Abstract
Two human genes, SFTPA1 (SP-A1) and SFTPA2 (SP-A2), encode surfactant protein A, a molecule of innate immunity and surfactant-related functions. Several genetic variants have been identified for both genes. These include nucleotide (nt) polymorphisms, as well as alternative splicing patterns at the 5′ untranslated region (5′UTR). Exon B (eB) is included in the 5′UTR of most SP-A2, but not SP-A1 splice variants. We investigated the role of eB in the regulation of gene expression and translation efficiency. A luciferase (Luc) reporter gene was cloned downstream of the entire (AeBD) or eB deletion mutants (del_mut) of the SP-A2 5′UTR, or heterologous 5′UTRs containing the eB sequence, or a random sequence of equal length. The del_mut constructs consisted in consecutive deletions of five nucleotides (n = 8) within eB and the exon-exon junctions in the AeBD 5′UTR. Luc activities and mRNA levels were compared after transfection of NCI-H441 cells. We found that 1) eB increased Luc mRNA levels when placed upstream of heterologous 5′UTR sequences or the promoter region, regardless of its position and orientation; 2) translation efficiency of in vitro-generated mRNAs containing eB was higher than that of mRNAs without eB; and 3) the integrity of eB sequence is crucial for transcription and translation of the reporter gene. Thus eB 1) is a transcription enhancer, because it increases mRNA content regardless of position and orientation, 2) enhances translation when placed in either orientation within its natural 5′UTR sequence and in heterologous 5′UTRs, and 3) contains potential regulatory elements for both transcription and translation. We conclude that eB sequence and length are determinants of transcription and translation efficiency.
Keywords: surfactant protein A, untranslated regions, secondary structure stability, trans-binding sites, cell-specific regulatory factors
surfactant protein A (SP-A) plays an important role in host defense and surfactant-related functions. Two functional genes, SFTPA1 (SP-A1), and SFTPA2 (SP-A2), encode this protein in humans. More than 30 gene variants have been described and characterized on the basis of nucleotide polymorphisms and amino acid sequences at the coding region (7, 8, 10). At the mRNA level, variants show differences at the untranslated regions (UTRs), resulting in sequence variants at the 3′UTR (16, 23, 42, 49) and splice variants at the 5′UTR (20). The genomic sequence of SP-A 5′UTR contains several exons (A, B, B′, C, C′, D, D′) (Fig. 1A) that are alternatively spliced to give rise to specific SP-A1 and SP-A2 patterns. The formation of SP-A 5′UTR splice variants is not a random process since there are major, minor, and rare splice variants for SP-A1 and SP-A2 transcripts (20). A study of SP-A transcripts found that the predominant splice variant configurations and their frequencies differ between the two genes, and these likely also differ among individuals (21). The major splice variant for SP-A1 is AD′ (81%), followed by ACD′ and AB′D′ (7%). The major SP-A2 variants are ABD′ (49%), and ABD (44%) (20).
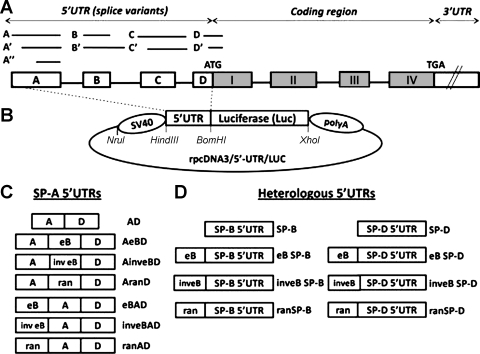
Fig. 1.
A: SP-A1 and SP-A2 common gene structure (not to scale). The 5′ untranslated region (5′UTR) exons A, B, C, and D differ in length, as represented by the lines and boxes. Alternative splicing of the 5′UTR exons gives rise to several SP-A1 and SP-A2 variants. All the SP-A2 mRNA variants described to date contain exon B (20). B: basic rpcDNA3/5′-UTR/LUC vector used to generate experimental plasmids. The rpcDNA3/5′-UTR/LUC was used to clone the experimental spliced 5′UTR sequences upstream of the luciferase reporter gene, as described in materials and methods. The SV40 promoter is located upstream of a 5′UTR sequence and a firefly luciferase cassette, followed by a polyadenylation sequence (polyA). The 5′UTR sequence is flanked by HindIII and BamHI restriction sites, and an XhoI site is located downstream of the luciferase coding sequence. C: diagrammatic representation of various SP-A 5′UTR constructs (n = 7). Boxes labeled with A, eB, and D represent the SP-A 5′UTR exons; inv eB and ran represent the SP-A2 exon B in inverted orientation and a 30-nt random sequence, respectively. D: heterologous 5′ UTR constructs (n = 8). The boxes labeled with SP-B and SP-D represent the heterologous 5′UTRs of the human SP-B and SP-D genes, respectively. Exon B or a random sequence were cloned upstream of the heterologous 5′UTRs. eB, exon B (30 nt); ran, random sequence (30 nt); SP-B, surfactant protein B 5′UTR (14 nt), SP-D, surfactant protein D 5′UTR (43 nt).
In humans, several diseases and complications are correlated with altered SP-A levels and specific SP-A genetic variants (4–6, 9, 22, 29, 30, 35, 37, 40, 41, 45, 53). Variations of the ratio of SP-A1 to total SP-A protein content have been observed as a function of pulmonary disease and aging (44). Therefore, the relative levels of SP-A1 and SP-A2 proteins could relate to disease susceptibility and/or function as predictors of disease specificity. Thus it is important to investigate the molecular mechanisms that govern SP-A1 and SP-A2 gene expression.
Gene regulation is a complex and multifaceted process, and both coding and noncoding sequences are capable of regulatory function. A genomic DNA sequence is considered a transcriptional enhancer when it has the ability to activate transcription independently of its location and orientation relative to a promoter (3). Transcriptional enhancers play numerous roles in gene regulation and cell signaling integration (2). In contrast, a repressor sequence is known to negatively affect the mRNA synthesis of a gene. Both positive and negative DNA regulatory elements exert their effects independently of orientation and position with regard to the transcriptional start site. On the other hand, several mRNA elements, such as the 5′CAP, poly(A) tail, and other structural elements, as well as the mRNA secondary structure, have been shown to modulate protein translation (17, 26). Translational enhancers are cis-acting factors that have the ability to stimulate the translational rate of a specific mRNA, usually by interaction with specific trans-acting factors (28). Numerous mRNA regulatory elements that participate in translational regulation have been identified and characterized in the UTRs of many eukaryotic genes (1, 13, 25, 26). The 5′UTRs play important roles in posttranscriptional RNA processing, localization, and stability, as well as translation initiation and ribosomal recycling (38). In addition, structural and functional communications between the 5′ and 3′ RNA termini, mediated by cis- or trans-acting factors, have been proposed in a number of gene expression studies. Trans-acting factors have been shown to interact with several RNA structures including the UTRs, and these interactions may positively affect the translation and mRNA stability of the majority of eukaryotic transcripts (14, 28).
In previous studies in which several SP-A gene variants were analyzed with regards to translation efficiency and mRNA stability, the translation efficiency of SP-A2 splice variants containing exon B (eB) was significantly higher than that of SP-A1 variants in both in vitro (42) and in lung epithelial cell culture systems (42, 48). The SP-A 5′UTR variant ABD (AeBD) displayed a lower rate of mRNA decay and a higher translation efficiency, compared with SP-A 5′UTR variants without eB (48). Furthermore, eB was shown to contain specific regions that mediate cap-independent translation (50), as well as potential binding sites for regulatory factors within its 30-nucleotide (nt) sequence. On the basis of these findings, it was hypothesized that eB is a cis-acting regulatory element for both transcription and translation (42, 48, 50). Moreover, eB confers SP-A 5′UTR variants with differential capabilities of modulating expression, and this in turn may contribute to differences observed in SP-A levels among individuals under several conditions (e.g., lung disease, aging) (44, 53). However, the location and properties of cis-acting elements in the UTRs of SP-A1 and SP-A2 transcripts have not been described.
In this work, we studied the ability of the SP-A2 5′UTR eB to modulate gene expression at the transcription and translation levels. We generated chimeric and deletion mutant mRNAs with eB in the natural and heterologous UTRs, and we studied the effects of eB position, orientation, and sequence in a reporter gene (luciferase) expression system. We found that eB 1) has the characteristics necessary to be considered a transcriptional enhancer, 2) increases translation when placed in its natural sequence environment (i.e., SP-A2 AeBD 5′UTR and poly(A) tail), independently of its orientation, as well as when it is placed upstream of heterologous 5′UTRs, and c) may contain potential regulatory sequence elements that modulate gene expression. Both length and sequence of eB, but not orientation, play a role in translation regulation.
MATERIALS AND METHODS
Recombinant Constructs
Experimental plasmids were derived from a modified pcDNA3 vector (Invitrogen), rpcDNA3/5′-UTR/LUC (48), that contains the SV40 promoter upstream the SP-A2 5′UTR (AeBD splice variant) flanked by HindIII and BamHI restriction sites, and the firefly luciferase (Luc) reporter gene downstream of the 5′UTR. A polyadenylation signal is located downstream of the Luc cassette (Fig. 1B). Four sets of experimental constructs were generated.
SP-A 5′ UTR constructs.
Seven different 5′UTRs were cloned upstream the Luc cassette, by digestion with HindIII and BamHI (Fig. 1, B and C): 1) the natural SP-A2 5′UTR splice variant AeBD (100 nt) (20); 2) the AeBD variant where the 30-nt eB sequence (eB: GTCGCTGATTTCTTGGAGCCTGAAAAGAAG) was replaced by a random sequence of the same length (ran: CTGCCTGGACCACCTCATCCTTGGCCTGTG) obtained from the coding region sequence of the human SP-B gene (AranD, 100 nt); 3) the AeBD variant without eB (AD, 70 nt); 4) eB placed upstream of the AD sequence (eBAD, 100 nt); 5) the random sequence upstream of AD (ranAD, 100 nt); 6) the inverted eB sequence (inveB: GAAGAAAAGTCCGAGGTTCTTTAGTCGCTG) upstream of AD (inveBAD), or 7) inveB between exons A and D (AinveBD). It should be noted that the natural SP-A1 AD′ splice-variant differs from the AD sequence by only three nucleotides at the 5′ end of exon D and is the most commonly observed SP-A1 splice-variant (20).
Heterologous 5′ UTR constructs.
For the heterologous study, eight additional constructs were obtained. The 14-nt 5′UTR of the surfactant protein B (SP-B) or the 43-nt 5′UTR of the surfactant protein D (SP-D) was cloned upstream of the Luc cassette (Fig. 1D) by digestion with HindIII and BamHI and site-directed mutagenesis. The SP-A eB, in its natural orientation (eB SP-B, eB SP-D) or inverted (inveB SP-B, inveB SP-D), or the 30-nt random sequence was cloned immediately upstream of the heterologous 5′UTR sequences (ran SP-B, ran SP-D). The SP-B and SP-D 5′UTRs without eB or random sequences were also studied.
Constructs with eB placed upstream of the promoter region.
To study the role of eB as a transcriptional enhancer, two new constructs in which eB was placed upstream of the SV40 promoter were obtained, by annealing of eB sequence oligonucleotides flanked by NruI restriction sites, followed by NruI digestion. eB was cloned in its natural or inverted orientation, upstream of SV40 and the luciferase reporter gene in the presence of the AD 5′UTR (eB-SV40-AD, inveB-SV40-AD).
Deletion mutant constructs.
The SP-A 5′UTR AeBD sequence was modified by site-directed mutagenesis with specific primers. The QuikChange II Site-Directed Mutagenesis Kit (Agilent) was used to generate a series of consecutive deletions of five nucleotides (n = 8) within eB, as well as in the exon-exon junctions (exons A-B, and exons B-D), following the manufacturer's protocol.
For all sets of experiments, a construct in which the complete 5′UTR was removed (LUC), by digestion with HindIII and BamHI followed by blunt-ending with T4 polymerase (Invitrogen) and ligation, was used as a negative control. The sequence of all the experimental constructs was verified by DNA sequencing (Molecular Genetics Core Facility at Pennsylvania State University College of Medicine).
Cell Culture
The lung adenocarcinoma cell line NCI-H441 (ATCC no. HTB-174) was obtained from the American Type Culture Collection (Manassas, VA). Cells were cultured and maintained in RPMI-1640 medium (GIBCO-Invitrogen) containing 1% antibiotic/antimycotic solution (GIBCO-Invitrogen), 1% l-glutamine solution (GIBCO-Invitrogen), and 10% heat-inactivated fetal bovine serum (FBS, GIBCO-Invitrogen) at 37°C in a 5% CO2 atmosphere and subcultured weekly.
Transient Transfection
Twenty-four hours before transfection, cells were trypsinized and transferred to six-well culture plates (80% confluency). Four hours before transfection, RPMI medium was replaced by serum-free, antibiotic-free Dulbecco's modified Eagle's medium (DMEM, GIBCO-Invitrogen). Transfection was performed with the Lipofectamine reagent kit (Invitrogen) and PLUS reagent (Invitrogen), following the manufacturer's protocol. Briefly, 1 μg of experimental DNA from large-scale plasmid preparations was delivered to the cells in the presence of pRL-SV40 Renilla luciferase control plasmid (0.05 μg), and cells were incubated at 37°C. After 4 h, medium was replaced by DMEM with 10% FBS and antibiotics/antimycotics, and cells were incubated until used for RNA extraction (30 h) or luciferase assay (36 h).
Total mRNA Preparation
RNA samples were obtained by harvesting NCI-H441 cells 30 h after transfection. After a washing with PBS (GIBCO-Invitrogen), 1 ml of RNA-Bee solution (Tel-Test) was added to each well, and cell lysates were transferred to tubes. Chloroform (0.2 ml) was added, and tubes were centrifuged at 12,000 g for 15 min at 4°C. The aqueous phase was transferred to a new tube and isopropanol (0.5 ml) was added. Tubes were incubated overnight at −20°C and centrifuged at 12,000 g for 30 min (4°C). Pellets were washed with 1 ml of 75% ethanol, and tubes were centrifuged 12,000 g for 5 min (4°C). Air-dried pellets were resuspended in 50 μl of nuclease-free water (GIBCO-Invitrogen) and stored at −80°C until used. RNA concentration was measured with a Nanodrop 1,000 spectrophotometer, and RNA quality was assayed with a 2100 bioanalyzer (Agilent Technologies) at the Functional Genomics Core Facility at the Penn State College of Medicine.
Quantitative Real-Time RT-PCR
RNA samples were treated with the DNA-free kit (Ambion) prior to real-time RT-PCR analysis, to remove any residual amount of genomic DNA. Primers were designed for both firefly (sense: GCCCGCGAACGACATTTA; antisense: TTTGCAACCCCTTTTTGGAA); and Renilla (sense: GCAGCATATCTTGAACCATTCAAA; antisense: CATCACTTGCACGTAGATAAGCATTATA) luciferase genes and obtained from the Macromolecular Core Facility at the Penn State College of Medicine. Fluorescent probes for firefly (FAM-CATTTCGCAGCCTACCGTAGTGTTT-TAMRA) and Renilla (HEX-TATCATGGCCTCGTGAAATCCCGTTAGTAA-TAMRA) were obtained from Biosearch Technologies and Sigma-Aldrich, respectively. The Taqman One-Step RT-PCR Master Mix Reagent kit (Applied Biosystems) was used to perform real-time RT-PCR reactions. Separate reactions were carried out for firefly and Renilla luciferase. Each reaction consisted of 100 ng of RNA, 600 nM sense primer, 900 nM antisense primer, and 300 nM probe in a final volume of 20 μl. Each sample was analyzed in triplicate along with specific standards and no-template controls. A standard curve of DNA concentrations ranging from 10−8 to 0.1 μg/μl was included for both the target gene (firefly) and the endogenous reference (Renilla). A reverse transcription was performed at 48°C for 30 min and 95°C for 10 min, followed by the amplification step, consisting of 43 cycles of 95°C for 15 s and 60°C for 1 min. Results were monitored and stored by the ABI PRISM 7900 sequence detection system (Applied Biosystems). RNA target quantity was calculated by the relative standard curve method (Applied Biosystems Guide to Performing Relative Quantitation of Gene Expression Using Real-Time Quantitative PCR).
Luciferase Activity Assay
Transfected cells were harvested after 36 h, and both firefly and Renilla luciferase activities were measured with the Dual-Luciferase reporter assay system kit (Promega), following the manufacturer's protocol, and an FB12 Luminometer (Zylux, Maryville, TN). Results are expressed by the ratio of firefly to Renilla luciferase activities (means ± SE).
In Vitro Transcription
Plasmids (n = 10) containing natural (AeBD), experimental (AD, AranD, eBAD, and ranAD), and heterologous (SP-B, SP-D, eB SP-B, eB SP-D) 5′UTRs, as well as the no-5′UTR control (LUC), were modified to replace the SV40 promoter by a T7 promoter. Each modified plasmid DNA was linearized by digestion with XhoI, and the Renilla luciferase plasmid (control) was linearized by digestion with BamHI and purified by QIAquick PCR purification kit (Qiagen). Transcription was performed with the mMESSAGE mMACHINE T7 Ultra kit (Ambion) in the presence of 15 mM NTPs, 60 mM ARCA (Anti-Reverse Cap Analog), and T7 RNA polymerase. Reactions were incubated at 37°C for 2 h and subsequently treated with TURBO DNAse. The cRNA products were purified by phenol-chloroform extraction and isopropanol precipitation, as described above. RNA quality was assessed by Nanodrop quantification and agarose denaturing gel electrophoresis. Two micrograms of each experimental transcript were incubated for 30 min at 37°C with 25 μM MnCl2, 10 μM ATP, E-PAP buffer, and 1 unit of E-PAP enzyme (Ambion) in a final volume of 20 μl, to add a poly(A) tail (poly(A)+ mRNA). The E-PAP enzyme was omitted in negative control reactions (poly(A)− mRNA).
In Vitro Translation
A rabbit reticulocyte lysate (RRL) kit (Promega) was used. Two-hundred ng (2 μl) of poly(A)+ or poly(A)− mRNA were incubated with 20 μM amino acid mixture, 40 units of RNAse inhibitor (RNAseOUT, Invitrogen), 45 ng of control Renilla luciferase poly(A)− mRNA, and 20 μl of RRL mix in a final volume of 40 μl, at 30°C for 90 min. Luc activity was measured in a 2-μl aliquot, as described above. In addition, translation reactions were performed with the RRL supplemented with 8 μl (40 μg of protein) of a whole cell lysate obtained from NCI-H441 cells. Results are shown as the ratio of firefly to Renilla luciferase activity (means ± SE).
NCI-H441 Whole Cell Lysate Extraction
The medium of twenty-four plates (90% confluency) was removed, and cells were washed with PBS. Cells were trypsinized and centrifuged, and the pellet (∼1 × 107 cells) was washed three times with 5 ml of washing solution (35 mM HEPES-KOH, 140 mM NaCl, 11 mM glucose, pH 7.5), and resuspended in one volume of extraction solution (20 mM HEPES-KOH, 135 mM potassium acetate, 30 mM potassium chloride, 1.2 mM magnesium acetate, pH 7.5). The cell suspension was transferred to 1.5-ml microtubes and lysed by five cycles of freezing on dry ice-ethanol bath and thawing at room temperature for 30 min. The suspension was centrifuged at 10,000 g for 5 min at 4°C, and the supernatant was desalted by using a PD-10 column (GE Healthcare), equilibrated with extraction solution diluted 1:10. Protein concentration was determined by the BCA protein assay kit (Thermo Fisher Scientific).
RNA Secondary Structure Predictions
We used the RNAfold software from the Vienna RNA websuite (15) (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) to perform mRNA secondary structure predictions. We considered default parameters for the analysis (i.e., no unpaired bases allowed, and GU pairs allowance at the end of helices).
Statistical Analysis
At least three independent experiments were carried out for transfection, mRNA quantification, luciferase assays, poly(A) tailing, and in vitro translation. Differences among treatment groups were estimated by one-way ANOVA followed by the Tukey's multiple-comparisons test using the SigmaStat version 2.0 Software. Statistically significant differences are indicated when P < 0.05.
RESULTS
To investigate the role of eB in gene expression, a luciferase reporter gene system was used, and a number of recombinant constructs were generated (Fig. 1). The lung adenocarcinoma cell line NCI-H441 was cotransfected with each of the experimental constructs and a Renilla luciferase control plasmid. In all cases, the reporter luciferase activity or mRNA levels are expressed after being normalized to Renilla luciferase activity or mRNA levels, respectively.
The Human SP-A 5′UTR eB Functions as a Transcription Enhancer
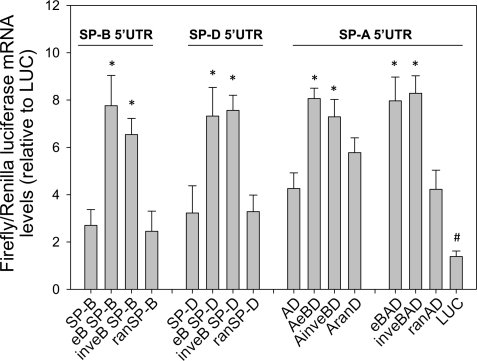
The ability of eB to modulate transcription was tested in the context of two independent heterologous 5′UTRs, from SP-B and SP-D (Fig. 1D). In both instances, the mRNA levels of NCI-H441 cells transfected with constructs containing the eB sequence (eB SP-B, eB SP-D) were higher by approximately threefold, compared with constructs without eB (SP-B, SP-D) or constructs in which eB was replaced by a random sequence of the same length (ranSP-B, ranSP-D) (Fig. 2). Moreover, when the eB sequence was placed in an inverted orientation with the heterologous 5′UTRs (inveB SP-B, inveB SP-D), a significant increase in transcription was also observed (Fig. 2).
Fig. 2.
SP-A eB enhances transcription in the natural and heterologous 5′UTR sequence context. Luciferase mRNA expression (expressed as firefly-to-Renilla ratio, relative to LUC control), measured by real-time RT-PCR after 30 h of transfection of NCI-H441 cells with experimental constructs containing eB upstream of the SP-B and SP-D 5′UTRs, or within the SP-A 5′UTR sequence. The sequence of eB was also placed upstream exons A and D (eBAD), or inverted (inveBAD). The presence of eB in the natural or heterologous 5′UTRs resulted in higher transcription, even when eB was inverted (*P < 0.05, vs. constructs without eB). This effect was not observed when eB was replaced by a random sequence of the same length (ran). The control construct without 5′UTR (LUC) exhibited the lowest mRNA levels (#P < 0.001 vs. all constructs). Real-time RT-PCR was done in triplicate for each sample. Each bar shows mean + SE (n = 3–6 independent experiments).
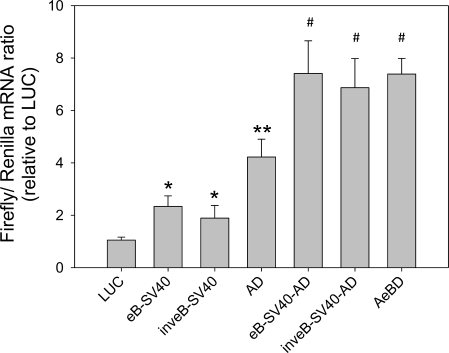
To test whether eB had all the characteristics of a transcription enhancer, its sequence was also cloned upstream of the SV40 promoter, in its natural or inverted orientation. After 30 h of transfection, luciferase mRNA levels were significantly higher when eB was placed in either orientation with respect to the promoter (Fig. 3). In addition, the presence of a 5′UTR (AD) resulted in higher mRNA levels than without the 5′UTR. Furthermore, even when eB was transcribed (i.e., in the context of the natural SP-A2 5′UTR sequence), regardless its position (AeBD or eBAD) or orientation (inveBAD, AinveBD), transcription was also increased (Fig. 2). Together, these findings indicate that eB exhibits characteristics of a transcription enhancer.
Fig. 3.
Exon B is a transcription enhancer. Luciferase mRNA levels (ratio of firefly to Renilla) were measured by real-time RT-PCR after 30 h of transfection with constructs containing the SP-A2 exon B in its natural (eB-SV40, eB-SV40-AD) or inverted (inveB-SV40, inveB-SV40-AD) orientation, upstream of the SV40 promoter and the AD 5′UTR. SV40-AeBD is used as positive control and SV40-LUC as a negative control. Transcription of luciferase was significantly higher in all cases regardless of eB orientation or position with respect to the promoter (*P < 0.05 vs. SV40-LUC; **,#P < 0.05 vs. all). Real-time RT-PCR was performed in triplicate for each sample. Each bar shows mean + SE (n = 3 independent experiments).
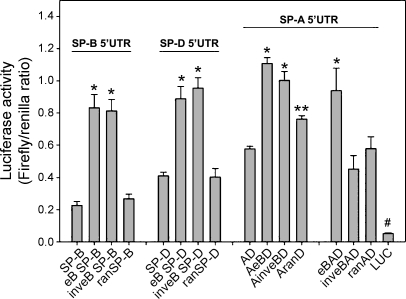
Luciferase Activity Is Enhanced When eB Is Placed Upstream of the Natural and Heterologous 5′UTR Sequences
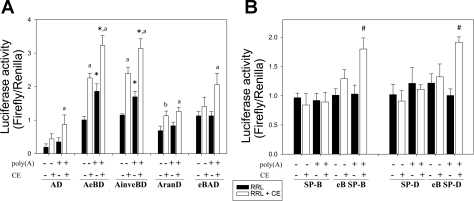
Luciferase activity was determined after 36 h of transfection of NCI-H441 cells with recombinant constructs of the intact SP-A 5′UTR (AeBD) and heterologous 5′UTRs (eB SP-B, inveB SP-B, eB SP-D, inveB SP-D). The latter contained eB upstream of their 5′UTR, in either the natural or inverted orientation of eB (Fig. 1, C and D). We found that the presence of eB resulted in a higher luciferase activity in eB SP-B, eB SP-D, AeBD, eBAD, and AinveBD when compared with AD, but this effect was not observed for inveBAD (Fig. 4). The luciferase activities observed for ranAD, ran SP-B, and ran SP-D did not differ from those obtained with AD, SP-B, or SP-D, respectively. These results indicate a role of eB sequence and position/orientation in the modulation of luciferase activity. The actual length of the UTR did not seem to play a role in luciferase activity, as evidenced by the luciferase levels of ranSP-B, ranSP-D, ranAD, and inveBAD constructs. However, in the natural sequence environment (AD), i.e., in the SP-A 5′UTR context (and not in the sequence environment of the heterologous 5′UTR) the 30-nt random sequence (AranD) displayed a significantly higher activity than AD, albeit significantly lower than AeBD and eBAD. This may indicate a contribution of either the length or the spatial distribution of exons A and D in SP-A2 AeBD splice variants. Furthermore, mRNA secondary structure predictions with RNAfold resulted in a more stable structure for AranD [Gibbs free energy (dG): −35.21 kcal/mol], when compared with ranAD (dG: −32.06 kcal/mol) or eBAD (dG: −33.18 kcal/mol) 5′UTRs. The free energy values for AeBD and AinveBD were dG: −37.79 kcal/mol and dG: −40.78 kcal/mol, respectively. To compare the stability of these 100-nt structures with that of the 70-nt AD (dG: −20.71 kcal/mol), we normalized the dG values to the sequence length in nucleotides. Our results confirmed that AD has a lower stability than AranD and the rest of the structures (normalized dG: AD: −296 cal/mol, AranD: −352 cal/mol, ranAD: −321 cal/mol, and eBAD: −332 cal/mol AeBD: −378 cal/mol, AinveBD: −408 cal/mol).
Fig. 4.
Role of eB in luciferase expression. Luciferase activity (expressed as the ratio of firefly to Renilla) measured after 36 h of transfection of NCI-H441 cells with experimental constructs containing eB within the heterologous SP-B and SP-D 5′UTRs, or the natural SP-A2 5′UTR sequence, in which eB was placed upstream (eBAD) or in between exons A and D (AeBD). Exon B increased luciferase activity in all cases (*P < 0.05 vs. constructs without eB), except when eB was placed inverted upstream of exons A and D (inveBAD), or replaced by a random sequence of the same length (ran). The AranD construct exhibited higher luciferase activity than AD (**P < 0.05), but lower than AeBD. The control construct without 5′UTR (LUC) exhibited the lowest translation compared with any other constructs shown in the figure (#P < 0.001). Luciferase activity was measured in triplicate for each sample. Each bar shows mean + SE (n = 3–6 independent experiments).
eB Increases Luciferase Translation Only When Placed Within Its Natural SP-A 5′UTR Sequence Environment
To further analyze the ability of eB to modulate translation when placed within its natural or heterologous context, as well as its orientation and position, and independently of factors such as mRNA stability, mRNA transport, etc., that may affect luciferase levels after transcription, we studied translation by starting from fixed amounts of in vitro synthesized mRNAs. We generated mRNA transcripts with several 5′UTRs (AD, AeBD, AinveBD, AranD, eBAD, SP-B, eB SP-B, SP-D, eB SP-D) placed upstream of the luciferase gene. A poly(A) tail was added or omitted, and transcripts were used for in vitro translation with a rabbit reticulocyte lysate (RRL) in the presence of a Renilla luciferase control transcript. The firefly-to-Renilla luciferase activity ratio was calculated as a measure of translation efficiency (Fig. 5). Furthermore, the translation reactions were carried out in the presence of NCI-H441 whole cell lysates, to provide potential factors important for translational regulation that may not be present in the RRL. The lowest translation efficiency was observed for transcripts containing a 5′UTR (AD) without eB, in the presence or absence of poly(A). In the absence of poly(A) tail, no differences were observed between wild type (AeBD) and constructs where the eB position (eBAD) or orientation (AinveBD) were modified, or when eB was replaced by the random sequence (AranD). However, in the presence of poly(A) tail (i.e., the natural mRNA context), the translation efficiency of SP-A2 AeBD and AinveBD 5′UTRs were significantly higher than the rest of the constructs in both translation systems. Moreover, translation efficiency was significantly increased in the presence of the human lung cell whole lysate, particularly in transcripts containing eB (Fig. 5A). On the other hand, for transcripts containing the heterologous 5′UTRs, translation was significantly higher only when transcripts contained eB and poly(A) tail (Fig. 5B) and were translated in the presence of whole cell lysates. Together, these data indicate a role of eB length and sequence context, as well as cellular factors on translation.
Fig. 5.
Effect of eB on translation. A: the in vitro translation (luciferase activity) was assessed for transcripts in which eB was placed within the SP-A2 5′UTRs context (AeBD), inverted (AinveBD), upstream of the 5′UTR (eBAD), replaced by a random sequence of the same length (AranD), or removed (AD). A poly(A) tail was added or omitted to all in vitro prepared mRNAs, and translation was measured in a rabbit reticulocyte lysate (RRL) system, with (white boxes) or without (black boxes) the addition of a NCI-H441 whole cell lysate (CE). The Renilla luciferase transcript was used as a normalization control. The presence of eB resulted in higher translation levels when the lung cell extract was added (*P < 0.05), and the poly(A) tail increased translation levels in all cases when the cell lysate was added (aP < 0.05). AranD resulted in higher translation than AD only in the presence of CE, but in the absence of poly(A) (bP < 0.05 vs. AD). B: reactions were also carried out for transcripts containing eB upstream of the heterologous 5′UTRs (SP-B, eB SP-B, SP-D, eB SP-D). Transcripts with eB and poly(A) reached significantly higher translation levels, only in the presence of the lung whole cell lysate (#P < 0.05). Luciferase activity was measured in triplicate for each sample. Each bar shows mean + SE (n = 4 independent experiments).
eB Sequence Contains Potential Transcription and Translation Regulatory Elements
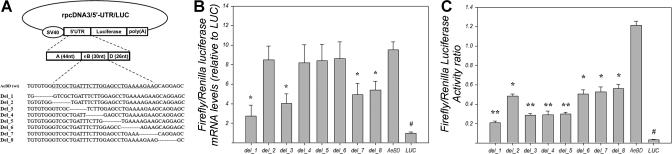
To determine whether the entire eB sequence was necessary for the observed transcription and translation enhancement, five consecutive base pairs at a time were deleted within eB of a complete SP-A2 AeBD 5′UTR luciferase construct, as well as five base pairs at each exon-exon junction sequence A-eB and eB-D (Fig. 6A). Luciferase mRNA levels after transient transfection of NCI-H441 cells were significantly lower for del_1, del_3, del_7, and del_8 compared with the intact AeBD sequence, indicating a potential role of eB and/or adjacent sequence elements in transcription (Fig. 6B). Of interest, when the luciferase activity was measured, none of the deletion constructs achieved a level of activity as high as the intact AeBD 5′UTR construct, indicating a potential role of the complete eB, and the immediate adjacent sequence, as modulators of translation (Fig. 6C). Moreover, the luciferase activity levels were remarkably lower for four deletion mutants (del_1, del_3, del_4, and del_5). These data indicate a potential translation-modulating role for eB nucleotides 6 through 20.
Fig. 6.
Exon B sequential sequence deletion analysis: effect on transcription and luciferase activity. A: experimental deletion-mutant (del_mut) constructs (n = 8), generated from the rpcDNA3/5′-UTR/LUC (AeBD) by site-directed mutagenesis. Del_2 to del_7 refer to 5-nucleotide consecutive deletions within eB. Del_1 corresponds to the removal of the last 5 nucleotides of exon A, whereas del_8 refers to deletion of the first 5 nucleotides of exon D. B: luciferase mRNA levels, expressed as the ratio of firefly to Renilla relative to LUC (no 5′UTR control), after 30 h of transfection of NCI-H441 with del_mut constructs. Del_1, del_3, del_7, and del_8 constructs exhibited lower transcription of luciferase than the natural AeBD 5′UTR (positive control) (*P < 0.05), but significantly higher transcription than the negative control construct (LUC) (#P < 0.01). C: luciferase activity (expressed as the firefly to Renilla ratio) after 36 h of transfection with control or del_mut constructs shown in A. All the del_mut constructs tested exhibited lower luciferase levels compared with the positive control AeBD (*P < 0.05; **P < 0.001). Luciferase activity levels were barely detectable for the construct without 5′UTR (LUC, #P < 0.001), relative to each of the other constructs. Luciferase activity was measured in triplicate for each sample. Each bar shows mean + SE (n = 3 independent experiments).
DISCUSSION
Human SP-A is encoded by two genes, and a number of genetic variants have been reported (7). Despite the high sequence similarity between SP-A1 and SP-A2, splice variants at the 5′UTR of SP-A1 and SP-A2 mRNA transcripts exist (20). We have shown previously that SP-A 5′UTR splice variants exhibit differences in activity and translation efficiency and indicated that eB may be involved in the modulation of gene transcription and/or translation efficiency (48). In this report, we used molecular and biochemical approaches to understand effects of eB on the expression of a reporter gene by investigating the eB-mediated activity within its natural SP-A2 AeBD 5′UTR splice variant and in two independent heterologous 5′UTRs (SP-B and SP-D), and by identifying regions within eB affecting mRNA and activity levels. We found that 1) there was a positive effect of eB in transcription and translation in both the natural and heterologous 5′UTR environments, independently of eB position and/or orientation; 2) specific sequences within eB were important for transcription, and the complete eB sequence was important for translation; 3) flanking sequences of eB appeared important for both transcription and translation; and 4) eB may interact with specific lung cell factors to exert its effect on translation. These observations indicate that eB can act as a transcription enhancer and that eB functions in a sequence-dependent manner to enhance translation. The enhancing effect of eB in transcription and translation may involve some of the same and/or different sequence motifs. Binding of trans-acting factors to eB sequence as well as stabilization of mRNA secondary structure by the presence of eB or by interactions with adjacent sequences are worthwhile future pursuits that may further explain these effects.
Gene regulation is an essential, dynamic, and complex process. Accurate regulation of gene function is achieved through a multitude of mechanisms, many of which are still unknown. The regulation of human SP-A is a complex biological process (19, 42, 48–50). The two human SP-A genes are highly similar at the sequence level, but both functional and structural differences between SP-A1 and SP-A2 products have been widely described (12, 20, 32–34, 46, 47, 51, 52) as well as differences in SP-A expression in model systems and/or among individuals have been observed (42, 44, 47–49). These differences, singly or together, may contribute to individual susceptibility to different insults, such as oxidative stress and disease (10, 44).
A DNA sequence is considered a transcription enhancer when it exhibits the ability to stimulate mRNA synthesis, independent of its orientation and position with respect to the promoter (3). The majority of these sequences are also able to enhance transcription when they are placed in a heterologous sequence environment. The results presented here show that the complete sequence of eB exhibits characteristics of a transcription enhancer. The present data show clearly that eB not only plays an important role in SP-A transcription, but also can act as a general enhancer for other genes if placed upstream of their 5′UTR, or the promoter. The sequence specificity by which eB exerts this function was identified by the deletion mutants study, in which the deletion of a five-nucleotide region (del_3, TGATT) resulted in a significantly reduced transcription, therefore indicating an important role of this region in mRNA synthesis (Fig. 6B). Interestingly, this sequence corresponds to the GATA-consensus motif (T/A)GATA(A/G) (27), a binding site element for the GATA family of transcription factors. These proteins are known to positively affect mRNA synthesis of several genes (43). Deletions carried out in the eB-D exon junction (del_7, del_8) within SP-A2 5′ UTR resulted in decreased transcription, indicating a potential participation of these elements in the modulation of mRNA synthesis. It is known that mRNA splicing and transcription are contemporaneous processes (39), and many factors of the exon junction complex (EJC) affect downstream gene expression events, such as polyadenylation, mRNA transport, and translation (36). Thus a participation of the A-eB (del_1) and/or eB-D (del_7, del_8) EJCs in the eB enhancer capabilities observed for the natural AeBD 5′UTR environment cannot be discarded.
On the other hand, translation enhancers are defined as mRNA sequences capable of increasing protein synthesis. These elements can act as binding sites for trans-acting factors that interact directly or indirectly with the translation machinery (24). In most eukaryotic mRNA transcripts, the UTRs exert regulatory effects at a variety of phases in the posttranscriptional stage, with the subsequent impact on the corresponding proteins levels. These phases include modulation of mRNA transport, stability, and translational efficiency, as well as subcellular localization (18, 31). Several kinds of sequences present in the UTRs could have a regulatory effect on gene expression (38), such as nucleotide sequence motifs that may interact with specific RNA binding proteins and modulate a variety of posttranscriptional events (18). In addition, biochemical and molecular evidence has indicated that the mRNA 5′ cap interacts synergistically with the poly(A) tail to stimulate translation initiation (11), and this communication is mediated by numerous RNA-RNA and RNA-protein interactions that result in the circularization of the mRNA (28). Previous studies suggested an involvement of SP-A 5′UTR eB in posttranscriptional regulation (42, 48, 50). The data presented here obtained by in vitro translation, are consistent with this postulate as these demonstrate that capped transcripts containing eB within the natural SP-A2 5′UTR context are translated more efficiently only in the presence of poly(A). Furthermore, transcripts with short 5′UTRs (exons A and D only) exhibited very low translation efficiency. However, the addition of eB (but not a random sequence) to these short 5′UTRs increased translation demonstrating a contribution of both length and sequence of eB, in translation. Moreover, in vitro translation of mRNAs containing eB in different position and/or orientation (AeBD, AinveBD, eBAD) resulted in higher translation efficiencies than AD or AranD (Fig. 5), particularly in the presence of poly(A) tail and when the translation reaction was supplemented with a lung cell lysate. On the basis of these observations, we speculate that mRNAs containing eB, such as the natural SP-A2 ABD splice variants, are translated more efficiently via 1) the binding of positive lung cell-specific regulatory factors to eB recognition sites and/or 2) interaction with the poly(A) tail via mRNA circularization, a mechanism already proposed (42), but not yet demonstrated, to be involved in the translation of SP-A variants. In addition, in silico comparison of predicted secondary structures resulted in more stable structures for RNAs containing AeBD (dG: −37.79 kcal/mol) and AinveBD (dG: −40.78 kcal/mol) 5′UTRs, relative to those that lacked eB (AD, dG: −20.71 kcal/mol) or had it replaced with a random sequence (AranD, dG: −35.21 kcal/mol). Together, these findings indicate that eB within its natural context may stabilize RNA transcripts or interact with translation factors to promote protein synthesis.
The deletion mutants study further confirmed that both length and sequence of eB play an important role in translation regulation, because the deletion of any five nucleotides within eB resulted in reduced activity when compared with the complete AeBD 5′UTR. It seems likely that the entire eB sequence is necessary and that exons A and/or D function in a synergistic way with eB to increase translatability. The deletion mutants displayed a more prominent reduction in luciferase levels than in mRNA levels, compared with that of AeBD (Fig. 6). The potential role of 5′UTR sequences in posttranscriptional events such as stabilization of mRNA secondary structures and/or binding of trans-acting factors that affect the translation rate cannot be discarded, although to date there is no available evidence of specific RNA-protein or RNA-RNA interactions identified in SP-A2 eB mRNA. For example, deletions del_4 and del_5 displayed high mRNA levels but low luciferase activities, indicating a potential role of these regions in the regulation of translation. However, further experimentation (i.e., in vitro translation) is needed to address this hypothesis.
In summary, both transcription and translation are complex and dynamic biological processes, regulated at several levels and by various means. The present findings indicate that eB of human SP-A plays an important role in gene expression, as assessed by the reporter gene approach and the cell culture and in vitro systems used. We demonstrated that the eB DNA sequence is an enhancer of transcription and the eB RNA sequence is a translation enhancer. All these observations together indicate that both eB sequence and length contribute to its enhancer properties. Future studies may investigate mechanisms of putative interactions with trans-acting factors and how these may affect eB enhancer activity.
GRANTS
This work was supported by the NIH HL-34788 grant.
DISCLOSURES
No conflicts of interest, financial or otherwise are declared by the author(s).
ACKNOWLEDGMENTS
The authors thank the Pennsylvania State University College of Medicine core facility for DNA sequencing, oligonucleotide synthesis, RNA quality control assay, and real-time PCR facilities.
Present address of G. Wang: Department of Surgery, SUNY Upstate Medical University, Syracuse, NY.
REFERENCES
- 1. Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol 19: 465–474, 2009 [DOI] [PubMed] [Google Scholar]
- 2. Arnosti D, Kulkarni M. Transcriptional enhancers: intelligent enhanceosomes or flexible billboards? J Cell Biochem 94: 890–898, 2005 [DOI] [PubMed] [Google Scholar]
- 3. Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 27: 299–308, 1981 [DOI] [PubMed] [Google Scholar]
- 4. Brasch F, Birzele J, Ochs M, Guttentag S, Schoch O, Boehler A, Beers M, Müller K, Hawgood S, Johnen G. Surfactant proteins in pulmonary alveolar proteinosis in adults. Eur Respir J 24: 426–435, 2004 [DOI] [PubMed] [Google Scholar]
- 5. Cheng G, Ueda T, Numao T, Kuroki Y, Nakajima H, Fukushima Y, Motojima S, Fukuda T. Increased levels of surfactant protein A and D in bronchoalveolar lavage fluids in patients with bronchial asthma. Eur Respir J 16: 831–835, 2000 [DOI] [PubMed] [Google Scholar]
- 6. deMello D, Heyman S, Phelps D, Floros J. Immunogold localization of SP-A in lungs of infants dying from respiratory distress syndrome. Am J Pathol 142: 1631–1640, 1993 [PMC free article] [PubMed] [Google Scholar]
- 7. DiAngelo S, Lin Z, Wang G, Phillips S, Ramet M, Luo J, Floros J. Novel, non-radioactive, simple and multiplex PCR-cRFLP methods for genotyping human SP-A and SP-D marker alleles. Dis Markers 15: 269–281, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Floros J. Human surfactant protein A (SP-A) variants: why so many, why such a complexity? Swiss Med Wkly 131: 87–90, 2001 [DOI] [PubMed] [Google Scholar]
- 9. Floros J, Fan R, Matthews A, DiAngelo S, Luo J, Nielsen H, Dunn M, Gewolb IH, Koppe J, van Sonderen L, Farri-Kostopoulos L, Tzaki M, Ramet M, Merrill J. Family-based transmission disequilibrium test (TDT) and case-control association studies reveal surfactant protein A (SP-A) susceptibility alleles for respiratory distress syndrome (RDS) and possible race differences. Clin Genet 60: 178–187, 2001 [DOI] [PubMed] [Google Scholar]
- 10. Floros J, Wang G, Mikerov A. Genetic complexity of the human innate host defense molecules, surfactant protein A1 (SP-A1) and SP-A2—impact on function. Crit Rev Eukaryot Gene Expr 19: 125–137, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gallie D, Tanguay R. Poly(A) binds to initiation factors and increases cap-dependent translation in vitro. J Biol Chem 269: 17166–17173, 1994 [PubMed] [Google Scholar]
- 12. García-Verdugo I, Wang G, Floros J, Casals C. Structural analysis and lipid-binding properties of recombinant human surfactant protein a derived from one or both genes. Biochemistry 41: 14041–14053, 2002 [DOI] [PubMed] [Google Scholar]
- 13. Gebauer F, Hentze M. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 5: 827–835, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Giri P, Kumar GS. Molecular aspects of small molecules-poly(A) interaction: an approach to RNA based drug design. Curr Med Chem 16: 965–987, 2009 [DOI] [PubMed] [Google Scholar]
- 15. Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res 36: W70–W74, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoover RR, Floros J. SP-A 3′-UTR is involved in the glucocorticoid inhibition of human SP-A gene expression. Am J Physiol Lung Cell Mol Physiol 276: L917–L924, 1999 [DOI] [PubMed] [Google Scholar]
- 17. Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jansen R. mRNA localization: message on the move. Nat Rev Mol Cell Biol 2: 247–256, 2001 [DOI] [PubMed] [Google Scholar]
- 19. Karinch A, Deiter G, Ballard P, Floros J. Regulation of expression of human SP-A1 and SP-A2 genes in fetal lung explant culture. Biochim Biophys Acta 1398: 192–202, 1998 [DOI] [PubMed] [Google Scholar]
- 20. Karinch A, Floros J. 5′ Splicing and allelic variants of the human pulmonary surfactant protein A genes. Am J Respir Cell Mol Biol 12: 77–88, 1995 [DOI] [PubMed] [Google Scholar]
- 21. Karinch AM, Floros J. Translation in vivo of 5′ untranslated-region splice variants of human surfactant protein-A. Biochem J 307: 327–330, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kida K, Oda H, Yamano Y, Kagawa J. Effects of cigarette smoking on the serum concentration of lung surfactant protein A (SP-A). Eur Respir J 10: 2124–2126, 1997 [DOI] [PubMed] [Google Scholar]
- 23. Krizkova L, Sakthivel R, Olowe S, Rogan P, Floros J. Human SP-A: genotype and single-strand conformation polymorphism analysis. Am J Physiol Lung Cell Mol Physiol 266: L519–L527, 1994 [DOI] [PubMed] [Google Scholar]
- 24. Kwon S, Barbarese E, Carson J. The cis-acting RNA trafficking signal from myelin basic protein mRNA and its cognate trans-acting ligand hnRNP A2 enhance cap-dependent translation. J Cell Biol 147: 247–256, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Le Quesne JP, Spriggs KA, Bushell M, Willis AE. Dysregulation of protein synthesis and disease. J Pathol 220: 140–151, 2010 [DOI] [PubMed] [Google Scholar]
- 26. Liu Y, Wimmer E, Paul AV. Cis-acting RNA elements in human and animal plus-strand RNA viruses. Biochim Biophys Acta 1789: 495–517, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maeda M, Kubo K, Nishi T, Futai M. Roles of gastric GATA DNA-binding proteins. J Exp Biol 199: 513–520, 1996 [DOI] [PubMed] [Google Scholar]
- 28. Martineau Y, Derry MC, Wang X, Yanagiya A, Berlanga JJ, Shyu AB, Imataka H, Gehring K, Sonenberg N. Poly(A)-binding protein-interacting protein 1 binds to eukaryotic translation initiation factor 3 to stimulate translation. Mol Cell Biol 28: 6658–6667, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McCormack FX, King TE, Jr, Bucher BL, Nielsen L, Mason RJ. Surfactant protein A predicts survival in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 152: 751–759, 1995 [DOI] [PubMed] [Google Scholar]
- 30. McCormack FX, Whitsett JA. The pulmonary collectins, SP-A and SP-D, orchestrate innate immunity in the lung. J Clin Invest 109: 707–712, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol 3: REVIEWS0004, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mikerov A, Umstead T, Gan X, Huang W, Guo X, Wang G, Phelps D, Floros J. Impact of ozone exposure on the phagocytic activity of human surfactant protein A (SP-A) and SP-A variants. Am J Physiol Lung Cell Mol Physiol 294: L121–L130, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mikerov A, Umstead T, Huang W, Liu W, Phelps D, Floros J. SP-A1 and SP-A2 variants differentially enhance association of Pseudomonas aeruginosa with rat alveolar macrophages. Am J Physiol Lung Cell Mol Physiol 288: L150–L158, 2005 [DOI] [PubMed] [Google Scholar]
- 34. Mikerov A, Wang G, Umstead T, Zacharatos M, Thomas N, Phelps D, Floros J. Surfactant protein A2 (SP-A2) variants expressed in CHO cells stimulate phagocytosis of Pseudomonas aeruginosa more than do SP-A1 variants. Infect Immun 75: 1403–1412, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Noah T, Murphy P, Alink J, Leigh M, Hull W, Stahlman M, Whitsett J. Bronchoalveolar lavage fluid surfactant protein-A and surfactant protein-D are inversely related to inflammation in early cystic fibrosis. Am J Respir Crit Care Med 168: 685–691, 2003 [DOI] [PubMed] [Google Scholar]
- 36. Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev 18: 210–222, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pavlovic J, Papagaroufalis C, Xanthou M, Liu W, Fan R, Thomas N, Apostolidou I, Papathoma E, Megaloyianni E, DiAngelo S, Floros J. Genetic variants of surfactant proteins A, B, C, and D in bronchopulmonary dysplasia. Dis Markers 22: 277–291, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pesole G, Mignone F, Gissi C, Grillo G, Licciulli F, Liuni S. Structural and functional features of eukaryotic mRNA untranslated regions. Gene 276: 73–81, 2001 [DOI] [PubMed] [Google Scholar]
- 39. Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell 108: 501–512, 2002 [DOI] [PubMed] [Google Scholar]
- 40. Rämet M, Haataja R, Marttila R, Floros J, Hallman M. Association between the surfactant protein A (SP-A) gene locus and respiratory-distress syndrome in the Finnish population. Am J Hum Genet 66: 1569–1579, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Seifart C, Lin H, Seifart U, Plagens A, DiAngelo S, von Wichert P, Floros J. Rare SP-A alleles and the SP-A1–6A(4) allele associate with risk for lung carcinoma. Clin Genet 68: 128–136, 2005 [DOI] [PubMed] [Google Scholar]
- 42. Silveyra P, Wang G, Floros J. Human SP-A1 (SFTPA1) variant-specific 3′ UTRs and poly(A) tail differentially affect the in vitro translation of a reporter gene. Am J Physiol Lung Cell Mol Physiol 299: L523–L534, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Staal FJ, Weerkamp F, Langerak AW, Hendriks RW, Clevers HC. Transcriptional control of t lymphocyte differentiation. Stem Cells 19: 165–179, 2001 [DOI] [PubMed] [Google Scholar]
- 44. Tagaram H, Wang G, Umstead T, Mikerov A, Thomas N, Graff G, Hess J, Thomassen M, Kavuru M, Phelps D, Floros J. Characterization of a human surfactant protein A1 (SP-A1) gene-specific antibody; SP-A1 content variation among individuals of varying age and pulmonary health. Am J Physiol Lung Cell Mol Physiol 292: L1052–L1063, 2007 [DOI] [PubMed] [Google Scholar]
- 45. Thomas N, Fan R, Diangelo S, Hess J, Floros J. Haplotypes of the surfactant protein genes A and D as susceptibility factors for the development of respiratory distress syndrome. Acta Paediatr 96: 985–989, 2007 [DOI] [PubMed] [Google Scholar]
- 46. Wang G, Bates-Kenney S, Tao J, Phelps D, Floros J. Differences in biochemical properties and in biological function between human SP-A1 and SP-A2 variants, and the impact of ozone-induced oxidation. Biochemistry 43: 4227–4239, 2004 [DOI] [PubMed] [Google Scholar]
- 47. Wang G, Guo X, Diangelo S, Thomas N, Floros J. Humanized SFTPA1 and SFTPA2 transgenic mice reveal functional divergence of SP-A1 and SP-A2: formation of tubular myelin in vivo requires both gene products. J Biol Chem 285: 11998–12010, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang G, Guo X, Floros J. Differences in the translation efficiency and mRNA stability mediated by 5′-UTR splice variants of human SP-A1 and SP-A2 genes. Am J Physiol Lung Cell Mol Physiol 289: L497–L508, 2005 [DOI] [PubMed] [Google Scholar]
- 49. Wang G, Guo X, Floros J. Human SP-A 3′-UTR variants mediate differential gene expression in basal levels and in response to dexamethasone. Am J Physiol Lung Cell Mol Physiol 284: L738–L748, 2003 [DOI] [PubMed] [Google Scholar]
- 50. Wang G, Guo X, Silveyra P, Kimball S, Floros J. Cap-independent translation of human SP-A 5′-UTR variants: a double-loop structure and cis-element contribution. Am J Physiol Lung Cell Mol Physiol 296: L635–L647, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang G, Myers C, Mikerov A, Floros J. Effect of cysteine 85 on biochemical properties and biological function of human surfactant protein A variants. Biochemistry 46: 8425–8435, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang G, Phelps D, Umstead T, Floros J. Human SP-A protein variants derived from one or both genes stimulate TNF-α production in the THP-1 cell line. Am J Physiol Lung Cell Mol Physiol 278: L946–L954, 2000 [DOI] [PubMed] [Google Scholar]
- 53. Wang J, Reid K. The immunoregulatory roles of lung surfactant collectins SP-A, and SP-D, in allergen-induced airway inflammation. Immunobiology 212: 417–425, 2007 [DOI] [PubMed] [Google Scholar]