IRE1 signaling robustly promoted P23H rhodopsin degradation by both proteasome and lysosome. When one degradation pathway was blocked, IRE1 signaling still promoted misfolded rhodopsin degradation using the remaining pathway. IRE1 signaling reduced levels of other misfolded rhodopsins, with lesser effects on misfolded CFTR.
Abstract
Endoplasmic reticulum (ER) is responsible for folding of secreted and membrane proteins in eukaryotic cells. Disruption of ER protein folding leads to ER stress. Chronic ER stress can cause cell death and is proposed to underlie the pathogenesis of many human diseases. Inositol-requiring enzyme 1 (IRE1) directs a key unfolded protein response signaling pathway that controls the fidelity of ER protein folding. IRE1 signaling may be particularly helpful in preventing chronic ER stress and cell injury by alleviating protein misfolding in the ER. To examine this, we used a chemical-genetic approach to selectively activate IRE1 in mammalian cells and tested how artificial IRE1 signaling affected the fate of misfolded P23H rhodopsin linked to photoreceptor cell death. We found that IRE1 signaling robustly promoted the degradation of misfolded P23H rhodopsin without affecting its wild-type counterpart. We also found that IRE1 used both proteasomal and lysosomal degradation pathways to remove P23H rhodopsin. Surprisingly, when one degradation pathway was compromised, IRE1 signaling could still promote misfolded rhodopsin degradation using the remaining pathway. Last, we showed that IRE1 signaling also reduced levels of several other misfolded rhodopsins with lesser effects on misfolded cystic fibrosis transmembrane conductance regulator. Our findings reveal the diversity of proteolytic mechanisms used by IRE1 to eliminate misfolded rhodopsin.
INTRODUCTION
Eukaryotic cells respond to protein misfolding in the endoplasmic reticulum (ER) by activating intracellular signaling pathways, collectively termed the unfolded protein response (UPR). Inositol-requiring enzyme 1 (IRE1) encodes an ER-resident transmembrane protein that controls the most highly conserved UPR signaling pathway (Hetz and Glimcher, 2009). IRE1 bears a luminal domain that monitors the fidelity of protein folding within the ER coupled to cytosolic kinase and endoribonuclease (RNase) domains (Ron and Walter, 2007). In response to protein misfolding in the ER, IRE1 undergoes oligomerization and activation of its kinase and RNase functions that then initiate the nonconventional splicing of Xbp-1 mRNA (Cox et al., 1993; Gonzalez et al., 1999; Yoshida et al., 2001; Calfon et al., 2002). Spliced Xbp-1 mRNA encodes a transcription activator that upregulates target genes, including ER chaperones and protein-folding enzymes, ER structural and transport proteins, and ER-associated degradation (ERAD) components (Lee et al., 2003). IRE1 signaling through the production of spliced X-box-binding protein 1 (XBP-1) transcription factor thereby enhances the protein folding environment of the ER by expanding the amount of ER and its constituent protein-folding machineries, as well as clearing misfolded ER proteins by targeting them for degradation through ERAD. In response to strong ER stress, IRE1 signaling also causes regulated IRE1-dependent mRNA decay (RIDD) and Jun N-terminal kinase (JNK) activation (Urano et al., 2000; Hollien and Weissman, 2006; Hollien et al., 2009; Oikawa et al., 2007). Many cells and tissues lacking IRE1 show profound defects in the generation of secreted and membrane proteins and undergo cell death (Zhang et al., 2005, 2011; Iwawaki et al., 2010), underscoring the importance of IRE1 in regulating ER protein folding and homeostasis.
Rhodopsin is a member of the G-protein–coupled receptor superfamily of membrane proteins that is expressed exclusively and robustly by photoreceptor cells in the eye in mammals, where it comprises >90% of all proteins in the photoreceptor outer segment disk membranes (Hargrave, 2001). Rhodopsin plays an essential role in vision by initiating the visual signal transduction cascade in the photoreceptor outer segment in response to light. More than 100 distinct mutations in rhodopsin have been identified that cause photoreceptor cell death and vision loss in autosomal-dominant forms of retinitis pigmentosa (RetNet—Retinal Information Network, www.sph.uth.tmc.edu/Retnet). Wild-type rhodopsin is normally synthesized and folded in the ER prior to delivery to the outer segment. By contrast, many mutant rhodopsins are defective in their folding and retained within the ER, where they cause ER stress and ultimately photoreceptor cell death (Sung et al., 1991; Kaushal and Khorana, 1994; Illing et al., 2002; Saliba et al., 2002; Lin et al., 2007; Gorbatyuk et al., 2010). Chemical and genetic chaperones have shown partial efficacy in restoring mutant rhodopsin protein folding, delivery out of the ER, and improving retinal function (Noorwez et al., 2003, 2004, 2009; Mendes and Cheetham, 2008; Kosmaoglou et al., 2009; Gorbatyuk et al., 2010). Alternatively, ribozyme-mediated degradation of P23H rhodopsin mRNA that spared wild-type rhodopsin has also shown efficacy in enhancing photoreceptor cell survival in transgenic animals expressing mutant and wild-type rhodopsins (Lewin et al., 1998; LaVail et al., 2000). Therefore finding mechanisms that enhance mutant rhodopsin folding or selectively remove mutant rhodopsin might offer new approaches to promote the survival of photoreceptors that are confronted with misfolded rhodopsin and ER stress.
Given IRE1's fundamental role in regulating ER function, we hypothesized that IRE1 activity might promote the folding and/or removal of misfolded proteins linked to disease such as mutant rhodopsins. To test this, we used a chemical-genetic strategy that enabled selective activation of IRE1's RNase function by the cell permeable small molecule 4-amino-1-tert-butyl-3-(1′-naphthylmethyl)pyrazolo[3,4-d]pyrimidine (1NM-PP1; Bishop et al., 2000). We previously showed that this system promoted robust splicing of Xbp-1 by IRE1 in the presence of 1NM-PP1 (Lin et al., 2007, 2009; Hollien et al., 2009). Of importance, this chemical-genetic system did not activate other UPR signaling pathways controlled by protein kinase RNA-like endoplasmic reticulum kinase (PERK) and activating transcription factor 6 (ATF6), did not activate RIDD or JNK, and did not cause apoptosis (Lin et al., 2007, 2009; Hollien et al., 2009). Here we used this approach to determine how selective activation of IRE1's RNase affected the fate of mutant rhodopsins and other misfolded proteins in mammalian cells.
RESULTS
ER stress reduces levels of wild-type and misfolded membrane proteins
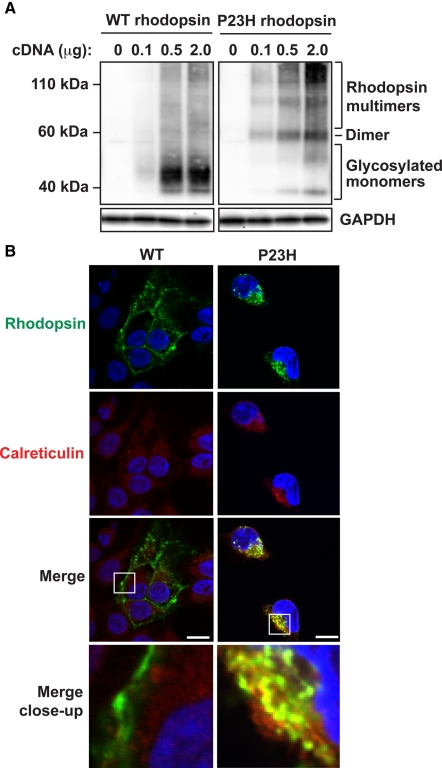
We focused on mutant rhodopsin bearing a proline-to-histidine missense mutation at amino acid residue 23 (P23H). The P23H rhodopsin mutation was first identified in families with autosomal-dominant retinitis pigmentosa (Dryja et al., 1990) and also caused photoreceptor cell death in mammalian models of retinal degeneration when expressed singly or as multicopy transgenes (Olsson et al., 1992; Lin et al., 2007; Sakami et al., 2011). Wild-type rhodopsin normally folds and undergoes N-linked glycosylation in the ER prior to export to the photoreceptor rod outer segment (Sung et al., 1991; Illing et al., 2002). These features are recapitulated in cell culture, where wild-type rhodopsin migrated predominantly as a diffuse 39- to 50-kDa molecular band on SDS gels and Western blot analysis corresponding to glycosylated monomeric rhodopsin and was efficiently exported from ER and localized at the plasma membrane (Figure 1, A and B, left). By contrast, mutant P23H rhodopsin protein primarily formed dimers and multimeric aggregates and was retained within the ER (Figure 1, A and B, right).
FIGURE 1:
Cellular and molecular defects induced by the P23H missense mutation in rhodopsin. (A) Wild-type or P23H rhodopsin cDNA was expressed in HEK293 cells as indicated, and rhodopsin protein levels and mobility were detected by SDS–PAGE separation and immunoblotting. GAPDH levels were assessed as a protein loading control. Positions of glycosylated monomeric, dimeric, and multimeric rhodopsin protein species are as indicated. (B) A 0.1-μg amount of wild-type or P23H rhodopsin cDNA was expressed in HEK293 cells, and the subcellular localization of rhodopsin was visualized by immunofluorescence labeling and confocal microscopy (shown in green). The endoplasmic reticulum was visualized by calreticulin immunostaining (shown in red). The nucleus was visualized by DAPI staining (shown in blue). Magnification bars, 10 μm.
First, we examined how activation of all UPR pathways including IRE1 affected P23H rhodopsin protein. We found sharps drop in P23H rhodopsin protein levels upon addition of tunicamycin (TM), an agent that blocks N-linked glycosylation, or thapsigargin (TG), an agent that blocks sarco/endoplasmic reticulum Ca2+ ATPase, to the medium of cells expressing P23H rhodopsin (Supplemental Figure S1A; reduced to 28% of levels in untreated cells by TM and 52% of levels in untreated cells by TG). This reduction in protein levels showed no specificity for P23H rhodopsin. Activation of UPR by TM or TG also caused sharp drops in the levels of wild-type rhodopsin protein (Supplemental Figure S1B; reduced to 67% of levels in untreated cells by TM and 33% of levels in untreated cells by TG) and another membrane protein, vascular cell adhesion molecule 1 (VCAM-1; Supplemental Figure S1C; reduced to 15% by TM and 17% by TG) in analogous experiments, even though neither protein bore missense misfolding mutations. This drop in levels of correctly folded proteins, as well as misfolded P23H rhodopsin, was consistent with the global translational attenuation observed after activation of the PERK branch of the UPR, concomitant with IRE1 activation, seen upon imposition of ER stress by these agents (Harding et al., 1999).
IRE1 selectively reduces levels of misfolded P23H rhodopsin
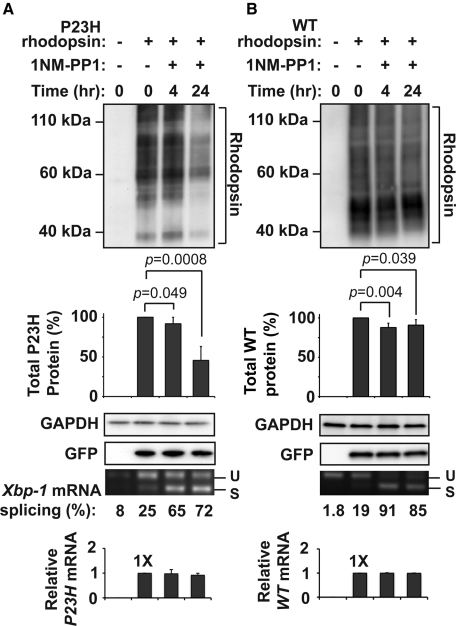
Next we examined how selective activation of IRE1 signaling affected P23H rhodopsin protein. When we expressed P23H rhodopsin in cells stably expressing IRE1[I642G], a genetically modified form of human IRE1 whose RNase activity can be toggled on/off by 1NM-PP1 without activating other UPR signaling pathways (Lin et al., 2007, 2009), we observed a sharp drop in P23H rhodopsin protein with addition of 1NM-PP1 compared with untreated cells (Figure 2A; down to ∼46% of protein levels in untreated cells by 24 h of drug exposure). This drop in P23H rhodopsin levels was highly specific for the misfolded protein, as wild-type rhodopsin protein levels showed minimal changes in analogous experiments (Figure 2B). Furthermore, this drop in P23H rhodopsin levels specifically required IRE1[I642G] because P23H rhodopsin protein levels were unchanged when 1NM-PP1 was added to wild-type HEK293 cells lacking IRE1[I642G] (Supplemental Figure S2).
FIGURE 2:
IRE1 selectively decreases levels of P23H rhodopsin protein. (A) P23H rhodopsin was expressed in HEK293 cells bearing IRE1[I642G], and 1NM-PP1 (5 μM) was applied for the indicated durations. (B) Wild-type rhodopsin was expressed in HEK293 cells bearing IRE1[I642G], and 1NM-PP1 (5 μM) was applied as indicated. (A, B) Total rhodopsin protein levels were detected by immunoblotting and quantified. GAPDH levels were assessed as a protein loading control. Cotransfected GFP levels were assessed to control for transfection efficiency. Xbp-1 mRNA splicing was assessed by reverse transcription-PCR, and the amount of spliced Xbp-1 mRNA was quantified as a percentage of the total amounts of unspliced (u) and spliced (s) amplicons. P23H and wild-type rhodopsin mRNA levels were measured by real-time PCR and are expressed relative to levels in cells that were not treated with 1NM-PP1. Immunoblots are representative of three independent experiments. Error bars represent SDs from three experiments. The p values were determined by Student's t test analysis.
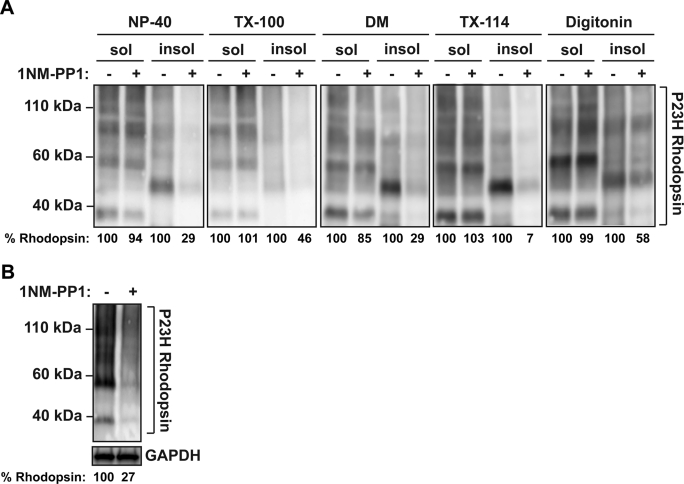
P23H rhodopsin is highly prone to form detergent-insoluble aggregates (Illing et al., 2002). Therefore we examined in more detail which species of P23H rhodopsin (detergent soluble or insoluble) was affected by IRE1 signaling. When we examined levels of P23H rhodopsin protein in 1% NP-40 soluble supernatant compared with detergent-insoluble pellet fractions, we found that the drop in P23H rhodopsin protein levels occurred almost entirely with the P23H rhodopsin present in the detergent-insoluble fraction after IRE1 activation by 1NM-PP1 (Figure 3A). Furthermore, we observed this selective drop in detergent-insoluble pellet fractions of P23H rhodopsin across a wide range of detergents, including 1% Triton X-100, 1% dodecyl-β-d-maltoside (DM), 1% Triton X-114, and 1% digitonin (Figure 3A). To rule out the possibility that selective IRE1 induction shifted the insoluble material into a form that was not reactive to 1D4 anti-rhodopsin antibody, recognizing the C-terminus of rhodopsin, we used a different rhodopsin antibody recognizing the N-terminus of rhodopsin, B630N anti-rhodopsin antibody. The selective drop in detergent-insoluble pellet fractions of P23H rhodopsin was also observed (Supplemental Figure S3). Taken together, our findings demonstrated that IRE1 signaling dramatically reduced misfolded P23H rhodopsin protein levels in cells, with minimal changes to the levels of its wild-type rhodopsin protein counterpart. Furthermore, IRE1 signaling selectively reduced levels of P23H rhodopsin protein in insoluble pellet forms, with minimal changes on levels of P23H rhodopsin in the soluble supernatant fraction.
FIGURE 3:
IRE1 signaling promotes P23H rhodopsin protein degradation. (A) P23H rhodopsin was expressed in cells bearing IRE1[I642G], and 1NM-PP1 (5 μM) was applied for 24 h. Cell lysates were solubilized in 1% of NP-40, Triton X-100, DM, Triton X-114, or digitonin. Total rhodopsin protein levels from detergent-soluble and -insoluble fractions were detected by immunoblotting and quantified. (B) P23H rhodopsin protein production was induced by application of doxycycline (1 μg/ml) for 30 min in cells bearing tetracycline-inducible P23H rhodopsin and IRE1[I642G]. Doxycycline was removed from media, and when P23H rhodopsin mRNA levels had returned to baseline (∼72 h), 1NM-PP1 (5 μM) was applied for 24 h. Total rhodopsin protein levels were detected by immunoblotting and quantified. GAPDH served as a protein loading control.
IRE1 promotes P23H rhodopsin protein degradation
Next we investigated how IRE1 caused the sharp drop seen in levels of P23H rhodopsin protein. Spliced XBP-1 protein transcriptionally upregulates a broad range of chaperones and enzymes that promote ERAD (Lee et al., 2003), and overexpression of ER-degradation enhancing α-mannosidase-like protein 1 (EDEM1), a lectin involved in ERAD (Yoshida et al., 2003; Eriksson et al., 2004; Olivari et al., 2005), has been shown to promote P23H rhodopsin protein degradation and enhance its delivery to membrane (Kosmaoglou et al., 2009). Therefore we investigated how IRE1 signaling affected P23H rhodopsin protein stability and degradation. Because the form of P23H rhodopsin protein most sensitive to IRE1 signaling was insoluble in detergents used for immunoprecipitation and metabolic labeling studies (Figure 3A and Supplemental Figure S3), we were unable to perform pulse-chase analysis to accurately measure how IRE1 signaling affected P23H rhodopsin protein half-life. Instead, we devised a strategy to measure the changes in P23H rhodopsin protein stability in response to IRE1 signaling by creating a cell line stably expressing both tetracycline-inducible P23H rhodopsin and IRE1[I642G]. In this system, P23H rhodopsin protein synthesis required the presence of doxycycline, and its synthesis could be induced with no toxicity for up to 10 d in vitro. When doxycycline was removed, P23H rhodopsin transcription ceased, and the remaining pool of P23H rhodopsin protein was eliminated via endogenous protein turnover. IRE1[I642G]'s ability to splice Xbp-1 after 1NM-PP1 addition was not changed in these cells (Supplemental Figure S4). Using this system, we briefly induced the expression of P23H rhodopsin protein by the addition of doxycycline. Doxycycline was then removed from media, and when P23H rhodopsin mRNA levels had returned to basal minimal levels, we activated IRE1 signaling by adding 1NM-PP1 for another 24 h. Under these conditions, we saw a pronounced decrease in the remaining pool of P23H rhodopsin protein with addition of 1NM-PP1 compared with untreated cells (Figure 3B; down to 27% of protein levels in untreated cells). These changes in P23H rhodopsin protein levels most likely arose from degradation, as there was minimal P23H rhodopsin mRNA and therefore little, if any, new P23H rhodopsin protein being synthesized under these conditions without doxycycline. These results provided evidence that IRE1 signaling directly and robustly promoted the degradation of P23H rhodopsin protein.
IRE1's RIDD activity also cleaves ER-targeted mRNAs, leading to drops in levels of some membrane and secreted proteins (Han et al., 2009; Hollien et al., 2009). Previous studies showed that 1NM-PP1 promotes Xbp-1 splicing by IRE1[I642G]'s RNase without activating RIDD (Hollien et al., 2009). Consistent with those findings, we observed robust Xbp-1 mRNA splicing without changes in P23H rhodopsin mRNA transcript levels after addition of 1NM-PP1 (Figure 2, A and B, bottom). These findings indicated that the drop in P23H rhodopsin protein levels seen after IRE1[I642G] activation did not arise from loss of transcription or decay of the P23H rhodopsin mRNA.
Proteasome and lysosome degrade P23H rhodopsin
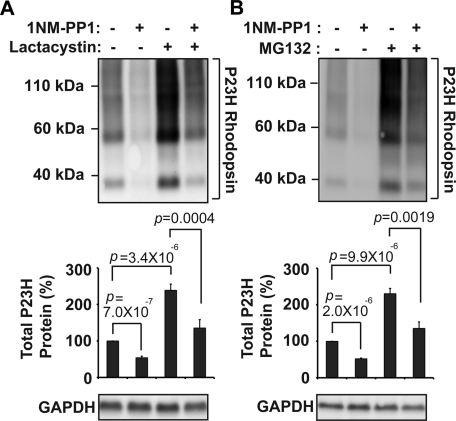
We next explored the proteolytic mechanisms responsible for degrading P23H rhodopsin. Similar to prior studies, we observed a significant increase in P23H rhodopsin protein levels, in both soluble and pellet fractions, after addition of proteasome inhibitors, lactacystin, or MG-132 (Figure 4A and 4B, top, and Supplemental Figure S5), indicating a role for the proteasome in P23H rhodopsin protein degradation (Illing et al., 2002; Saliba et al., 2002). However, to our surprise, when we activated IRE1 signaling with 1NM-PP1 in the presence of proteasome inhibitors, total P23H rhodopsin protein levels still decreased, dropping from ∼240 to ∼140% of untreated levels after IRE1 activation in the presence of lactacystin or MG-132 (Figure 4, A and B). We saw similar findings when we examined soluble and pellet fractions of P23H rhodopsin treated with lactacystin and 1NM-PP1 (Supplemental Figure S5). We confirmed that lactacystin and MG-132 remained effective in these studies by demonstrating increased protein levels of GFPu, an unstable variant of green fluorescent protein (GFP) previously used as a reporter of proteasome activity (Bence et al., 2001; Supplemental Figure S6). We also confirmed that 1NM-PP1 remained effective in activating IRE1[I642G]'s RNase activity under conditions of proteasome inhibition by demonstrating robust Xbp-1 mRNA splicing after addition of lactacystin or MG-132 (Supplemental Figure S7, A and B). Taken together, our findings indicated that P23H rhodopsin protein degradation depended upon proteasome function, but, of interest, our findings suggested that additional proteolytic mechanisms cleared P23H rhodopsin after IRE1 activation.
FIGURE 4:
P23H rhodopsin protein degradation depends on proteasome function. (A) P23H rhodopsin was expressed in cells bearing IRE1[I642G], and lactacystin (1 μM) and/or 1NM-PP1 (5 μM) was added for 24 h as indicated. Rhodopsin protein levels were detected by immunoblotting and quantified. (B) P23H rhodopsin was expressed in cells bearing IRE1[I642G]. MG132 (1 μM) and/or 1NM-PP1 (5 μM) were applied for 24 h as indicated. (A, B) GAPDH protein levels were assessed as a protein loading control. Immunoblots are representative of three independent experiments. Error bars represent SDs from three experiments. The p values were determined by Student's t test analysis.
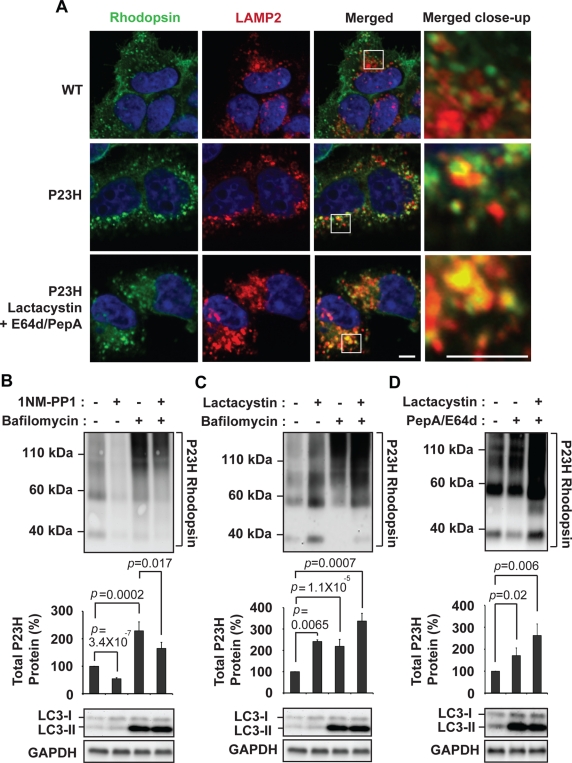
Besides the proteasome, the autophagy–lysosome system is another cellular mechanism that degrades proteins, and rapamycin was previously reported to promote P23H rhodopsin degradation via induction of autophagy (Kaushal, 2006). When we expressed P23H rhodopsin in cells, we saw increased colocalization between P23H rhodopsin and lysosomal-associated membrane protein 2 (LAMP2) in immunofluorescence studies compared with cells expressing wild-type rhodopsin (Figure 5A, top two rows), and this colocalization was even more apparent when cells were treated with proteasomal and lysosomal protease inhibitors (Figure 5A, bottom row). These findings suggested that P23H rhodopsin was subjected to lysosomal protein degradation. When we treated cells expressing P23H rhodopsin with bafilomycin, a V-ATPase inhibitor that interrupts autophagosome fusion with lysosomes (Yamamoto et al., 1998), P23H rhodopsin protein levels increased to ∼220% of levels seen in untreated cells (Figure 5B). Levels of the lipidated form of light-chain 3 (LC3-II) also increased after bafilomycin treatment (Mizushima and Yoshimori, 2007), confirming the efficacy of bafilomycin in these studies (Figure 5B). These studies indicated that P23H rhodopsin protein degradation also depended on lysosome function through autophagy. Of interest, P23H rhodopsin protein levels decreased in the presence of bafilomycin when IRE1 was activated by1NM-PP1 (Figure 5B and Supplemental Figure S7C). This finding likely reflected the continued proteasomal degradation of P23H rhodopsin when autophagic delivery to lysosome was inhibited.
FIGURE 5:
P23H rhodopsin protein degradation depends on lysosome function. (A) Wild-type or P23H rhodopsin was expressed in cells, and the subcellular localization of rhodopsin was visualized by immunofluorescence labeling and confocal microscopy (shown in green). The lysosome was visualized by LAMP2 immunostaining (shown in red). The nucleus was visualized by DAPI staining (shown in blue). Magnification bar, 5 μm. (B) P23H rhodopsin was expressed in cells bearing IRE1[I642G], and bafilomycin (1 μM) and/or 1NM-PP1 (5 μM) was added for 20 h as indicated. (C) P23H rhodopsin was expressed in cells, and lactacystin (1 μM) and/or bafilomycin (1 μM) was added for 20 h as indicated. (D) P23H rhodopsin was expressed in cells, and lactacystin (1 μM) and/or E64d (20 μg/ml)/pepstatin A (20 μg/ml) was added for 20 h as indicated. (B–D) Rhodopsin protein levels, LC3-I, and LC3-II protein isoforms were assessed by immunoblotting. GAPDH protein levels were assessed as a protein loading control. Immunoblots are representative of three independent experiments. Error bars represent SDs from three experiments. The p values were determined by Student's t test analysis.
To test the possibility that P23H rhodopsin protein degradation depended on both lysosome and proteasome, we simultaneously inhibited proteasome and lysosome/autophagy functions and examined effects on P23H rhodopsin protein levels. When we combined lactacystin with bafilomycin, we saw a greater increase in P23H rhodopsin protein levels than with either inhibitor alone (Figure 5C). Similarly, when we combined lactacystin with E64d and pepstatin A, agents that inhibit several lysosomal proteases (McGowan et al., 1989; Mizushima and Yoshimori, 2007), we saw a greater increase in P23H rhodopsin protein levels than with E64d and pepstatin A alone (Figure 5D). Taken together, our results provided evidence that P23H rhodopsin was a substrate for both lysosomal and proteasomal degradation.
IRE1 does not enhance P23H rhodopsin folding and delivery to surface membrane
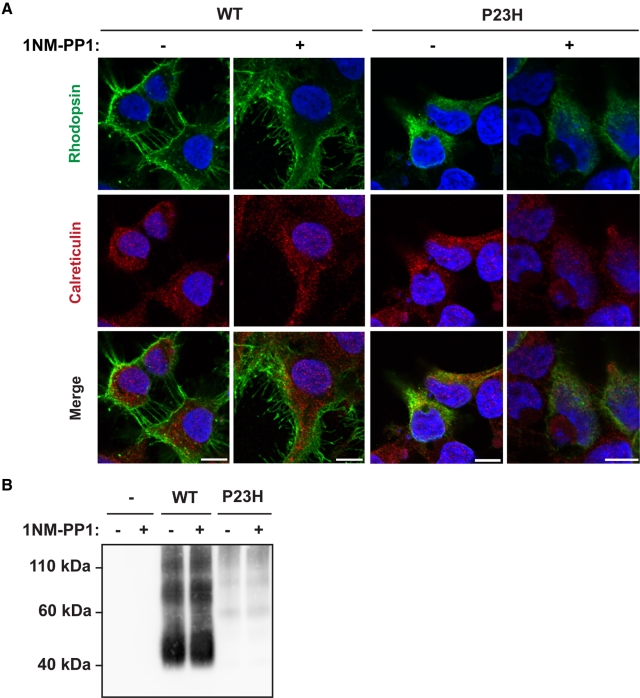
Chemical and genetic chaperones have been demonstrated to promote P23H rhodopsin exit from the ER by partially ameliorating the misfolding defect (Noorwez et al., 2003, 2004, 2009; Mendes and Cheetham, 2008; Kosmaoglou et al., 2009). The IRE1 signaling pathway transcriptionally upregulates numerous ER chaperones. Therefore we examined whether selective activation of IRE1 could partially restore delivery of P23H rhodopsin to the plasma membrane as another function in addition to promoting P23H rhodopsin protein degradation. When we expressed wild-type rhodopsin in cells bearing IRE1[I642G], we found that rhodopsin was predominantly at the plasma membrane in immunofluorescence studies, and this subcellular localization was not altered upon activation of IRE1 signaling by addition of 1NM-PP1 (Figure 6A). When we expressed P23H rhodopsin in analogous experiments, we found that P23H rhodopsin was retained within the ER, and activation of IRE1[I642G] by addition of 1NM-PP1 did not increase the amount of P23H rhodopsin observed at the plasma membrane in these studies (Figure 6A). Consistent with these imaging studies, when we examined the amount of wild-type and P23H rhodopsin present at the plasma membrane by cell surface biotinylation, we detected large amounts of biotinylated wild-type rhodopsin and minimal P23H rhodopsin at the plasma membrane, and the levels of biotinylated wild-type and P23H rhodopsin were not altered by the addition of 1NM-PP1 (Figure 6B). Therefore our findings demonstrated that IRE1 signaling does not restore P23H rhodopsin delivery to the plasma membrane. Instead, the main effect of IRE1 on P23H rhodopsin in our system was to promote its degradation via the proteasome and lysosome.
FIGURE 6:
IRE1 does not restore the delivery of P23H rhodopsin to the plasma membrane. (A) Wild-type or P23H rhodopsin was expressed in cells bearing IRE1[I642G], and 1NM-PP1 (5 μM) was applied for 24 h as indicated. The subcellular localization of rhodopsin was visualized by immunofluorescence labeling and confocal microscopy (shown in green). The endoplasmic reticulum was visualized by calreticulin immunostaining (shown in red). The nucleus was visualized by DAPI staining (shown in blue). Magnification bars, 10 μm. (B) Wild-type or P23H rhodopsin were expressed in cells bearing IRE1[I642G] with or without application of 1NM-PP1 (5 μM) for 24 h. Surface membrane proteins were biotinylated, and rhodopsin protein levels in the biotinylated fraction were assessed by immunoblotting.
Effects of IRE1 on class II rhodopsins and CFTR∆508
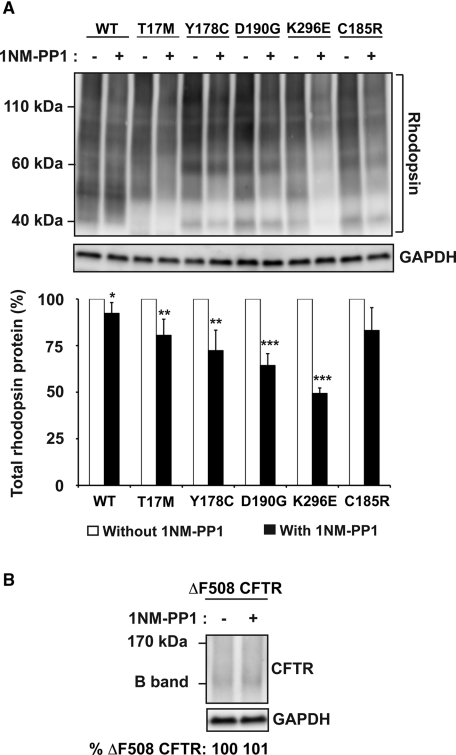
Besides P23H rhodopsin, many additional missense mutations in rhodopsin have been identified that cause photoreceptor cell death and vision loss (Berson, 1993). We investigated how IRE1 signaling affected protein levels of five other mutant rhodopsins. When we expressed T17M, Y178C, D190G, K296E, or C185R mutant rhodopsins in cells expressing IRE1[I642G], these mutant rhodopsins formed higher-order multimers and were retained within the ER (Figure 7A; Sung et al., 1991; Saliba et al., 2002; Liu et al., 2010). With addition of 1NM-PP1, we observed reduction in the protein levels of all these mutant rhodopsin proteins that was not seen with wild-type rhodopsin (Figure 7A; reduced to 81% for T17M rhodopsin, 72% for Y178C rhodopsin, 64% for D190G rhodopsin, 49% for K296E rhodopsin, and 83% for C185R rhodopsin after 24 h of drug exposure compared with protein levels in untreated cells). Furthermore, we also observed that these mutant rhodopsins remained predominantly localized in the ER (Supplemental Figure S8). Therefore our results suggested that rhodopsin mutants that cause misfolding were selectively targeted by IRE1 for degradation. Of interest, the magnitude of the reduction in protein levels varied between different rhodopsin mutants for unclear reasons but could reflect differences in the level of protein misfolding and aggregation induced by a particular mutation.
FIGURE 7:
(A) Wild-type or mutant T17M, Y178C, D190G, K296E, or C185R rhodopsin was expressed in cells bearing IRE1[I642G], and 1NM-PP1 (5 μM) was applied for 24 h. Total rhodopsin protein levels were detected by immunoblotting and quantified. Error bars represent SDs from three experiments. The p values were determined by Student's t test analysis. *p < 0.05, **p < 0.01, ***p < 0.001. (B) CFTR∆F508 was expressed in cells bearing IRE1[I642G], and 1NM-PP1 (5 μM) was applied for 24 h. CFTR∆F508 protein levels were detected by immunoblotting and quantified. (A, B) GAPDH levels were assessed as a protein loading control.
We also asked how IRE1 signaling affected misfolded cystic fibrosis transmembrane conductance regulator (CFTR), a 12-transmembrane ion channel whose loss of function leads to cystic fibrosis. Point mutations in CFTR, such as the CFTR∆F508 mutant, lead to its misfolding and rapid degradation by ERAD (Ward and Kopito, 1994; Farinha and Amaral, 2005; Sun et al., 2006; Younger et al., 2006). By contrast to P23H rhodopsin, CFTR∆508 does not accumulate as dimers or higher order multimers and instead, is typically found at very low steady-state levels when expressed in cells (Ward and Kopito, 1994; Farinha and Amaral, 2005; Sun et al., 2006; Younger et al., 2006). When we activated IRE1, we did not observe a decrease in CFTR∆F508 protein levels, most likely because its steady-state level was already very low (Figure 8B). These findings indicated that misfolded CFTR∆508 protein levels were not altered significantly, if at all, by IRE1 signaling in our cell culture system. More broadly, our analysis of multiple mutant rhodopsins and CFTR∆508 indicated that IRE1 signaling had markedly varying effects on different misfolded membrane proteins.
FIGURE 8:
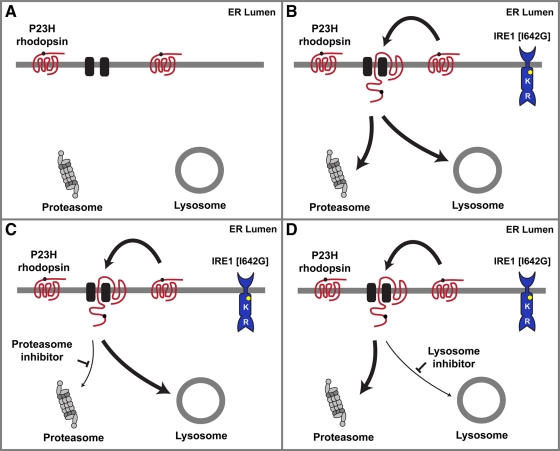
Proposed model of P23H rhodopsin protein degradation by IRE1 signaling. (A) P23H rhodopsin is degraded by proteasome and lysosome. (B) IRE1[I642G] activation by 1NM-PP1 enhances P23H rhodopsin degradation. (C) When proteasome function is blocked, IRE1 promotes P23H rhodopsin extraction and degradation by lysosome. (D) When lysosome function is blocked, IRE1 promotes P23H rhodopsin extraction and degradation by proteasome.
DISCUSSION
The goal of the present study was to decipher the effects of IRE1 signaling on the fate of the misfolded P23H rhodopsin. A striking effect of IRE1 activation was the sharp drop in the protein levels of misfolded P23H rhodopsin. Intriguingly, the greatest drop was seen with P23H rhodopsin protein present in detergent-insoluble pellet fractions. P23H rhodopsin is highly prone to aggregation, and this aggregated form is likely to comprise much of the P23H rhodopsin protein found in the detergent-insoluble pellet. We envisage several mechanisms to account for the selective reduction in P23H rhodopsin protein levels in pellet fractions after IRE1 activation. First, IRE1 could directly target P23H rhodopsin aggregates for degradation. Second, IRE1 could induce chaperones and enzymes that “solubilize” P23H rhodopsin aggregates, and then this soluble form is degraded. Third, IRE1 could prevent newly synthesized P23H rhodopsin from aggregating and, instead, steer the misfolded protein toward degradation. Many of these mechanisms might be at play, given IRE1's ability to induce numerous genes involved in ER protein folding and degradation. Future studies will identify the factors that selectively recognize and target P23H rhodopsin for degradation.
The present study provided evidence that P23H rhodopsin is a substrate for both proteasomal and lysosomal degradation and suggested that the pathways compensated for one another to degrade P23H rhodopsin when one pathway was inhibited (Figure 8). The cellular and molecular events that identify, retrieve, and deliver misfolded membrane proteins for degradation are best understood for ERAD via the proteasome. Recent studies in yeast have proposed a mechanism for ERAD of misfolded membrane proteins that initially entails the recognition of a “bipartite signal” on a misfolded membrane protein comprised of chaperones bound to hydrophobic polypeptides abnormally exposed on the surface of the misfolded protein coupled to lectins bound to modified glycans added through N-linked glycosylation (Kosmaoglou et al., 2008; Buchberger et al., 2010). Then, if the protein-folding problem cannot be corrected, the irretrievably misfolded membrane protein is handed off to a retrotranslocation complex that extracts it from ER membrane for ubiquitinylation and proteasomal degradation in the cytosol (Kawaguchi and Ng, 2007). ERAD components such as EDEM1 and VCP/ter94 have been shown to play important roles in misfolded rhodopsin degradation (Kang and Ryoo, 2009; Kosmaoglou et al., 2009; Griciuc et al., 2010). Of interest, other lectins and ERAD components, such as calnexin, ninaA/peptidyl-prolyl-isomerase, and ninaG/oxidoreductase, are necessary for folding of some rhodopsins but not others (Ahmad et al., 2006; Rosenbaum et al., 2006; Kosmaoglou and Cheetham, 2008; Kosmaoglou et al., 2008). These findings suggest that mutation-specific ERAD cofactors work together with a conserved core ERAD machinery to target P23H rhodopsin for proteasomal degradation. Identifying the full complement of ERAD cofactors specifically targeting human P23H rhodopsin is an important next step.
The role of the lysosome in processing P23H rhodopsin is poorly understood. The present study provided multiple lines of evidence that P23H rhodopsin is a bona fide substrate for lysosomal degradation. However, it is unclear how the cell determines whether to use the proteasome or the lysosome for P23H rhodopsin degradation. P23H rhodopsin readily forms higher-order multimers and aggregates (Figure 1; Illing et al., 2002; Saliba et al., 2002). These aggregates, when arising in the cytosol, could be highly resistant to unfolding and translocation into the 20S proteasome core particle for proteolysis. Furthermore, if P23H rhodopsin aggregates formed while still in the ER membrane, they could overwhelm the ability of ERAD to extract and retrotranslocate the misfolded protein for proteasomal degradation. Autophagic recognition of P23H rhodopsin aggregates and delivery to lysosome could enable degradation under these circumstances. Indeed, the autophagy–lysosome system has been implicated in the degradation of a growing number of cytosolic aggregates of misfolded proteins found in neurodegenerative diseases (Rubinsztein, 2006; Winslow and Rubinsztein, 2008; Wong and Cuervo, 2010). More recently, ER-phagy—selective autophagy of ER—has been observed after ER stress and IRE1 activation (Bernales et al., 2006, 2007). ER-phagy could be a mechanism to encapsulate regions of ER containing aggregated P23H rhodopsin for ultimate delivery to the lysosome for degradation. Future studies to identify how IRE1 promotes the degradation of P23H rhodopsin through autophagy/lysosome are required.
Chaperones have shown efficacy in correcting P23H rhodopsin misfolding and enhancing photoreceptor cell survival. Pharmacologic chaperones, such as 11-cis-7-ring retinal and 9-cis-retinal, can partially correct the P23H rhodopsin misfolding by enhancing its ability to form pigment, acquire mature glycosylation, and transit to the cell surface (Li et al., 1998; Saliba et al., 2002; Noorwez et al., 2003, 2004). Genetic overexpression of EDEM1 in cell culture enhanced P23H rhodopsin degradation and also promoted delivery of P23H rhodopsin to cell surface (Kosmaoglou et al., 2009). The IRE1 signaling pathway upregulates numerous ER protein-folding chaperones, yet we saw no restoration of P23H rhodopsin delivery to the plasma membrane in our studies. This could arise from the concomitant enhancement of P23H rhodopsin protein degradation after IRE1 activation, preventing any “escape” of P23H rhodopsin to plasma membrane. Intriguingly, overexpression of the immunoglobulin-binding protein (BiP)/Grp78 chaperone did not correct P23H rhodopsin misfolding, yet BiP/Grp78 overexpression still enhanced visual function in transgenic animals expressing P23H rhodopsin, in part, by suppressing ER stress-induced proapoptotic signaling pathways in photoreceptors (Gorbatyuk et al., 2010). Chemical-genetic activation of IRE1 in photoreceptors expressing P23H rhodopsin could have similar beneficial effects.
The present findings emphasize the striking ability of IRE1 signaling to remove misfolded P23H rhodopsin from cells while sparing the wild-type counterpart. These findings raise an intriguing question of whether failure of endogenous IRE1 signaling accounts for the toxicity of P23H rhodopsin by allowing its levels to build up to a critical lethal threshold. Genetic studies in lower organisms indicated that the fidelity of protein quality control breaks down as the organism ages (Cuervo and Dice, 2000; David et al., 2010), and recent studies demonstrated that chronic ER stress can render refractive IRE1 signaling required for ER proteostasis (Lin et al., 2007; Li et al., 2010). Determining how chronic stress and/or aging affect IRE1 may reveal why photoreceptors ultimately fail to cope with P23H rhodopsin and die.
Retinitis pigmentosa arising from P23H rhodopsin is emblematic of numerous heritable proteinopathies in which a mutant misfolded protein coexists with the wild-type protein. In many cases, blocking the abnormal protein encoded by the mutated allele is sufficient to restore cell viability because the remaining normal allele encodes sufficient wild-type protein for organismal functions (Lewin et al., 1998; LaVail et al., 2000). Modulation of IRE1 signaling could be particularly useful in treating these types of proteinopathies by preferentially removing the misfolded protein encoded by the mutated allele while sparing the normal protein encoded by the wild-type allele.
MATERIALS AND METHODS
Cell culture and transfection
HEK293 cells were maintained at 37°C and 5% CO2 in DMEM (Mediatech, Manassas, VA) supplemented with 10% fetal calf serum (Invitrogen, Carlsbad, CA) and 1% penicillin/streptomycin (Invitrogen). For transient transfection of wild-type rhodopsin, mutant rhodopsins, CFTR, CFTR∆508, or GFPu, 0.5 μg or indicated amount of plasmids were transfected using Lipofectamine 2000 reagent (Invitrogen) following the manufacturer's instruction. The generation and use of 1NM-PP1–sensitized HEK293 stable cell lines expressing IRE1[I642G] was previously described (Lin et al., 2007, 2009). For the construction and selection of tetracycline-inducible wild-type rhodopsin or P23H rhodopsin cell lines, we used the Flp-In T-Rex-293 cell line system (Invitrogen). Briefly, wild-type rhodopsin and the P23H mutant were cloned into the pcDNA5/FRT/TO TOPO TA expression vector (Invitrogen). These plasmids were cotransfected with pCI-neo vector using Lipofectamine 2000 reagent into Flp-In T-Rex-293 cells that were already stably expressing IRE1[I642G]. Two days after transfection, 0.8 mg/ml of G418 (Mediatech) was added to the culture media, and surviving colonies of cells were isolated. To induce the expression of rhodopsin, 1 μg/ml of doxycycline was added to media.
Chemicals
1NM-PP1 was generously provided by C. Zhang and K. Shokat (University of California, San Francisco, CA; Bishop et al., 2000). Doxycycline and pepstatin A was obtained from Sigma-Aldrich (St. Louis, MO). Tunicamycin, thapsigargin, bafilomycin, and MG132 were obtained from Calbiochem EMD Bioscience (Darmstadt, Germany). Lactacystin was obtained from Cayman Chemical (Ann Arbor, MI). (2S,3S)-trans-Epoxysuccinyl-l-leucylamido-3-methylbutane ethyl ester (E64d) was obtained from Enzo Life Sciences (Plymouth Meeting, PA).
Molecular biology
Cells were lysed and total RNA was collected (RNeasy, Qiagen, Hilden, Germany). PolyA mRNA was reverse transcribed using the SuperScript-RT system (Invitrogen). cDNA was used as template for PCR amplification across the fragment of the Xbp-1 cDNA bearing the intron target of IRE1 RNAse activity. Primers used included human Xbp-1, 5′-TTACGAGAGAAAACTCATGGC-3′ and 5′-GGGTCCAAGTTGTCCAGAATGC-3′. PCR conditions were as follows: 95°C for 5 min; 95°C for 1 min; 58°C for 30 s; 72°C for 30 s; and 72°C for 5 min, with 35 cycles of amplification. PCR products were resolved on a 2.5% agarose/1× Tris-acetate-EDTA gel. Quantification of Xbp-1 mRNA splicing as a percentage of total Xbp-1 mRNA was performed using VisionWork LS Software (UVP, Upland, CA).
For quantitative PCR, total RNA was collected from lysed cells (RNeasy). PolyA mRNA was reverse transcribed using the SuperScript-RT system, and aliquots of cDNA were used as template for quantitative PCR. Primers used included human Rpl19 mRNA, 5′-ATGTATCACAGCCTGTACCTG–3′ and 5′-TTCTTGGTCTCTTCCTCCTTG-3′; and human rhodopsin mRNA, 5′- CTTCACCGTCAAGGAGGC-3′ and 5′- GCAAAGAACGCTGGGATG-3′. Quantitative PCR was performed using a Chromo4 DNA Engine (Bio-Rad, Hercules, CA). Rpl19 mRNA levels, a transcript whose levels are not altered by ER stress, served as an internal normalization standard. PCR conditions were as follows: 95°C for 5 min; 95°C for 10 s; 56.5°C for 10 s; 72°C for 10 s, with 40 cycles of amplification.
Protein biochemistry and quantification
Cells were lysed and homogenized by sonication in ice-cold lysis buffer (1% NP-40, 50 mM Tris-HCl, pH 8, and 150 mM NaCl, containing protease inhibitors [Roche, Basel, Switzerland]). For solubilizing rhodopsin, cells were lysed in ice-cold lysis buffer (1% NP-40, 50 mM Tris-HCl, pH 8, and 150 mM NaCl, containing protease inhibitors [Roche]) and incubated on ice for 30 min. Cell lysates were mixed vigorously every 5 min. Insoluble material was recovered by centrifugation at 15,000 × g for 30 min and solubilized in phosphate-buffered saline (PBS) containing 1% SDS for 10 min at room temperature, followed by sonication. Additional lysis buffers used for solubilizing rhodopsin included 1% Triton X-100 (Sigma-Aldrich) in 50 mM Tris-HCl, pH 8, and 150 mM NaCl; 1% DM (Calbiochem EMD Bioscience) in PBS, pH 7.4, 1% Triton X-114 (Sigma-Aldrich) in PBS, pH 7.4, and 1% digitonin (Sigma-Aldrich) in PBS, pH 7.4. Protein concentrations of the total cell lysates, soluble rhodopsin, and insoluble material were determined by BCA Protein Assay Kit (Pierce, Rockford, IL). Five micrograms or the indicated amount of total protein was loaded onto 4–12% SDS–PAGE minigels (NuPAGE, Invitrogen) or 4–15% Mini-PROTEAN TGX precast gels (Bio-Rad) and analyzed by Western blot. The following antibodies and dilutions were used for immunoblotting: anti-rhodopsin at 1:1000 (Santa Cruz Biotechnologies, Santa Cruz, CA); B630N anti-rhodopsin at 1:1000 (a generous gift from W. C. Smith, University of Florida, Gainesville, FL); anti-VCAM1 at 1:1000 (Santa Cruz Biotechnologies); anti–glyceraldehyde 3-phosphate dehydrogenase (GAPDH) at 1:5000 (Santa Cruz Biotechnologies); anti-GFP at 1:5000 (Santa Cruz Biotechnologies); anti-LC3 antibody at 1:1000 (Cell Signaling, Danvers, MA); and anti-CFTR at 1:1000 (Millipore, Billerica, MA). After incubation with primary antibody, membranes were washed in TBS with 0.1% Tween-20 and incubated in horseradish peroxidase–coupled secondary antibody (Amersham, Piscataway, NJ) diluted at 1:5000 in washing buffer with 5% nonfat milk for 1 h. Immunoreactivity was detected using the enhanced chemiluminescence assay (Pierce).
PNGase (New England Biolabs, Ipswich, MA) digestion was performed on total cell lysate for 6 h at 37°C in the buffer supplied by the manufacturer (Supplemental Figure S9).
Protein quantification was measured using VisionWork LS Software. Rhodopsin protein levels were determined by measuring the area density within the bracket (including glycosylated monomers, dimer, and multimers) indicated in the figures after normalizing with the equivalent area density from the control lane.
Protein biotinylation
For cell surface biotinylation, cells were grown in 10-cm dishes coated with poly-d-lysine and transfected with 3 μg of wild-type rhodopsin or mutant P23H rhodopsin. Four hours after transfection, cells were treated with or without 1NM-PP1 for 24 h. The cells were then washed with ice-cold PBS containing 1 mM MgCl2 and 0.1 mM CaCI2. After washing, 5 ml of NaIO4 (Pierce; 10 mM in ice-cold PBS containing 1 mM MgCl2 and 0.1 mM CaCI2) was added to the dishes and agitated for 30 min at 4°C in the dark. After washing (three times) with PBS containing MgCl2 and CaCI2, the cells were labeled with biotin hydrazide (Pierce; 2 mM in ice-cold PBS containing MgCl2 and CaCI2) at 4°C in the dark. Biotinylated cells were collected and pelleted by brief centrifugation and solubilized in 0.5 ml of ice-cold lysis buffer (1% NP-40, 50 mM Tris-HCl, pH 8, and 150 mM NaCl, containing protease inhibitors [Roche]). Three hundred μl of a 50% slurry of neutrAvidin-agarose beads (Pierce) was incubated with biotinylated cell lysates. The neutrAvidin-agarose beads were washed three times with 0.5 ml of PBS containing protease inhibitor, resuspended in 200 μl of 1× SDS sample buffer, and boiled for 5 min. Aliquots of recovered biotinylated proteins were used for Western blot analysis.
Immunofluorescence and confocal microscopy
Cells were grown on poly-d-lysine-coated glass coverslips and transfected with wild-type or P23H rhodopsin. For immunofluorescence analysis, cells were fixed for 20 min at room temperature in 4% paraformaldehyde in PBS, pH 7.4, washed briefly with PBS, and permeabilized with 0.1% Triton X-100 in PBS. Cells were then washed two times with 1% bovine serum albumen (BSA) in PBS and blocked with 5% goat serum in 1% BSA/PBS for 20 min. The coverslips were then incubated at room temperature for 1 h with the mouse monoclonal anti-rhodopsin antibody (1D4; Santa Cruz Biotechnologies) used at a dilution of 1:2000 and the rabbit polyclonal anti-calreticulin antibody (Assay Designs, Plymouth Meeting, PA) used at a dilution of 1:500 or the rabbit polyclonal anti-LAMP2 antibody (a gift from Minoru Fukuda, Burnham Institute, La Jolla, CA; Carlsson et al., 1988) at a dilution of 1:500. After washing in 0.1% BSA in PBS three times, cells were incubated with secondary antibodies. Secondary antibodies included Alexa 488 goat anti-mouse (green) antibody (Molecular Probes, Invitrogen) and Alexa 546 goat anti-rabbit (red) antibody (Molecular Probes), and these were used at a dilution of 1:500. After washing in PBS three times, the cover slips were mounted in ProLong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen), and images were collected with a FluoView-1000 confocal microscope (Olympus, Tokyo, Japan) and processed using Olympus FluoView, version 2.0a, Viewer software at the University of California, San Diego, microscopy facility (http://microscopy.ucsd.edu).
Statistical analysis
All results are expressed as mean ± SD of at least three independent experiments. Two-tailed t tests were performed to determine p values for paired samples. A value of p < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank W. C. Smith, R. Ayyagari, M. Fukuda, M. Kamps, R. Kopito, K. Shokat, L. Wiseman, and C. Zhang for helpful comments and generous provision of reagents. These studies were supported by the American Federation for Aging Research, the Hope for Vision Foundation, the Karl Kirchgessner Foundation, the University of California, San Diego, Neuroscience Shared Microscopy Facility (P30-NS047101), and National Institutes of Health Grants EY018313 and EY020846. W.-C.C. received postdoctoral support from the Fight-for-Sight Foundation.
Abbreviations used:
- 1NM-PP1
4-amino-1-tert-butyl-3-(1′-naphthylmethyl)pyrazolo[3,4-d]pyrimidine; BiP, immunoglobulin-binding protein
- CFTR
cystic fibrosis transmembrane conductance regulator
- EDEM1
ER-degradation enhancing α-mannosidase-like protein 1
- ER
endoplasmic reticulum
- ERAD
ER-associated protein degradation
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GFP
green fluorescent protein
- IRE1
inositol-requiring enzyme 1
- PERK
PKR-like endoplasmic reticulum kinase
- UPR
unfolded protein response
- XBP-1
X-box-binding protein 1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-08-0663) on January 4, 2012.
REFERENCES
- Ahmad ST, Natochin M, Barren B, Artemyev NO, O’Tousa JE. Heterologous expression of bovine rhodopsin in Drosophila photoreceptor cells. Invest Ophthalmol Vis Sci. 2006;47:3722–3728. doi: 10.1167/iovs.06-0281. [DOI] [PubMed] [Google Scholar]
- Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, Schuck S, Walter P. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy. 2007;3:285–287. doi: 10.4161/auto.3930. [DOI] [PubMed] [Google Scholar]
- Berson EL. Retinitis pigmentosa. Invest Ophthalmol Vis Sci. 1993;34:1659–1676. [PubMed] [Google Scholar]
- Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol Cell. 2010;40:238–252. doi: 10.1016/j.molcel.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Carlsson SR, Roth J, Piller F, Fukuda M. Isolation and characterization of human lysosomal membrane glycoproteins, h-lamp-1 and h-lamp-2Major sialoglycoproteins carrying polylactosaminoglycan. J Biol Chem. 1988;263:18911–18919. [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, Kenyon C. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 2010;8:e1000450. doi: 10.1371/journal.pbio.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryja TP, McGee TL, Reichel E, Hahn LB, Cowley GS, Yandell DW, Sandberg MA, Berson EL. A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature. 1990;343:364–366. doi: 10.1038/343364a0. [DOI] [PubMed] [Google Scholar]
- Eriksson KK, Vago R, Calanca V, Galli C, Paganetti P, Molinari M. EDEM contributes to maintenance of protein folding efficiency and secretory capacity. J Biol Chem. 2004;279:44600–44605. doi: 10.1074/jbc.M407972200. [DOI] [PubMed] [Google Scholar]
- Farinha CM, Amaral MD. Most F508del-CFTR is targeted to degradation at an early folding checkpoint and independently of calnexin. Mol Cell Biol. 2005;25:5242–5252. doi: 10.1128/MCB.25.12.5242-5252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez TN, Sidrauski C, Dorfler S, Walter P. Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J. 1999;18:3119–3132. doi: 10.1093/emboj/18.11.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbatyuk MS, Knox T, LaVail MM, Gorbatyuk OS, Noorwez SM, Hauswirth WW, Lin JH, Muzyczka N, Lewin AS. Restoration of visual function in P23H rhodopsin transgenic rats by gene delivery of BiP/Grp78. Proc Natl Acad Sci USA. 2010;107:5961–5966. doi: 10.1073/pnas.0911991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griciuc A, Aron L, Roux MJ, Klein R, Giangrande A, Ueffing M. Inactivation of VCP/ter94 suppresses retinal pathology caused by misfolded rhodopsin in Drosophila. PLoS Genet. 2010;6(pii):e1001075. doi: 10.1371/journal.pgen.1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Hargrave PA. Rhodopsin structure, function, and topography. Invest Ophthalmol Vis Sci. 2001;42:3–9. [PubMed] [Google Scholar]
- Hetz C, Glimcher LH. Fine-tuning of the unfolded protein response: assembling the IRE1alpha interactome. Mol Cell. 2009;35:551–561. doi: 10.1016/j.molcel.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Illing ME, Rajan RS, Bence NF, Kopito RR. A rhodopsin mutant linked to autosomal dominant retinitis pigmentosa is prone to aggregate and interacts with the ubiquitin proteasome system. J Biol Chem. 2002;277:34150–34160. doi: 10.1074/jbc.M204955200. [DOI] [PubMed] [Google Scholar]
- Iwawaki T, Akai R, Kohno K. IRE1alpha disruption causes histological abnormality of exocrine tissues, increase of blood glucose level, and decrease of serum immunoglobulin level. PLoS One. 2010;5:e13052. doi: 10.1371/journal.pone.0013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MJ, Ryoo HD. Suppression of retinal degeneration in Drosophila by stimulation of ER-associated degradation. Proc Natl Acad Sci USA. 2009;106:17043–17048. doi: 10.1073/pnas.0905566106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal S. Effect of rapamycin on the fate of P23H opsin associated with retinitis pigmentosa (an American Ophthalmological Society thesis) Trans Am Ophthalmol Soc. 2006;104:517–529. [PMC free article] [PubMed] [Google Scholar]
- Kaushal S, Khorana HG. Structure and function in rhodopsin. 7. Point mutations associated with autosomal dominant retinitis pigmentosa. Biochemistry. 1994;33:6121–6128. doi: 10.1021/bi00186a011. [DOI] [PubMed] [Google Scholar]
- Kawaguchi S, Ng DT. SnapShot: ER-associated protein degradation pathways. Cell. 2007;129:1230. doi: 10.1016/j.cell.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Kosmaoglou M, Cheetham ME. Calnexin is not essential for mammalian rod opsin biogenesis. Mol Vis. 2008;14:2466–2474. [PMC free article] [PubMed] [Google Scholar]
- Kosmaoglou M, Kanuga N, Aguila M, Garriga P, Cheetham ME. A dual role for EDEM1 in the processing of rod opsin. J Cell Sci. 2009;122:4465–4472. doi: 10.1242/jcs.055228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmaoglou M, Schwarz N, Bett JS, Cheetham ME. Molecular chaperones and photoreceptor function. Prog Retin Eye Res. 2008;27:434–449. doi: 10.1016/j.preteyeres.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVail MM, Yasumura D, Matthes MT, Drenser KA, Flannery JG, Lewin AS, Hauswirth WW. Ribozyme rescue of photoreceptor cells in P23H transgenic rats: long-term survival and late-stage therapy. Proc Natl Acad Sci USA. 2000;97:11488–11493. doi: 10.1073/pnas.210319397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin AS, Drenser KA, Hauswirth WW, Nishikawa S, Yasumura D, Flannery JG, LaVail MM. Ribozyme rescue of photoreceptor cells in a transgenic rat model of autosomal dominant retinitis pigmentosa. Nat Med. 1998;4:967–971. doi: 10.1038/nm0898-967. [DOI] [PubMed] [Google Scholar]
- Li H, Korennykh AV, Behrman SL, Walter P. Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proc Natl Acad Sci USA. 2010;107:16113–16118. doi: 10.1073/pnas.1010580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Sandberg MA, Pawlyk BS, Rosner B, Hayes KC, Dryja TP, Berson EL. Effect of vitamin A supplementation on rhodopsin mutants threonine-17 –> methionine and proline-347 –> serine in transgenic mice and in cell cultures. Proc Natl Acad Sci USA. 1998;95:11933–11938. doi: 10.1073/pnas.95.20.11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, LaVail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Zhang Y, Ron D, Walter P. Divergent effects of PERK and IRE1 signaling on cell viability. PLoS ONE. 2009;4:e4170. doi: 10.1371/journal.pone.0004170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang M, Xia CH, Du X, Flannery JG, Ridge KD, Beutler B, Gong X. Severe retinal degeneration caused by a novel rhodopsin mutation. Invest Ophthalmol Vis Sci. 2010;51:1059–1065. doi: 10.1167/iovs.09-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan EB, Becker E, Detwiler TC. Inhibition of calpain in intact platelets by the thiol protease inhibitor E-64d. Biochem Biophys Res Commun. 1989;158:432–435. doi: 10.1016/s0006-291x(89)80065-8. [DOI] [PubMed] [Google Scholar]
- Mendes HF, Cheetham ME. Pharmacological manipulation of gain-of-function and dominant-negative mechanisms in rhodopsin retinitis pigmentosa. Hum Mol Genet. 2008;17:3043–3054. doi: 10.1093/hmg/ddn202. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- Noorwez SM, Kuksa V, Imanishi Y, Zhu L, Filipek S, Palczewski K, Kaushal S. Pharmacological chaperone-mediated in vivo folding and stabilization of the P23H-opsin mutant associated with autosomal dominant retinitis pigmentosa. J Biol Chem. 2003;278:14442–14450. doi: 10.1074/jbc.M300087200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noorwez SM, Malhotra R, McDowell JH, Smith KA, Krebs MP, Kaushal S. Retinoids assist the cellular folding of the autosomal dominant retinitis pigmentosa opsin mutant P23H. J Biol Chem. 2004;279:16278–16284. doi: 10.1074/jbc.M312101200. [DOI] [PubMed] [Google Scholar]
- Noorwez SM, Sama RR, Kaushal S. Calnexin improves the folding efficiency of mutant rhodopsin in the presence of pharmacological chaperone 11-cis-retinal. J Biol Chem. 2009;284:33333–33342. doi: 10.1074/jbc.M109.043364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa D, Tokuda M, Iwawaki T. Site-specific cleavage of CD59 mRNA by endoplasmic reticulum-localized ribonuclease, IRE1. Biochem Biophys Res Commun. 2007;360:122–127. doi: 10.1016/j.bbrc.2007.06.020. [DOI] [PubMed] [Google Scholar]
- Olivari S, Galli C, Alanen H, Ruddock L, Molinari M. A novel stress-induced EDEM variant regulating endoplasmic reticulum-associated glycoprotein degradation. J Biol Chem. 2005;280:2424–2428. doi: 10.1074/jbc.C400534200. [DOI] [PubMed] [Google Scholar]
- Olsson JE, Gordon JW, Pawlyk BS, Roof D, Hayes A, Molday RS, Mukai S, Cowley GS, Berson EL, Dryja TP. Transgenic mice with a rhodopsin mutation (Pro23His): a mouse model of autosomal dominant retinitis pigmentosa. Neuron. 1992;9:815–830. doi: 10.1016/0896-6273(92)90236-7. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rosenbaum EE, Hardie RC, Colley NJ. Calnexin is essential for rhodopsin maturation, Ca2+ regulation, and photoreceptor cell survival. Neuron. 2006;49:229–241. doi: 10.1016/j.neuron.2005.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Sakami S, Maeda T, Bereta G, Okano K, Golczak M, Sumaroka A, Roman AJ, Cideciyan AV, Jacobson SG, Palczewski K. Probing mechanisms of photoreceptor degeneration in a new mouse model of the common form of autosomal dominant retinitis pigmentosa due to P23H opsin mutations. J Biol Chem. 2011;286:10551–10567. doi: 10.1074/jbc.M110.209759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba RS, Munro PM, Luthert PJ, Cheetham ME. The cellular fate of mutant rhodopsin: quality control, degradation and aggresome formation. J Cell Sci. 2002;115:2907–2918. doi: 10.1242/jcs.115.14.2907. [DOI] [PubMed] [Google Scholar]
- Sun F, Zhang R, Gong X, Geng X, Drain PF, Frizzell RA. Derlin-1 promotes the efficient degradation of the cystic fibrosis transmembrane conductance regulator (CFTR) and CFTR folding mutants. J Biol Chem. 2006;281:36856–36863. doi: 10.1074/jbc.M607085200. [DOI] [PubMed] [Google Scholar]
- Sung CH, Schneider BG, Agarwal N, Papermaster DS, Nathans J. Functional heterogeneity of mutant rhodopsins responsible for autosomal dominant retinitis pigmentosa. Proc Natl Acad Sci USA. 1991;88:8840–8844. doi: 10.1073/pnas.88.19.8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- Ward CL, Kopito RR. Intracellular turnover of cystic fibrosis transmembrane conductance regulatorInefficient processing and rapid degradation of wild-type and mutant proteins. J Biol Chem. 1994;269:25710–25718. [PubMed] [Google Scholar]
- Winslow AR, Rubinsztein DC. Autophagy in neurodegeneration and development. Biochim Biophys Acta. 2008;1782:723–729. doi: 10.1016/j.bbadis.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci. 2010;13:805–811. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Hosokawa N, Kaufman RJ, Nagata K, Mori K. A time-dependent phase shift in the mammalian unfolded protein response. Dev Cell. 2003;4:265–271. doi: 10.1016/s1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Younger JM, Chen L, Ren HY, Rosser MF, Turnbull EL, Fan CY, Patterson C, Cyr DM. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- Zhang K, et al. The unfolded protein response transducer IRE1alpha prevents ER stress-induced hepatic steatosis. EMBO J. 2011;30:1357–1375. doi: 10.1038/emboj.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest. 2005;115:268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.