Abstract
Chemosensory receptors (CR) are crucial for animals to sense the environmental changes and survive on earth. The emergence of whole-genome sequences provides us an opportunity to identify the entire CR gene repertoires. To completely gain more insight into the evolution of CR genes in vertebrates, we identified the nearly all CR genes in 25 vertebrates using homology-based approaches. Among these CR gene repertoires, nearly half of them were identified for the first time in those previously uncharacterized species, such as the guinea pig, giant panda and elephant, etc. Consistent with previous findings, we found that the numbers of CR genes vary extensively among different species, suggesting an extreme form of ‘birth-and-death’ evolution. For the purpose of facilitating CR gene analysis, we constructed a database with the goals to provide a resource for CR genes annotation and a web tool for exploring their evolutionary patterns. Besides a search engine for the gene extraction from a specific chromosome region, an easy-to-use phylogenetic analysis tool was also provided to facilitate online phylogeny study of CR genes. Our work can provide a rigorous platform for further study on the evolution of CR genes in vertebrates.
Introduction
In vertebrate, chemosensory receptors (CR) can detect different odorant and taste molecules in the environment and play important roles for the survival of animals because it can help them to find food, identify mates and avoid danger, etc. [1], [2], [3], [4]. Vertebrate CRs are all G protein-coupled receptors (GPCRs) characterized by their seven trans-membrane regions, and CR genes are diversely encoded in animal genomes. CRs mainly contain six different multigene families in vertebrate animals: olfactory receptor (OR), vomeronasal receptor type 1 and type 2 (V1R and V2R), trace amine-associated receptor (TAAR) and taste receptor type 1 and type 2 (T1R and T2R). OR, V1R, V2R and TAAR genes encode pheromone receptors, among which OR and TAAR genes are mainly expressed in the main olfactory epithelium (MOE), and V1R and V2R are encoded in the vomeronasal organ (VNO). The taste receptors, T1R and T2R, are mainly expressed in the taste buds of the tongue [2].
Previous studies have identified many CR genes using PCR-based approaches [5]–[12]. However, these methods have many limitations which cannot identify the entire set of CR genes and might overestimate the proportion of potentially functional genes. With the emergence of whole-genome sequences, it is possible for us to identify the nearly complete CR gene repertoires using data-mining methods [3], [4], [13]–[27]. Up to date, myriads of works have identified many CR gene repertoires and reported that the numbers of CR genes vary extensively among different species [2]. For examples, rats have nearly ∼1,800 OR genes, whereas humans only have ∼800 OR genes [20]; Herbivorous (such as cows and horses) and omnivorous animals (such as humans and mice) have larger T2R gene repertoires than that of carnivores (such as dogs) [14], [23]; Zebrafishes have more than 100 TAAR genes, whereas humans have not more than 10 TAAR genes [26]. Moreover, the faction of pseudogenes also varies extensively among different species [2]. These findings suggested that CR genes are subject to an extreme form of birth-and-death evolution, and are closely related to the survival environments and dietary habits of different species [14], [20]–[23], [25]–[27].
These CR genes provide us an opportunity to better understand their evolution among vertebrate animals. In this work, we identified and annotated nearly the entire sets of CR genes in 25 vertebrate species whose high-coverage genome sequences are available. Furthermore, we constructed a database of CR genes in vertebrates (CRDB, http://zldev.ccbr.utoronto.ca/CRDB/) with the purpose of facilitating CR gene analysis. The product of this effort might become an important repository for CR genes in vertebrates.
Results and Discussion
Identification of CR genes in vertebrates
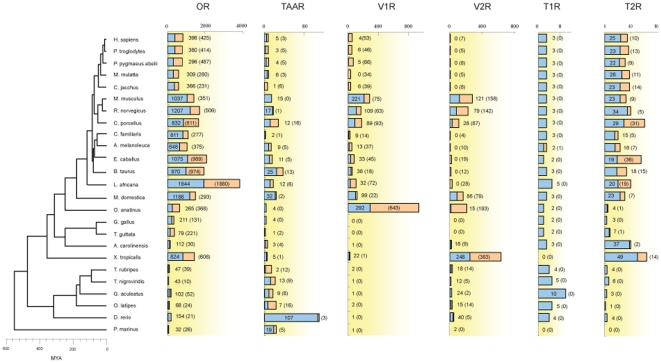
Using previously published CR proteins as query sequences, we identified the CR genes from 25 vertebrate genome sequences, which can be classified into six different multigene families (see Materials and methods ). Figure 1 summarizes the numbers of identified CR intact genes and pseudogenes in each species. It contains a total of 28,596 CR genes, including 23,217 OR genes, 449 TAAR genes, 2,288 V1R genes, 1,902 V2R genes, 80 T1R genes and 660 T2R genes, respectively. The number of CR genes are quite different among these species, which reinforces the concept of an extreme form of ‘birth-and-death’ evolution of CR genes, especially in OR gene family [2]. Many works have reported the extreme form of ‘birth-and-death’ evolution in olfactory and bitter taste receptor genes [2], [13], [14], [20]. To further detect this evolution pattern of CR genes, we performed a further analysis using simulation method. Briefly, we randomly selected 50 genes from 10 species, and then built the phylogenetic tree to detect whether these genes underwent ‘birth-and-death’ evolution. This work performed 1,000 times, and then we found significant ‘birth-and-death’ mechanisms at each time (100% bootstrap value support).
Figure 1. Number of chemosensory receptor genes in vertebrates.
The numbers of each bar represent the number of functional (intact) and pseudogenes of OR, TAAR, V1R, V2R, T1R and T2R genes. The blue and orange bars represent the number of functional genes and pseudogenes, respectively.
(i) OR genes
OR genes form one of the largest multigene families in vertebrate animals. As shown in Figure 1 , OR genes are present in all vertebrate species, among which 13,396 intact OR genes and 9,821 pseudogenes were identified. The number of OR genes is not equally distributed in each species. It is obviously that mammals contain larger OR gene repertories (400–3,800 genes) than that of fishes and birds (30–100 genes). For example, mice and dogs are considered as macrosmatic animals that rely on the sense of smell, and they contain 1,388 and 1,088 OR genes [20], respectively. In contrast, fishes have only ∼100 OR genes which are much smaller than that of tetrapod animals. Consistent with the concept that birds often have poor sense of smell, limited numbers of OR genes were found in the chicken and zebra finch (342 and 300 genes, respectively). In addition to the previously identified OR genes, we also detected OR genes from some previously uncharacterized species. We found 1,643 OR genes from the guinea pig genome, which is nearly similar to the numbers of OR genes in the mouse and rat. Surprisingly, nearly half of OR genes in the guinea pig are pseudogenes (49% pseudogenes), which are significantly higher than those of the mouse (25%) and rat (29%) [20]. Cows and horses are domesticated herbivores, and they both contain large OR gene repertoires (1,944 and 2,064 genes) and high fractions of pseudogenes (50% and 48%). It was documented that horses and cows are all have strong sense of smell [28], [29], [30]. It raised an interesting question why these animals have large numbers of OR genes and nearly the same number of intact gene and pseudogenes. As pseudogenes can be regarded as relics under the relaxed natural selection, it is possible that some of functional genes have become nonfunctional during the domestication. The giant panda, as a ‘vegetarian carnivore’, contains 1,023 OR genes (648 intact genes and 375 pseudogenes), and indicates that the giant panda still has a strong sense of smell which is concerted with previous findings [31]. Notably, we found that the elephant contains the largest OR gene repertoires (3,824 genes) among vertebrate species, indicating its keener sense of smell than other species.
(ii) TAAR genes
TAAR can respond to trace amines. In contrast to OR genes, the number of TAAR genes is smaller ( Figure 1 ). For examples, primates have no more than 10 TAAR genes; in the chicken and zebra finch, only three and four TAAR genes were identified. Interestingly, the number of TAAR genes highly diverse in fishes, and an expansion of TAAR genes were found in the zebrafish [26]. The real reason is still unclear, and it is possible that biogenic amines are more important odorants in fishes than that of tetrapod animals.
(iii) VR genes
The vomeronasal organ can detect pheromones and is responsible for various behavioral and neuroendocrine responses [25]. Two multigene families of vomeronasal pheromone receptors, V1R and V2R, differ extensively in expression locations and gene structures. Many VR genes have been identified and it has been suggested that V1Rs detect airborne chemicals, whereas V2Rs detect water-soluble molecules [25]. For example, teleost fishes lack V1R genes, but have expanded V2R gene repertoires (20–50 genes) [25]. In this work, we found the guinea pig also has relatively expanded VR gene families (182 V1Rs and 115 V2Rs), which is consistent with the finding that rodents have strong vomeronasal pheromone perceptions. The horse, cow and elephant have small sizes of V1R families (78, 56 and 104 genes respectively), whereas V2R genes totally degenerated in these species. It has been documented that the lizard vomeronasal organ is very poorly supplied with secretion [32], and we only found one V1R genes and 25 V2R genes in the lizard. We did not find any VR genes in the chicken and zebra finch genomes indicating the lack of the vomeronasal system in birds.
(iv) T1R genes
Taste perception is essential to diet selection in animals. In vertebrates, T1R and T2R are characterized as sweet/umami and bitter taste receptors, respectively. T1R family contains three members: T1R1 and T1R3 are receptors for sensing umami taste, and T1R2 is receptors for sensing sweet taste [23]. Most mammals contain three T1R genes, except in the horse, platypus and elephant. For examples, the elephant contains five T1R genes, including three T1R2 genes; T1R2 gene is not present in the horse and T1R3 is lost in the platypus. Interestingly, T1R1 gene in the panda has become a pseudogene, which can partly explain why the panda is a ‘vegetarian carnivore’ [33]. Furthermore, T1R2 gene was not found in avian genomes. In teleost fishes, many T1R2 genes were found, indicating the importance of T1R2 genes in teleost fishes.
(v) T2R genes
It was reported that many poisonous substances tend to be bitter taste, and the sense of bitter taste can prevent animals from ingesting harmful food [34], [35]. Glendinning and co-workers have reported that plants contain more bitter constituents [35]. Therefore, herbivorous and omnivorous mammals would be expected to need a greater level of bitter taste rejection compared with carnivores. We found that the guinea pig, horse and cow have larger T2R gene repertoires (60, 54 and 33 genes, respectively), whereas the panda and dog have relative smaller T2R gene repertoires (23 and 20 genes, respectively). Moreover, the lizard and frog have large number of lineage-specific T2R genes. By contrast, T2R genes birds and fishes have no more than 10 T2R genes.
Database of CR genes
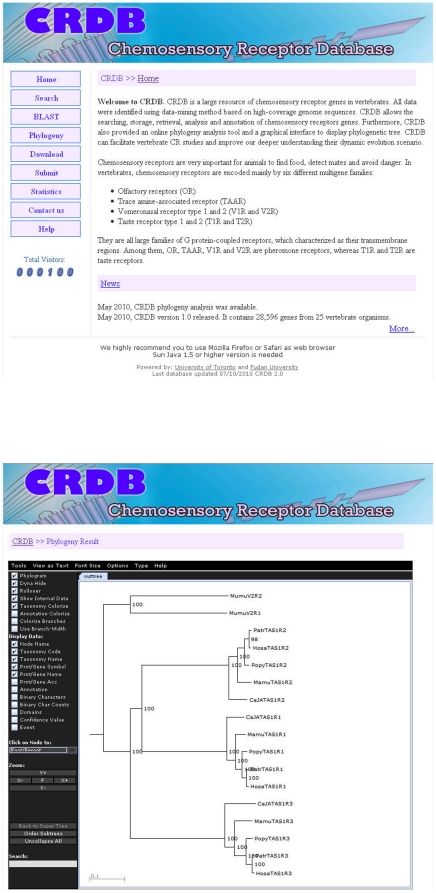
To our knowledge, the CR genes we identified are the largest curated dataset identified from high-coverage genome sequences. To facilitate CR gene annotation and evolutionary analysis, we subsequently constructed a database (CRDB, http://zldev.ccbr.utoronto.ca/CRDB/). The current version of CRDB ( Figure 2A ) provides a user friendly search engine, which allows to search the content of CR genes using ‘Basic search’ or ‘Advanced search’. In ‘Basic search’, search options for CR genes include by gene name, common name or aliases. Many species have lineage specific CR genes generated from gene duplication. Therefore, these CR genes tend to be located into clusters on specific regions of the same chromosome. For example, T2R genes in the mouse and rat are mainly located on chromosomes 6 and 4 [14], respectively. CRDB allows users to search CR genes at a specific chromosome region of interests in ‘Advanced search’. The system provides details of a specific CR gene including gene annotation, sequence information and cross-reference links. Moreover, we predicted the transmembrane topology of these CR proteins using TMHMM [36], and the transmembrane helix information is provided. Furthermore, to facilitate researchers to submit their identified CR genes to our database, we have implemented a tool for researchers to directly submit their data.
Figure 2. The web interface of CRDB.
A) Snapshot of the CRDB home page. B) Result of phylogenetic tree of primate T1R genes.
To better understand the phylogenetic relationship among CR genes, CRDB provides an online phylogeny tool. Here, we only confined the phylogenetic analysis to intact CR genes, because most pseudogenes contained deletions and were much shorter than intact genes. Considering the accuracy and efficacy, NJ method was implemented. We have adapted phyloXML [37] for the online phylogenetic tree visualization. Figure 2B illustrates an example of phylogenetic tree of T1R genes in five primate species. Mouse V2R sequences were used as the outgroup of the phylogenetic tree. The result showed that primates T1Rs form three monophyletic clades (T1R1, T1R2 and T1R3 clades) with high bootstrap values.
Conclusion
In this study, we identified nearly the entire CR genes based on 25 vertebrate high-coverage genome sequences. Our results strongly suggest the numbers of CR genes are diverse in vertebrates. In addition, it can also provide us a preliminary scenario for the extensive expansions and contractions of CR genes in vertebrate evolution. The extensively varying numbers of CR genes might be responsible for the species-specific ability of odor and taste perceptions [2]. Our work can be very useful for several reasons. First, we comprehensively identified CR genes in 25 vertebrates, and illustrated a dynamic fluctuating numbers of CR genes. Second, these identified CR genes provide a large resource for further analysis. At last, our CR gene database provides a platform for CR genes, and can facilitate CR gene evolutionary researches in vertebrates.
Materials and Methods
Genome sequences
The high coverage genome sequences of 25 vertebrate were downloaded from UCSC genome browser (http://genome.ucsc.edu/), which include human (Homo sapiens, hg19), chimpanzee (Pan troglodytes, panTro2), orangutan (Pongo pygmaeus abelii, ponAbe2), rhesus (Macaca mulatta, rheMac2), marmoset (Callithrix jacchus, calJac1), mouse (Mus musculus, mm9), rat (Rattus norvegicus, rn4), guinea Pig (Cavia porcellus, cavPor3), dog (Canis lupus familiaris, canFam2), panda (Ailuropoda melanoleuca, ailMel1), horse (Equus caballus, equCab2), cow (Bos Taurus, bosTau4), elephant (Loxodonta africana, loxAfr3), opossum (Monodelphis domestica, monDom5), platypus (Ornithorhynchus anatinus, ornAna1), chicken (Gallus gallus, galGal3), zebra finch (Taeniopygia guttata, taeGut1), lizard (Anolis carolinensis, anoCar1), x. tropicalis (Xenopus tropicalis, xenTro2), zebrafish (Danio rerio, danRer6), tetraodon (Tetraodon nigroviridis, tetNig2), fugu (Takifugu rubripes, fr2), stickleback (Gasterosteus aculeatus, gasAcu1), medaka (Oryzias latipes, oryLat2), sea lamprey (Petromyzon marinus, petMar1).
Identification of CR genes
Semi-automatic data-mining methods were used to identify CR genes from these whole-genome sequences. At first, we collected previously published CR proteins as query sequences [14], [20], [21], [23], [25], [26], [27]. Then, we conducted TBLASTN searches using the E-value 1e-10 against each genome sequence for different types of CR proteins, respectively. Many TBLASTN query hits might located at the same genomic regions, and we extracted non-overlapping sequences showing the lowest E-value among the hits to a given region. Each of the blast hit sequence was extended in both 3′ and 5′ directions along the genome sequences. Among CR genes, the OR, TAAR, V1R and T2R genes lack introns in their coding regions. The coding sequences with proper ATG and the stop codon were directly extracted, and they were regarded as intact CR genes; Sequences that contain interrupting stop codons or frameshifts were regarded as pseudogenes; Remaining sequences containing either initiation codons or stop codons were not considered. T1R and V2R genes contain many introns, and we conducted a cDNA-to-genomic alignment using Spidey to identify the exon/intron genomic structure [38]. At last, all identified sequences were confirmed by BLASTP searches against the NCBI database to ensure that genuine CR sequences were obtained.
Database construction
We developed a web interface (CRDB) to facilitate online search and phylogeny study of CR genes. CRDB was designed using Java language that provides access to a local MySQL database. To perform the online phylogenetic analysis of CR genes, MAFFT 6.717 [39] was used to generate multiple sequence alignment. The Neighbor-Joining (NJ) method, implemented by QuickTree [40], was used to generate phylogenetic result due to its good balance between accuracy and efficiency. Then, phyloXML were implemented for online phylogenetic tree visualization [37].
Acknowledgments
We appreciate Dr. Niimura and Dr. Peng Shi for providing their identified genes of olfactory receptors and vomeronasal receptors. We thank Dr. Yufang Zheng for her helpful comments and discussion.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work is supported by Key Construction Program of the National ‘985’ project of East China Normal University to Dong Dong, and the funding No. is 79633006. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Glusman G, Yanai I, Rubin I, Lancet D. The complete human olfactory subgenome. Genome Res. 2001;11:685–702. doi: 10.1101/gr.171001. [DOI] [PubMed] [Google Scholar]
- 2.Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet. 2008;9:951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- 4.Zozulya S, Echeverri F, Nguyen T. The human olfactory receptor repertoire. Genome Biol. 2001;2:RESEARCH0018. doi: 10.1186/gb-2001-2-6-research0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilad Y, Man O, Paabo S, Lancet D. Human specific loss of olfactory receptor genes. Proc Natl Acad Sci U S A. 2003;100:3324–3327. doi: 10.1073/pnas.0535697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilad Y, Przeworski M, Lancet D. Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. PLoS Biol. 2004;2:E5. doi: 10.1371/journal.pbio.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rouquier S, Blancher A, Giorgi D. The olfactory receptor gene repertoire in primates and mouse: evidence for reduction of the functional fraction in primates. Proc Natl Acad Sci U S A. 2000;97:2870–2874. doi: 10.1073/pnas.040580197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rouquier S, Friedman C, Delettre C, van den Engh G, Blancher A, et al. A gene recently inactivated in human defines a new olfactory receptor family in mammals. Hum Mol Genet. 1998;7:1337–1345. doi: 10.1093/hmg/7.9.1337. [DOI] [PubMed] [Google Scholar]
- 9.Rouquier S, Giorgi D. Olfactory receptor gene repertoires in mammals. Mutat Res. 2007;616:95–102. doi: 10.1016/j.mrfmmm.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 10.Rouquier S, Stubbs L, Gaillard-Sanchez I, Giorgi D. Sequence and chromosomal localization of the mouse ortholog of the human olfactory receptor gene 912–93. Mamm Genome. 1999;10:1172–1174. doi: 10.1007/s003359901185. [DOI] [PubMed] [Google Scholar]
- 11.Steiger SS, Fidler AE, Valcu M, Kempenaers B. Avian olfactory receptor gene repertoires: evidence for a well-developed sense of smell in birds? Proc Biol Sci. 2008;275:2309–2317. doi: 10.1098/rspb.2008.0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer A, Gilad Y, Man O, Paabo S. Evolution of bitter taste receptors in humans and apes. Mol Biol Evol. 2005;22:432–436. doi: 10.1093/molbev/msi027. [DOI] [PubMed] [Google Scholar]
- 13.Dong D, He G, Zhang S, Zhang Z. Evolution of olfactory receptor genes in primates dominated by birth-and-death process. Genome Biol Evol. 2009;2009:258–264. doi: 10.1093/gbe/evp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong D, Jones G, Zhang S. Dynamic evolution of bitter taste receptor genes in vertebrates. BMC Evol Biol. 2009;9:12. doi: 10.1186/1471-2148-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Go Y. Proceedings of the SMBE Tri-National Young Investigators' Workshop 2005. Lineage-specific expansions and contractions of the bitter taste receptor gene repertoire in vertebrates. Mol Biol Evol. 2006;23:964–972. doi: 10.1093/molbev/msj106. [DOI] [PubMed] [Google Scholar]
- 16.Niimura Y, Nei M. Evolution of olfactory receptor genes in the human genome. Proc Natl Acad Sci U S A. 2003;100:12235–12240. doi: 10.1073/pnas.1635157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niimura Y, Nei M. Comparative evolutionary analysis of olfactory receptor gene clusters between humans and mice. Gene. 2005;346:13–21. doi: 10.1016/j.gene.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 18.Niimura Y, Nei M. Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc Natl Acad Sci U S A. 2005;102:6039–6044. doi: 10.1073/pnas.0501922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niimura Y, Nei M. Evolutionary dynamics of olfactory and other chemosensory receptor genes in vertebrates. J Hum Genet. 2006;51:505–517. doi: 10.1007/s10038-006-0391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niimura Y, Nei M. Extensive gains and losses of olfactory receptor genes in Mammalian evolution. PLoS ONE. 2007;2:e708. doi: 10.1371/journal.pone.0000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niimura Y. On the origin and evolution of vertebrate olfactory receptor genes: comparative genome analysis among 23 chordate species. Genome Biol Evol. 2009;2009:34–44. doi: 10.1093/gbe/evp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steiger SS, Kuryshev VY, Stensmyr MC, Kempenaers B, Mueller JC. A comparison of reptilian and avian olfactory receptor gene repertoires: species-specific expansion of group gamma genes in birds. BMC Genomics. 2009;10:446. doi: 10.1186/1471-2164-10-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi P, Zhang J. Contrasting modes of evolution between vertebrate sweet/umami receptor genes and bitter receptor genes. Mol Biol Evol. 2006;23:292–300. doi: 10.1093/molbev/msj028. [DOI] [PubMed] [Google Scholar]
- 24.Shi P, Zhang J, Yang H, Zhang YP. Adaptive diversification of bitter taste receptor genes in Mammalian evolution. Mol Biol Evol. 2003;20:805–814. doi: 10.1093/molbev/msg083. [DOI] [PubMed] [Google Scholar]
- 25.Shi P, Zhang J. Comparative genomic analysis identifies an evolutionary shift of vomeronasal receptor gene repertoires in the vertebrate transition from water to land. Genome Res. 2007;17:166–174. doi: 10.1101/gr.6040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashiguchi Y, Nishida M. Evolution of trace amine associated receptor (TAAR) gene family in vertebrates: lineage-specific expansions and degradations of a second class of vertebrate chemosensory receptors expressed in the olfactory epithelium. Mol Biol Evol. 2007;24:2099–2107. doi: 10.1093/molbev/msm140. [DOI] [PubMed] [Google Scholar]
- 27.Young JM, Trask BJ. V2R gene families degenerated in primates, dog and cow, but expanded in opossum. Trends Genet. 2007;23:212–215. doi: 10.1016/j.tig.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 28.Bignetti E, Cavaggioni A, Pelosi P, Persaud KC, Sorbi RT, et al. Purification and characterisation of an odorant-binding protein from cow nasal tissue. Eur J Biochem. 1985;149:227–231. doi: 10.1111/j.1432-1033.1985.tb08916.x. [DOI] [PubMed] [Google Scholar]
- 29.Blazquez NB, French JM, Long SE, Perry GC. A pheromonal function for the perineal skin glands in the cow. Vet Rec. 1988;123:49–50. doi: 10.1136/vr.123.2.49. [DOI] [PubMed] [Google Scholar]
- 30.Yamate J, Izawa T, Ogata K, Kobayashi O, Okajima R, et al. Olfactory neuroblastoma in a horse. J Vet Med Sci. 2006;68:495–498. doi: 10.1292/jvms.68.495. [DOI] [PubMed] [Google Scholar]
- 31.Endo H, Komiya T, Narushima E, Suzuki N. Three-dimensional image analysis of a head of the giant panda by the cone-beam type CT. J Vet Med Sci. 2002;64:1153–1155. doi: 10.1292/jvms.64.1153. [DOI] [PubMed] [Google Scholar]
- 32.Kratzing JE. The fine structure of the olfactory and vomeronasal organs of a lizard (Tiliqua scincoides scincoides). Cell Tissue Res. 1975;156:239–252. doi: 10.1007/BF00221807. [DOI] [PubMed] [Google Scholar]
- 33.Li R, Fan W, Tian G, Zhu H, He L, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glendinning JI. Is the bitter rejection response always adaptive? Physiol Behav. 1994;56:1217–1227. doi: 10.1016/0031-9384(94)90369-7. [DOI] [PubMed] [Google Scholar]
- 35.Glendinning JI, Tarre M, Asaoka K. Contribution of different bitter-sensitive taste cells to feeding inhibition in a caterpillar (Manduca sexta). Behav Neurosci. 1999;113:840–854. [PubMed] [Google Scholar]
- 36.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 37.Han MV, Zmasek CM. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics. 2009;10:356. doi: 10.1186/1471-2105-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang H, Shi P, Zhang YP, Zhang J. Composition and evolution of the V2r vomeronasal receptor gene repertoire in mice and rats. Genomics. 2005;86:306–315. doi: 10.1016/j.ygeno.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 39.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howe K, Bateman A, Durbin R. QuickTree: building huge Neighbour-Joining trees of protein sequences. Bioinformatics. 2002;18:1546–1547. doi: 10.1093/bioinformatics/18.11.1546. [DOI] [PubMed] [Google Scholar]