Abstract
Our understanding of the composition of multi-clonal malarial infections and the epidemiological factors which shape their diversity remain poorly understood. Traditionally within-host diversity has been defined in terms of the multiplicity of infection (MOI) derived by PCR-based genotyping. Massively parallel, single molecule sequencing technologies now enable individual read counts to be derived on genome-wide datasets facilitating the development of new statistical approaches to describe within-host diversity. In this class of measures the FWS metric characterizes within-host diversity and its relationship to population level diversity. Utilizing P. falciparum field isolates from patients in West Africa we here explore the relationship between the traditional MOI and FWS approaches. FWS statistics were derived from read count data at 86,158 SNPs in 64 samples sequenced on the Illumina GA platform. MOI estimates were derived by PCR at the msp-1 and -2 loci. Significant correlations were observed between the two measures, particularly with the msp-1 locus (P = 5.92×10−5). The FWS metric should be more robust than the PCR-based approach owing to reduced sensitivity to potential locus-specific artifacts. Furthermore the FWS metric captures information on a range of parameters which influence out-crossing risk including the number of clones (MOI), their relative proportions and genetic divergence. This approach should provide novel insights into the factors which correlate with, and shape within-host diversity.
Introduction
Particularly in high transmission regions, individuals may carry multiple genetically distinct parasite clones in a single malaria infection (multi-clonal infection). This dynamic poses numerous challenges to malaria control. Recombination between genetically distinct parasite clones (out-crossing) is a major risk factor for generating novel parasite variants with clinically important phenotypes such as virulence, drug resistance or immune evasion. Parasite diversity dynamics also have important implications for host immune acquisition, and influence the selection and spread of drug resistance [1], [2], [3], [4], [5]. Furthermore, the intense competition enabled between distinct clones within an infection appears to promote the evolution of highly virulent Plasmodium parasites [6], [7], [8], [9]. Numerous studies have sought correlations between parasite multi-clonality and clinical phenotypes such as patient age, immune status, clinical outcome, drug sensitivity, gametocyte production and gametocyte infectivity [10], [11], [12], [13], [14], [15], [16], [17]. However, these studies have been constrained by the limited availability of tools to effectively gauge the genetic composition of multi-clonal infections. Constrained in part by these limitations we still have only a poor understanding of how within-host diversity is shaped in different epidemiological settings.
Our current understanding is that multi-clonal infections may be generated by super-infection (the composition of multiple infectious bites with genetically distinct clones), and that this may be a frequent occurrence in high transmission areas. However, it has also been demonstrated that a single mosquito inoculation may carry a large amount of parasite diversity [18], and that this again may be dependent on the population level diversity. In addition, within-host dynamics may be shaped by factors such as host immunity and density-dependent control mechanisms [19]. In order to better understand the dynamics of multiple-clone infections and parasite out-crossing, effective tools to address within-host diversity within the context of the population-level diversity are essential.
Traditionally, within-host parasite diversity has been described by the number of distinct clones within an infection, or, multiplicity of infection (MOI). A common approach is the use of PCR to derive the number of repeat length variants observed at a few highly diverse loci, most frequently variable number tandem repeats (VNTRs) in the genes encoding antigens such as the merozoite surface protein 1 (MSP1) and 2 (MSP2) [20], [21]. The large repertoire of alleles at a single VNTR locus is practical for finger-printing malaria parasites, particularly for drug surveillance. However, limitations on the number and “nature” of the loci constrain effective characterization of within-host diversity. Furthermore, as the clone counting approach does not account for the population level of diversity, limited insight is gained into the risks of out-crossing in a given population.
Massively parallel sequencing technologies now enable characterization of parasite diversity at high depth and molecular resolution [22], [23], [24]. Individually, SNPs do not capture as much diversity as VNTRs, and so the clone counting MOI method is not effective for SNP data. Alternatively, the single molecule sequencing approach of platforms such as the Illumina Genome Analyzer enables individual allele counts to be derived at each SNP position, and the opportunity for new statistical approaches to describe within-host parasite diversity based on population genetics such as the FWS metric (Manske, Miotto et al., in preparation). This metric characterizes not just within-host diversity, but also its relationship to local population diversity, essentially measuring the risk of out-crossing/inbreeding.
Here, using PCR at the msp1 and 2 loci, we explore the relationship between the traditional VNTR-based MOI and genome-wide, SNP-based FWS metric in clinical (non-cultured) Plasmodium falciparum samples from malaria-endemic regions in West Africa.
Results and Discussion
PCR-based estimation of MOI
The traditional method for characterizing within-host diversity in P. falciparum entails MOI estimation using genotype data from the msp-1 and -2 loci [20]. This approach utilizes family specific and repeat length variants in each gene to differentiate parasite clones (see Methods). In the West African samples, genotyping success rates at the msp-1 and -2 loci were 100% (64/64) and 98% (63/64), respectively. The distributions of MOI estimates for each assay/population are presented in Figure S1. A summary of these distributions is presented in Table 1. MOI estimates were slightly higher in Burkina Faso (mean = 2.95) than Mali (mean = 2.57), but the difference was not significant (t = 1.28, P = 0.209). These estimates are similar to previous observations in Mali and Burkina Faso [25], [26]. In the combined population (West Africa), the mean MOI estimate was 2.83. The gene-specific MOI estimates demonstrated similar distributions to the maximum MOI (Figures S2, 3), although msp-2 estimates (mean = 2.54) were moderately higher than msp-1 (mean = 2.19) (t = −1.82; P = 0.071). Moderate gene-specific differences were also observed between populations. The msp-1 MOI estimates were higher in Burkina Faso (mean = 2.33) than Mali (mean = 1.90), with borderline significance (t = 1.94, P = 0.059). The msp-2 MOI estimates were only moderately higher in Burkina Faso (mean = 2.64) than Mali (mean = 2.33), and the difference was not significant (t = 0.91, P = 0.368). In summary, the MSP MOI data indicated high multi-clonality in West Africa, with slightly higher levels in Burkina Faso than Mali, and subtle differences in the estimates derived from the msp-1 and -2 loci.
Table 1. Summary of MOI and FWS estimates in Burkina Faso, Mali and the combined (West Africa) population.
| Burkina Faso | Mali | All (West Africa) | |
| Mean Maximum MOI (range) | 2.95 (1–5) | 2.57 (1–4) | 2.82 (1–5) |
| Mean MSP1 MOI (range) | 2.33 (1–4) | 1.91 (1–4) | 2.19 (1–4) |
| Mean MSP2 MOI (range) | 2.64 (1–5) | 2.33 (1–4) | 2.54 (1–5) |
| Mean FWS (range) | 0.69 (0.19–1.00) | 0.81 (0.32–1.00) | 0.73 (0.19–1.00) |
Genome-wide characterization of within-host diversity (FWS)
The FWS metric describes the relationship between the diversity observed within a patient to that of the population using estimations of heterozygosity (i.e. the probability that two randomly selected parasites carry different alleles at a given locus). In any given population, heterozygosity is dependent on the allele frequencies at that locus, as described by the Hardy-Weinberg principle, and is therefore influenced by the level of out-crossing in the population. By combining allele counts across all samples in the population, we can derive an estimate of population allele frequencies at each locus, and consequently estimate heterozygosity at the population level (HS). Similarly, for each sample, using allele counts at each locus, we can estimate within-host heterozygosity (HW), enabling computation of the FWS measure. Essentially, the FWS metric provides a measure of the risk of out-crossing between the parasites within an individual to generate new genotypes during recombination in the mosquito host. Thus, the metric captures information on all of the sample parameters which influence the risk of new parasite genotypes being generated at meiosis. These include not just the overall diversity within an individual but also the level of similarity between the parasites and their relative proportions. A low FWS reflects a low risk of inbreeding/high risk of out-crossing and thus high within-host diversity. Whilst simple calculations of the heterozygosity at each locus within a sample provide some level of information on within-host diversity, the ability to describe this diversity in the context of the level of diversity in the population is critical to capturing information on the risk of out-crossing. Consequently, population level heterozygosity estimates are essential for effective computation and interpretation of the FWS metric.
We previously described FWS distributions in a range of P. falciparum populations, demonstrating global patterns of within-host diversity consistent with our knowledge of the epidemiology and human (and parasite) migration patterns in the regions studied (Manske, Miotto et al., in preparation). In the West African samples addressed here, a mean FWS of 0.73 was observed, with 28% samples exhibiting “high” FWS estimates (i.e. ≥0.95: see Figure S2). In each of Mali and Burkina Faso, the mean FWS scores were 0.81 (29% ≥0.95) and 0.69 (30% ≥0.95) respectively. The low proportion of samples with high FWS scores is indicative of a large degree of panmixis (low sub-structure) amongst the parasites in the populations sampled here. Presumably, super-infection with genetically distinct parasites is a relatively frequent occurrence. It follows that the risks of parasite out-crossing, and consequent threat to malaria control efforts, are moderately high in these populations.
In accordance with the msp-based results, greater within-host diversity (lower FWS) was observed in Burkina Faso (mean = 0.69) than Mali (mean = 0.81) (t = −2.08, P = 0.042). This observation may reflect subtle differences in the epidemiology of the sites studied here, such as rural (Mali) versus urban (Burkina Faso) influences on transmission dynamics [27], [28]. Further exploration of the FWS profile in different epidemiological settings is required to address this and other putative determinants of within-host parasite diversity. This should also enable more effective interpretation of the relative out-crossing risks associated with different FWS scores.
A wide range of FWS scores were observed in the West African populations (Figure S2). Thus, even after gauging within-host diversity estimates in terms of the out-crossing risk and related parameters (number, relative proportion, and degree of divergence between clones), there appears to be heterogeneity in within-host diversity in the West African populations. It remains to be determined how well this heterogeneity reflects the heterogeneity in clinical outcomes such as disease severity, gametocyte prevalence and infectivity, patient age and immune status, in West Africa and other populations. To date, the parasite genetic basis of these outcomes has generally only been addressed with regard to clone count measures of within-host diversity (review in [29]). The extent to which the clone counting MOI approach reflected the additional diversity parameters captured by the FWS metric remained uncertain. We therefore explored this relationship further.
Relationship between genome-wide FWS metric and MSP1+MSP2 MOI estimates
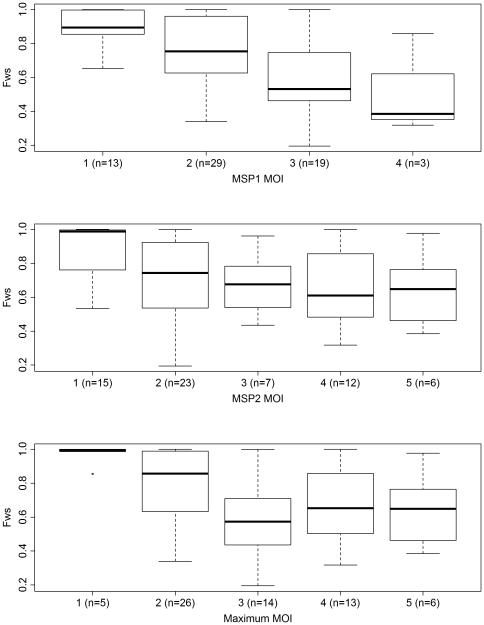
Using the FWS score as a proxy to genome-wide within-host SNP diversity, we assessed the correlation between this metric and the MSP-based MOI estimates. A significant negative correlation was observed between the MSP1 MOI and FWS (rho = −0.480, P = 5.92×10−5), as illustrated in Figure 1. The FWS correlations with each of the MSP2 and maximum MOI estimates were less strong but both remained significant (rho = −0.366, P = 0.003; rho = −0.384, P = 0.002, respectively) (Figure 1). These correlations demonstrated that the MOI and FWS metric broadly agreed on their interpretations of within-host diversity. However, differences in two key areas, 1) the analytical approach, and 2) the number and nature of loci examined, may underlie the remaining deviations between the two measures.
Figure 1. FWS against MSP1 MOI, MSP2 MOI, and maximum MOI.
A moderate proportion of samples (13/64 [20.3%]) demonstrated multiple clones by MSP genotyping but appeared to be largely “clonal” with respect to the FWS (≥0.95). These results presumably reflect infections with multiple clones exhibiting limited divergence and thus limited within-host diversity. As detailed below, any of a number of differences, both technical and analytical, between the PCR-based MOI and FWS approaches may underlie these discrepancies. Fewer samples (4/64 [6.25%]) exhibited a clonal MSP outcome concurrent with high diversity FWS (≤0.70). The ability to capture the relatedness between the clones within an infection should facilitate our understanding of the impact of within-host regulatory effects such as inter-parasite competition and host immunity on parasite diversity dynamics.
In addition to the core analytical approach, the PCR-based MOI approach may be constrained in ability to capture the complexity of an individual infection owing to sensitivity to the number of loci examined, the level of polymorphism at each locus, family-specific versus intra-family variation and selective pressures at one or more loci. In these respects, the genome-wide, SNP-based, FWS approach is more robust. In general, even with highly diverse loci, more markers ensure a more accurate representation of diversity [30]. Indeed, a larger number of multiple clone infections were detected by the maximum MOI (92% [59/64]) than with the msp-1 (79% [50/63]) and -2 (77% [49/64]) loci individually. However, practicalities limit the number of loci that can be examined by PCR.
Owing to differences in the level and nature of polymorphism, inter-locus variation in diversity is not unexpected within a multi-clonal malaria infection. Indeed, the msp-1 and -2 genes exhibited modest differences in polymorphism, reflected in their different strengths of correlation with the FWS metric (Figure 1). Inter-locus differences may also result from locus-specific selective pressures which are not representative of the genome as a whole. This is a potential limitation of MSP1 and 2, which appear to be under selective pressure from the host immune system [31].
The distinction of alleles at VNTR loci by electrophoretic mobility is another potential limitation of the PCR-based approach, as differences in sequence composition between equally migrating fragments may be missed. Furthermore, it is generally accepted that VNTRs evolve at different rates and under different mechanisms from SNPs [32], [33], [34], [35]. Thus, VNTRs may not provide an effective representation of the SNP diversity in the genome. Rather, as observed with the moderately high frequency (20.3%) of isolates demonstrating largely clonal dynamics with FWS estimates but polyclonal dynamics at the MSP loci, VNTR-based estimates may overestimate the true genome-wide within-host diversity.
A potential shortfall of the FWS approach is the integrity of the genome-wide dataset. A highly stringent SNP discovery and allele-calling process was employed here to ensure high confidence in the dataset (see Methods). In addition, only good quality samples with high sequence depth were included (Figure S3). The comparable sensitivity of the PCR-based and genome-sequencing approach has yet to be determined. However, in contrast to the potential ambiguity in detecting and distinguishing bands on gels, the ability to count individual reads in Illumina sequence data enables more objective allele-calling.
Juliano and colleagues achieved extensive sequence depth at the msp-1 and -2 loci by focusing 454 sequencing on amplicons of these two genes alone [36]. Using the clone-counting approach, facilitated by haplotype information, an average of ∼5 more msp-1 and -2 variants were detected relative to the genotyping data. As with the PCR-based MOI method, this approach is powerful for distinguishing clones in clinical drug trials. For characterization of within-host diversity however, the use of just 2 VNTR loci under potential selective pressure renders this approach sensitive to the features discussed above. With continual improvements in read length and sequence yield on high-throughput, single-molecule sequencing platforms, haplotype-based clone counting approaches similar to those described by Juliano and colleagues should be feasible on a broader, genome-wide, scale.
Conclusion
The traditional approach to estimating MOI using VNTRs at a handful of loci such as msp-1 and -2 is a simple and moderately cheap method with demonstrated practicality for finger-printing clones in drug trials. However, genome-wide measures such as the FWS statistic, by capturing information not just on the number of clones in a sample but also on their respective ratios and the degree of inter-clone variation at hundreds of thousands of polymorphic positions, provide a greater wealth of information on within-host diversity which is highly robust against potential locus-specific artifacts. With rapidly advancing progress in whole genome sequencing technologies, including ever increasing read lengths and continual reductions in cost, this approach is now feasible in hundreds of samples. The ability to assess within-host diversity in genome-wide datasets from non-cultured (clinical) samples sourced from low resource field settings, as demonstrated here, should revolutionize clinical and epidemiological studies of Plasmodium.
Methods
Samples
Samples were collected from field sites in Mali and Burkina Faso within the framework of a large, multi-center P. falciparum genome sequencing project to facilitate SNP discovery and population genetic characterization (Manske, Miotto et al., in preparation). Consenting patients of all ages and ethnic groups presenting at the clinic with symptoms of uncomplicated malaria and P. falciparum parasitaemia were recruited to the study. In Mali, samples were collected from clinics in two rural villages (Kolle and Faladje). In Burkina Faso, samples were collected from permanent health dispensaries in three suburbs of Bobo-Dioulasso (Colsamma, Ouezzinville, and Sakaby). Details of the sample processing procedures in the laboratory are described elsewhere [37]. Briefly, venous blood (2–8 ml) was processed to deplete the human white blood cell fraction, and DNA extraction was undertaken using Qiagen QIAamp blood extraction kits as per the manufacturer's instructions.
Ethics
Informed, written consent was obtained from patients over 18 years of age and from a parent or guardian for younger patients. The study was approved by the Comite d'Ethique de la Faculté de Médecine de Pharmacie et d'Odontostomatologie, Bamako, Mali, and Comite d'Ethique Institutionnel du Centre Muraz, Bobo- Dioulasso, Burkina Faso.
Sequencing and SNP Calling
Sixty-four samples (21 Mali, 43 Burkina Faso) with >500 ng total DNA and <60% human DNA contamination were sequenced on the Illumina Genome Analyzer platform. Details of the sequencing, SNP discovery and genotype calling process are described elsewhere (Manske, Miotto et al., in preparation). Briefly, up to 6 lanes were sequenced per sample, with 37, 54 or 76 bp paired-end reads. Coverage distributions are presented in Figure S3. The “raw” sequence data for the samples can be accessed in the European Nucleotide Archive (www.ebi.ac.uk/ena/data/search/?query=plasmodium). For each sample, sequence data from multiple lanes was merged, and mapped to the P. falciparum reference genome (3D7 version 2.1.5; ftp://ftp.sanger.ac.uk/pub/pathogens/Plasmodium/falciaprum/3D7/3D7.version2.1.5) using the bwa program (available from http://bio-bwa.sourceforge.net) with the default parameters [38]. The resulting alignments were processed in samtools (http://samtools.sourceforge.net/cns0.shtml) using the default parameters to identify positions with one or more bases differing from the reference sequence (putative SNPs). Genotypes were called at 86,158 high confidence SNP positions (typable SNPs) identified by a stringent quality-filtering SNP discovery process detailed elsewhere (Manske, Miotto et al., in preparation). This SNP list is referred to as version 1gamma and is available on the MapSeq database (www.mapseq.net/pf). For the purpose of assigning confident genotype calls to the dataset, all genotypes in the version 1 gamma dataset with less than 5 reads were assigned a status of missing data.
Calculation of FWS
Details of the underlying basis for calculations used to derive the Fws metric are described elsewhere (Manske, Miotto et al., in preparation). The FWS metric was calculated for each individual sample, using the formula FWS = 1−(Hw/Hs), where Hw is the within-individual heterozygosity and Hs is the within-population heterozygosity. At each biallelic SNP, heterozygosity was estimated using the formula H = 1−(p 2+q 2), where p and q are the frequencies of the two alleles (p = 1−q). At each SNP, p and q were estimated for each individual as the proportions of sequencing reads that carried each allele in the individual sample. At population level, the allele frequencies at the SNP were estimated as the mean of the allele frequencies in the individuals comprising the population sample; the frequency of the least common allele at that SNP in the West African population was noted as the minor allele frequency (MAF). Since heterozygosity depends on allele frequencies, each of the 86,158 SNPs was assigned to one of ten equally-sized MAF intervals ([0.0–0.05], [0.05–0.1] … [0.45–0.5]). Estimates of Hw and Hs were calculated for each MAF interval, as the means of the Hw and Hs across all SNPs in that interval. For each individual, Hw estimates were plotted against the corresponding Hs estimates for all MAF intervals, and the slope of the resulting straight-line plot was used to evaluate the Hw/Hs ratio, and thus compute FWS for the individual.
Genotype-based MOI Estimation
The traditional method of genotyping in the P. falciparum merozoite surface protein - 1 (msp-1) and -2 (msp-2) genes was undertaken on 64 of the typable samples using a nested PCR approach [20]. This method utilises family specific variation and variation in repeat length in each gene to distinguish parasite clones. In general, each parasite will exhibit one of three family-specific sequences, K1, R033 or MAD20, in the msp-1 gene, and one of two family-specific sequences, FC27 or IC/3D7, in the msp-2 gene. Within each family-specific sequence, regions comprising repeat-length polymorphisms enable further distinction of clones. The number of distinct bands observed in each sample was summed across the families in each of the msp-1 and -2 genes separately, providing gene-specific MOI estimates. The maximum MOI estimate was then derived as the maximum gene-specific MOI estimate for each sample. The standard Welch two-sample t-test was used to measure the significance of difference in MOI estimates between populations and assays using R software.
Supporting Information
MSP-based MOI Estimates.
(TIFF)
Distribution of Fws scores in the West African samples. Dashed lines indicate thresholds for highly diverse (Fws≤0.7) and moderately “clonal” (Fws≥0.95) samples.
(TIFF)
Read depth distribution at the typable SNP positions in the West African samples. Distribution of SNP coverage at read depth (number of sequenced nucleotides covering a given locus) thresholds of 1, 5, 10, 15, 20 and 25.
(TIFF)
Acknowledgments
We wish to thank the patients who donated samples, the numerous individuals who facilitated sample collections in the field sites, and the Wellcome Trust Sanger Institute library preparation and sequencing teams.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The genetic component of this research was funded by the Wellcome Trust, the Bill and Melinda Gates Foundation, and the Medical Research Council. SA, SC, OM, MM, GM, DA, BM, KAA, TGC and DPK were funded by the Wellcome Trust, the Bill and Melinda Gates Foundation, and the Medical Research Council. AAD is supported by a European and Developing Countries Clinical Trial Partnership Senior Fellowship (grant 2004.2.C.f1) and a Howard Hughes Medical Institution International Scholarship (grant 55005502). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hastings IM, D'Alessandro U. Modelling a predictable disaster: the rise and spread of drug-resistantmalaria. Parasitol Today. 2000;16:340–347. doi: 10.1016/s0169-4758(00)01707-5. [DOI] [PubMed] [Google Scholar]
- 2.Contamin H, Fandeur T, Rogier C, Bonnefoy S, Konate L, et al. Different genetic characteristics of Plasmodium falciparum isolates collected during successive clinical malaria episodes in Senegalese children. Am J Trop Med Hyg. 1996;54:632–643. doi: 10.4269/ajtmh.1996.54.632. [DOI] [PubMed] [Google Scholar]
- 3.Forsyth KP, Philip G, Smith T, Kum E, Southwell B, et al. Diversity of antigens expressed on the surface of erythrocytes infected with mature Plasmodium falciparum parasites in Papua New Guinea. Am J Trop Med Hyg. 1989;41:259–265. [PubMed] [Google Scholar]
- 4.Marsh K, Otoo L, Hayes RJ, Carson DC, Greenwood BM. Antibodies to blood stage antigens of Plasmodium falciparum in rural Gambians and their relation to protection against infection. Trans R Soc Trop Med Hyg. 1989;83:293–303. doi: 10.1016/0035-9203(89)90478-1. [DOI] [PubMed] [Google Scholar]
- 5.Newbold CI, Pinches R, Roberts DJ, Marsh K. Plasmodium falciparum: the human agglutinating antibody response to the infected red cell surface is predominantly variant specific. Exp Parasitol. 1992;75:281–292. doi: 10.1016/0014-4894(92)90213-t. [DOI] [PubMed] [Google Scholar]
- 6.Bell AS, de Roode JC, Sim D, Read AF. Within-host competition in genetically diverse malaria infections: parasite virulence and competitive success. Evolution. 2006;60:1358–1371. [PubMed] [Google Scholar]
- 7.de Roode JC, Culleton R, Bell AS, Read AF. Competitive release of drug resistance following drug treatment of mixed Plasmodium chabaudi infections. Malar J. 2004;3:33. doi: 10.1186/1475-2875-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Roode JC, Pansini R, Cheesman SJ, Helinski ME, Huijben S, et al. Virulence and competitive ability in genetically diverse malaria infections. Proc Natl Acad Sci U S A. 2005;102:7624–7628. doi: 10.1073/pnas.0500078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mackinnon MJ, Read AF. Virulence in malaria: an evolutionary viewpoint. Philos Trans R Soc Lond B Biol Sci. 2004;359:965–986. doi: 10.1098/rstb.2003.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.al-Yaman F, Genton B, Reeder JC, Anders RF, Smith T, et al. Reduced risk of clinical malaria in children infected with multiple clones of Plasmodium falciparum in a highly endemic area: a prospective community study. Trans R Soc Trop Med Hyg. 1997;91:602–605. doi: 10.1016/s0035-9203(97)90046-8. [DOI] [PubMed] [Google Scholar]
- 11.Engelbrecht F, Togel E, Beck HP, Enwezor F, Oettli A, et al. Analysis of Plasmodium falciparum infections in a village community in Northern Nigeria: determination of msp2 genotypes and parasite-specific IgG responses. Acta Trop. 2000;74:63–71. doi: 10.1016/s0001-706x(99)00044-3. [DOI] [PubMed] [Google Scholar]
- 12.Felger I, Smith T, Edoh D, Kitua A, Alonso P, et al. Multiple Plasmodium falciparum infections in Tanzanian infants. Trans R Soc Trop Med Hyg. 1999;93(Suppl 1):29–34. doi: 10.1016/s0035-9203(99)90324-3. [DOI] [PubMed] [Google Scholar]
- 13.Mayor A, Saute F, Aponte JJ, Almeda J, Gomez-Olive FX, et al. Plasmodium falciparum multiple infections in Mozambique, its relation to other malariological indices and to prospective risk of malaria morbidity. Trop Med Int Health. 2003;8:3–11. doi: 10.1046/j.1365-3156.2003.00968.x. [DOI] [PubMed] [Google Scholar]
- 14.Owusu-Agyei S, Smith T, Beck HP, Amenga-Etego L, Felger I. Molecular epidemiology of Plasmodium falciparum infections among asymptomatic inhabitants of a holoendemic malarious area in northern Ghana. Trop Med Int Health. 2002;7:421–428. doi: 10.1046/j.1365-3156.2002.00881.x. [DOI] [PubMed] [Google Scholar]
- 15.Smith T, Beck HP, Kitua A, Mwankusye S, Felger I, et al. Age dependence of the multiplicity of Plasmodium falciparum infections and of other malariological indices in an area of high endemicity. Trans R Soc Trop Med Hyg. 1999;93(Suppl 1):15–20. doi: 10.1016/s0035-9203(99)90322-x. [DOI] [PubMed] [Google Scholar]
- 16.Taylor LH, Walliker D, Read AF. Mixed-genotype infections of malaria parasites: within-host dynamics and transmission success of competing clones. Proc Biol Sci. 1997;264:927–935. doi: 10.1098/rspb.1997.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zwetyenga J, Rogier C, Tall A, Fontenille D, Snounou G, et al. No influence of age on infection complexity and allelic distribution in Plasmodium falciparum infections in Ndiop, a Senegalese village with seasonal, mesoendemic malaria. Am J Trop Med Hyg. 1998;59:726–735. doi: 10.4269/ajtmh.1998.59.726. [DOI] [PubMed] [Google Scholar]
- 18.Druilhe P, Daubersies P, Patarapotikul J, Gentil C, Chene L, et al. A primary malarial infection is composed of a very wide range of genetically diverse but related parasites. J Clin Invest. 1998;101:2008–2016. doi: 10.1172/JCI119890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Portugal S, Carret C, Recker M, Armitage AE, Goncalves LA, et al. Host-mediated regulation of superinfection in malaria. Nat Med. 2011;17:732–737. doi: 10.1038/nm.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snounou G, Beck HP. The use of PCR genotyping in the assessment of recrudescence or reinfection after antimalarial drug treatment. Parasitol Today. 1998;14:462–467. doi: 10.1016/s0169-4758(98)01340-4. [DOI] [PubMed] [Google Scholar]
- 21.Viriyakosol S, Siripoon N, Petcharapirat C, Petcharapirat P, Jarra W, et al. Genotyping of Plasmodium falciparum isolates by the polymerase chain reaction and potential uses in epidemiological studies. Bull World Health Organ. 1995;73:85–95. [PMC free article] [PubMed] [Google Scholar]
- 22.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 25.Bereczky S, Dolo A, Maiga B, Hayano M, Granath F, et al. Spleen enlargement and genetic diversity of Plasmodium falciparum infection in two ethnic groups with different malaria susceptibility in Mali, West Africa. Trans R Soc Trop Med Hyg. 2006;100:248–257. doi: 10.1016/j.trstmh.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 26.Soulama I, Nebie I, Ouedraogo A, Gansane A, Diarra A, et al. Plasmodium falciparum genotypes diversity in symptomatic malaria of children living in an urban and a rural setting in Burkina Faso. Malar J. 2009;8:135. doi: 10.1186/1475-2875-8-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coene J. Malaria in urban and rural Kinshasa: the entomological input. Med Vet Entomol. 1993;7:127–137. doi: 10.1111/j.1365-2915.1993.tb00665.x. [DOI] [PubMed] [Google Scholar]
- 28.Omumbo JA, Guerra CA, Hay SI, Snow RW. The influence of urbanisation on measures of Plasmodium falciparum infection prevalence in East Africa. Acta Trop. 2005;93:11–21. doi: 10.1016/j.actatropica.2004.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiwanuka GN. Genetic diversity in Plasmodium falciparum merozoite surface protein 1 and 2 coding genes and its implications in malaria epidemiology: a review of published studies from 1997–2007. J Vector Borne Dis. 2009;46:1–12. [PubMed] [Google Scholar]
- 30.Havryliuk T, Ferreira MU. A closer look at multiple-clone Plasmodium vivax infections: detection methods, prevalence and consequences. Mem Inst Oswaldo Cruz. 2009;104:67–73. doi: 10.1590/s0074-02762009000100011. [DOI] [PubMed] [Google Scholar]
- 31.Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, et al. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- 32.Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, et al. High resolution of human evolutionary trees with polymorphic microsatellites. Nature. 1994;368:455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- 33.Forbes SH, Hogg JT, Buchanan FC, Crawford AM, Allendorf FW. Microsatellite evolution in congeneric mammals: domestic and bighorn sheep. Mol Biol Evol. 1995;12:1106–1113. doi: 10.1093/oxfordjournals.molbev.a040284. [DOI] [PubMed] [Google Scholar]
- 34.Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 35.Schlotterer C. Genome evolution: are microsatellites really simple sequences? Curr Biol. 1998;8:R132–134. doi: 10.1016/s0960-9822(98)70989-3. [DOI] [PubMed] [Google Scholar]
- 36.Juliano JJ, Porter K, Mwapasa V, Sem R, Rogers WO, et al. Exposing malaria in-host diversity and estimating population diversity by capture-recapture using massively parallel pyrosequencing. Proc Natl Acad Sci U S A. 2010;107:20138–20143. doi: 10.1073/pnas.1007068107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auburn S, Campino S, Clark TG, Djimde AA, Zongo I, et al. An Effective Method to Purify Plasmodium falciparum DNA Directly from Clinical Blood Samples for Whole Genome High-Throughput Sequencing. PLoS One. 2011;6:e22213. doi: 10.1371/journal.pone.0022213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MSP-based MOI Estimates.
(TIFF)
Distribution of Fws scores in the West African samples. Dashed lines indicate thresholds for highly diverse (Fws≤0.7) and moderately “clonal” (Fws≥0.95) samples.
(TIFF)
Read depth distribution at the typable SNP positions in the West African samples. Distribution of SNP coverage at read depth (number of sequenced nucleotides covering a given locus) thresholds of 1, 5, 10, 15, 20 and 25.
(TIFF)