Abstract
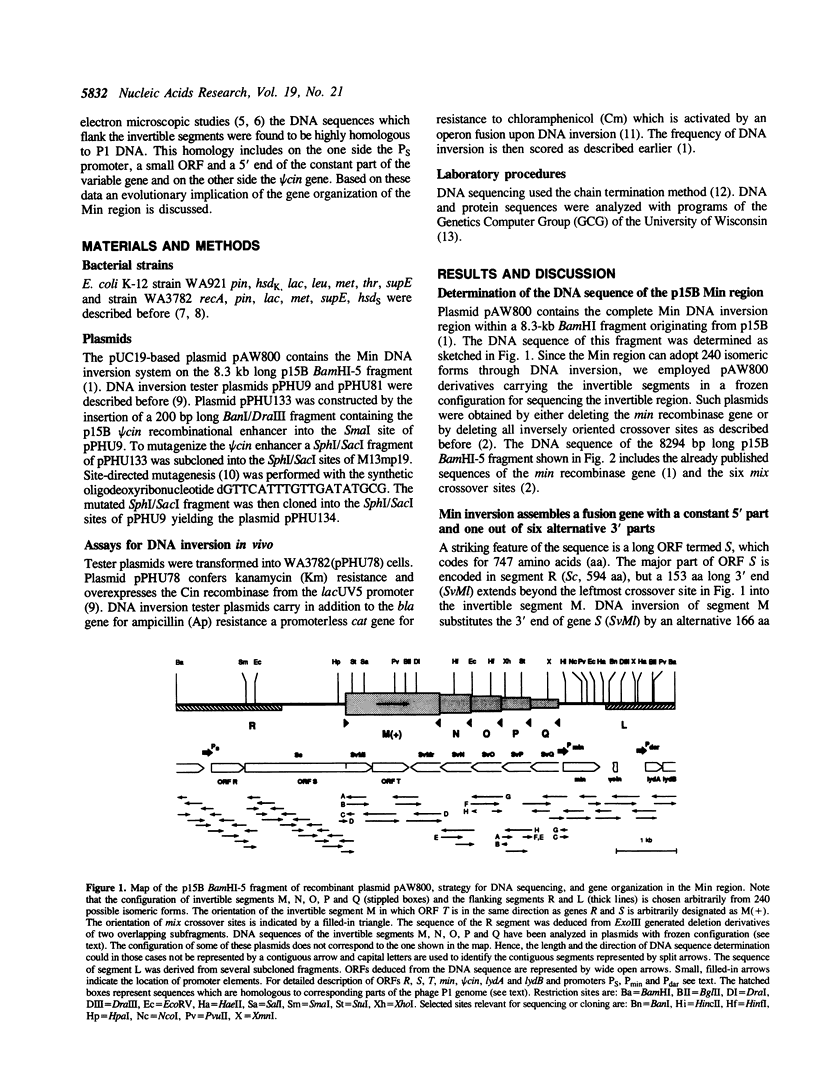
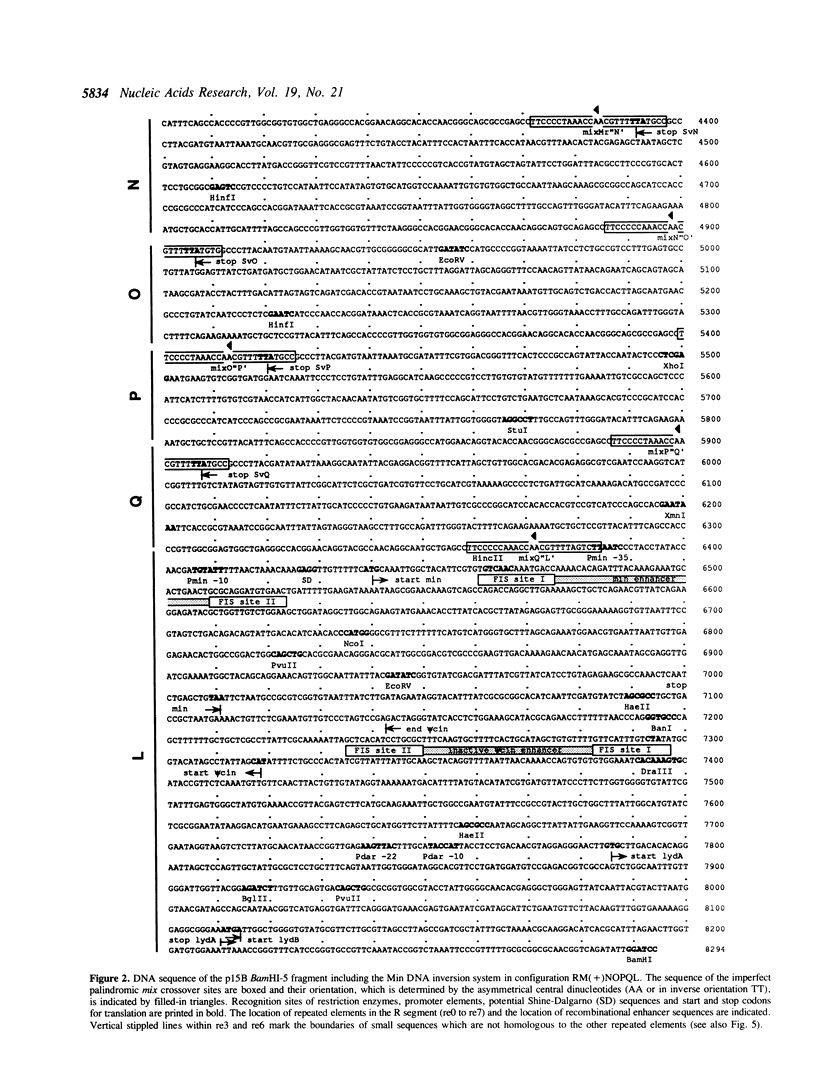
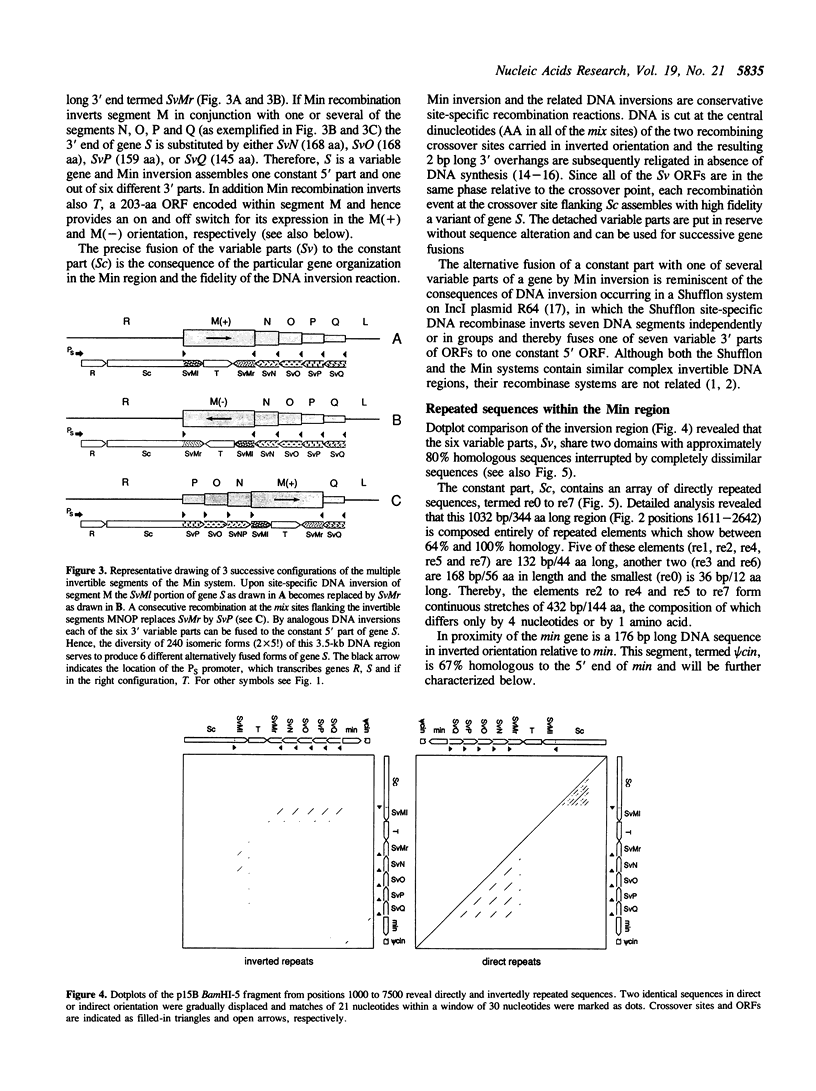
The bacteriophage P1-related plasmid p15B of E. coli 15T- contains a 3.5 kb long region which frequently undergoes complex rearrangements by DNA inversion. Site-specific recombination mediated by the Min DNA invertase occurs at six crossover sites and it eventually results in a population of 240 isomeric configurations of this region. We have determined 8.3-kb sequences of the invertible DNA and its flanking regions. The result explains how DNA inversion fuses variable 3' parts to a constant 5' part, thereby alternatively assembling one out of six different open reading frames (ORF). The resulting variable gene has a coding capacity of between 739 and 762 amino acids. A large portion of its constant part is composed of repeated sequences. The p15B sequences in front of the variable fusion gene encode a small ORF and a phage-specific late promoter and are highly homologous to P1 DNA. Adjacent to the DNA invertase gene min, we have found a truncated 5' region of a DNA invertase gene termed psi cin which is highly homologous to the phage P1 cin gene. Its recombinational enhancer segment is inactive, but it can be activated by the substitution of two nucleotides.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidolin A., Zingg J. M., Arber W. Organization of the bacteriophage P1 tail-fibre operon. Gene. 1989;76(2):239–243. doi: 10.1016/0378-1119(89)90164-9. [DOI] [PubMed] [Google Scholar]
- Guidolin A., Zingg J. M., Lehnherr H., Arber W. Bacteriophage P1 tail-fibre and dar operons are expressed from homologous phage-specific late promoter sequences. J Mol Biol. 1989 Aug 20;208(4):615–622. doi: 10.1016/0022-2836(89)90152-6. [DOI] [PubMed] [Google Scholar]
- Haffter P., Bickle T. A. Purification and DNA-binding properties of FIS and Cin, two proteins required for the bacteriophage P1 site-specific recombination system, cin. J Mol Biol. 1987 Dec 20;198(4):579–587. doi: 10.1016/0022-2836(87)90201-4. [DOI] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiestand-Nauer R., Iida S. Sequence of the site-specific recombinase gene cin and of its substrates serving in the inversion of the C segment of bacteriophage P1. EMBO J. 1983;2(10):1733–1740. doi: 10.1002/j.1460-2075.1983.tb01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber H. E., Iida S., Arber W., Bickle T. A. Site-specific DNA inversion is enhanced by a DNA sequence element in cis. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3776–3780. doi: 10.1073/pnas.82.11.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübner P., Arber W. Mutational analysis of a prokaryotic recombinational enhancer element with two functions. EMBO J. 1989 Feb;8(2):577–585. doi: 10.1002/j.1460-2075.1989.tb03412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübner P., Haffter P., Iida S., Arber W. Bent DNA is needed for recombinational enhancer activity in the site-specific recombination system Cin of bacteriophage P1. The role of FIS protein. J Mol Biol. 1989 Feb 5;205(3):493–500. doi: 10.1016/0022-2836(89)90220-9. [DOI] [PubMed] [Google Scholar]
- Iida S., Hiestand-Nauer R. Localized conversion at the crossover sequences in the site-specific DNA inversion system of bacteriophage P1. Cell. 1986 Apr 11;45(1):71–79. doi: 10.1016/0092-8674(86)90539-8. [DOI] [PubMed] [Google Scholar]
- Iida S., Huber H., Hiestand-Nauer R., Meyer J., Bickle T. A., Arber W. The bacteriophage P1 site-specific recombinase cin: recombination events and DNA recognition sequences. Cold Spring Harb Symp Quant Biol. 1984;49:769–777. doi: 10.1101/sqb.1984.049.01.087. [DOI] [PubMed] [Google Scholar]
- Iida S., Sandmeier H., Hübner P., Hiestand-Nauer R., Schneitz K., Arber W. The Min DNA inversion enzyme of plasmid p15B of Escherichia coli 15T-: a new member of the Din family of site-specific recombinases. Mol Microbiol. 1990 Jun;4(6):991–997. doi: 10.1111/j.1365-2958.1990.tb00671.x. [DOI] [PubMed] [Google Scholar]
- Iida S., Streiff M. B., Bickle T. A., Arber W. Two DNA antirestriction systems of bacteriophage P1, darA, and darB: characterization of darA- phages. Virology. 1987 Mar;157(1):156–166. doi: 10.1016/0042-6822(87)90324-2. [DOI] [PubMed] [Google Scholar]
- Ikeda H., Inuzuka M., Tomizawa J. I. P1-like plasmid in Escherichia coli 15. J Mol Biol. 1970 Jun 14;50(2):457–470. doi: 10.1016/0022-2836(70)90204-4. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Bruist M. F. Intermediates in Hin-mediated DNA inversion: a role for Fis and the recombinational enhancer in the strand exchange reaction. EMBO J. 1989 May;8(5):1581–1590. doi: 10.1002/j.1460-2075.1989.tb03542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp D., Kardas E., Ritthaler W., Sandulache R., Schmucker R., Stern B. Comparative analysis of invertible DNA in phage genomes. Cold Spring Harb Symp Quant Biol. 1984;49:301–311. doi: 10.1101/sqb.1984.049.01.036. [DOI] [PubMed] [Google Scholar]
- Klippel A., Cloppenborg K., Kahmann R. Isolation and characterization of unusual gin mutants. EMBO J. 1988 Dec 1;7(12):3983–3989. doi: 10.1002/j.1460-2075.1988.tb03286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komano T., Kubo A., Nisioka T. Shufflon: multi-inversion of four contiguous DNA segments of plasmid R64 creates seven different open reading frames. Nucleic Acids Res. 1987 Feb 11;15(3):1165–1172. doi: 10.1093/nar/15.3.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J., Iida S., Arber W. Physical analysis of the genomes of hybrid phages between phage P1 and plasmid p15B. J Mol Biol. 1983 Mar 25;165(1):191–195. doi: 10.1016/s0022-2836(83)80250-2. [DOI] [PubMed] [Google Scholar]
- Meyer J., Stålhammar-Carlemalm M., Streiff M., Iida S., Arber W. Sequence relations among the IncY plasmid p15B, P1, and P7 prophages. Plasmid. 1986 Sep;16(2):81–89. doi: 10.1016/0147-619x(86)90066-1. [DOI] [PubMed] [Google Scholar]
- Oliphant A. R., Struhl K. Defining the consensus sequences of E.coli promoter elements by random selection. Nucleic Acids Res. 1988 Aug 11;16(15):7673–7683. doi: 10.1093/nar/16.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier H., Iida S., Meyer J., Hiestand-Nauer R., Arber W. Site-specific DNA recombination system Min of plasmid p15B: a cluster of overlapping invertible DNA segments. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1109–1113. doi: 10.1073/pnas.87.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streiff M. B., Iida S., Bickle T. A. Expression and proteolytic processing of the darA antirestriction gene product of bacteriophage P1. Virology. 1987 Mar;157(1):167–171. doi: 10.1016/0042-6822(87)90325-4. [DOI] [PubMed] [Google Scholar]
- Vandeyar M. A., Weiner M. P., Hutton C. J., Batt C. A. A simple and rapid method for the selection of oligodeoxynucleotide-directed mutants. Gene. 1988 May 15;65(1):129–133. doi: 10.1016/0378-1119(88)90425-8. [DOI] [PubMed] [Google Scholar]
- Wood W. B. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966 Mar;16(1):118–133. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]



