Abstract
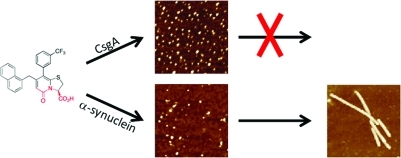
Small organic molecules that inhibit functional bacterial amyloid fibers, curli, are promising new antibiotics. Here we investigated the mechanism by which the ring-fused 2-pyridone FN075 inhibits fibrillation of the curli protein CsgA. Using a variety of biophysical techniques, we found that FN075 promotes CsgA to form off-pathway, non-amyloidogenic oligomeric species. In light of the generic properties of amyloids, we tested whether FN075 would also affect the fibrillation reaction of human α-synuclein, an amyloid-forming protein involved in Parkinson’s disease. Surprisingly, FN075 stimulates α-synuclein amyloid fiber formation as measured by thioflavin T emission, electron microscopy (EM), and atomic force microscopy (AFM). NMR data on 15N-labeled α-synuclein show that upon FN075 addition, α-synuclein oligomers with 7 nm radius form in which the C-terminal 40 residues remain disordered and solvent exposed. The polypeptides in these oligomers contain β-like secondary structure, and the oligomers are detectable by AFM, EM, and size-exclusion chromatography (SEC). Taken together, FN075 triggers oligomer formation of both proteins: in the case of CsgA, the oligomers do not proceed to fibers, whereas for α-synuclein, the oligomers are poised to rapidly form fibers. We conclude that there is a fine balance between small-molecule inhibition and templation that depends on protein chemistry.
Introduction
Amyloid fibrils are β-sheet-rich protein structures associated with human neurodegenerative diseases, such as Alzheimer’s and Parkinson’s.1 The common structural element of these fibrils is the cross-β conformation, that is, β-sheets that are packed perpendicular to the fiber axis. Most amyloid fibrils form via nucleation-dependent pathways that involve oligomeric, prefibrillar structures.2 α-Synuclein is a 140-residue protein involved in Parkinson’s disease, a condition that affects 2% of the population over 60 years of age.3 The function of α-synuclein is unknown; it is an intrinsically unstructured protein although α-helical structure is observed in the presence of vesicles or membranes.4 Residues 50–100 in the synuclein sequence appear to be the primary aggregation-promoting region.4 In Parkinson’s disease, the process of aggregation of α-synuclein from monomers, via oligomeric intermediates, into amyloid fibrils is considered to be the disease-causative mechanism. The mature amyloid fibers may not be the source of α-synuclein mediated cytotoxicity, however; the transient oligomeric structures have been shown to be toxic in vivo.3 Synuclein fibrils accumulate in cytosolic inclusions called Lewy bodies in the brain of Parkinson’s disease patients.5
Amyloid formation is associated not only with disease: nature employs the amyloid structure for a number of functions.6 These functional amyloid fibrils have been described in a number of organisms spanning the diversity of cellular life. In particular, microbial functional amyloids are major components of the extracellular matrix that promotes biofilm formation and other community behaviors.6 Among these, the amyloid-forming properties of Escherichia coli CsgA are among the most studied.7 The extracellular curli fibrils, a common component of bacterial biofilms, are composed of mainly CsgA and are attached to the surface of the bacteria through the membrane-bound CsgB protein, which acts as a “seed” for polymerization of CsgA.8 CsgA is an unstructured monomeric protein that rapidly forms amyloid fibrils both in vivo and in vitro.9,10 The expression of CsgA and CsgB is tightly regulated, and specific chaperones prevent fibril formation in the cell prior to export.7
Inhibition of the bacteria’s ability to attach to the host and establish bacterial biofilms is emerging as an attractive new way to prevent bacterial infections.11−13 A number of substituted dihydro thiazolo ring-fused 2-pyridone peptidomimetics14 (general structure 1, Figure 1) have been synthesized and tested both in vivo and in vitro for their ability to inhibit bacterial biofilm formation. One of these compounds, FN075 (also referred to as a curlicide) inhibits CsgA fibril formation both in vitro and in curli-dependent biofilm assays with uropathogenic E. coli.15 It has also been found that FN075 inhibits in vitro fibril formation of the human Aβ peptide.16 However, the molecular mechanism governing FN075-mediated inhibition of amyloid fibers remains unclear. Here, we reveal the mechanism by which FN075 inhibits CsgA fibrillation in vitro, and surprisingly, we discover that FN075 increases the rate of α-synuclein fibrillation. Thus FN075 displays cross-reactivity with diverging activity for two different amyloidogenic proteins. This finding illustrates the importance of assessing cross-reactivity when small molecules are designed to inhibit amyloid-fiber formation.
Figure 1.
Substituted dihydro thiazolo ring fused 2-pyridones (1) have been designed and synthesized to mimic small rigid C-terminal peptides. The peptidomimetic backbone is highlighted in red and the possibilities to introduce substituents on this central fragment via established methods17 is indicated with R1–R4. This study is focused around the compound FN075 known to be an inhibitor of functional amyloids, curli, in uropathogenic E. coli and the analog FN071, with a blocked C-terminal.
Materials and Methods
Compounds
Synthetic schemes of FN075 and FN071 are described in ref (15).
Protein Preparation and NMR Experiments
CsgA and α-Synuclein Fibrillation Assays
FN075 and FN071 stock solutions were prepared at 10–100 mM in dimethyl sulfoxide (DMSO) and diluted to appropriate concentrations in 50 mM potassium phosphate. Purified soluble CsgA was diluted to 10 μM in 50 mM potassium phosphate. Equal volumes of compound and protein were mixed in a 96-well black plate and incubated at 20 or 37 °C. The α-synuclein experiments were performed with continuous agitation (using 2 mm glass beads; pH 7.4, 20 mM Tris, 140 mM NaCl); for CsgA there was agitation every 15 min. In both cases, thioflavin T was added to a final concentration of 20 μM, and fluorescence was measured at 480 nm (excitation 440 nm) on Tecan Infinite or BioTek Hybrid H4 plate reader incubators.18 Control experiments were performed with 0.5% or 0.05% DMSO. Background fluorescence (samples without protein) was subtracted from the signals at each time point. Origin 8.0 was used to fit the data to standard sigmoidal equations to extract lag times.19 Fibrillation curves shown are representative from at least four replicates. Fiber seeds were prepared as described in ref (20).
Atomic Force Microscopy (AFM)
AFM measurements were performed on a PICO PLUS 5500 microscope in tapping mode at room temperature.21 A silicon probe was oscillated at about 330 kHz, and images were collected at a scan rate of 1 Hz. Samples were diluted to approximately 5 μM protein concentration with sterile filtered Milli-Q water and applied to freshly cleaved mica surface, incubated for 5 min, washed 3 times with Milli-Q water and dried with air flow.
Electron Microscopy (EM)
Ten microliter samples from the fibrillation studies were applied to Formvar coated 200 mesh size copper grids, incubated for 5 min, washed with Milli-Q water, and stained with 1% phosphotungstic acid or 1% sodium silicon tungstate. The grids were examined with a Jeol (Tokyo, Japan) model 1230 electron microscope operated at 80 kV. Images were obtained using a Gatan MultiScan 600W camera.
Circular Dichroism
Spectra were recorded on Jasco J-710 and J-810 spectropolarimeters equipped with peltiers (0.1 cm quartz cuvette, 190–300 nm). Buffer was 10 mM (α-synuclein) or 2 mM (CsgA) phosphate at pH 7.4 at 20 and 10 °C.
Size Exclusion Chromatography (SEC)
Protein samples were loaded on a Superdex 200 GL gel filtration column, which was connected to an ÄKTA purifier at 10 °C. The column was equilibrated with 20 mM Tris, 140 mM NaCl, pH 7.4 (α-synuclein), or with 50 mM phosphate buffer (pH 7.4) for CsgA. Elution was followed by absorption at 220, 280, and 320 nm.
Results and Discussion
FN075 (Figure 1) is a strategically designed small molecule with a dihydro thiazolo ring-fused 2-pyridone central fragment containing a peptide-like segment with a free carboxylic acid (highlighted in red in Figure 1). The compounds within this class are designed to mimic a small C-terminal peptide with an extended β-sheet conformation. The ring-fused central fragment provides stability to the β-sheet-like conformation, and substituents can be introduced onto this central fragment to mimic peptide side chain properties. FN075, which is substituted with a large naphthyl substituent and a CF3-phenyl substituent (i.e., mimicking hydrophobic side chains), is designed to target hydrophobic β-sheet regions in proteins. In the related compound FN071 (Figure 1), the free carboxylic acid substituent is exchanged for a neutral methyl ester thus keeping the peptide-like backbone but not having a free and charged C-terminal. FN075 potently inhibits CsgA aggregation in vitro and curli formation in vivo.15 To better understand how FN075 interferes with CsgA amyloid formation, we assessed CsgA biophysical properties in the presence and absence of FN075. Since amyloid fibers made of bacterial and human proteins display generic properties, we also tested the interaction of FN075 with the human amyloid-forming protein α-synuclein.
CsgA Fibrillation Involves an Intermediate
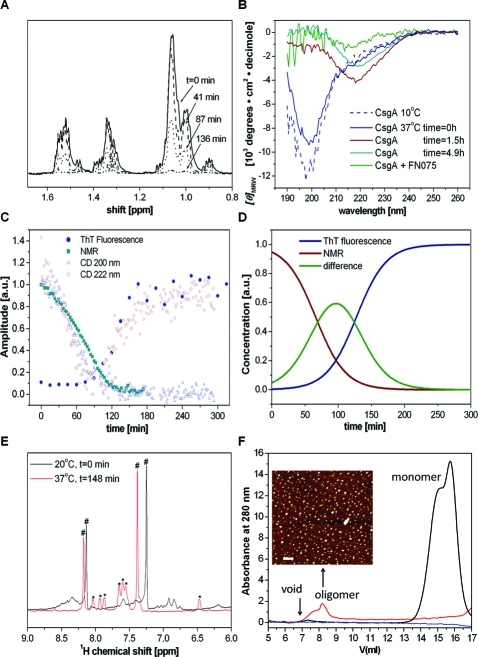
Fibril formation is readily followed by the amyloid-specific dye thioflavin T (ThT): the fluorescence of ThT is increased in the presence of amyloid fibrils.22 A typical fibrillation curve has a sigmoid shape composed of a lag phase, a growth phase and a stationary phase. We studied the intrinsic polymerization reaction of CsgA at 37 °C by a combination of far-UV circular dichroism (CD), one-dimensional NMR, and ThT signals (Figure 2). The depletion of monomers was followed by the disappearance of NMR peaks in 1H spectra. Amyloid formation was probed by ThT emission. Finally, changes in secondary structure were followed by far-UV CD. At 200 nm, the random coil signal dominates, and it is found to disappear with time. In contrast, the signal at 220 nm is found to increase with time. There is a striking overlap in the kinetic CD traces (at 200 and 220 nm, respectively) with the depletion of monomer (from NMR data) and fiber appearance (from ThT data). Combining CD, NMR, and ThT data as a function of time reveals that the monomer disappears prior to fiber appearance and an intermediate species is present at transitional time points (Figure 2C,D).
Figure 2.
Time-dependent structural conversion of CsgA from monomer to β-rich polymer. (A) Depletion of monomeric CsgA during the fibrillation reaction probed by 1H NMR at 37 °C. The figure is an expansion of the aliphatic region in one-dimensional spectra. (B) Time-dependent far-UV CD acquired at 37 °C. The spectrum at 10 °C is a reference of monomeric CsgA; fibrillation is induced by a temperature jump to 37 °C (4.5 μM CsgA and in one sample also 45 μM FN075). (C) Overlay of kinetic traces from NMR, CD, and ThT fluorescence. (D) Sigmodial fits of monomer depletion (from NMR) and fiber build-up (from ThT). The analysis reveals the presence of an intermediate during the reaction. (E) 1H NMR expanded in the amide region upon FN075 addition to CsgA (450 μM FN075, 45 μM CsgA): (∗) FN075 peaks, (#) imidazole (small amount leftover from purification). (F) Size exclusion chromatograms of CsgA in the absence and presence of FN075. Ten micromolar freshly isolated (black) or overnight incubated protein (at room temperature) with 100 μM (red) or without (blue) FN075. Inset: AFM of the oligomer fraction (red trace) (bar 200 nm).
FN075 Stabilizes CsgA Oligomers
Upon addition of FN075 to CsgA at 37 °C, as expected from earlier work, fiber formation is delayed signficantly (Figure S1, Supporting Information).15 However, NMR reveals that upon FN075 addition, there is rapid formation of large soluble oligomeric assemblies as all the CsgA NMR peaks disappear (due to their high molecular weight and slow tumbling rate) (Figure 2E). The formation of CsgA oligomers was also confirmed by size-exclusion chromatography (SEC) where CsgA in the presence of FN075 migrated as molecular weights much larger than that of monomeric CsgA (Figure 2F). Gel electrophoresis of soluble and insoluble fractions reveals that CsgA remains in the soluble fraction, even after long incubation times, in the presence of FN075 (Figure S1, Supporting Information). In contrast, after prolonged incubation in the absence of FN075, CsgA is localized in the insoluble fraction. Analysis of the absorption properties of the content in the oligomeric peak in the SEC profile indicates the presence of both protein and FN075. Atomic force microscopy (AFM) images of this oligomer fraction revealed the presence of oligomeric spherical particles (Figure 2F, inset). In contrast, fresh CsgA elutes at 15–16 mL, which corresponds to the elution volume of a small unstructured protein. CsgA incubated overnight does not exhibit any SEC peaks likely due to formation of fibers during the long incubation (Figure 2F, blue line); fibers will be removed in the filtering procedure performed prior to gel filtration. Thus, we conclude that inhibition of fiber formation by FN075 is through the formation of inert soluble CsgA oligomers. In further support of an off-pathway species, addition of preformed FN075–CsgA oligomers to fresh CsgA monomers did not affect CsgA fibrillation kinetics as probed by ThT (Figure S1, Supporting Information). Faster rates of CsgA fibrillation upon seeding with preformed CsgA fibers have been reported.9,10
FN075 Promotes α-Synuclein Fibrillation
Next we tested whether FN075 could also inhibit α-synuclein fibrillation. In contrast to CsgA polymerization, α-synuclein fibers form slowly with a long lag time (i.e., >20 h at 37 °C, pH 7, with agitation). Surprisingly, we found that incubation of unfolded α-synuclein monomers at 37 °C with FN075 results in faster appearance of the increased ThT fluorescence. The lag time for fibrillation decreased from 23.3 ± 2.5 h to 7.3 ± 3.5 h for 35 μM α-synuclein upon including 1:1 molar ratio of FN075 (Figure 3A). Thus, FN075 stimulates α-synuclein fiber formation. The effect of FN075 on α-synuclein fibrillation is dose-dependent, and the lag time decreases as a function of FN075 concentration (Figure 3B). The shortest lag time measured for fibrillation was 2.5 ± 0.3 h for the highest FN075 concentration.
Figure 3.
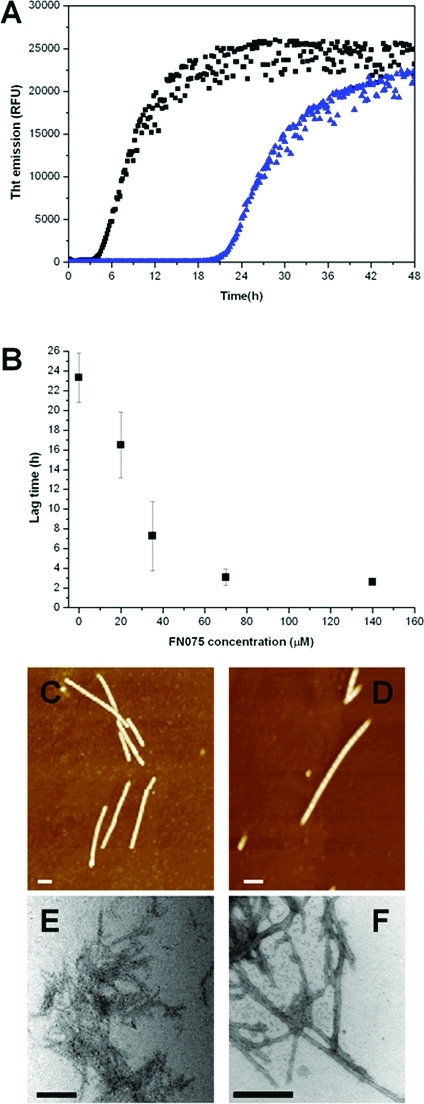
α-Synuclein fibrillation reaction. (A) α-Synuclein (35 μM) fibrillation at 37 °C in the presence of 0.05% DMSO alone (blue) and with 1:1 molar ratio FN075 (black) followed by ThT fluorescence. (B) Lag times of α-synuclein fibrillation without and with various amounts of FN075. Error bars come from multiple experiments (at least four for each point); the error bar is smaller than the symbol for the point at the highest FN075 concentration. (C, D) AFM images of structures formed during fibrillation in the absence (C) and presence (D) of FN075. (E, F) EM image of structures formed during fibrillation in the absence (E) and presence (F) of FN075. Bars designate 100 nm on the AFM and 200 nm on the EM images.
The formation of fibrils was verified by AFM and electron microscopy (EM): samples taken from the stationary phase contain fibrils (Figure 3C–F). Next, we tested whether preformed α-synuclein fibers could seed α-synuclein fibrillation. Fibers formed both in absence and in presence of FN075 were used to seed new α-synuclein fibers (Figure S2, Supporting Information). In these experiments, the effect of FN075 is negligible since the final FN075 concentration is less than 1/20th of the α-synuclein concentration (control in Figure S2, Supporting Information). We found that seeds from fibers formed in the presence and absence of FN075 increase the rate of fiber formation to the same extent. Moreover, this enhanced speed of fibrillation is similar to that found upon addition of 1 equiv of FN075.
FN075 Triggers α-Synuclein Oligomer Formation
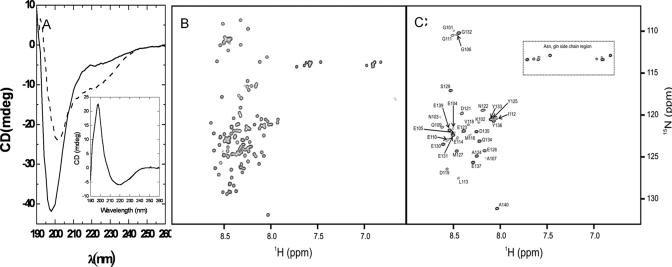
The interaction between FN075 and α-synuclein was studied by far-UV CD and NMR at 10 °C. The CD spectra of α-synuclein in the presence of 0.05% DMSO corresponds to that of an unstructured polypeptide with a negative peak at 195 nm. Upon addition of FN075, the negative peak at 195 nm decreases and shifts toward 210 nm; at the same time, a distinct negative shoulder at 220 nm appears (Figure 4A). These results suggest that when FN075 interacts with α-synuclein, the protein becomes more ordered. The same far-UV CD changes as here were reported for α-synuclein alone upon temperature increase; these changes were interpreted as formation of an aggregation-competent partially folded intermediate with significant β-structure.19
Figure 4.
α-Synuclein–FN075 interaction. (A) CD spectra of 20 μM α-synuclein with 0.05% DMSO (solid line) and with 30 μM FN075 (dashed line) 5 min after mixing. (Inset) The difference CD spectrum resembles that of β-sheet structure. (B) 1H–15N HSQC spectra of 70 μM α-synuclein at 10 °C. (C) 1H–15N HSQC spectra of 70 μM α-synuclein with 200 μM FN075 at 10 °C. Cross peaks corresponding to residues 1–100 are missing due to formation of a large oligomeric species. Assignments23 of residues 101–140 that remain visible in presence of FN075 are indicated in the figure.
1H–15N HSQC spectra acquired for 15N-labeled α-synuclein at 10 °C show that upon FN075 addition to α-synuclein, the cross peaks for residues 1–100 disappear (using assignments from ref (23)), while the cross peaks for residues 100–140 remain visible (Figure 4B,C). NMR diffusion experiments using the remaining cross peaks (at 10 and 37 °C) reveal that a large assembly with a hydrodynamic radius of about 7 nm is formed upon FN075 addition (Table 1). This value matches the dimensions (19 nm × 9 nm × 4.5 nm) reported for regular α-synuclein oligomers in SAXS experiments.24 Our data is thus consistent with the formation of a large oligomeric species where the intramolecular interactions are mediated with the N-terminal 100 residues, whereas the C-terminal 40 residues are unstructured and solvent-exposed. The N-terminal residues are missing in the spectrum as a result of the large molecular weight of the oligomer, whereas the C-terminal unstructured residues tumble independently of the oligomer and gives rise to observable resonances. It has been proposed earlier using site-directed spin labeling methods that residues 36–98 are involved in the α-synuclein fibril core25 and, in the presence of membranes, residues 1–100 interact with these and adopt a helical structure.4
Table 1. Dependence of Stokes Hydrodynamic Radii of α-Synuclein on FN075 Concentration and Temperature Estimated from NMR Diffusion Experiments at the Different Conditions.
| T (°C) | [αSyn] (μM) | [FN075] (μM) | r (nm) |
|---|---|---|---|
| 10 | 70 | 2.96 | |
| 10 | 20 | 60 | 6.49 |
| 10 | 20 | 120 | 7.46 |
| 37 | 70 | 2.16 | |
| 37 | 70 | 200 | 7.21 |
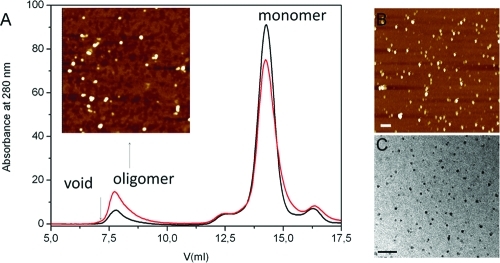
Size-exclusion chromatography (SEC) of α-synuclein–FN075 mixtures reveals the presence of α-synuclein oligomeric species (Figure 5A). AFM analysis of the oligomer peak reveals the presence of spherical particles (Figure 5A, inset). Although the NMR data indicates in essence 100% oligomeric assemblies in the presence of FN075, SEC reveals a large fraction of monomers in addition to the oligomer peak. Contact with the matrix of the gel-filtration column and actual separation of individual species can cause shifts in the equilibrium between species.24 It was proposed that synuclein oligomers are metastable and convert to monomers upon gel filtration.24,26 Thus, the FN075-triggered α-synuclein oligomer may to some extent revert to monomers upon gel filtration. The same oligomeric peak is found upon prolonged incubation of α-synuclein in the absence of FN075, albeit the peak is then smaller (Figure 5A). In the FN075-treated α-synuclein sample, the oligomer peak contains FN075 according to absorption measurements. The exact ratio is not fully reproducible (between 5 and 25 proteins per FN075 molecule are found in oligomers), but there are always substoichiometric amounts of FN075 in the oligomer peak.
Figure 5.
α-Synuclein oligomers. (A) SEC analysis of α-synuclein: 140 μM protein was incubated in presence of 0.5% DMSO at 37 °C for 20 h and injected onto the column (black trace); the same amount of protein was incubated with 300 μM FN075 for 1 h (red trace). Inset shows an AFM image of oligomer peak from the sample with FN075 (bar 200 nm). (B) AFM of a FN075−α-synuclein mixture equivalent to the NMR sample (Figure 4C). (C) EM of FN075−α-synuclein mixture equivalent to the NMR sample (Figure 4C). Bars correspond to 100 nm.
FN075-Triggered Oligomers Appear Similar to Regular α-Synuclein Oligomers
Upon prolonged incubation of α-synuclein alone at 37 °C, but prior to fiber appearance, spherical soluble oligomers (height of 3–6 nm) are observed by microscopy (Figure S3, Supporting Information). AFM and EM data on α-synuclein–FN075 mixtures demonstrate the immediate appearance of similar, on a macroscopic level, oligomers with heights around 3–6 nm in these samples (Figure 5B,C, see also inset in panel A).
If FN075 causes faster appearance of synuclein fibers via the formation of synuclein oligomers, the addition of preformed FN075–synuclein oligomers (as seeds) to monomeric synuclein should speed up synuclein fibrillation as probed by ThT. As expected, and in contrast to the FN075–CsgA oligomer, the addition of seed amount (10% of total protein concentration) of FN075–synuclein oligomers to synuclein monomers resulted in a shorter lag time in the ThT fibrillation curve compared with synuclein alone (Figure S4, Supporting Information). This result suggests that the FN075-induced synuclein oligomer is on the pathway toward fibers.
FN075 Is Monomeric in Solution
When a methyl ester is added to the position of the peptide-like C-terminal end in FN075 (i.e., FN071, Figure 1), the compound loses its effect on both proteins (Figure S5, Supporting Information, NMR synuclein FN071; Figure S6, Supporting Information, ThT data). Thus, the peptide-like C-terminal part of FN075 is important for its activity. Since small organic molecules often aggregate in aqueous solution,27 we tested this using 1D NMR for both molecules. We found that FN075 is present as a monomer under the conditions used for NMR measurements between 10 and 37 °C (Table S1, Supporting Information), whereas FN071 is highly aggregated at all conditions and no NMR resonances from the compound could be observed. This observation can be explained by the lack of charge in FN071 that will result in a higher tendency for aggregation compared with the charged FN075 molecule. For FN075, the radius extracted from NMR diffusion measurements is 0.77 nm, which is in good agreement with the predicted molecular size (0.72 nm).
Studies have been pursued in order to find small-molecule inhibitors of α-synuclein fibrillation, and a range of binding modes have been reported. For example, the anti-Parkinsonian drug selegiline causes the formation of nontoxic amorphous aggregates of α-synuclein.28 Like FN075, the flavonoid compound baicalein induces spherical α-synuclein oligomers, but in contrast to the FN075 case, these oligomers cannot proceed to fibers.29 Moreover, Congo red and lacmoid molecules interact as aggregates with monomeric α-synuclein and thereby inhibit fibrillation,27 while some natural polyphenolic compounds were found to disaggregate preformed α-synuclein oligomers.30 To our knowledge, there is no other small molecule reported with analogous effects on α-synuclein as FN075; however, the formation of toxic α-synuclein oligomers is accelerated by the concerted action of the 20S proteasome and liposomes.31
Conclusion
We discovered that a small molecule, a ring-fused 2-pyridone, designed to inhibit functional amyloids like the bacterial curli fibers, templates the formation of human α-synuclein fibers associated with Parkinson’s disease. Analysis of the reaction mechanism reveals that FN075 accelerates α-synuclein oligomer formation, and this in turn results in faster assembly of fibers. The FN075-induced oligomers contain polypeptides that are structured in the N-termini but the C-terminal 40 residues remain flexible and solvent exposed. Analysis of the effect of FN075 on the curli protein CsgA reveals that also here the small molecule stabilizes an oligomeric form; however, for CsgA, these FN075-induced oligomers cannot assemble into fibers, and an inhibitory effect is observed. Our in vitro findings imply that, despite FN075 triggering oligomers in both cases, the output effect is either inhibition or templation. Moreover, since FN075 has a peptide-like backbone, one may speculate that small peptide metabolites present in vivo may have similar (templating/inhibiting) properties. In accord, non-natural amino acid peptides have been found to interfere with amyloid fiber formation.32
Acknowledgments
We gratefully thank the Swedish Research Council (2011-6259 P.W.-S., 2010-4730 F.A., 2010-5247 M.W.-W., 2011-2619 A.O.), the Knut and Alice Wallenberg Foundation (P.W.-S., F.A.), Göran Gustafsson Foundation (P.W.-S.), JC Kempe Foundation (P.W.-S., M.W.-W., F.A., A.O.), the Swedish Alzheimer Foundation (A.O.), the FAMY organization (A.O.), Umeå University Young Researcher Awards (P.W.-S., M.W.-W.), and National Institutes of Health (Grant RO1 AI073847 MC and ROI AI048689 SH) for financial support.
Glossary
Abbreviations
- AFM
atomic force microscopy
- EM
electron microscopy
- CD
circular dichroism
- NMR
nuclear magnetic resonance
- SEC
size exclusion chromatography
- ThT
thioflavin T
Supporting Information Available
Experimental details, Table S1, and Figures S1–S6. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Fink A. L. Acc. Chem. Res. 2006, 39 (9), 628–34. [DOI] [PubMed] [Google Scholar]
- Kayed R.; Head E.; Thompson J. L.; McIntire T. M.; Milton S. C.; Cotman C. W.; Glabe C. G. Science 2003, 300 (5618), 486–9. [DOI] [PubMed] [Google Scholar]
- Winner B.; Jappelli R.; Maji S. K.; Desplats P. A.; Boyer L.; Aigner S.; Hetzer C.; Loher T.; Vilar M.; Campioni S.; Tzitzilonis C.; Soragni A.; Jessberger S.; Mira H.; Consiglio A.; Pham E.; Masliah E.; Gage F. H.; Riek R. Proc. Natl. Acad. Sci. U.S.A. 2011, 108 (10), 4194–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliezer D.; Kutluay E.; Bussell R. Jr.; Browne G. J. Mol. Biol. 2001, 307 (4), 1061–73. [DOI] [PubMed] [Google Scholar]
- Galvin J. E.; Lee V. M.; Schmidt M. L.; Tu P. H.; Iwatsubo T.; Trojanowski J. Q. Adv. Neurol. 1999, 80, 313–24. [PubMed] [Google Scholar]
- Larsen P.; Nielsen J. L.; Dueholm M. S.; Wetzel R.; Otzen D.; Nielsen P. H. Environ. Microbiol. 2007, 9 (12), 3077–90. [DOI] [PubMed] [Google Scholar]
- Chapman M. R.; Robinson L. S.; Pinkner J. S.; Roth R.; Heuser J.; Hammar M.; Normark S.; Hultgren S. J. Science 2002, 295 (5556), 851–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer N. D.; Schmidt J. C.; Chapman M. R. Proc. Natl. Acad. Sci. U.S.A. 2007, 104 (30), 12494–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Smith D. R.; Jones J. W.; Chapman M. R. J. Biol. Chem. 2007, 282 (6), 3713–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueholm M. S.; Nielsen S. B.; Hein K. L.; Nissen P.; Chapman M.; Christiansen G.; Nielsen P. H.; Otzen D. E. Biochemistry 2011, 50 (39), 8281–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. M.; Almqvist F.; Hultgren S. J. Curr. Opin. Pharmacol. 2003, 3 (5), 513–9. [DOI] [PubMed] [Google Scholar]
- Aberg V.; Almqvist F. Org. Biomol. Chem. 2007, 5 (12), 1827–34. [DOI] [PubMed] [Google Scholar]
- Pinkner J. S.; Remaut H.; Buelens F.; Miller E.; Aberg V.; Pemberton N.; Hedenstrom M.; Larsson A.; Seed P.; Waksman G.; Hultgren S. J.; Almqvist F. Proc. Natl. Acad. Sci. U.S.A. 2006, 103 (47), 17897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emtenas H.; Alderin L.; Almqvist F. J. Org. Chem. 2001, 66 (20), 6756–61. [DOI] [PubMed] [Google Scholar]
- Cegelski L.; Pinkner J. S.; Hammer N. D.; Cusumano C. K.; Hung C. S.; Chorell E.; Aberg V.; Walker J. N.; Seed P. C.; Almqvist F.; Chapman M. R.; Hultgren S. J. Nat. Chem. Biol. 2009, 5 (12), 913–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberg V.; Norman F.; Chorell E.; Westermark A.; Olofsson A.; Sauer-Eriksson A. E.; Almqvist F. Org. Biomol. Chem. 2005, 3 (15), 2817–23. [DOI] [PubMed] [Google Scholar]
- Pemberton N.; Aberg V.; Almstedt H.; Westermark A.; Almqvist F. J. Org. Chem. 2004, 69 (23), 7830–5. [DOI] [PubMed] [Google Scholar]
- Giehm L.; Otzen D. E. Anal. Biochem. 2011, 400 (2), 270–81. [DOI] [PubMed] [Google Scholar]
- Uversky V. N.; Li J.; Fink A. L. J. Biol. Chem. 2001, 276 (14), 10737–44. [DOI] [PubMed] [Google Scholar]
- Nonaka T.; Watanabe S. T.; Iwatsubo T.; Hasegawa M. J. Biol. Chem. 2010, 285 (45), 34885–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanamandra K.; Gruden M. A.; Casaite V.; Meskys R.; Forsgren L.; Morozova-Roche L. A. PLoS One 2011, 6 (4), e18513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiki H.; Higuchi K.; Hosokawa M.; Takeda T. Anal. Biochem. 1989, 177 (2), 244–9. [DOI] [PubMed] [Google Scholar]
- Rao J. N.; Kim Y. E.; Park L. S.; Ulmer T. S. J. Mol. Biol. 2009, 390 (3), 516–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giehm L.; Svergun D. I.; Otzen D. E.; Vestergaard B. Proc. Natl. Acad. Sci. U.S.A. 2011, 108 (8), 3246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.; Margittai M.; Chen J.; Langen R. J. Biol. Chem. 2007, 282 (34), 24970–9. [DOI] [PubMed] [Google Scholar]
- Bhak G.; Lee J. H.; Hahn J. S.; Paik S. R. PLoS One 2009, 4 (1), e4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendel C.; Bertoncini C. W.; Cremades N.; Waudby C. A.; Vendruscolo M.; Dobson C. M.; Schenk D.; Christodoulou J.; Toth G. Biochemistry 2009, 48 (35), 8322–34. [DOI] [PubMed] [Google Scholar]
- Braga C. A.; Follmer C.; Palhano F. L.; Khattar E.; Freitas M. S.; Romao L.; Di Giovanni S.; Lashuel H. A.; Silva J. L.; Foguel D. J. Mol. Biol. 2011, 405 (1), 254–73. [DOI] [PubMed] [Google Scholar]
- Hong D. P.; Fink A. L.; Uversky V. N. J. Mol. Biol. 2008, 383 (1), 214–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruana M.; Hogen T.; Levin J.; Hillmer A.; Giese A.; Vassallo N. FEBS Lett. 2011, 585 (8), 1113–20. [DOI] [PubMed] [Google Scholar]
- Lewis K. A.; Yaeger A.; DeMartino G. N.; Thomas P. J. J. Bioenerg. Biomembr. 2010, 42 (1), 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers S. A.; Karanicolas J.; Chang H. W.; Zhao A.; Jiang L.; Zirafi O.; Stevens J. T.; Munch J.; Baker D.; Eisenberg D. Nature 2011, 475 (7354), 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.