Abstract
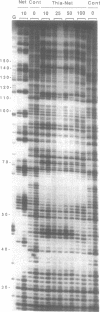
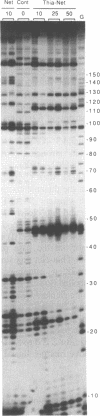
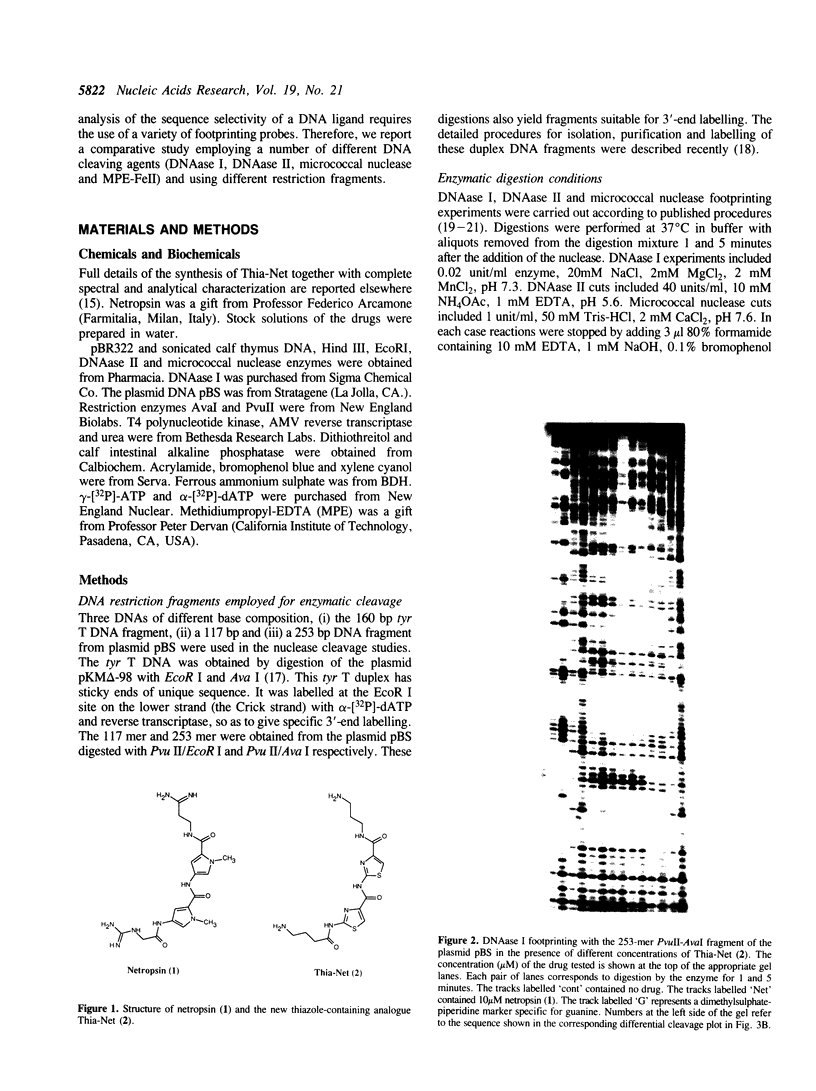
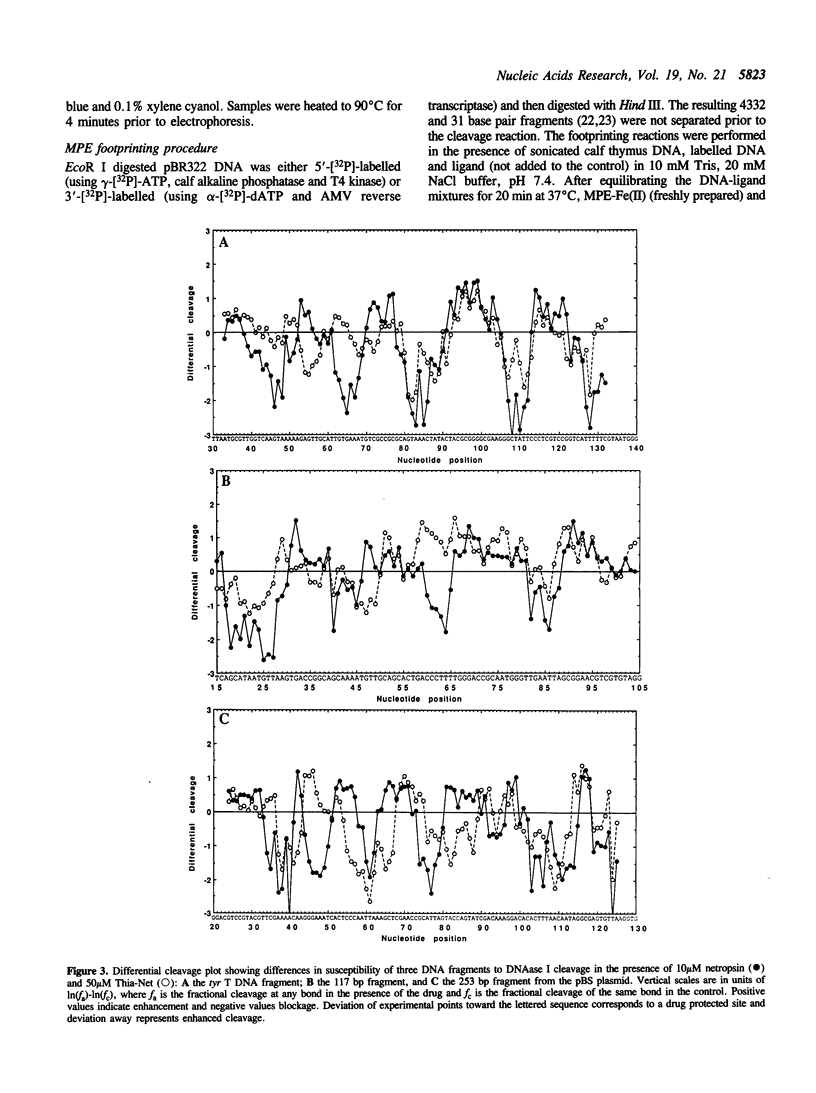
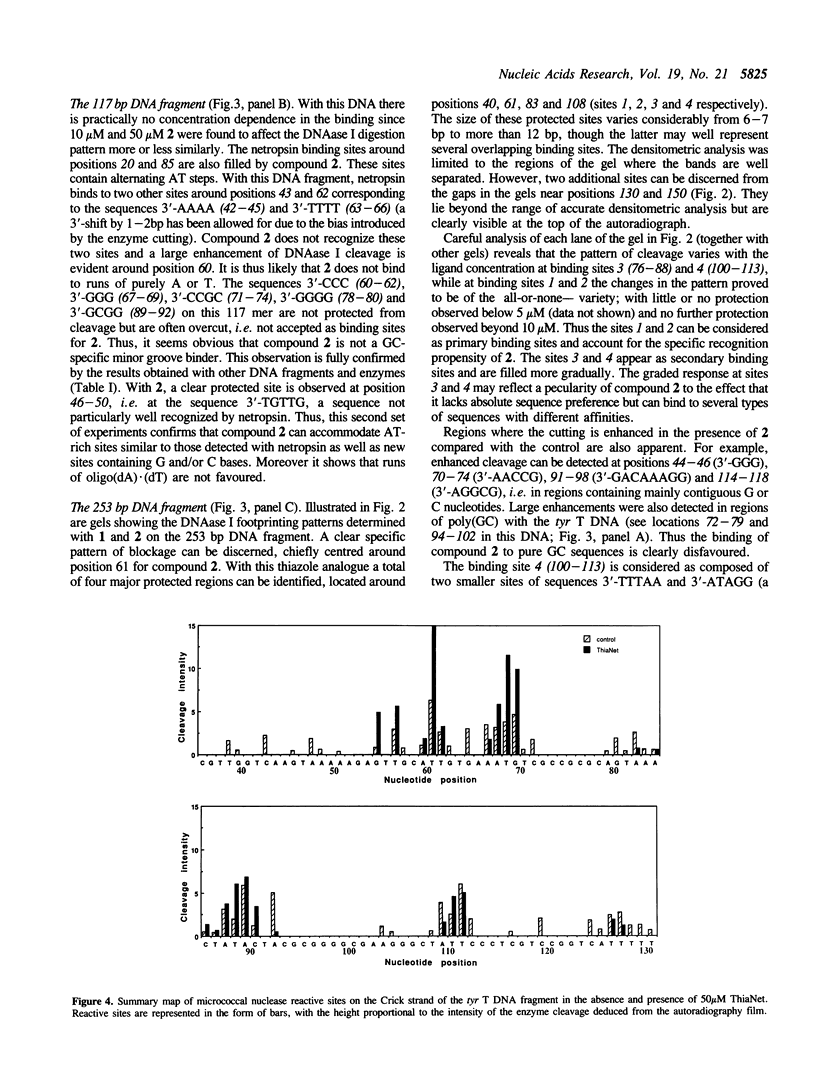
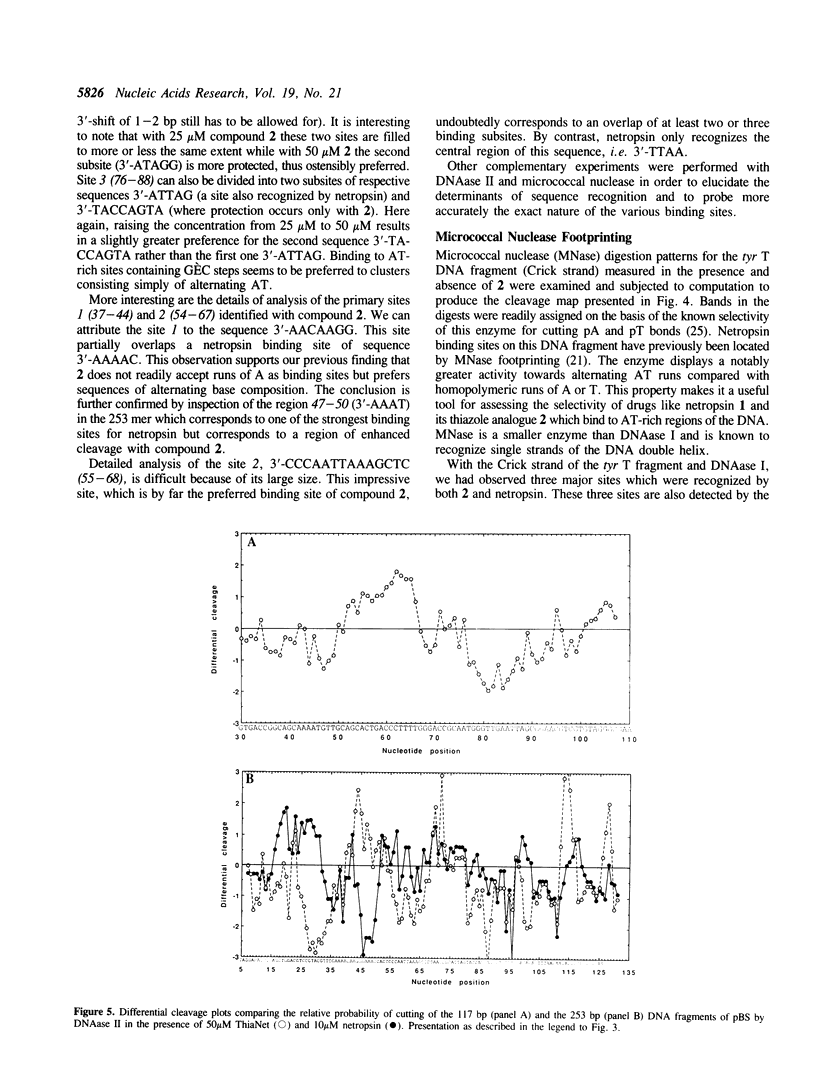
Four different footprinting techniques have been used to probe the DNA sequence selectivity of Thia-Net, a bis-cationic analogue of the minor groove binder netropsin in which the N-methylpyrrole moieties are replaced by thiazole groups. In Thia-Net the ring nitrogen atoms are directed into the minor groove where they could accept hydrogen bonds from the exocyclic 2-amino group of guanine. Three nucleases (DNAase I, DNAase II, and micrococcal nuclease) were employed to detect binding sites on the 160bp tyr T fragment obtained from plasmid pKM delta-98, and further experiments were performed with 117mer and 253mer fragments cut out of the plasmid pBS. MPE.Fe(II) was used to footprint binding sites on an EcoRI/HindIII fragment from pBR322. Thia-Net binds to sites in the minor groove containing 4 or 5 base pairs which are predominantly composed of alternating A and T residues, but with significant acceptance of intrusive GC base pairs. Unlike the parent antibiotic netropsin, Thia-Net discriminates against homooligomeric runs of A and T. The evident preference of Thia-Net for AT-rich sites, despite its containing thiazole nitrogens capable of accepting GC sites by hydrogen bonding, supports the view that the biscationic nature of the ligand imposes a bias due to the electrostatic potential differences in the receptor which favour the ligand reading alternating AT sequences.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bailly C., OhUigin C., Rivalle C., Bisagni E., Hénichart J. P., Waring M. J. Sequence-selective binding of an ellipticine derivative to DNA. Nucleic Acids Res. 1990 Nov 11;18(21):6283–6291. doi: 10.1093/nar/18.21.6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill M. E., Travers A. A. Protein motifs that recognize structural features of DNA. Trends Biochem Sci. 1991 Mar;16(3):92–97. doi: 10.1016/0968-0004(91)90040-3. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA structural variations in the E. coli tyrT promoter. Cell. 1984 Jun;37(2):491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- Flick J. T., Eissenberg J. C., Elgin S. C. Micrococcal nuclease as a DNA structural probe: its recognition sequences, their genomic distribution and correlation with DNA structure determinants. J Mol Biol. 1986 Aug 20;190(4):619–633. doi: 10.1016/0022-2836(86)90247-0. [DOI] [PubMed] [Google Scholar]
- Fox K. R., Sansom C. E., Stevens M. F. Footprinting studies on the sequence-selective binding of pentamidine to DNA. FEBS Lett. 1990 Jun 18;266(1-2):150–154. doi: 10.1016/0014-5793(90)81527-u. [DOI] [PubMed] [Google Scholar]
- Fox K. R., Waring M. J. DNA structural variations produced by actinomycin and distamycin as revealed by DNAase I footprinting. Nucleic Acids Res. 1984 Dec 21;12(24):9271–9285. doi: 10.1093/nar/12.24.9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K. R., Waring M. J. The use of micrococcal nuclease as a probe for drug-binding sites on DNA. Biochim Biophys Acta. 1987 Jul 14;909(2):145–155. doi: 10.1016/0167-4781(87)90036-4. [DOI] [PubMed] [Google Scholar]
- Gago F., Reynolds C. A., Richards W. G. The binding of nonintercalative drugs to alternating DNA sequences. Mol Pharmacol. 1989 Feb;35(2):232–241. [PubMed] [Google Scholar]
- Goodsell D., Dickerson R. E. Isohelical analysis of DNA groove-binding drugs. J Med Chem. 1986 May;29(5):727–733. doi: 10.1021/jm00155a023. [DOI] [PubMed] [Google Scholar]
- Harshman K. D., Dervan P. B. Molecular recognition of B-DNA by Hoechst 33258. Nucleic Acids Res. 1985 Jul 11;13(13):4825–4835. doi: 10.1093/nar/13.13.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. Q., Rehfuss R. P., LaPlante S. R., Boudreau E., Borer P. N., Lane M. J. Actinomycin D induced DNase I cleavage enhancement caused by sequence specific propagation of an altered DNA structure. Nucleic Acids Res. 1988 Dec 9;16(23):11125–11139. doi: 10.1093/nar/16.23.11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissinger K., Krowicki K., Dabrowiak J. C., Lown J. W. Molecular recognition between oligopeptides and nucleic acids. Monocationic imidazole lexitropsins that display enhanced GC sequence dependent DNA binding. Biochemistry. 1987 Sep 8;26(18):5590–5595. doi: 10.1021/bi00392a002. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Yoon C., Goodsell D., Pjura P., Dickerson R. E. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1376–1380. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Jaseja M., Zimmermann J., Yadagiri B., Pon R. T., Sapse A. M., Lown J. W. Molecular recognition and binding of a GC site-avoiding thiazole-lexitropsin to the decadeoxyribonucleotide d-[CGCAATTGCG]2: 1H-NMR evidence for thiazole intercalation. J Biomol Struct Dyn. 1990 Aug;8(1):99–121. doi: 10.1080/07391102.1990.10507792. [DOI] [PubMed] [Google Scholar]
- Laughton C. A., Jenkins T. C., Fox K. R., Neidle S. Interaction of berenil with the tyrT DNA sequence studied by footprinting and molecular modelling. Implications for the design of sequence-specific DNA recognition agents. Nucleic Acids Res. 1990 Aug 11;18(15):4479–4488. doi: 10.1093/nar/18.15.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Chang D. K., Hartley J. A., Pon R. T., Krowicki K., Lown J. W. Structural and dynamic aspects of binding of a prototype lexitropsin to the decadeoxyribonucleotide d(CGCAATTGCG)2 deduced from high-resolution 1H NMR Studies. Biochemistry. 1988 Jan 12;27(1):445–455. doi: 10.1021/bi00401a066. [DOI] [PubMed] [Google Scholar]
- Lee M., Coulter D. M., Pon R. T., Krowicki K., Lown J. W. Sequence specific molecular recognition and binding of a monocationic bis-imidazole lexitropsin to the decadeoxyribonucleotide d-[(GATCCGTATG).(CATACGGATC)]: structural and dynamic aspects of intermolecular exchange studied by 1H-NMR. J Biomol Struct Dyn. 1988 Apr;5(5):1059–1087. doi: 10.1080/07391102.1988.10506449. [DOI] [PubMed] [Google Scholar]
- Lee M., Hartley J. A., Pon R. T., Krowicki K., Lown J. W. Sequence specific molecular recognition by a monocationic lexitropsin of the decadeoxyribonucleotide d-[CATGGCCATG]2: structural and dynamic aspects deduced from high field 1H-NMR studies. Nucleic Acids Res. 1988 Jan 25;16(2):665–684. doi: 10.1093/nar/16.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Krowicki K., Shea R. G., Lown J. W., Pon R. T. Molecular recognition between oligopeptides and nucleic acids. Specificity of binding of a monocationic bis-furan lexitropsin to DNA deduced from footprinting and 1H NMR studies. J Mol Recognit. 1989 Sep;2(2):84–93. doi: 10.1002/jmr.300020206. [DOI] [PubMed] [Google Scholar]
- Low C. M., Drew H. R., Waring M. J. Sequence-specific binding of echinomycin to DNA: evidence for conformational changes affecting flanking sequences. Nucleic Acids Res. 1984 Jun 25;12(12):4865–4879. doi: 10.1093/nar/12.12.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lown J. W., Krowicki K., Balzarini J., De Clercq E. Structure-activity relationship of novel oligopeptide antiviral and antitumor agents related to netropsin and distamycin. J Med Chem. 1986 Jul;29(7):1210–1214. doi: 10.1021/jm00157a016. [DOI] [PubMed] [Google Scholar]
- Lown J. W., Krowicki K., Bhat U. G., Skorobogaty A., Ward B., Dabrowiak J. C. Molecular recognition between oligopeptides and nucleic acids: novel imidazole-containing oligopeptides related to netropsin that exhibit altered DNA sequence specificity. Biochemistry. 1986 Nov 18;25(23):7408–7416. doi: 10.1021/bi00371a024. [DOI] [PubMed] [Google Scholar]
- Lown J. W. Lexitropsins: rational design of DNA sequence reading agents as novel anti-cancer agents and potential cellular probes. Anticancer Drug Des. 1988 Jun;3(1):25–40. [PubMed] [Google Scholar]
- Peden K. W. Revised sequence of the tetracycline-resistance gene of pBR322. Gene. 1983 May-Jun;22(2-3):277–280. doi: 10.1016/0378-1119(83)90112-9. [DOI] [PubMed] [Google Scholar]
- Portugal J., Waring M. J. Assignment of DNA binding sites for 4',6-diamidine-2-phenylindole and bisbenzimide (Hoechst 33258). A comparative footprinting study. Biochim Biophys Acta. 1988 Feb 28;949(2):158–168. doi: 10.1016/0167-4781(88)90079-6. [DOI] [PubMed] [Google Scholar]
- Portugal J., Waring M. J. Comparison of binding sites in DNA for berenil, netropsin and distamycin. A footprinting study. Eur J Biochem. 1987 Sep 1;167(2):281–289. doi: 10.1111/j.1432-1033.1987.tb13334.x. [DOI] [PubMed] [Google Scholar]
- Rao K. E., Shea R. G., Yadagiri B., Lown J. W. Molecular recognition between oligopeptides and nucleic acids: DNA sequence specificity and binding properties of thiazole-lexitropsins incorporating the concepts of base site acceptance and avoidance. Anticancer Drug Des. 1990 Feb;5(1):3–20. [PubMed] [Google Scholar]
- Reinert K. E. Adenosine-thymidine cluster-specific elongation and stiffening of DNA induced by the oligopeptide antibiotic netropsin. J Mol Biol. 1972 Dec 30;72(3):593–607. doi: 10.1016/0022-2836(72)90178-7. [DOI] [PubMed] [Google Scholar]
- Struhl K. Helix-turn-helix, zinc-finger, and leucine-zipper motifs for eukaryotic transcriptional regulatory proteins. Trends Biochem Sci. 1989 Apr;14(4):137–140. doi: 10.1016/0968-0004(89)90145-X. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Methidiumpropyl-EDTA.Fe(II) and DNase I footprinting report different small molecule binding site sizes on DNA. Nucleic Acids Res. 1983 Aug 25;11(16):5555–5567. doi: 10.1093/nar/11.16.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke M. W., Hertzberg R. P., Dervan P. B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II). Proc Natl Acad Sci U S A. 1982 Sep;79(18):5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward B., Rehfuss R., Goodisman J., Dabrowiak J. C. Rate enhancements in the DNase I footprinting experiment. Nucleic Acids Res. 1988 Feb 25;16(4):1359–1369. doi: 10.1093/nar/16.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewska K., Randrianarivelo M., Pullman B. Drug recognition of DNA. Proposal for GC minor groove specific ligands: vinylexins. J Biomol Struct Dyn. 1988 Oct;6(2):331–344. doi: 10.1080/07391102.1988.10507716. [DOI] [PubMed] [Google Scholar]
- Zimmer C., Wähnert U. Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Prog Biophys Mol Biol. 1986;47(1):31–112. doi: 10.1016/0079-6107(86)90005-2. [DOI] [PubMed] [Google Scholar]