SUMMARY
Rapid and reversible methods for perturbing the function of specific proteins are desirable tools for probing complex biological systems. We have developed a general technique to regulate the stability of specific proteins in mammalian cells using cell-permeable, synthetic molecules. We engineered mutants of the human FKBP12 protein that are rapidly and constitutively degraded when expressed in mammalian cells, and this instability is conferred to other proteins fused to these destabilizing domains. Addition of a synthetic ligand that binds to the destabilizing domains shields them from degradation, allowing fused proteins to perform their cellular functions. Genetic fusion of the destabilizing domain to a gene of interest ensures specificity, and the attendant small-molecule control confers speed, reversibility, and dose-dependence to this method. This general strategy for regulating protein stability should enable conditional perturbation of specific proteins with unprecedented control in a variety of experimental settings.
INTRODUCTION
Techniques that target gene function at the level of DNA and mRNA are general and powerful strategies for perturbing the protein products encoded by specific genes. The tet/dox and Cre/lox systems have been widely used to target various genes at the transcriptional level (Ryding et al., 2001), and RNA interference is rapidly being adopted as a method to achieve posttranscriptional gene silencing (Fire et al., 1998; Medema, 2004). However, experimental approaches to regulate proteins directly are limited, especially in mammalian cells. In certain cases, inhibitors or activators of specific proteins have been found in nature, and these reagents are often cell-permeable small molecules. Many of these molecules have found widespread use as biological probes, often because the speed, dose-dependence, and reversibility of their activities provide a useful complement to genetic techniques (Schreiber, 2003). However, the question of specificity remains of the utmost importance; in many cases, proteomic analysis reveals that a small-molecule regulator of protein function targets at least one, if not many, off-target proteins (Davies et al., 2000; Bain et al., 2003; Godl et al., 2003).
Shokat and coworkers have developed a method by which a specific kinase can be inhibited using a small-molecule modulator (Shah et al., 1997; Bishop et al., 1998). This method involves genetic manipulation of the protein of interest, typically replacing a large conserved residue in the active site with a smaller glycine or alanine. Specificity is achieved by chemically modifying a previously promiscuous inhibitor with a large substituent, which prevents binding to kinases lacking the cavity-forming mutation. This approach has been successful both in cultured cells and in mice (Bishop et al., 2000; Wang et al., 2003, Chen et al., 2005); however, it is limited to ATPases and GTPases. Although the relatively large size of the kinase family makes this approach fairly general, additional methods are required in order to probe the functions of a wider array of proteins.
To this end, investigators have devised alternative strategies to perturb protein function by taking advantage of existing cellular processes (Banaszynski and Wandless, 2006). Varshavsky and coworkers’ recognition that a protein’s intrinsic stability is in part dependent upon its N-terminal residue (Bachmair et al., 1986) resulted in the genesis of several methods to control the function of a protein of interest in a general manner. Szostak and coworkers showed that a small peptide sequence could be fused to the N terminus of a protein of interest, and that fusion of this degron resulted in decreased stability of that protein in yeast (Park et al., 1992). Varshavsky and coworkers then isolated a temperature-sensitive dihydrofolate reductase degron with a greatly reduced half-life at nonpermissive temperatures (Dohmen et al., 1994), enabling studies of essential proteins in yeast (Labib et al., 2000; Kanemaki et al., 2003). More recently, several researchers have engineered systems in which dimeric small molecules are used to conditionally target fusion proteins for degradation through induced localization to either an E3 ligase complex or to the proteasome itself (Schneekloth et al., 2004; Janse et al., 2004). However, these systems either require a prior knowledge of high-affinity ligands for the protein of interest or are restricted to engineered yeast strains.
An alternative approach for controlling protein function is to perturb subcellular localization. Several technologies achieve small-molecule regulation of protein localization by taking advantage of the FKBP•rapamycin•FRB ternary complex (Kohler and Bertozzi, 2003; Inoue et al., 2005). Fusions of proteins of interest can be made to either FKBP or a small domain of the mTOR protein called FRB, and colocalization is induced upon addition of the small molecule rapamycin. Because of rapamycin’s inherent biological activity, researchers have developed a “bump-hole” strategy similar to that employed by Shokat and coworkers. Rapamycin derivatives possessing large substituents at the FRB binding interface bind poorly to wild-type FRB and in turn bind poorly to the biologically relevant target mTOR, with binding restored upon introduction of compensatory cavity-forming mutations in FRB. Specifically, a C20-methallyl-rapamycin derivative (MaRap) binds to a triple mutant of FRB called FRB* (Liberles et al., 1997). We recently fused GSK-3β to FRB* with the goal of using MaRap to conditionally mislocalize GSK-3β from the nucleus (Stankunas et al., 2003). Interestingly, we noticed decreased levels of the GSK-3β-FRB* fusion relative to an otherwise identical fusion with wild-type FRB. Levels of the FRB* fusion protein were rescued upon addition of MaRap.
Although fusion to FRB* confers instability to multiple different proteins in the absence of MaRap, this chance observation of conditional stabilization is less than ideal. First, two proteins (FKBP and FRB) are required to stabilize the protein of interest. A second and more troubling problem is that of the ligand itself. MaRap is expensive, difficult to synthesize and formulate, and exhibits poor pharmacokinetics in vivo. The inaccessibility of the stabilizing ligand makes widespread implementation of this technology unlikely. Nevertheless, FRB* serves as proof-of-concept that the ligand-dependent stability of one protein can predictably affect the stability of a fused partner protein.
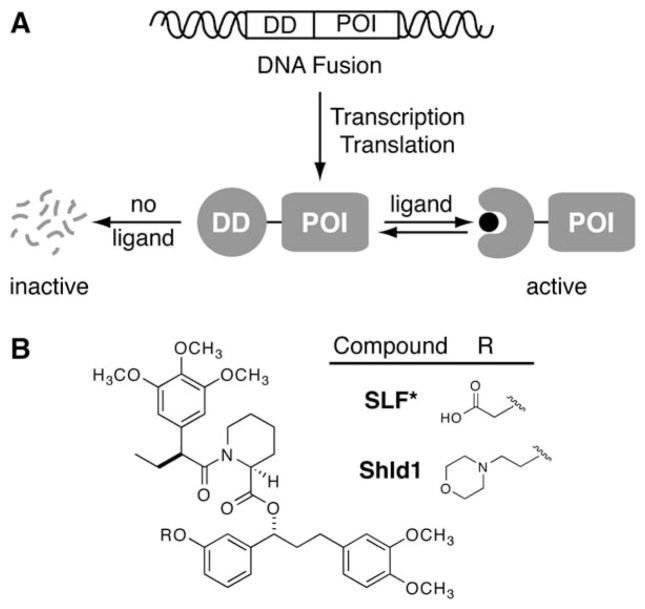
We thus set out to develop a “single ligand-single domain” system that would allow conditional small-molecule control of protein stability. We envisioned the fusion of any protein of interest to a ligand binding domain that is engineered to be unstable, and thus degraded, in the absence of its ligand. Binding of the ligand to this destabilizing domain would stabilize the fusion protein and shield it from degradation, thus restoring function to the protein of interest (Figure 1A). Ideally, this destabilizing domain would be capable of conferring ligand-dependent stability to a wide variety of proteins, thus achieving generality.
Figure 1. A General Method to Conditionally Control Protein Stability.
(A) Genetic fusion of a destabilizing domain (DD) to a protein of interest (POI) results in degradation of the entire fusion. Addition of a ligand for the destabilizing domain protects the fusion from degradation.
(B) Synthetic ligands for FKBP12 F36V.
We chose the FK506- and rapamycin-binding protein (FKBP12) as a candidate destabilizing domain. This 107 residue protein has been widely studied, often in the context of fusion proteins, and dozens of high-affinity ligands for FKBP12 have been developed (Pollock and Clackson, 2002). In one study, ligands that possess a synthetic “bump” in the FKBP12 binding domain were shown to bind more tightly to the cavity-forming F36V mutant relative to the wild-type protein by almost three orders of magnitude (Clackson et al., 1998). Importantly, this family of ligands does not elicit any undesired responses when administered to cultured cells or animals including humans (Iuliucci et al., 2001).
RESULTS
Identification of Ligand-Responsive Destabilizing Domains
To identify mutants that display the desired ligand-dependent behavior, we implemented a cell-based screen in which the fluorescence of yellow fluorescent protein (YFP) served as an indicator of FKBP12 stability. A library based on the FKBP12 F36V gene sequence (hereafter FKBP) was generated using error-prone PCR and then cloned in-frame in front of YFP. A Moloney murine leukemia retroviral expression system was used to stably integrate this library of FKBP-YFP fusions into NIH3T3 fibroblasts, and the transduced cells were subjected to three rounds of sorting using flow cytometry. In the first round, cells were treated with 5 μM of the FKBP ligand SLF* (Figure 1B) for 24 hr prior to sorting. Fluorescent cells were collected and further cultured in the absence of ligand for 60 hr. Reanalysis revealed that approximately 5% of the cell population exhibited decreased fluorescence levels, indicating that the majority of the sequences were either unmutated or contained mutations that did not affect stability of the fusion protein. This small population of cells exhibiting decreased fluorescence was collected and cultured once more in the presence of 5 μM SLF* for 24 hr, at which time YFP-expressing cells were collected and the genomic DNA was isolated.
Sequence analysis of 72 FKBP clones (see Table S1 in the Supplemental Data) revealed several frequently recurring mutations. Mutations were distributed fairly evenly over the primary amino acid sequence, and localized clustering on the tertiary structure was not observed. All observed sequences maintained the F36V mutation and the majority were spatially separated from the ligand binding site, suggesting that ligand binding was crucial for selection.
Before analyzing the behavior of the individual mutants, we synthesized a derivative of SLF* in which the carboxylic acid is replaced with a morpholine group (Figure 1B). This functional group is commonly appended to drug-like molecules to improve their pharmacokinetic properties, and we hypothesized that its addition to SLF* in a position known not to interfere with FKBP binding would enhance intracellular availability and improve the potency of the stabilizing ligand. This cell-permeable FKBP ligand is designed to protect an otherwise unstable protein domain from degradation, so we call the morpholine-containing ligand Shield-1 (Shld1).
Characterization of Shld1-Responsive Destabilizing Domains
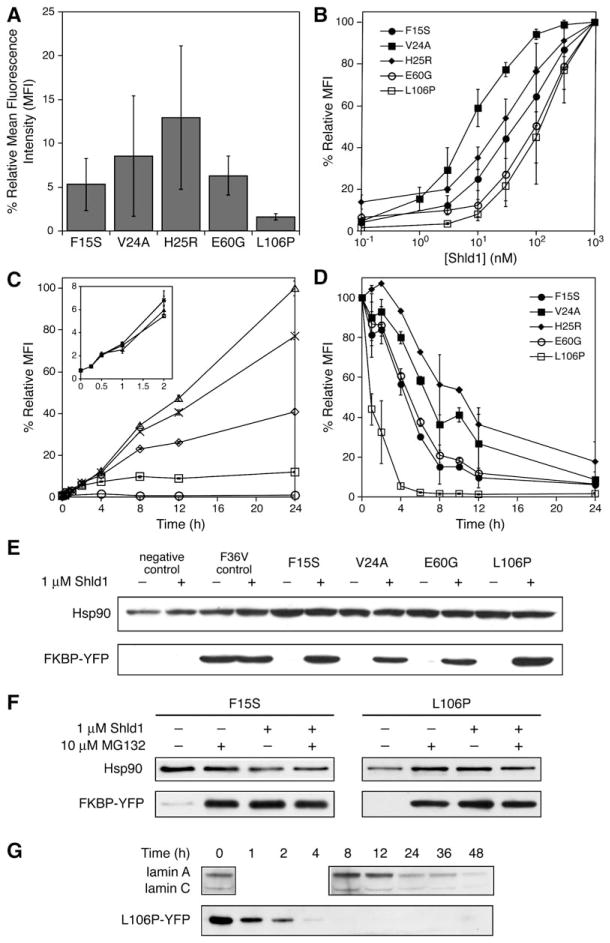
To validate the screening method and to further characterize ligand-responsive destabilizing domains, we chose five mutants (F15S, V24A, H25R, E60G, and L106P) for further analysis. Each mutant was separately transduced into NIH3T3 cells, and YFP fluorescence levels were measured in the absence of Shld1 (Figure 2A). All five mutants showed decreased fluorescence levels with respect to a positive control, indicating that the mutants identified from the library screen are indeed destabilizing. The mutants exhibit varying degrees of destabilization, with the most destabilizing mutant, L106P, expressing YFP fluorescence at a level of only 1%–2% relative to the positive control. All mutants showed increased fluorescence upon addition of Shld1 (see Figure S1 in the Supplemental Data), with observed efficiencies of rescue varying by over an order of magnitude (Figure 2B). Mutant V24A showed the most efficient rescue (EC50 ~5 nM), whereas the more destabilizing L106P required higher concentrations of Shld1 (EC50 ~100 nM) to stabilize the YFP fusion protein.
Figure 2. Characterization of FKBP Mutants that Display Shld1-Dependent Stability.
(A) Fluorescence of FKBP-YFP fusions expressed in NIH3T3 cells in the absence of Shld1 as determined by flow cytometry. (B) NIH3T3 cells stably expressing FKBP-YFP fusions were treated with 3-fold dilutions of Shld1 (1 μM to 0.1 nM) and monitored by flow cytometry. (C) NIH3T3 cells stably expressing FKBP-YFP fusions were either mock-treated (circles) or treated with 30 nM (squares), 100 nM (diamonds), 300 nM (crosses), or 1 μM (triangles) Shld1. Increases in fluorescence were monitored over time using flow cytometry. Mean fluorescence intensity (MFI) was normalized to 100% at 24 hr, 1 μM Shld1. (D) NIH3T3 cells stably expressing FKBP-YFP fusions were treated with 1 μM Shld1 for 24 hr, at which point the cells were washed with media to remove Shld1, and decreases in fluorescence were monitored using flow cytometry. Data for panels (A) through (D) are presented as the average MFI ± SEM relative to that of the maximum fluorescence intensity observed for the individual mutant. Experiments were performed in triplicate.
(E) FKBP-YFP fusions were either mock-treated or treated with 1 μM Shld1 for 24 hr and immunoblotted with an anti-FKBP antibody.
(F) NIH3T3 cells stably expressing F15S-YFP and L106P-YFP were treated with 1 μM Shld1 for 24 hr. Cells were then washed with media and treated with 10 μM MG132 in the presence or absence of 1 μM Shld1 for 4 hr. Immunoblotting was performed with an anti-YFP antibody.
(G) HeLa cells were transfected with siRNA against lamin A/C and monitored over time. Time required for knockdown of lamin A/C is compared against time required for degradation of L106P-YFP upon removal of Shld1 from NIH3T3 cells stably expressing the fusion.
In a kinetic study of NIH3T3 cells stably expressing each destabilizing domain, we observed that YFP fluorescence for all five mutants increased at approximately the same rate upon addition of Shld1, with maximum fluorescence achieved at 24 hr and stably maintained for at least an additional 48 hr without further dosing of Shld1 (Figure S2). These results imply that, upon addition of Shld1, these FKBP mutants are able to adopt a conformation that approximates the stability of the wild-type protein, and that increases in fluorescence are mainly a function of the rate of protein synthesis and/or YFP maturation within the cell. In a related experiment, NIH3T3 cells transduced with the FKBP L106P-YFP fusion (hereafter L106P-YFP) were treated with various concentrations of Shld1, and YFP fluorescence was monitored as a function of time (Figure 2C). YFP expression is observed within 15 min, and we observe that when cells are incubated with lower concentrations of Shld1 they achieve steady state expression levels more rapidly than cells that have been incubated with higher concentrations of Shld1.
We next assayed the five destabilizing domains for kinetics of protein degradation. Upon withdrawal of Shld1, we observed distinct differences in fluorescence decay profiles among the destabilizing domains (Figure 2D). This study revealed a correlation between the rate of degradation and the degree of destabilization conferred by each mutation. Mutant H25R, which is the least destabilizing of this group, showed the slowest rate of degradation, whereas L106P, the most destabilizing of the five, was degraded most quickly, with protein levels becoming negligible within 4 hr.
To correlate YFP fluorescence with intracellular protein levels, and to look for evidence of partial proteolysis, cells stably expressing each destabilizing domain fused to YFP were either mock-treated or treated with Shld1. Antibodies against either FKBP12 (Figure 2E) or YFP (data not shown) were used to immunoblot cell lysates. Neither antibody was capable of detecting protein in lysates from mock-treated cells expressing the mutant FKBP-YFP fusions, whereas Shld1-treated cells showed strong expression of the expected fusion proteins, which correlated with the observed fluorescence levels. F15S and L106P fusions to YFP were also monitored using fluorescence microscopy, and the predicted Shld1-dependent fluorescence is observed (Figure S3).
To gain additional insight into this inducible degradation system and to understand possible limitations thereof, we examined the mechanism of degradation for the F15S and L106P mutants. The ubiquitin-proteasome system is a major mediator of intracellular protein degradation (Pickart, 2004), so we treated cells expressing either F15S or L106P fusions to YFP with either MG132 (Figure 2F) or lactacystin (Figure S4). Following withdrawal of Shld1, the inability of cells to degrade the fusions in the presence of proteasome inhibitors suggests that the degradation of the YFP fusion proteins is mediated, at least in part, by the proteasome.
RNA interference (RNAi) has become a widely used tool for reducing intracellular levels of a protein of interest, so we wanted to compare the rate of RNAi-mediated silencing of an endogenous gene to the rate of degradation achieved through fusion of a protein of interest to a destabilizing domain. Lamin A/C is a nonessential cytoskeletal protein commonly used as a control in RNAi experiments. Previous studies have shown more than 90% reduction in lamin A/C expression in HeLa cells assayed 40 to 45 hr after transfection with a cognate siRNA duplex (Elbashir et al., 2001). This suggests that the half-life of the lamin A/C proteins is no more than 10 to 12 hr, which is significantly shorter than that of green fluorescent protein (t1/2 = 26 hr, Corish and Tyler-Smith, 1999). When HeLa cells were transfected with siRNA against lamin A/C, we began to observe a decrease in protein levels after 24 hr, with a significant reduction in lamin A/C observed by 48 hr (Figure 2G, Figure S5). In contrast, cells stably expressing L106P-YFP show nearly complete degradation of the fusion within 4 hr of removal of Shld1, illustrating that fusion of a destabilizing domain to a protein of interest dramatically reduces its stability in cultured cells.
Predictable Regulation of Intracellular Protein Levels
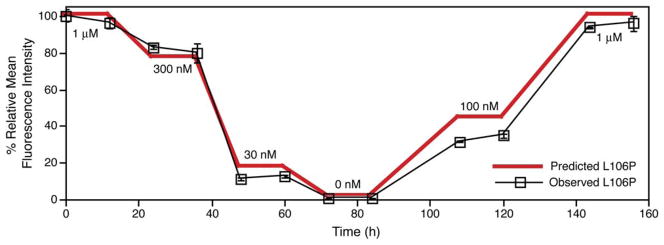
Taken together, these data show that we have identified ligand-sensitive mutants of FKBP, and they further suggest that we may be able to predictably regulate YFP levels with excellent temporal control. To test this theory, we subjected a population of NIH3T3 cells stably expressing L106P-YFP to various concentrations of Shld1 over the course of 1 week (Figure 3). The dose-dependent control that this technology offers is exemplified by the proximity of the observed fluorescence levels to values predicted from the dose-response experiments shown in Figure 2B. This level of control could prove invaluable when the biological function of a protein of interest depends upon its intracellular concentration (Niwa et al., 2000; Pan et al., 2005).
Figure 3. Fusion of an FKBP Destabilizing Domain to the N Terminus of YFP Results in Predictable and Reversible Small-Molecule Regulation of Intracellular Protein Levels.
A population of NIH3T3 cells stably expressing L106P-YFP was treated with varying concentrations of Shld1 over the course of one week, and samples of the population were assayed by flow cytometry at the indicated time points. Data are presented as the average mean fluorescence intensity ± SEM relative to that of the maximum fluorescence intensity observed for L106P-YFP. Predicted fluorescence is based upon the dose response experiment shown in Figure 2B. The experiment was performed in triplicate.
Identification and Characterization of C-Terminal Destabilizing Domains
Many proteins can accommodate fusions at their N termini without loss of function; however, in some cases the intrinsic protein structure or requirements for posttranslational modifications may prohibit N-terminal fusions. Reversing the orientation of FKBP and YFP, we performed a screen of a YFP-FKBP library to identify several candidate C-terminal destabilizing domains (Table S2). From these candidate domains, we chose six FKBP mutants (M66T, R71G, D100G, D100N, E102G, and K105I) for further analysis. At the same time, we tested the ability of the L106P destabilizing domain to confer ligand-dependent stability when placed at the C terminus of a protein of interest. Overall, destabilizing domains fused to the C terminus of YFP are less destabilizing than their N-terminal counterparts (Table S3). For example, when the L106P mutant is fused to the N terminus of YFP (L106P-YFP), fluorescence is only ~1%–2% of that observed in the presence of Shld1; however, when the orientation is reversed (YFP-L106P), fluorescence in the absence of Shld1 is ~10% of that observed in its presence. Interestingly, L106P at the C terminus of YFP is as destabilizing as any mutant identified through our screening process.
Both C-terminal and N-terminal destabilizing domains respond similarly to Shld1, with EC50s ranging from 10 nM to 100 nM (Figure S6). As observed with N-terminal destabilizing domains, all mutants exhibit nearly identical rates of increase in fluorescence upon addition of Shld1, regardless of the degree of instability conferred (Figure S7). Again, rates of fluorescence decay upon removal of Shld1 could be correlated with the relative degree of destabilization conferred by each mutant (Figure S8), with levels of the most destabilizing domains (D100G and L106P) becoming negligible within 8 hr.
Destabilizing Domains Confer Shld1-Dependent Stability in Multiple Cells Lines
These destabilizing domains appear to be quite effective in the context of transduced fibroblasts, so we wanted to ensure that the same behavior would be observed upon transient introduction of the fusions into a variety of different cell types. We therefore tested our destabilizing domains fused to either the N or C terminus of YFP in several commonly used cell lines (NIH3T3, HEK 293T, HeLa, and COS-1) using transient transfection to introduce the chimeric gene. Shld1-dependent fluorescence is observed (Table 1), demonstrating that ligand-dependent stability is not restricted to one cell type. Additionally, these FKBP-derived destabilizing domains can be stabilized by commercially available ligands such as FK506 (Figure S9), keeping in mind that FK506, unlike Shld1, will perturb the cellular environment by inhibiting calcineurin.
Table 1.
Fluorescence of FKBP-YFP Fusions in the Absence of Shld1 in Transiently Transfected Cell Lines
| Percent residual YFP fluorescence* | ||||
|---|---|---|---|---|
| FKBP-YFP | YFP-FKBP | |||
| F15S | L106P | D100G | L106P | |
| NIH3T3 | 7 | 8 | 16 | 16 |
| HEK 293T | 7 | 5 | 15 | 19 |
| HeLa | 8 | 6 | 9 | 12 |
| COS-1 | 12 | 19 | 22 | 26 |
Data are presented as the average mean fluorescence intensity relative to that of the maximum fluorescence intensity observed for the individual mutant. The experiment was performed in duplicate.
Destabilizing Domains Confer Shld1-Dependent Stability to a Variety of Proteins
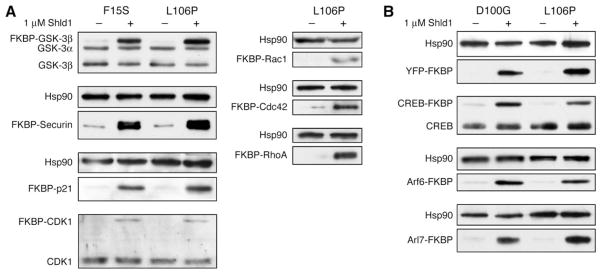
Although the FKBP mutants are efficient destabilizing domains for YFP, it was unclear if this behavior could be used to target other proteins of interest that perform more relevant cellular functions. In choosing candidates, we aimed to target proteins of various characteristics (e.g., size, fold, function, and cellular localization). Using the F15S and L106P destabilizing domains fused at the N termini, Shld1-dependent stability is conferred to the kinases GSK-3β and CDK1, the cell cycle regulatory proteins securin and p21, and three small GTPases, Rac1, RhoA, and Cdc42 (Figure 4A). Interestingly, we were able to induce degradation of an otherwise stable protein (CDK1) and stabilize relatively short-lived cell cycle regulators (p21 and securin), proteins that are normally targeted for degradation by the APC complex (Nigg, 2001). When either the D100G or L106P destabilizing domain was fused to the C terminus of the transcription factor CREB or the small GTPases Arf6 and Arl7, we observed Shld1-dependent stability of these fusion proteins (Figure 4B). To date, we have tested 14 proteins and all have shown ligand-dependent stability when expressed in NIH3T3 cells.
Figure 4. FKBP Destabilizing Domains Confer Shld1-Dependent Stability to a Variety of Proteins.
(A) FKBP mutants F15S and L106P were fused to the N termini of several different proteins and transduced into NIH3T3 cells. Cell populations stably expressing the fusions were then either mock-treated or treated with 1 mM Shld1, and cell lysates were immunoblotted with antibodies against the protein of interest. Endogenous proteins are shown as loading controls when detected, and Hsp90 serves this purpose in cases where they are not detected.
(B) FKBP mutants D100G and L106P were fused to the C termini of several different proteins of interest and treated as above.
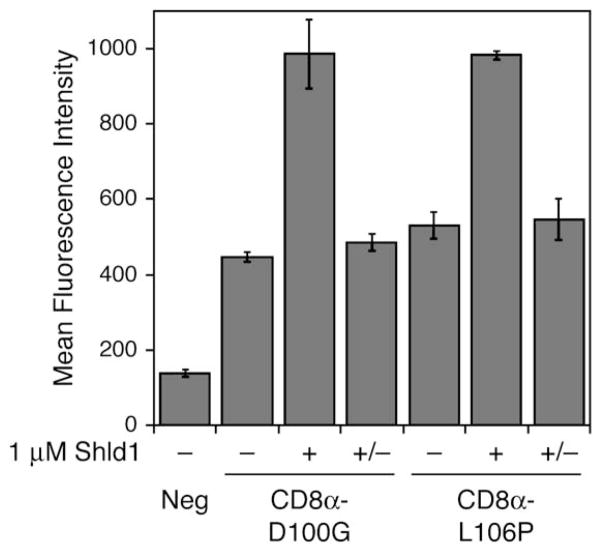
The ability to regulate the function of membrane-bound proteins would allow greater understanding of a range of physiological processes. When CD8α, a transmembrane glycoprotein found on the surface of T cells, was fused at its C terminus to either the D100G or L106P destabilizing domain and expressed in NIH3T3 cells, we were able to elicit Shld1-dependent expression as assayed by flow cytometry (Figure 5). We observed a decrease in CD8α levels at the cell surface upon removal of Shld1, suggesting that the FKBP destabilizing domains possess the ability to recruit the cellular proteins necessary for internalization of membrane-bound proteins (Hicke and Dunn, 2003), presumably leading to degradation of the CD8α-FKBP fusion.
Figure 5. Destabilizing Domains Confer Shld1-Dependent Stability to a Transmembrane Protein.
FKBP mutants D100G and L106P were fused to the C terminus of CD8α, and NIH3T3 cells stably expressing the fusions were split into three pools. The first population (−) was mock-treated and the second population (+) was treated with 1 mM Shld1 for 24 hr. The third population (+/−) was treated with 1 mM Shld1 for 24 hr, then washed with media and cultured for 24 hr in the absence of Shld1. Live cells were then probed with a FITC-conjugated anti-CD8a antibody and assayed by flow cytometry. Data are presented as the average mean fluorescence intensity ± SEM from an experiment performed in triplicate.
Shld1-Dependent Control of Cellular Phenotypes
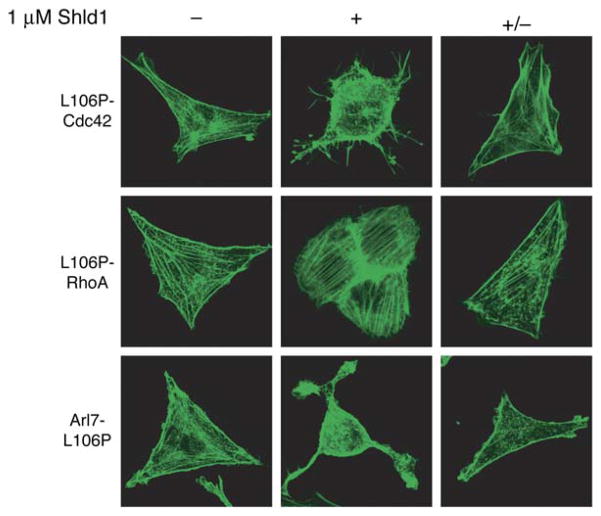
We next sought to correlate changes in cellular behavior with the Shld1-dependent stabilization of a specific protein. Expression of constitutively active small GTPases causes well-characterized changes in cellular morphology (Heo and Meyer, 2003), and intracellular levels of small GTPases fused to destabilizing domains are Shld1-dependent (Figures 4A and 4B). NIH3T3 cells were individually transduced with L106P-RhoA, L106P-Cdc42, or Arl7-L106P, mock-treated or treated with Shld1, and visualized using confocal microscopy (Figure 6). Shld1-treated populations displayed the predicted morphologies. Expression of RhoA induces the formation of stress fibers, expression of Cdc42 results in filopodia formation, and expression of Arl7 induces the shrunken cell phenotype (Heo and Meyer, 2003). These GTPase-dependent morphology changes were reversible, as treatment with Shld1 followed by removal of Shld1 resulted in fibroblast-like morphologies in transduced cells that were indistinguishable from the morphologies observed for mock-treated transduced cells. The penetrance of the observed phenotype was high, with a large percentage of cells (>90%) exposed to a given experimental condition displaying the predicted behavior (Figure S10).
Figure 6. Stabilization of Specific Proteins with Shld1 Results in Predictable Changes in Cellular Morphologies.
NIH3T3 cells stably expressing fusions of a constitutively active small GTPase to the L106P destabilizing domain were split into three pools. The first population (−) was mock-treated and the second population (+) was treated with 1 mM Shld1 for 24 hr. The third population (+/−) was treated with 1 mM Shld1 for 24 hr, then washed with media and cultured in the absence of Shld1 for 24 hr (RhoA Q63L) or 48 hr (Cdc42 Q61L, Arl7 Q72L). Cells were serum-starved for 12 hr, fixed, stained with Alexa Fluor 488-conjugated phalloidin, and visualized using confocal microscopy.
DISCUSSION
Modern experimental biology often relies on the perturbation of a gene followed by observation of the resulting phenotype to elucidate gene function. The success of a given experimental approach is often a function of the quality of the perturbation as well as the richness of the technique used for observation. RNAi has become an integral tool for biologists for probing the functions of various proteins and pathways (Medema, 2004). One feature that makes RNAi so attractive is its relative ease of application. Theoretically, one need only know the gene sequence encoding a protein of interest to design short RNA sequences capable of catalyzing the degradation of the mRNA encoding that protein. After its initial discovery in C. elegans, implementation of RNAi in cultured mammalian cells proved difficult due to the challenges of introducing the RNA sequences capable of entering the RNAi pathway. However, a variety of techniques designed to introduce RNAi effectors into mammalian cells have emerged (e.g., synthetic siRNA, plasmid-encoded shRNA, enzymatically diced pools of RNA), allowing RNAi to become widely used in mammalian cells (Medema, 2004).
Despite its general utility, RNAi is not ideal. Some aspects of the silencing mechanism are poorly understood, making the design of appropriate RNA silencing elements a nontrivial task. The success rate for synthetic siRNAs is typically one in four, with some genes proving more difficult to silence, perhaps due to the accessibility or stability of the messenger RNA. Diced pools improve the “hit rate” of silencing, but they also increase the occurrence of off-target effects. Once an effective RNA sequence has been identified, the extent of mRNA degradation can be variable, and in many cases significant amounts of protein expression are maintained. Reliably introducing RNA into cells is not trivial, and populations of cells that have been transfected with a silencing RNA typically show heterogeneous responses as the extent of RNA delivery can vary significantly between members of a population. Perhaps the greatest disadvantage of RNAi is the time required to reduce protein levels below a functional threshold. The efficacy of the chosen RNA silencing element toward its target message plays a role in this equation; however, the major determinant affecting the rate of knockdown is the half-life of the protein of interest, with 48 hr being a typical timeframe required for significant knockdown of protein levels (Raab and Stephanopoulos, 2004).
An ideal technique to perturb biological macromolecules would be specific, fast, reversible, and tunable. Cell-permeable small molecules often deliver the latter three characteristics, but apart from a few well-known exceptions, they are not typically specific for one biological target. The ideal perturbation technology combines the specificity of reverse genetics (i.e., well-defined DNA changes in a large genomic background) with the conditionality of cell-permeable small molecules.
Using a small library of FKBP mutants (20,000 to 30,000 members) and a cell-based screen, we have identified several small (107 residues) FKBP-derived destabilizing domains that, when fused to their partners, are capable of conferring ligand-dependent stability to a variety of other proteins. Stability, and therefore function, of the resulting fusion protein is induced upon addition of a cell-permeable high-affinity ligand. When the most destabilizing mutant from our screen, FKBP L106P, is fused to YFP, the fusion protein is expressed at only ~1%–2% of its maximum level in the absence of the stabilizing ligand, and this fusion protein is fully stabilized by 1 μM Shld1. However, lower concentrations of Shld1 may be sufficient to restore expression levels that would allow a physiologically relevant protein of interest to perform its cellular function (Figure 2C).
Turnover is quite rapid upon removal of Shld1, with levels of the L106P mutant becoming negligible within 4 hr. We have shown that the FKBP-derived destabilizing domains confer ligand-dependent stability to cytoplasmic proteins, nuclear proteins, and a transmembrane protein, indicating that this might be a general method with which to perturb protein function. One of the biophysical revelations from this study is the size of the sequence space for protein domains that exhibit the desired ligand-dependent stability. The abundance of mutants that display ligand-dependent stability suggests that further refinements in screening may lead to additional destabilizing domains selected for various properties (e.g., rate of degradation, potency of stabilization, and subcellular localization).
The destabilizing domains confer ligand-dependent stability when fused to either the N or the C terminus of a protein of interest, although the N-terminal fusions appear to exhibit a stronger destabilizing effect on the fusion proteins. This observation may reflect a context-dependent ability of the degradation machinery to recognize unstable protein domains. Alternatively, the observed discrepancies in the degree of destabilization conferred by N- and C-terminal destabilizing domains may indicate independent mechanisms of recognition and/or degradation. Additional mechanistic studies should be able to discriminate between these and other alternatives. The proteasome-mediated degradation process appears to be processive, as we have not observed any evidence of partial degradation of any fusion proteins.
Destabilizing domains not only function in virally transduced NIH3T3 fibroblasts, but they also confer Shld1-dependent stability to fusion proteins in a variety of cell lines, including human, upon transient introduction of the genetic fusions. We did, however, observe slight increases in residual fluorescence in the absence of Shld1, which might be attributed to the broader range of expression levels observed upon transient transfection versus viral transduction. It is possible that a small percentage of cells are expressing high levels of a constitutively unstable fusion that may in turn compromise function of the proteasome (Bence et al., 2001).
The use of a small-molecule regulator of protein stability allows one to rapidly and predictably regulate protein levels within a cell, allowing unprecedented control of protein function. The excellent dose and temporal control this technology offers is illustrated by our ability to regulate L106P-YFP stability over an extended period of time (Figure 3). The predicted expression levels were inferred from the simple dose-response curve shown in Figure 2B. When the Shld1 concentration is changed, the rates at which the predicted YFP levels are achieved are probably nonlinear and faster than those shown in Figure 3.
One of the most labor-intensive but minimally perturbing applications of this technology would be to create knockin mice expressing Shld1-dependent alleles of a protein of interest. Expression of the fusion protein would be driven by the endogenous promoter, ideally reproducing the spatial and temporal expression patterns of the unmodified gene. The ligand could be given regularly to stabilize the fusion protein until the mice achieved the age of experimental interest. Withdrawal of Shld1 should result in rapid but reversible loss of the fusion protein. Unlike Cre-mediated gene disruption, this method is reversible. Re-addition of Shld1 stabilizes the fusion protein and reverses the effects of ligand withdrawal, allowing rapid, reversible, and conditional control of protein function in a complex system.
In its most simple implementation, this strategy appears to be a “drug-on” strategy. The stabilizing ligand must be present for expression of the desired fusion protein. However, if one expresses a protein that exhibits a dominant-negative phenotype, the system can be implemented in a “drug-off” manifold. In this configuration, addition of Shld1 results in stabilization of the fusion protein and loss of function of the target protein. A similar situation could be imagined if a constitutively active variant of a protein (e.g., oncogene) was placed under the control of a ligand-responsive fusion protein (Figure 6). In these experimental configurations, the addition of ligand rather than its withdrawal triggers the experimental event.
Recently, investigators screening libraries of synthetic small molecules have discovered inhibitors for several proteins of interest (Mayer et al., 1999; Tan, 2005). In at least one respect, destabilizing domains are not as portable as a library-derived small molecule or RNAi, which, at least in theory, can be applied to any cell type or organism of interest without molecular biological intervention. In order to implement our approach, investigators must determine if the protein of interest retains its intrinsic function(s) in the context of a fusion protein. Then, the destabilizing domain must be either knocked in to an endogenous gene or expressed as a transgene, with the possibility that the endogenous alleles of the protein of interest, if present, may complicate interpretation of the studies. However, the dividend of these genetic interventions is specificity. The discovery of a small-molecule activator or inhibitor from a large pool of candidates in itself is a significant accomplishment; however, proving the specificity of the observed perturbation is an even more formidable task. In contrast, the genetic fusion of our destabilizing domain to any protein of interest ensures the specificity of our approach while maintaining the speed, reversibility, and tunability inherent to small-molecule control but lacking in RNAi.
EXPERIMENTAL PROCEDURES
FKBP Library Generation
Diversity in the FKBP sequence was generated using a combination of error-prone PCR and nucleotide analog mutagenesis. Primers for mutagenic PCR were designed to anneal upstream of the 5′ restriction site to be used for cloning the mutagenesis products into the pBMN iHcRed-tandem retroviral expression vector and to anneal downstream of the 3′ restriction site. Three independent condition sets were used to generate diversity. Condition set A utilized 4 ng template, 0.5 μM of each oligonucleotide primer, 5 units Taq polymerase, 5 mM MgCl2, 0.2 mM MnCl2, 0.4 mM dNTPs in equal ratio, and an excess of 0.2 mM dATP and dCTP. Condition set B was identical to A, except that dGTP and dTTP were present in excess. Condition set C utilized the nonnatural nucleotides 8-oxo-dGTP and dPTP to encourage nucleotide misincorporation (Zaccolo et al., 1996). The FKBP libraries were pooled and ligated into the pBMN iHcRed-t retroviral expression vector, affording a library containing ~3 × 104 members.
FKBP Synthetic Ligands
SLF* and Shld1 were synthesized essentially as described (Holt et al., 1993; Yang et al., 2000). Reagent requests should be directed to the corresponding author.
Cell Culture, Transfections, and Transductions
The NIH3T3 cell line was cultured in DMEM supplemented with 10% heat-inactivated donor bovine serum (Invitrogen), 2 mM glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. All other cell lines were cultured with 10% heat-inactivated fetal bovine serum (Invitrogen), 2 mM glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin.
The ΦNX ecotropic packaging cell line was transfected using standard Lipofectamine 2000 protocols. Viral supernatants were harvested 48 hr posttransfection, filtered, and concentrated 10-fold using an Amicon Ultra centrifugal filter device (Millipore, 100 kDa cutoff). NIH3T3 cells were incubated with the concentrated retroviral supernatants supplemented with 4 μg/mL polybrene for 4 hr at 37°C. Cells were washed once with PBS and cultured in growth media for 24 to 36 hr to allow for viral integration, then assayed as described.
HeLa cells were plated at 7 × 104 cells per well of a 24-well plate 12 hr prior to transfection. Cells were transfected with either 200 ng Silencer Lamin A/C siRNA (Ambion) or a negative control siRNA using the GeneSilencer protocol. Cell lysates were immunoblotted with an antilamin A/C antibody (Clone 14, BD Transduction Laboratories).
Flow Cytometry
Twenty-four hours prior to analysis, transduced NIH3T3 cells were plated at 1 × 105 cells per well of a 12-well plate and treated as described. Cells were removed from the plate using PBS + 2 mM EDTA, washed once with PBS, and resuspended in 200 μl PBS. Cells were analyzed at the Stanford Shared FACS Facility using FlasherII with 10,000 events represented.
Protein of Interest Origin and Antibodies
Proteins tested as fusions to destabilizing domains were of the following origin, and the following antibodies were used for immunoblotting: Arf6 Q67L (human, 3A-1, Santa Cruz Biotechnology); Arl7 Q72L (human, BC001051, Protein Tech Group, Inc.); Cdc42 Q61L (human, P1, Santa Cruz Biotechnology); CD8α (mouse, 5H10, Caltag Laboratories); CDK1 (human, H-297, Santa Cruz Biotechnology); CREB (mouse, 86B10, Cell Signaling Technology); FKBP (human, 2C1-97, BD Phar-Mingen); GSK-3β (mouse, 0011-A, Santa Cruz Biotechnology); Hsp90 (mouse, 68, BD Transduction Laboratories); p21 (human, H-164, Santa Cruz Biotechnology); Rac1 Q61L (human, C-11, Santa Cruz Biotechnology); RhoA Q63L (human, 26C4, Santa Cruz Biotechnology); Securin (human, Z23.YU, Zymed Laboratories); YFP, Aequorea victoria (JL-8, Clontech).
Phalloidin Staining and Microscopy
NIH3T3 cells stably expressing constitutively active GTPases fused to destabilizing domains were treated with 1 μM Shld1 for 24 hr. At this time, cells were washed once with PBS and plated at 8 × 103 cells in 4-well LabTek Chambered coverglass (NUNC) coated with 1 mg/ml poly-D-lysine (Sigma). Mock-treated transduced cells and transduced cells treated with 1 μM Shld1 were plated likewise as negative and positive controls, respectively. Cells were cultured for 24 hr in 10% DBS, then cultured in serum-free media for 12 hr. Cells were then washed with PBS, fixed in 4% paraformaldehyde for 15 min, permeabilized in 0.2% Triton X-100 for 5 min, stained with 1 μg/ml Alexa Fluor 488-conjugated phalloidin (Invitrogen; A12379) in PBS for 20 min, and washed with PBS. Fixed cells were imaged using a Bio-Rad Radiance 2100 confocal microscope.
Supplementary Material
Acknowledgments
We thank the Crabtree, Felsher, Ferrell, Jackson, Kopito, Meyer, and Nolan labs for reagents and advice. We thank H. Bayle, J. Gestwicki, and W. D. Heo for helpful discussions. This work was supported by the NIH (GM068589 and GM073046).
Footnotes
Supplemental Data include three tables and ten figures and can be found with this article online at http://www.cell.com/cgi/content/full/126/5/995/DC1/.
References
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Bain J, McLauchlan H, Elliott M, Cohen P. The specificities of protein kinase inhibitors: an update. Biochem J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski LA, Wandless TJ. Conditional control of protein function. Chem Biol. 2006;13:11–21. doi: 10.1016/j.chembiol.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- Bishop AC, Shah K, Liu Y, Witucki L, Kung CY, Shokat KM. Design of allele-specific inhibitors to probe protein kinase signaling. Curr Biol. 1998;8:257–266. doi: 10.1016/s0960-9822(98)70198-8. [DOI] [PubMed] [Google Scholar]
- Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Chen X, Ye H, Kuruvilla R, Ramanan N, Scangos KW, Zhang C, Johnson NM, England PM, Shokat KM, Ginty DD. A chemical-genetic approach to studying neurotrophin signaling. Neuron. 2005;46:13–21. doi: 10.1016/j.neuron.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Clackson T, Yang W, Rozamus LW, Hatada M, Amara JF, Rollins CT, Stevenson LF, Magari SR, Wood SA, Courage NL, et al. Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc Natl Acad Sci USA. 1998;95:10437–10442. doi: 10.1073/pnas.95.18.10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corish P, Tyler-Smith C. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng. 1999;12:1035–1040. doi: 10.1093/protein/12.12.1035. [DOI] [PubMed] [Google Scholar]
- Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen RJ, Wu P, Varsahvsky A. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science. 1994;263:1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in C. elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Godl K, Wissing J, Kurtenbach A, Habenberger P, Blencke S, Gutbrod H, Salassidis K, Stein-Gerlach M, Missio A, Cotton M, Daub H. An efficient proteomics method to identify the cellular targets of protein kinase inhibitors. Proc Natl Acad Sci USA. 2003;100:15434–15439. doi: 10.1073/pnas.2535024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo WD, Meyer T. Swith-of-function mutants based on morphology classification of Ras superfamily small GTPases. Cell. 2003;113:315–328. doi: 10.1016/s0092-8674(03)00315-5. [DOI] [PubMed] [Google Scholar]
- Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- Holt DA, Luengo JI, Yamashita DS, Oh H-J, Konialian AL, Yen H-K, Rozamus LW, Brandt M, Bossard MJ, Levy MA, et al. Design, synthesis, and kinetic evaluation of high-affinity FKBP ligands and the X-ray crystal structures of their complexes with FKBP12. J Am Chem Soc. 1993;115:9925–9938. [Google Scholar]
- Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. Inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuliucci JD, Oliver SD, Morley S, Ward C, Ward J, Dalgarno D, Clackson T, Berger HJ. Intravenous safety and pharmacokinetics of a novel dimerizer drug, AP1903, in healthy volunteers. J Clin Pharmacol. 2001;41:870–879. doi: 10.1177/00912700122010771. [DOI] [PubMed] [Google Scholar]
- Janse DM, Crosas B, Finley D, Church GM. Localization to the proteasome is sufficient for degradation. J Biol Chem. 2004;279:21415–21420. doi: 10.1074/jbc.M402954200. [DOI] [PubMed] [Google Scholar]
- Kanemaki M, Sanchez-Diaz A, Gambus A, Labib K. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature. 2003;423:720–724. doi: 10.1038/nature01692. [DOI] [PubMed] [Google Scholar]
- Kohler JJ, Bertozzi CR. Regulating cell surface glycosylation by small molecule control of enzyme location. Chem Biol. 2003;10:1303–1331. doi: 10.1016/j.chembiol.2003.11.018. [DOI] [PubMed] [Google Scholar]
- Labib K, Tercero JA, Diffley JFX. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 2000;288:1643–1646. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- Liberles SD, Diver ST, Austin DJ, Schreiber SL. Inducible gene expression and protein translocation using nontoxic ligands identified by a mammalian three-hybrid screen. Proc Natl Acad Sci USA. 1997;94:7825–7830. doi: 10.1073/pnas.94.15.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer TT, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitors of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- Medema RH. Optimizing RNA interference for application in mammalian cells. Biochem J. 2004;380:593–603. doi: 10.1042/BJ20040260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Pan X, Ohneda O, Ohneda K, Lindeboom F, Iwata F, Shimizu R, Nagano M, Suwabe N, Philipsen S, Lim K-C, et al. Graded levels of GATA-1 expression modulate survival, proliferation, and differentiation of erythroid progenitors. J Biol Chem. 2005;280:22385–22394. doi: 10.1074/jbc.M500081200. [DOI] [PubMed] [Google Scholar]
- Park EC, Finley D, Szostak JW. A strategy for the generation of conditional mutations by protein destabilization. Proc Natl Acad Sci USA. 1992;89:1249–1252. doi: 10.1073/pnas.89.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Back to the future with ubiquitin. Cell. 2004;116:181–190. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- Pollock R, Clackson T. Dimerizer-regulated gene expression. Curr Opin Biotechnol. 2002;13:459–467. doi: 10.1016/s0958-1669(02)00373-7. [DOI] [PubMed] [Google Scholar]
- Raab RM, Stephanopoulos G. Dynamics of gene silencing by RNA interference. Biotechnol Bioeng. 2004;88:121–132. doi: 10.1002/bit.20216. [DOI] [PubMed] [Google Scholar]
- Ryding ADS, Sharp MGF, Mullins JJ. Conditional transgenic technologies. J Endocrinol. 2001;171:1–14. doi: 10.1677/joe.0.1710001. [DOI] [PubMed] [Google Scholar]
- Schneekloth JS, Fonseca FN, Koldobskiy M, Mandal A, De-shaies R, Sakamoto K, Crews CM. Chemical genetic control of protein levels: selective in vivo targeted degradation. J Am Chem Soc. 2004;126:3748–3754. doi: 10.1021/ja039025z. [DOI] [PubMed] [Google Scholar]
- Schreiber SL. The small-molecule approach to biology: Chemical genetics and diversity-oriented organic synthesis make possible the systematic exploration of biology. Chem Eng News. 2003;81:51–61. [Google Scholar]
- Shah K, Liu Y, Deirmengian C, Shokat KM. Engineering unnatural nucleotide specificity for Rous sarcoma virus tyrosine kinase to uniquely label its direct substrates. Proc Natl Acad Sci USA. 1997;94:3565–3570. doi: 10.1073/pnas.94.8.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankunas K, Bayle JH, Gestwicki JE, Lin YL, Wandless TJ, Crabtree GR. Conditional protein alleles using knockin mice and a chemical inducer of dimerization. Mol Cell. 2003;12:1615–1624. doi: 10.1016/s1097-2765(03)00491-x. [DOI] [PubMed] [Google Scholar]
- Tan DS. Diversity-oriented synthesis: exploring the intersections between chemistry and biology. Nat Chem Biol. 2005;1:74–84. doi: 10.1038/nchembio0705-74. [DOI] [PubMed] [Google Scholar]
- Wang H, Shimizu E, Tang YP, Cho M, Kyin M, Zuo W, Robinson DA, Alaimo PJ, Zhang C, Morimoto H, et al. Inducible protein knockout reveals temporal requirement of CaMK11 reactivation memory consolidation in the brain. Proc Natl Acad Sci USA. 2003;100:4287–4292. doi: 10.1073/pnas.0636870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Rozamus LW, Narula S, Rollins CT, Yuan R, Andrade LJ, Ram MK, Phillips TB, van Schravendijk MR, Dalgarno D, et al. Investigating protein-ligand interactions with a mutant FKBP possessing a designed specificity pocket. J Med Chem. 2000;43:1135–1142. doi: 10.1021/jm9904396. [DOI] [PubMed] [Google Scholar]
- Zaccolo M, Williams DM, Brown DM, Gherardi E. An approach to random mutagenesis of DNA using mixtures of triphosphate derivatives of nucleoside analogues. J Mol Biol. 1996;255:589–603. doi: 10.1006/jmbi.1996.0049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.