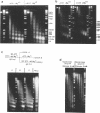
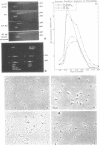
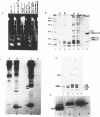
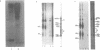Abstract
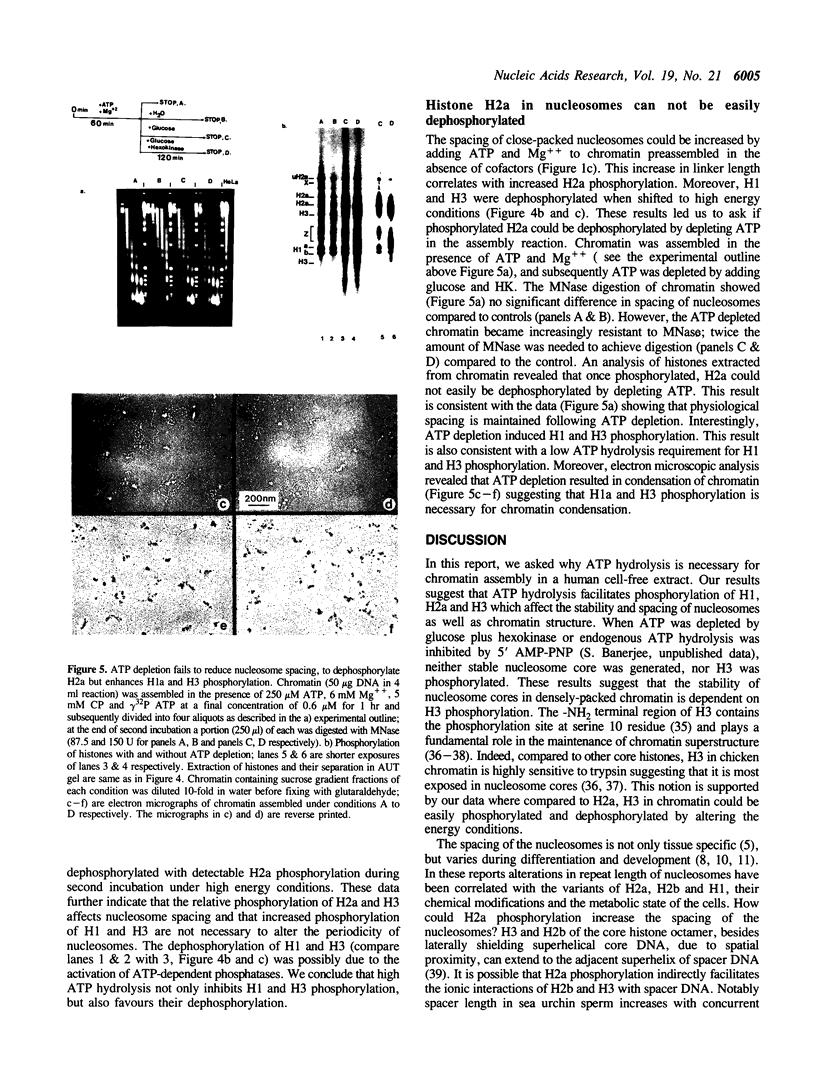
Physiologically spaced nucleosome formation in HeLa cell extracts is ATP dependent. ATP hydrolysis is required for chromatin assembly on both linear and covalently closed circular DNA. The link between the phosphorylation state of histones and nucleosome formation has been examined and we demonstrate that in the absence of histone phosphorylation no stable and regularly spaced nucleosomes are formed. Phosphorylated H3 stabilizes the nucleosome core; while phosphorylation of histone H2a is necessary to increase the linker length between nucleosomes from 0 to approximately 45 bp. Histone H1 alone, whether phosphorylated or unphosphorylated, does not increase the nucleosome repeat length in the absence of core histone phosphorylation. Phosphorylations of H1 and H3 correlate with condensation of chromatin. Maximum ATP hydrolysis which is necessary to increase the periodicity of nucleosomes from approximately 150 to approximately 185 bp, not only inhibits H1 and H3 phosphorylation but facilitates their dephosphorylation.
Full text
PDF

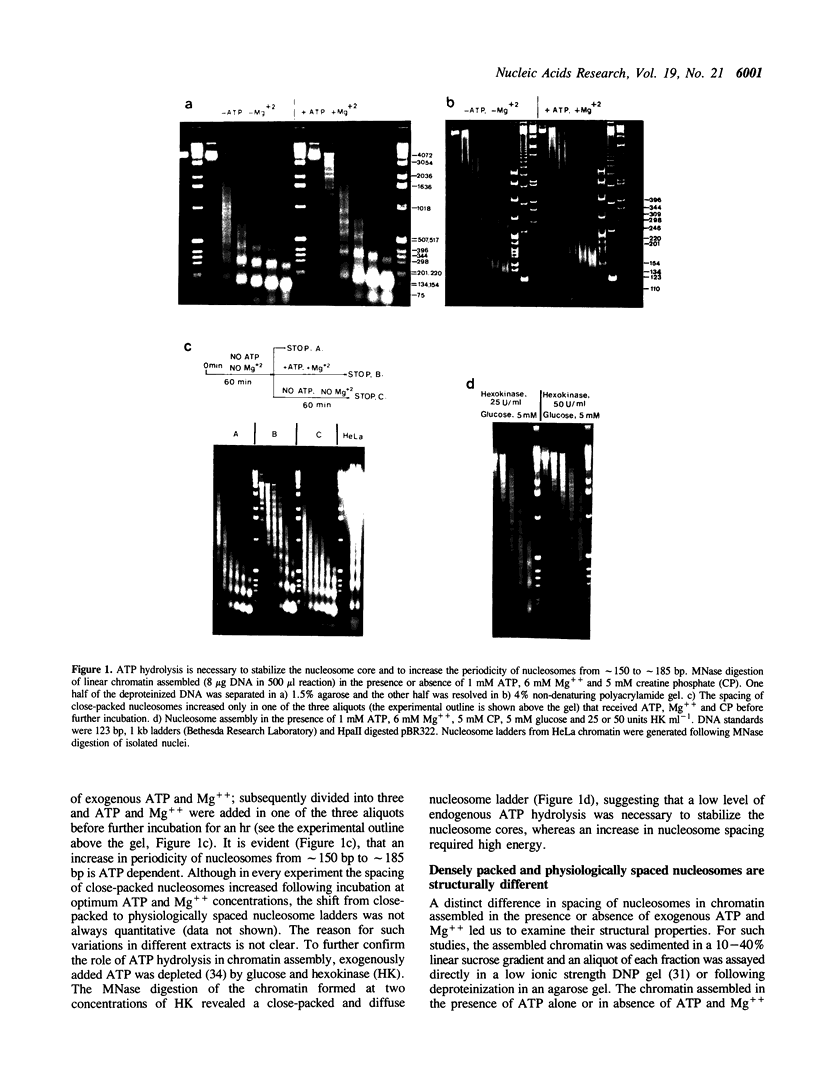

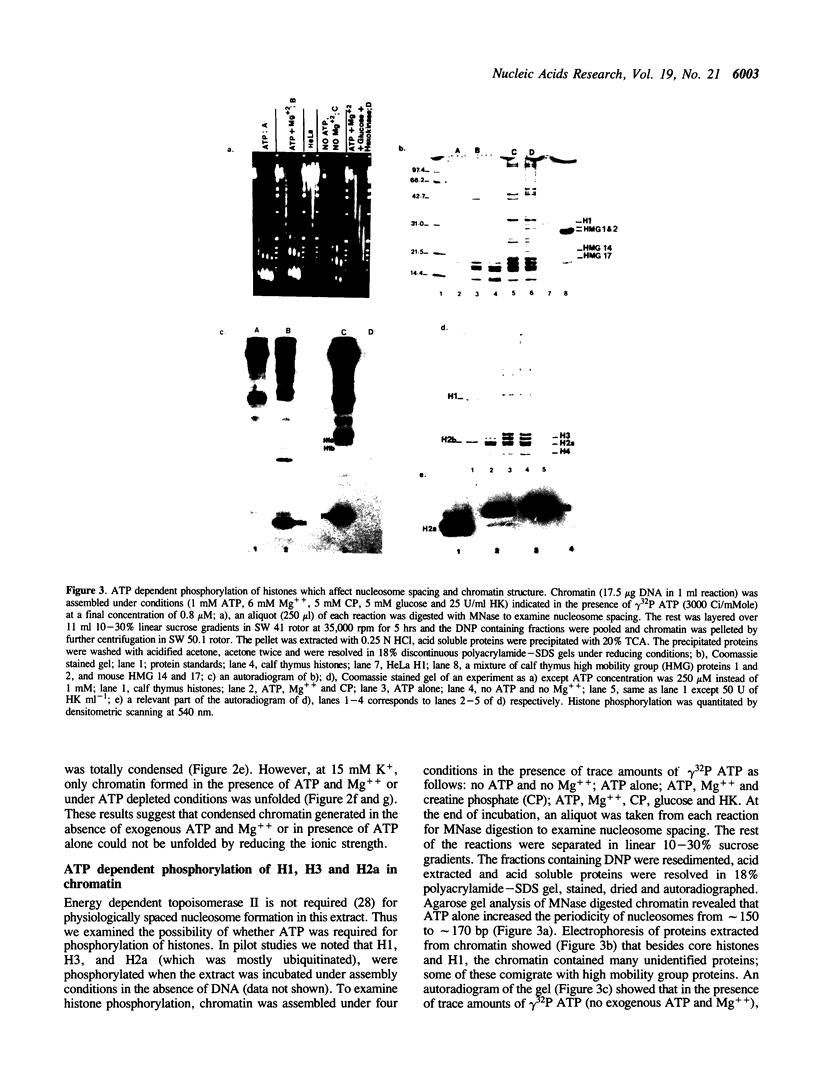
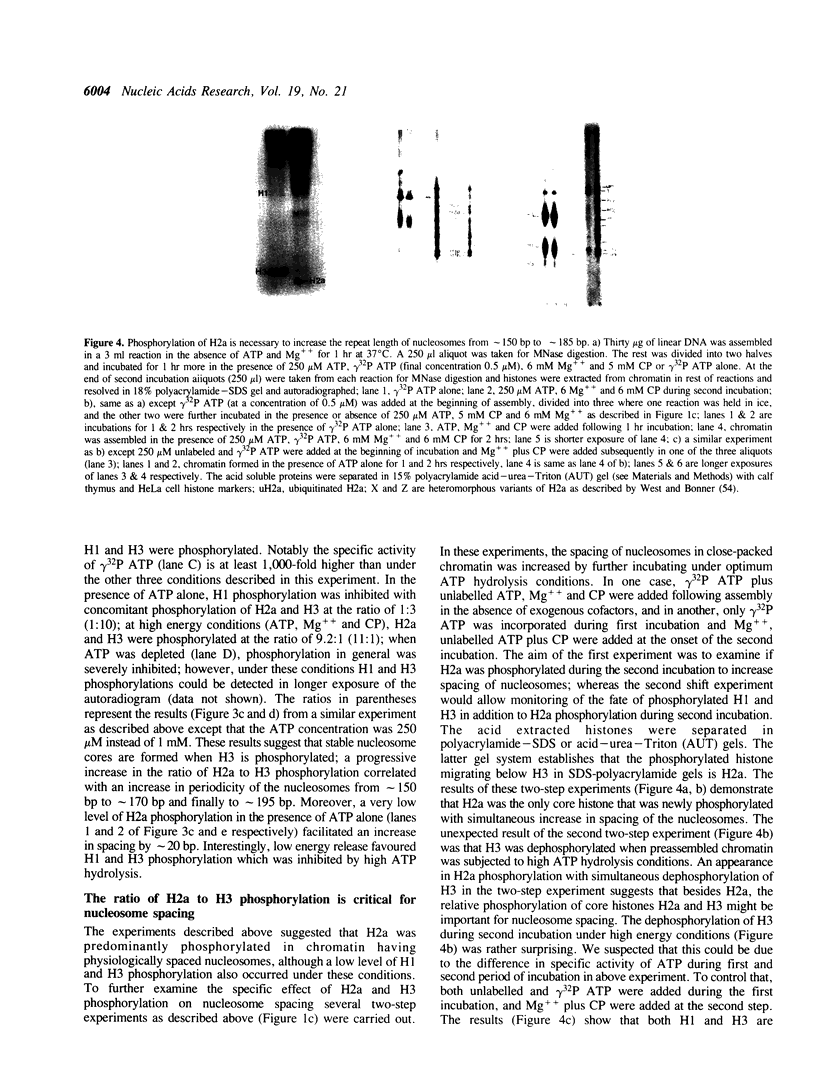

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ajiro K., Borun T. W., Cohen L. H. Phosphorylation states of different histone 1 subtypes and their relationship to chromatin functions during the HeLa S-3 cell cycle. Biochemistry. 1981 Mar 17;20(6):1445–1454. doi: 10.1021/bi00509a007. [DOI] [PubMed] [Google Scholar]
- Almouzni G., Méchali M. Assembly of spaced chromatin involvement of ATP and DNA topoisomerase activity. EMBO J. 1988 Dec 20;7(13):4355–4365. doi: 10.1002/j.1460-2075.1988.tb03334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arceci R. J., Gross P. R. Histone variants and chromatin structure during sea urchin development. Dev Biol. 1980 Nov;80(1):186–209. doi: 10.1016/0012-1606(80)90508-4. [DOI] [PubMed] [Google Scholar]
- Banerjee S., Cantor C. R. Nucleosome assembly of simian virus 40 DNA in a mammalian cell extract. Mol Cell Biol. 1990 Jun;10(6):2863–2873. doi: 10.1128/mcb.10.6.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavykin S. G., Usachenko S. I., Zalensky A. O., Mirzabekov A. D. Structure of nucleosomes and organization of internucleosomal DNA in chromatin. J Mol Biol. 1990 Apr 5;212(3):495–511. doi: 10.1016/0022-2836(90)90328-J. [DOI] [PubMed] [Google Scholar]
- Berlin R. D., Oliver J. M., Walter R. J. Surface functions during Mitosis I: phagocytosis, pinocytosis and mobility of surface-bound Con A. Cell. 1978 Oct;15(2):327–341. doi: 10.1016/0092-8674(78)90002-8. [DOI] [PubMed] [Google Scholar]
- Bonner W. M., West M. H., Stedman J. D. Two-dimensional gel analysis of histones in acid extracts of nuclei, cells, and tissues. Eur J Biochem. 1980 Aug;109(1):17–23. doi: 10.1111/j.1432-1033.1980.tb04762.x. [DOI] [PubMed] [Google Scholar]
- Bradbury E. M., Inglis R. J., Matthews H. R. Control of cell division by very lysine rich histone (F1) phosphorylation. Nature. 1974 Feb 1;247(5439):257–261. doi: 10.1038/247257a0. [DOI] [PubMed] [Google Scholar]
- Burke B., Gerace L. A cell free system to study reassembly of the nuclear envelope at the end of mitosis. Cell. 1986 Feb 28;44(4):639–652. doi: 10.1016/0092-8674(86)90273-4. [DOI] [PubMed] [Google Scholar]
- Chambers S. A., Vaughn J. P., Shaw B. R. Shortest nucleosomal repeat lengths during sea urchin development are found in two-cell embryos. Biochemistry. 1983 Nov 22;22(24):5626–5631. doi: 10.1021/bi00293a026. [DOI] [PubMed] [Google Scholar]
- Gautier J., Matsukawa T., Nurse P., Maller J. Dephosphorylation and activation of Xenopus p34cdc2 protein kinase during the cell cycle. Nature. 1989 Jun 22;339(6226):626–629. doi: 10.1038/339626a0. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glikin G. C., Ruberti I., Worcel A. Chromatin assembly in Xenopus oocytes: in vitro studies. Cell. 1984 May;37(1):33–41. doi: 10.1016/0092-8674(84)90298-8. [DOI] [PubMed] [Google Scholar]
- Gottschling D. E., Palen T. E., Cech T. R. Different nucleosome spacing in transcribed and non-transcribed regions of the ribosomal RNA gene in Tetrahymena thermophila. Nucleic Acids Res. 1983 Apr 11;11(7):2093–2109. doi: 10.1093/nar/11.7.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziani Y., Erikson E., Erikson R. L. Characterization of the Rous sarcoma virus transforming gene product. J Biol Chem. 1983 May 25;258(10):6344–6351. [PubMed] [Google Scholar]
- Green G. R., Poccia D. L. Interaction of sperm histone variants and linker DNA during spermiogenesis in the sea urchin. Biochemistry. 1988 Jan 26;27(2):619–625. doi: 10.1021/bi00402a019. [DOI] [PubMed] [Google Scholar]
- Gurley L. R., D'Anna J. A., Barham S. S., Deaven L. L., Tobey R. A. Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur J Biochem. 1978 Mar;84(1):1–15. doi: 10.1111/j.1432-1033.1978.tb12135.x. [DOI] [PubMed] [Google Scholar]
- Gurley L. R., Walters R. A., Tobey R. A. Cell cycle-specific changes in histone phosphorylation associated with cell proliferation and chromosome condensation. J Cell Biol. 1974 Feb;60(2):356–364. doi: 10.1083/jcb.60.2.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. S., Packman L. C., Thomas J. O. Phosphorylation at clustered -Ser-Pro-X-Lys/Arg- motifs in sperm-specific histones H1 and H2B. EMBO J. 1990 Mar;9(3):805–813. doi: 10.1002/j.1460-2075.1990.tb08177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt J. A., Steinbeisser H. DNA-dependent phosphorylation of histone H2A.X during nucleosome assembly in Xenopus laevis oocytes: involvement of protein phosphorylation in nucleosome spacing. EMBO J. 1991 Oct;10(10):3043–3050. doi: 10.1002/j.1460-2075.1991.tb07855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal G. W., Poccia D. Control of chromosome condensation in the sea urchin egg. Exp Cell Res. 1979 Oct 15;123(2):207–219. doi: 10.1016/0014-4827(79)90461-0. [DOI] [PubMed] [Google Scholar]
- Langan T. A., Gautier J., Lohka M., Hollingsworth R., Moreno S., Nurse P., Maller J., Sclafani R. A. Mammalian growth-associated H1 histone kinase: a homolog of cdc2+/CDC28 protein kinases controlling mitotic entry in yeast and frog cells. Mol Cell Biol. 1989 Sep;9(9):3860–3868. doi: 10.1128/mcb.9.9.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey R. A., Earnshaw W. C. Nucleosome assembly. Nature. 1980 Aug 21;286(5775):763–767. doi: 10.1038/286763a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D., Morris N. R. Assembly of SV40 chromatin in a cell-free system from Xenopus eggs. Cell. 1977 Feb;10(2):237–243. doi: 10.1016/0092-8674(77)90217-3. [DOI] [PubMed] [Google Scholar]
- Lohr D., Tatchell K., Van Holde K. E. On the occurrence of nucleosome phasing in chromatin. Cell. 1977 Nov;12(3):829–836. doi: 10.1016/0092-8674(77)90281-1. [DOI] [PubMed] [Google Scholar]
- Marion C., Roux B., Coulet P. R. Role of histones H1 and H3 in the maintenance of chromatin in a compact conformation. Study with an immobilized enzyme. FEBS Lett. 1983 Jul 4;157(2):317–321. doi: 10.1016/0014-5793(83)80568-7. [DOI] [PubMed] [Google Scholar]
- Marion C., Roux B., Pallotta L., Coulet P. R. Study of chromatin organization with trypsin immobilized on collagen membranes. Biochem Biophys Res Commun. 1983 Aug 12;114(3):1169–1175. doi: 10.1016/0006-291x(83)90685-x. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y., Yasuda H., Mita S., Marunouchi T., Yamada M. Evidence for the involvement of H1 histone phosphorylation in chromosome condensation. Nature. 1980 Mar 13;284(5752):181–183. doi: 10.1038/284181a0. [DOI] [PubMed] [Google Scholar]
- Miake-Lye R., Kirschner M. W. Induction of early mitotic events in a cell-free system. Cell. 1985 May;41(1):165–175. doi: 10.1016/0092-8674(85)90071-6. [DOI] [PubMed] [Google Scholar]
- Morris N. R. A comparison of the structure of chicken erythrocyte and chicken liver chromatin. Cell. 1976 Dec;9(4 Pt 1):627–632. doi: 10.1016/0092-8674(76)90045-3. [DOI] [PubMed] [Google Scholar]
- Müller U., Zentgraf H., Eicken I., Keller W. Higher order structure of simian virus 40 chromatin. Science. 1978 Aug 4;201(4354):406–415. doi: 10.1126/science.208155. [DOI] [PubMed] [Google Scholar]
- Nelson T., Hsieh T. S., Brutlag D. Extracts of Drosophila embryos mediate chromatin assembly in vitro. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5510–5514. doi: 10.1073/pnas.76.11.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J., Spann T. Disassembly of the nucleus in mitotic extracts: membrane vesicularization, lamin disassembly, and chromosome condensation are independent processes. Cell. 1987 Jan 30;48(2):219–230. doi: 10.1016/0092-8674(87)90425-9. [DOI] [PubMed] [Google Scholar]
- Noll M. Differences and similarities in chromatin structure of Neurospora crassa and higher eucaryotes. Cell. 1976 Jul;8(3):349–355. doi: 10.1016/0092-8674(76)90146-x. [DOI] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Paulson J. R., Taylor S. S. Phosphorylation of histones 1 and 3 and nonhistone high mobility group 14 by an endogenous kinase in HeLa metaphase chromosomes. J Biol Chem. 1982 Jun 10;257(11):6064–6072. [PubMed] [Google Scholar]
- Pearson E. C., Bates D. L., Prospero T. D., Thomas J. O. Neuronal nuclei and glial nuclei from mammalian cerebral cortex. Nucleosome repeat lengths, DNA contents and H1 contents. Eur J Biochem. 1984 Oct 15;144(2):353–360. doi: 10.1111/j.1432-1033.1984.tb08471.x. [DOI] [PubMed] [Google Scholar]
- Poccia D., Greenough T., Green G. R., Nash E., Erickson J., Gibbs M. Remodeling of sperm chromatin following fertilization: nucleosome repeat length and histone variant transitions in the absence of DNA synthesis. Dev Biol. 1984 Aug;104(2):274–286. doi: 10.1016/0012-1606(84)90083-6. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Campos A., Shimamura A., Worcel A. Assembly and properties of chromatin containing histone H1. J Mol Biol. 1989 Sep 5;209(1):135–150. doi: 10.1016/0022-2836(89)90177-0. [DOI] [PubMed] [Google Scholar]
- Ruiz-Carrillo A., Jorcano J. L., Eder G., Lurz R. In vitro core particle and nucleosome assembly at physiological ionic strength. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3284–3288. doi: 10.1073/pnas.76.7.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone G. T., Skinner J. D., Burgoyne L. A. Resistance of chromatin superstructure to tryptic digestion modulated by conjugated polyacrylamide. FEBS Lett. 1983 Jun 27;157(1):111–114. doi: 10.1016/0014-5793(83)81126-0. [DOI] [PubMed] [Google Scholar]
- Savić A., Richman P., Williamson P., Poccia D. Alterations in chromatin structure during early sea urchin embryogenesis. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3706–3710. doi: 10.1073/pnas.78.6.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura A., Tremethick D., Worcel A. Characterization of the repressed 5S DNA minichromosomes assembled in vitro with a high-speed supernatant of Xenopus laevis oocytes. Mol Cell Biol. 1988 Oct;8(10):4257–4269. doi: 10.1128/mcb.8.10.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. D., Seale R. L., Yu J. Transcribed chromatin exhibits an altered nucleosomal spacing. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5505–5509. doi: 10.1073/pnas.80.18.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadafora C., Bellard M., Compton J. L., Chambon P. The DNA repeat lengths in chromatins from sea urchin sperm and gastrule cells are markedly different. FEBS Lett. 1976 Oct 15;69(1):281–285. doi: 10.1016/0014-5793(76)80704-1. [DOI] [PubMed] [Google Scholar]
- Stein A., Bina M. A model chromatin assembly system. Factors affecting nucleosome spacing. J Mol Biol. 1984 Sep 15;178(2):341–363. doi: 10.1016/0022-2836(84)90148-7. [DOI] [PubMed] [Google Scholar]
- Stein A., Mitchell M. Generation of different nucleosome spacing periodicities in vitro. Possible origin of cell type specificity. J Mol Biol. 1988 Oct 20;203(4):1029–1043. doi: 10.1016/0022-2836(88)90127-1. [DOI] [PubMed] [Google Scholar]
- Sun J. M., Ali Z., Lurz R., Ruiz-Carrillo A. Replacement of histone H1 by H5 in vivo does not change the nucleosome repeat length of chromatin but increases its stability. EMBO J. 1990 May;9(5):1651–1658. doi: 10.1002/j.1460-2075.1990.tb08285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Thompson R. J. Variation in chromatin structure in two cell types from the same tissue: a short DNA repeat length in cerebral cortex neurons. Cell. 1977 Apr;10(4):633–640. doi: 10.1016/0092-8674(77)90096-4. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Bakayev V. V., Chumackov P. M., Georgiev G. P. Minichromosome of simian virus 40: presence of histone HI. Nucleic Acids Res. 1976 Aug;3(8):2101–2113. doi: 10.1093/nar/3.8.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G., Davoust J., Cockcroft A. Recycling of transferrin receptors in A431 cells is inhibited during mitosis. EMBO J. 1984 Oct;3(10):2217–2225. doi: 10.1002/j.1460-2075.1984.tb02119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H. The nucleosome repeat length increases during erythropoiesis in the chick. Nucleic Acids Res. 1978 Apr;5(4):1179–1188. doi: 10.1093/nar/5.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M. H., Bonner W. M. Histone 2A, a heteromorphous family of eight protein species. Biochemistry. 1980 Jul 8;19(14):3238–3245. doi: 10.1021/bi00555a022. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P., Andrews M. T., Crawford E., Losa R., Brown D. D. Negative supercoiling is not required for 5S RNA transcription in vitro. Cell. 1987 May 8;49(3):301–303. doi: 10.1016/0092-8674(87)90279-0. [DOI] [PubMed] [Google Scholar]