL5 mRNA was shown to accumulate in SA-treated Arabidopsis (Ostergaard et al., 2002) as well as in the Arabidopsis mpk4 mutant, which exhibits constitutive systemic acquired resistance and high levels of SA (Petersen et al., 2000). Nishimura et al. (2003) hypothesised that rapid callose deposition during the early stages of infection initiated by SA may then in turn repress later defense responses that may potentially be harmful for the plant. However, our data suggest that callose deposition may not be the primary mechanism of defense gene supression by GSL5. We conclude this because in uninfected edr1 mutant plants, little to no callose is detectable by aniline blue staining, and there is very low level expression of PR-1, yet loss of GSL5 function in the edr1 mutant background leads to high level expression of PR-1 even in the absence of pathogen infection (Fig. 4). Since there was no detectable callose in the edr1 mutant leaves prior to infection, it seems unlikely that callose could have been responsible for suppressing PR-1 expression in these same leaves. If callose is not directly responsible for suppression of SA signaling in the edr1 mutant, it would suggest that GSL5 has functions in addition to stress-induced callose biosynthesis. As mentioned above, GSL5 likely works in concert with GSL1 during normal plant development and likely contributes to normal cellular homeostasis (Enns et al., 2005). We speculate that it is this disruption in normal cellular homeostasis, when combined with loss of EDR1 function, that leads to massive induction of SA signaling.
Surprisingly, we also observed highly elevated expression of the JA-inducible gene AtERF1 in the edr1eed3 double mutant (Fig. 4). This is surprising because SA and JA signaling pathways often function antagonistically to each other (Kunkel and Brooks, 2002), and the pmr4-1 mutant has been shown to produce less JA than wild-type plants during infection by Alterneria brassicicola (Flors et al., 2008). However, we have previously observed that edr1 single mutant plants have modestly elevated expression of AtERF1 as well as enhanced responsiveness to exogenous abscisic acid (Wawrzynska et al., 2008), suggesting that EDR1 negatively regulates multiple hormone signaling pathways. We hypothesize that loss of GSL5 somehow induces SA and JA signaling that in the absence of EDR1 function, reaches deleterious levels. In this context, it is noteworthy that overexpression of AtERF1 is known to cause dwarfing in Arabidopsis (Pre et al., 2008), as does constitutive elevation of SA signaling (Bowling et al., 1994), thus the severely dwarfed stature of the edr1eed3 mutant may be a consequence of elevating both pathways.
MATERIALS AND METHODS
Plant growth conditions
Arabidopsis thaliana plants were grown in growth rooms under a 9-h light/15-h dark cycle at 23°C as described previously (Frye and Innes, 1998).
Pathogen infections
Golovinomyces cichoracearum strain UCSC1 was maintained on hyper-susceptible A. thaliana pad4-2 mutant plants. Plants were inoculated between 4 and 6 weeks of age by gently brushing the leaves of diseased plants and healthy plants together to pass the conidia (asexual spores).
Genetic and physical mapping of eed3
Genetic mapping was accomplished using an F2 population derived from a cross between the eed3 mutant (Columbia genotype, Col-0) and Landsberg erecta (Ler). F2 seeds were planted and scored for disease resistance to G. cichoracearum as described above. Plants displaying an edr1 phenotype were confirmed to be homozygous for the edr1 mutation using a PCR-based assay, then allowed to self to generate F3 families. Progeny from 37 F3 families were planted and scored for the eed3 phenotype (dwarfed plants with lesions). Genomic DNA was isolated from a total of 218 dwarfed F3 plants and scored with published microsatellite markers. This initial mapping localized the eed3 mutation between molecular markers CIW5 and T7M24 on chromosome 4. New molecular markers at intervals between these two markers were developed using the Monsanto Col-0 and Ler polymorphism database (http://www.arabidopsis.org/Cereon/index.jsp; primer sequences available upon request). These analyses localized the eed3 mutation to a 160 kb interval on chromosome 4, spanning Arabidopsis BAC clones F9H3 and T5L23.
DNA sequencing
The GSL5 gene was amplified from the eed3 mutant by PCR and directly sequenced. All sequencing reactions were performed using BigDye Terminator Kits (Applied Biosystems, Foster City, CA, USA) and separated on an ABI 3730 automated DNA sequencer (Applied Biosystems).
Analysis of fungal infection, callose deposition, and dead cells in Arabidopsis leaves
Fungal structures and dead plant cells were stained using alcoholic trypan blue (Koch and Slusarenko, 1990). Callose deposition was detected by aniline blue staining as described by (Adam and Somerville, 1996). Stained leaves were mounted under coverslips with 50% glycerol and observed with a Nikon e800 microscope.
qRT-PCR analysis
Plants were grown under 9 hr days and RNA was isolated from leaves of WT Col-0, edr1, pmr4-1 and edr1eed3 at 4.5 weeks. Total RNA was isolated using the Qiagen RNeasy kit and treated with DNase (Invitrogen) to remove DNA contamination. The High Capacity Reverse Transcriptase Kit (Applied Biosystems) was utilized to obtain cDNA and the samples purified with Qiagen QIAquick PCR Purification Kit. qRT-PCR was performed using primers for PR-1 (At2g14610; 5′-CATGGGACCTACGCCTACC-3′ and 5′-TTCTTCCCTCGAAAGCTCAA-3′), PBS3 (At5g13320; 5′-ACTGGATTCTTGCTAAGTTCTG-3′ and 5′-CACACCTTTCACATGCTTGGTT-3′), ICS1 (At1g74710; 5′-AAACACGCCTGAGAGACTATT-3′ and 5′-TCTTTCGGACTGGTTAGTAAGT-3′), AtERF1 (At4g17500; 5′-GAGATTTGCCGTTGAAAGAA-3′ and 5′-GTCGGAAGACGAAGAAGACG-3′), SRG1-like (At4g10500; 5′-GACCAAATGCAGGTCATAAG-3′ and 5′-GGGAAATAGAAAGTCGGAAT-3′), and endochitinase (At2g43570; 5′-AAGAAACAGGGTTCATGTGT-3′ and 5′-ACTCTGGTTTCTCTGTGTCG-3′). An alpha-tubulin gene, TUA3 (At5g19770; 5′-GTATTGAACGCATCGTGTG-3′ and 5′-TGGGAGCTTTACTGTCTCGAA-3′) was used as a control for normalizing the amount of cDNA. Takara SYBR Premix Extaq was used for all qRT-PCR reactions and samples analyzed on an Mx3000P qPCR instrument (Stratagene).
Acknowledgments
We thank the Arabidopsis Biological Resource Center at Ohio State for providing pmr4-1 mutant seed (ABRC stock number CS3858). This work was supported by a grant from the National Institute of General Medical Sciences of the NIH (grant number R01 GM063761 to R.W. I.).
LITERATURE CITED
- Adam L, Somerville SC. Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana. Plant J. 1996;9:341–356. doi: 10.1046/j.1365-313x.1996.09030341.x. [DOI] [PubMed] [Google Scholar]
- Andreasson E, Jenkins T, Brodersen P, Thorgrimsen S, Petersen NH, Zhu S, Qiu JL, Micheelsen P, Rocher A, Petersen M, Newman MA, Bjorn Nielsen H, Hirt H, Somssich I, Mattsson O, Mundy J. The MAP kinase substrate MKS1 is a regulator of plant defense responses. Embo J. 2005;24:2579–2589. doi: 10.1038/sj.emboj.7600737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X. A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell. 1994;6:1845–1857. doi: 10.1105/tpc.6.12.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- Conrath U, Pieterse CM, Mauch-Mani B. Priming in plant-pathogen interactions. Trends Plant Sci. 2002;7:210–216. doi: 10.1016/s1360-1385(02)02244-6. [DOI] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, Ryals J. A central role of salicylic acid in plant disease resistance. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL. Arabidopsis mutants simulating disease resistance response. Cell. 1994;77:565–577. doi: 10.1016/0092-8674(94)90218-6. [DOI] [PubMed] [Google Scholar]
- Enns LC, Kanaoka MM, Torii KU, Comai L, Okada K, Cleland RE. Two callose synthases, GSL1 and GSL5, play an essential and redundant role in plant and pollen development and in fertility. Plant Mol Biol. 2005;58:333–349. doi: 10.1007/s11103-005-4526-7. [DOI] [PubMed] [Google Scholar]
- Flors V, Ton J, van Doorn R, Jakab G, Garcia-Agustin P, Mauch-Mani B. Interplay between JA, SA and ABA signalling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. Plant J. 2008;54:81–92. doi: 10.1111/j.1365-313X.2007.03397.x. [DOI] [PubMed] [Google Scholar]
- Frye CA, Innes RW. An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell. 1998;10:947–956. doi: 10.1105/tpc.10.6.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye CA, Tang D, Innes RW. Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc Natl Acad Sci U S A. 2001;98:373–378. doi: 10.1073/pnas.98.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J, Chen W, Estes B, Chang HS, Nawrath C, Metraux JP, Zhu T, Katagiri F. Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 2003;34:217–228. doi: 10.1046/j.1365-313x.2003.01717.x. [DOI] [PubMed] [Google Scholar]
- Inoue SB, Qadota H, Arisawa M, Anraku Y, Watanabe T, Ohya Y. Signaling toward yeast 1,3-beta-glucan synthesis. Cell Struct Funct. 1996;21:395–402. doi: 10.1247/csf.21.395. [DOI] [PubMed] [Google Scholar]
- Jacobs AK, Lipka V, Burton RA, Panstruga R, Strizhov N, Schulze-Lefert P, Fincher GB. An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell. 2003;15:2503–2513. doi: 10.1105/tpc.016097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL. Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 2002;129:440–450. doi: 10.1104/pp.003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D, Zhou N, Cooper B, Clarke JD, Dong X, Glazebrook J. Constitutive salicylic acid-dependent signaling in cpr1 and cpr6 mutants requires PAD4. Plant J. 2001;26:395–407. doi: 10.1046/j.1365-313x.2001.2641040.x. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Koch E, Slusarenko A. Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell. 1990;2:437–445. doi: 10.1105/tpc.2.5.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol. 2002;5:325–331. doi: 10.1016/s1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET. The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell. 2004;16:319–331. doi: 10.1105/tpc.016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo A, Funck D, Muhlenbock P, Kular B, Mullineaux PM, Karpinski S. Controlled levels of salicylic acid are required for optimal photosynthesis and redox homeostasis. J Exp Bot. 2006;57:1795–1807. doi: 10.1093/jxb/erj196. [DOI] [PubMed] [Google Scholar]
- Mauch F, Mauch-Mani B, Gaille C, Kull B, Haas D, Reimmann C. Manipulation of salicylate content in Arabidopsis thaliana by the expression of an engineered bacterial salicylate synthase. Plant J. 2001;25:67–77. doi: 10.1046/j.1365-313x.2001.00940.x. [DOI] [PubMed] [Google Scholar]
- Nawrath C, Metraux JP. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell. 1999;11:1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- Nishimura MT, Stein M, Hou BH, Vogel JP, Edwards H, Somerville SC. Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science. 2003;301:969–972. doi: 10.1126/science.1086716. [DOI] [PubMed] [Google Scholar]
- Nobuta K, Okrent RA, Stoutemyer M, Rodibaugh N, Kempema L, Wildermuth MC, Innes RW. The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis. Plant Physiol. 2007;144:1144–1156. doi: 10.1104/pp.107.097691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard L, Petersen M, Mattsson O, Mundy J. An Arabidopsis callose synthase. Plant Mol Biol. 2002;49:559–566. doi: 10.1023/a:1015558231400. [DOI] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, Sharma SB, Klessig DF, Martienssen R, Mattsson O, Jensen AB, Mundy J. Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell. 2000;103:1111–1120. doi: 10.1016/s0092-8674(00)00213-0. [DOI] [PubMed] [Google Scholar]
- Pre M, Atallah M, Champion A, De Vos M, Pieterse CM, Memelink J. The AP2/ERF-domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008;147:1347–1357. doi: 10.1104/pp.108.117523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TA, Somerville CR. The cellulose synthase superfamily. Plant Physiol. 2000;124:495–498. doi: 10.1104/pp.124.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K, Nakajima H, Rajasekhar VK, Dixon RA, Lamb C. Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell. 1997;9:261–270. doi: 10.1105/tpc.9.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell. 2006;18:3415–3428. doi: 10.1105/tpc.106.046532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Christiansen KM, Innes RW. Regulation of plant disease resistance, stress responses, cell death, and ethylene signaling in Arabidopsis by the EDR1 protein kinase. Plant Physiol. 2005;138:1018–1026. doi: 10.1104/pp.105.060400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J, Mauch-Mani B. Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 2004;38:119–130. doi: 10.1111/j.1365-313X.2004.02028.x. [DOI] [PubMed] [Google Scholar]
- Ton J, Jakab G, Toquin V, Flors V, Iavicoli A, Maeder MN, Metraux JP, Mauch-Mani B. Dissecting the beta-aminobutyric acid-induced priming phenomenon in Arabidopsis. Plant Cell. 2005;17:987–999. doi: 10.1105/tpc.104.029728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Glazebrook J, Cohen JD, Katagiri F. Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 2008;53:763–775. doi: 10.1111/j.1365-313X.2007.03369.x. [DOI] [PubMed] [Google Scholar]
- van Hulten M, Pelser M, van Loon LC, Pieterse CM, Ton J. Costs and benefits of priming for defense in Arabidopsis. Proc Natl Acad Sci U S A. 2006;103:5602–5607. doi: 10.1073/pnas.0510213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma DP, Hong Z. Plant callose synthase complexes. Plant Mol Biol. 2001;47:693–701. doi: 10.1023/a:1013679111111. [DOI] [PubMed] [Google Scholar]
- Vernooij B, Uknes S, Ward E, Ryals J. Salicylic acid as a signal molecule in plant-pathogen interactions. Curr Opin Cell Biol. 1994;6:275–279. doi: 10.1016/0955-0674(94)90147-3. [DOI] [PubMed] [Google Scholar]
- Vogel J, Somerville S. Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc Natl Acad Sci U S A. 2000;97:1897–1902. doi: 10.1073/pnas.030531997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzynska A, Christiansen KM, Lan Y, Rodibaugh NL, Innes RW. Powdery mildew resistance conferred by loss of the ENHANCED DISEASE RESISTANCE1 protein kinase is suppressed by a missense mutation in KEEP ON GOING, a regulator of abscisic acid signaling. Plant Physiol. 2008;148:1510–1522. doi: 10.1104/pp.108.127605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]