This is the first report on the role of Fer kinase in down-regulating the expression of laminin-binding glycans that suppress cell migration. The data show a novel biochemical interaction between glycan-based adhesion and cell migration, mediated by a tyrosine kinase.
Abstract
Glycans of α-dystroglycan (α-DG), which is expressed at the epithelial cell–basement membrane (BM) interface, play an essential role in epithelium development and tissue organization. Laminin-binding glycans on α-DG expressed on cancer cells suppress tumor progression by attenuating tumor cell migration from the BM. However, mechanisms controlling laminin-binding glycan expression are not known. Here, we used small interfering RNA (siRNA) library screening and identified Fer kinase, a non–receptor-type tyrosine kinase, as a key regulator of laminin-binding glycan expression. Fer overexpression decreased laminin-binding glycan expression, whereas siRNA-mediated down-regulation of Fer kinase increased glycan expression on breast and prostate cancer cell lines. Loss of Fer kinase function via siRNA or mutagenesis increased transcription levels of glycosyltransferases, including protein O-mannosyltransferase 1, β3-N-acetylglucosaminyltransferase 1, and like-acetylglucosaminyltransferase that are required to synthesize laminin-binding glycans. Consistently, inhibition of Fer expression decreased cell migration in the presence of laminin fragment. Fer kinase regulated STAT3 phosphorylation and consequent activation, whereas knockdown of STAT3 increased laminin-binding glycan expression on cancer cells. These results indicate that the Fer pathway negatively controls expression of genes required to synthesize laminin-binding glycans, thus impairing BM attachment and increasing tumor cell migration.
INTRODUCTION
During cancer metastasis, interactions between epithelial cells and the basement membrane (BM) are disrupted by loss of adhesion molecules expressed on solid tumors after the epithelial–mesenchymal transition (EMT), similar to EMT mechanisms that occur during BM disassembly in normal development (White et al., 2004; Levayer and Lecuit, 2008; Polyak and Weinberg, 2009). Thus molecules functioning in epithelial cell–BM interaction are important markers of tumor progression.
Dystroglycan complex is a major mediator of interactions between cells and the BM in many tissues, including brain, mammary gland, muscle, and prostate (Barresi and Campbell, 2006). In the dystroglycan complex, α-dystroglycan (α-DG) expressed on the cell surface is a major receptor for several BM proteins, such as laminin, perlecan, and agrin, through binding to laminin G–like domain (Barresi and Campbell, 2006; Larsen et al., 2006), whereas β-dystroglycan (β-DG) links the complex to the actin cytoskeleton via dystrophin and α-dystrobrevin (Grady et al., 1999). α-DG is highly glycosylated and contains both N-linked glycans and mucin type O-glycans, all of which are clustered in a mucin-like domain at the N-terminus of mature α-DG protein (Chiba et al., 1997; Barresi and Campbell, 2006). Laminin-binding glycans contain phosphorylated O-mannose in the core (Yoshida-Moriguchi et al., 2010), but the structure of the long side chain, which is responsible for the laminin binding, has not been elucidated. The glycans can be synthesized by coordinated action of protein O-mannosyltransferase 1 (POMT1; Manya et al., 2004), protein O-mannosyltransferase 2 (POMT2), protein O-mannose β1, 2-N-acetylglucosaminyltransferase 1 (POMGnT1; Yoshida et al., 2001), β3-N-acetylglucosaminyltransferase 1 (β3GnT1; Sasaki et al., 1997; Bao et al., 2009), like-acetylglucosaminyltransferase protein (LARGE; Peyrard et al., 1999; Barresi et al., 2004; Kanagawa et al., 2004), fukutin (Kobayashi et al., 1998), and fukutin-related protein (FKRP) (Brockington et al., 2001). Both analysis of mouse mutants and studies of human disease such as congenital muscular dystrophies indicate that laminin-binding glycans synthesized by these glycosyltransferases are crucial for interaction of epithelial cells with the BM (Grewal et al., 2001; Yoshida et al., 2001; Longman et al., 2003). Recent findings demonstrate that cancer progression of tumors of epithelial origin, including prostate, breast, colon, and oral cancers, is associated with loss of α-DG and/or its laminin-binding glycans (Muschler et al., 2002; Jing et al., 2004; Sgambato et al., 2007), indicating that they function as a tumor suppressor (Bao et al., 2009). In fact, signaling activated by cell adhesion through laminin-binding glycans on α-DG can compete with mitogen-activated protein kinase signaling activated by integrin (Spence et al., 2004; Bao et al., 2009).
We and others recently found that the activity of glycosyltransferases required for synthesis of laminin-binding glycans plays key roles in tumor cell behavior (Bao et al., 2009; de Bernabe et al., 2009). In mouse models, PC3-H cells highly expressing laminin-binding glycans formed smaller prostate tumors than did PC3-L, which express minimal amounts of laminin-binding glycans due to lack of β3GnT1 (Bao et al., 2009). β3GnT1 overexpression rescued laminin-mediated adhesion, making cells less tumorigenic. Interactions between laminin and laminin-binding glycans on α-DG antagonize migration signals initiated by integrin-mediated adhesion, suppressing tumor cell migration (Ferletta et al., 2003; Spence et al., 2004; Bao et al., 2009). Expression of LARGE, which contains two distinct structural domains homologous to UDP-glucose protein glucosyltransferase (Patnaik and Stanley, 2005) and β3GnT1 (Sasaki et al., 1997), is also lost in highly metastatic epithelial cell–derived cancer cell lines (de Bernabe et al., 2009), promoting tumor progression due to decreased tumor cell adhesion to BM. Because increases in laminin-binding glycans on the cell surface suppress tumor progression, it is critical to define pathways that control expression levels of α-DG and glycosyltransferases required for glycan synthesis. However, it is not known which signaling pathways function in that process.
In the present study, we used siRNA screening and found that Fer, a non–receptor-type tyrosine kinase (Greer, 2002), plays a critical role in synthesis of the laminin-binding glycans on α-DG. Overexpression of Fer increased tyrosine phosphorylation and decreased expression of laminin-binding glycans, whereas short interfering RNA (siRNA)–mediated down-regulation of Fer increased expression of laminin-binding glycans. RT-PCR analysis demonstrated that siRNA-mediated knockdown of Fer increased transcription of β3GnT1 and LARGE. Increases in laminin-binding glycans by siRNA-mediated knockdown of Fer resulted in reduced cell migration, indicating that the Fer-mediated signaling is a valuable target for tumor suppression.
RESULTS
Fer kinase down-regulates laminin-binding glycan expression in prostate cancer cells
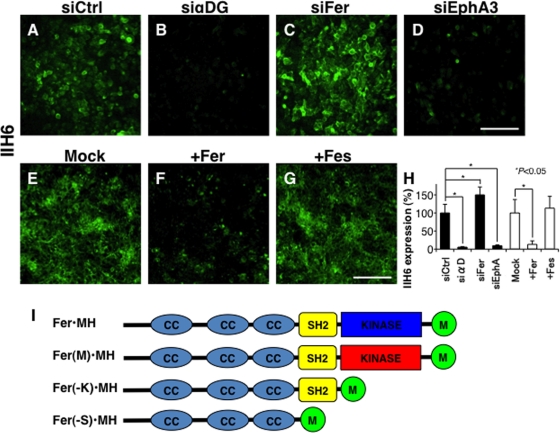
We showed that overexpression of LARGE or β3GnT1 blocks migration of many carcinoma cells (Bao et al., 2009). However, the same treatment did not reduce cell migration of some cell lines, probably due to potential low abundance of other glycosyltransferases such as POMT1, POMGnT1, fukutin, and FKRP in these lines. Alternatively, other, unidentified genes may be responsible for this effect (Godfrey et al., 2007; Muntoni et al., 2007). To reveal mechanisms underlying expression of laminin-binding glycans in tumor cells, we undertook an siRNA-based, high-throughput screening of the human prostate DU145 cell line, as these cells express moderate levels of laminin-binding glycans and are less heterogeneous than PC3 cells (Bao et al., 2009). For the screening (Konig et al., 2008, 2010), we tested a druggable kinase siRNA library (704 genes × 3 siRNAs) to identify the intracellular signaling molecules regulating laminin-binding glycans recognized by IIH6 antibody. After analyzing three different siRNAs in duplicate, we identified 10 genes whose knockdown promoted the glycan expression, defined as a 150% increase over controls, and 26 genes whose knockdown was associated with decreased expression, defined as <50% of control (Supplemental Table S1). Among the former, we found that knockdown of Fer kinase, a non–receptor-type tyrosine kinase, significantly increased expression of laminin-binding glycans (Figure 1, C and H, 141–365% over control, p = 0.002; Table 1). Consistently, overexpression of Fer dramatically reduced the laminin-binding glycans (Figure 1, F and H), whereas Fes kinase, ∼50% identical to Fer kinase, did not affect the glycan expression (Figure 1, G and H). We also found that siRNAs targeting fibroblast growth factor (FGF) receptor 1 (FGFR1), FGF receptor 2 (FGFR2), and FGF receptor 4 (FGFR4) also increased laminin-binding glycan expression (100–385% over control, p = 0.004, 0.014, and 0.006, respectively), whereas knockdown of Eph receptor A3 decreased laminin-binding glycan expression (10–30% of control). The results were individually verified by using three different siRNAs for each gene. These results indicated that expression of laminin-binding glycans on α-DG is regulated by several different signaling pathways.
FIGURE 1:
Fer-siRNA increases laminin-binding glycan expression on DU145 cells. Expression of laminin-binding glycans detected by IIH6 antibody was determined after transfection with (A) siRNA control (Sc-5701), (B) siRNA for α-DG, (C) siRNA targeting Fer, or (D) siRNA targeting EphA3. siRNA targeting α-DG blocked laminin-binding glycan expression, siRNA-Fer increased it, and siRNA-EphA3 decreased it. (E–G) Laminin-binding glycans after transfection of Mock (E), Fer (F), or Fes cDNA (G). (H) Summary of expression of IIH6 is summarized, represented by mean fluorescence intensity per cell. Experiments were repeated in triplicate, and error bars represent the SE of the mean. (I) Schematic representation of Fer and Fer mutants. Fer(M)•MH, Fer(-K)•MH, and Fer(–S)•MH correspond to Fer with Arg743Asp mutation, kinase domain-deleted mutant, and kinase domain and SH2-domain deleted mutant, respectively. Myc and histidine tag (MH) is added to the COOH-terminal. CC, coiled-coil domain. Scale bar in D and G, 100 μm.
TABLE 1:
Relative expression of laminin-binding glycans that showed higher (>150%) or lower expression (<50%) after siRNA treatments.
| Kinase siRNA librarya | Relative expression (%) | |||
|---|---|---|---|---|
| Gene name | Gene symbol | siRNA1 | siRNA2 | siRNA3 |
| Fer (fps/fes related) tyrosine kinase (NCP94) | Fer | 306.0 | 233.5 | 240.5 |
| Fibroblast growth factor receptor 1 | FGFR1 | 189.5 | 257.0 | 343.0 |
| Fibroblast growth factor receptor 2 | FGFR2 | 331.5 | 143.5 | 187.0 |
| Fibroblast growth factor receptor 4 | FGFR4 | 301.0 | 153.0 | 327.0 |
| Eph receptor A3 | EphA3 | 13.5 | 25.0 | 23.0 |
| Fes (feline sarcoma oncogene) | Fes | 119.0 | 172.5 | 179.0 |
a704 genes/3 siRNAs per gene.
Only glycans that showed a prominent change are listed. Expression of the glycans was assessed by IIH6 antibody staining, followed by fluorescein isothiocyanate–conjugated secondary antibody and evaluated by high-throughput microscopy.
Down-regulation of Fer increases the laminin-binding glycans in breast carcinoma cells
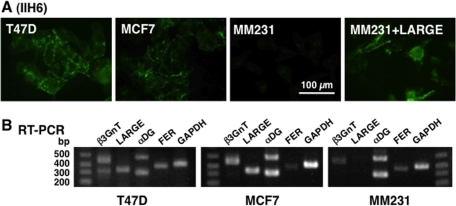
To examine whether the regulation of laminin-binding glycan by Fer is general for different cancer cells, we tested the effects of siRNA-Fer in T47D, MCF-7, and MDA-MB-231 breast cancer cells. The cells express high, moderate, and minimal laminin-binding glycan, respectively, at cell surface examined by immunostaining and flow cytometry (Figure 2; Bao et al., 2009).
FIGURE 2:
Laminin-binding glycan expression in breast cancer cells. (A) Breast cancer cell lines T47D, MCF-7, and MDA-MB-231 (MM231) were stained with IIH6 for laminin-binding glycans. MDA-MB-231 cells were transfected with LARGE cDNA (right). (B) RT-PCR analysis of the three breast cancer lines for α-DG, β3GnT1, and LARGE. Note that LARGE was not detected in parent MDA-MB-231 (MM231) cells.
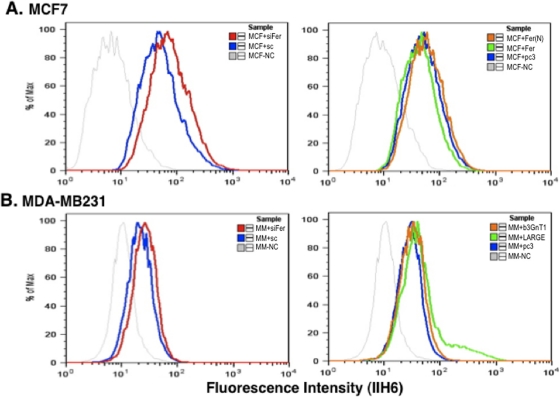
On the basis of previous analysis of laminin-binding glycan expression (Bao et al., 2009), we undertook siRNA-mediated down-regulation of α-DG, β3GnT1, and LARGE in T47D cells and observed decreased glycan levels (data not shown), confirming common proteins required for the glycan synthesis. Among them, LARGE was not expressed well in MDA-MB-231 cells, whereas all the other genes examined were expressed in other two cell lines (Figure 2B; Bao et al., 2009). Significantly, transfection of MDA-MB-231 cells with LARGE, which are more mobile than T47D and MCF-7 cells, rescued expression of laminin-binding glycans (Figures 2A and 3B), confirming that LARGE loss underlies lack of glycan expression in this line. Because Fer kinase is expressed in all three breast cancer lines, albeit at different levels, we asked whether transfection of siRNA-Fer would increase laminin-binding glycans in these breast cancer cells. Transfection of siRNA targeting Fer increased expression of laminin-binding glycans in MCF-7 and MDA-MB-231 cells, suggesting that Fer inhibits laminin-binding glycan synthesis in epithelium-derived cancers (Figure 3).
FIGURE 3:
Fer knockdown induces laminin-binding glycan expression in breast cancer cells. (A) MCF-7 cells were transfected with siRNA-Fer (left) and Fer cDNA or a kinase mutant Fer cDNA (right), followed by FACS analysis with IIH6 antibody. (B) MDA-MB-231 cells were transfected with siRNA-Fer (left), and LARGE cDNA (right) and analyzed in a similar way. Control experiment omitted the primary antibody.
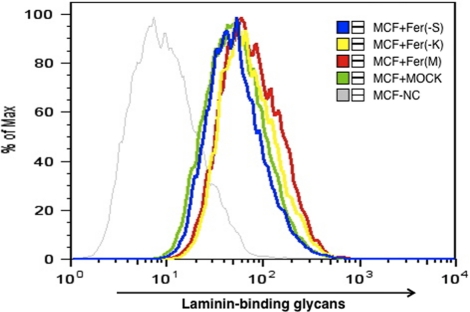
Fer kinase contains three coiled-coil (CC) domains, an SH2 domain, and a tyrosine kinase domain (Figure 1I; Greer, 2002). To undertake functional analysis, we constructed a dominant-negative form of Fer, substituting Asp-743 for Arg, which was tagged with Myc and hexahistidine (Fer(M)•MH in Figure 1I; Cole et al., 1999). To identify which Fer domain functions to control laminin-binding glycan expression, we also generated deletion mutants lacking the kinase domain (Fer(−K)•MH) or lacking both the SH2 and kinase domains (Fer(−S)•MH; Figure 1I). Transfection of MCF-7 cells with tagged wild-type Fer cDNA decreased glycan expression, whereas transfection with Fer(M) increased it (Figure 4). Transfection of MCF-7 cells with Fer(−K) construct, which lacks the kinase domain, but not with Fer(−S) construct, which lacks both the SH2 and kinase domains, increased glycan expression (Figure 4). These results indicate that inhibition of glycan expression by Fer likely requires its tyrosine kinase activity toward target molecules interacting through SH2 domain of Fer.
FIGURE 4:
Mutated Fer increases laminin-binding glycan expression detected by IIH6 staining in MCF-7 breast cancer cells. MCF cells transfected with mutated Fer or Fer deletion mutants expressed higher levels of laminin-binding glycans than did mock-transfected cells (MCF + MOCK, green). Fer cDNA constructs are shown in Figure 1I. Negative control (NC) without the first antibody is indicated by a thin gray line. Fer(M), Fer(−K), and Fer(−S) correspond to Fer kinase with Arg743Asp mutant, kinase domain-deleted Fer mutant, and kinase and SH2 domains–deleted Fer mutant, respectively.
Fer down-regulates glycosyltransferase expression required for laminin-binding glycan synthesis
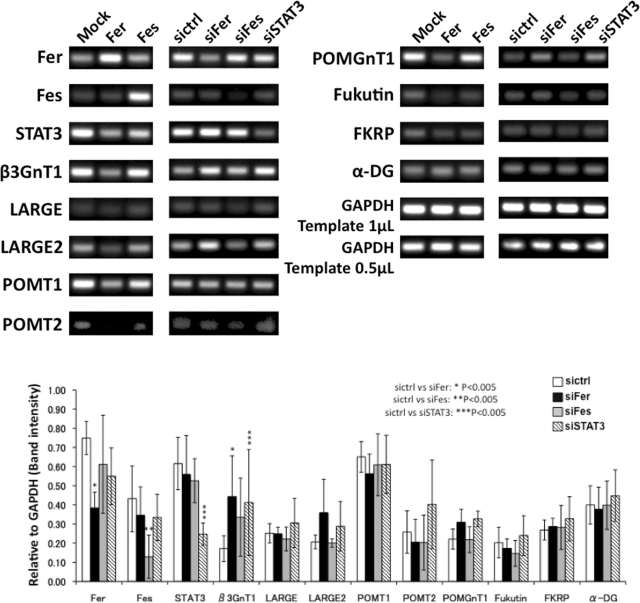
We first hypothesized that Fer either down-regulates transcription of genes required for laminin-binding glycan synthesis or blocks activity of proteins such as α-DG and β3GnT1. siRNA-Fer switched on the expression of LARGE and laminin-binding glycan in MDA-MB-231 cells, and LARGE transfection also increased the glycans (Supplemental Figures S1 and 2A). These results suggested that Fer downregulates transcription of genes for the laminin-binding glycans synthesis rather than inactivation of translated proteins such as α-DG and β3GnT1. Semiquantitative RT-PCR on prostate cancer DU145 cells showed that siRNA-Fer increased the transcripts for β3GnT1 and LARGE, whereas overexpression of Fer significantly decreased the transcripts for β3GnT1 and LARGE2 (Figure 5 and Supplemental Figure S2). It is noteworthy that transcription of α-DG was not perturbed by these treatments. Similarly, siRNA-Fer increased β3GnT1, LARGE, POMT1, and α-DG expression on breast cancer cells, MCF-7. and MDA-MB-231 (Supplemental Figure S1). These results strongly suggest that increased laminin-binding glycan expression mediated by siRNA-Fer results from increased transcription of glycosyltransferases required for their synthesis.
FIGURE 5:
Fer down-regulates mRNAs functioning in laminin-binding glycan synthesis. (Top) Prostate cancer DU145 cells were transfected with Fer, Fes cDNA (left), or siRNA for Fer, Fes, or STAT3 (right). Control siRNA (sictrl) or empty vector (Mock) served as controls for RT-PCR analysis. GAPDH was used to normalize mRNA levels. (Bottom) Expression levels of laminin-binding glycan synthesis–related proteins in siRNA-transfected DU145 cells were semiquantitatively estimated by RT-PCR. mRNA levels were normalized to that of GAPDH. Expression levels were assessed by ImageJ. Experiments were repeated in triplicate, and error bars represent the SE of the mean. β3GnT1 and LARGE2 mRNA levels were increased by siRNA-Fer but decreased by Fer cDNA transfection. Statistical difference (p < 0.005) between control siRNA and each siRNA is indicated by asterisk(s).
STAT3 down-regulates laminin-binding glycans
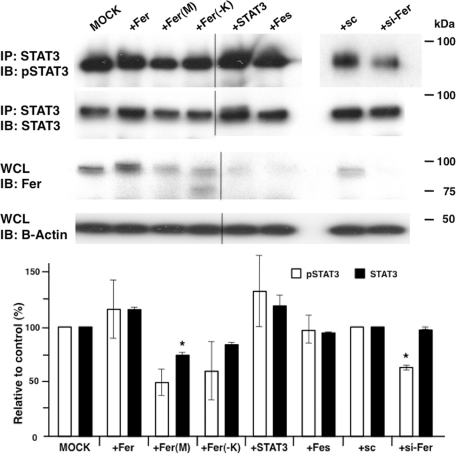
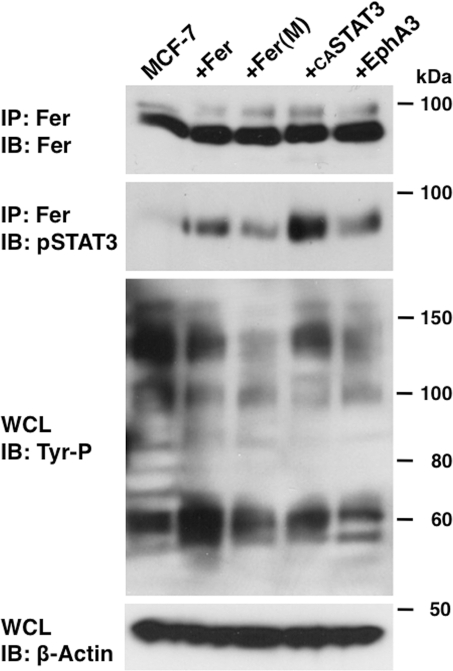
Fer kinase functions in cell growth, adhesion, and migration through activating factors such as cortactin (CTTN), p120Cas, β-catenin, and STAT3 (Greer, 2002; Piedra et al., 2003; El Sayegh et al., 2005; Pasder et al., 2006; Lee et al., 2008). When we performed Western blot analysis using an anti-phosphotyrosine antibody of MCF-7 cells transfected with Fer•MH, we found that overall tyrosine phosphorylation, as well as phosphorylation of Fer itself, was increased (Supplemental Figure S4), whereas expression of the dominant-negative form of Fer, Fer(M)•MH, slightly suppressed tyrosine phosphorylation (Supplemental Figure S4). Transfection of MCF-7 cells with siRNA-Fer decreased Fer protein levels, but tyrosine phosphorylation levels of whole protein were similar to those seen in control cells, likely due to the relatively low tyrosine phosphorylation activity by endogenous Fer kinase in those cells. Fer has been shown to bind to p120-catenin (Piedra et al., 2003), and Fer down-regulation resulted in a decrease in E-cadherin protein (Supplemental Figure S4). Previously Fer was shown to activate STAT3 and promote human prostate cancer cell growth (Zoubeidi et al., 2009). Thus we asked whether STAT3 interacts with Fer in breast cancer cell lines. RT-PCR analysis detected STAT3 and Fer mRNAs in MCF-7 and DU-145 cell lines (Figure 5 and Supplemental Figure S1). We then asked whether STAT3 phosphorylation is regulated by Fer. Transfection of Fer kinase into those cells increased STAT3 phosphorylation (Figure 6, second lane from left), indicating that STAT3 is activated by Fer. Of importance, siRNA-Fer significantly decreased phosphorylation of STAT3 without affecting STAT3 amount (Figure 6). Furthermore, Fer-dependent STAT3 phosphorylation was more elevated by Fer overexpression than mutated Fer overexpression (Figure 7).
FIGURE 6:
Immunoblot analysis of tyrosine phosphorylation of STAT3. (Top) MCF-7 cells were transfected with cDNAs encoding Fer, mutated Arg743Asp Fer (Fer(M)), Fer lacking kinase domain (Fer(−K)), STAT3, Fes, siRNA control (sc), or siRNA-Fer (si-Fer). STAT3 was immunoprecipitated, and blots were probed with anti–phosphorylated STAT3 (pSTA3) followed by STAT3. Whole-cell lysate (WCL) was also blotted and probed with anti-Fer, followed by anti–β-actin. (Bottom) Each pan STAT3 (STAT3) and pSTAT3 band intensity was quantified by ImageJ and normalized to that of β-actin. Averages of independent experiments are indicated by white columns (pSTAT3) or black columns (STAT3), and error bars denote SD. Comparison to MOCK or control siRNA transfection after Student's t test demonstrated that Fer(M) and si-Fer significantly decreased STAT3 and pSTAT3 levels, respectively. *p < 0.02.
FIGURE 7:
Fer affects tyrosine phosphorylation of STAT3. MCF-7 cells were transfected with Fer cDNA, mutated Fer cDNA, constitutively active (CA) STAT3 cDNA, or EphA3 cDNA. Proteins immunoprecipitated by anti-Fer antibody were subjected to Western blotting with Fer or phosphorylated STAT3 antibodies. A portion of the whole-cell lysate (WCL) was also probed with phosphorylated tyrosine. β-Actin was probed for loading control.
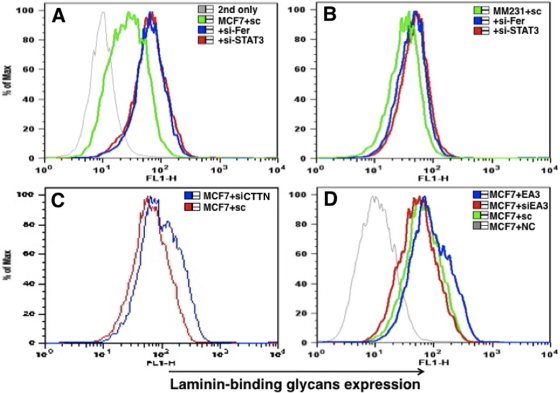
Then we analyzed the STAT3 function in regulating laminin-binding glycan expression. Knockdown of either Fer or STAT3 increased levels of laminin-binding glycans in MCF-7 and MDA-MB-231 breast carcinoma lines (Figure 8, A and B). It is noteworthy that siRNA-mediated down-regulation of STAT3 slightly increased β3GnT1 transcription (Figure 5). Overall, these results suggest that Fer-mediated down-regulation of laminin-binding glycans requires Fer kinase activity and that STAT3 may be one of effectors of Fer kinase.
FIGURE 8:
Laminin-binding glycan expression is regulated by STAT3. STAT3 siRNA increased expression of laminin-binding glycans, as did siRNA-Fer in MCF-7 (A) and MDA-MB-231 (B) cells. (C) CTTN siRNA also increased laminin-binding glycan expression in MCF-7 cells. (D) EphA3 (EA3) cDNA increased laminin-binding glycan expression in MCF-7 cells, whereas EphA3 siRNA decreased laminin-binding glycans expression. SC, siRNA control; NC, nonprimary antibody; 2nd only, second antibody only. The results are shown after FACS analysis.
In parallel, we asked whether gain- or loss-of-function of tyrosine kinase EphA3 altered glycan expression. Transfection of MCF-7 cells with an EphA3 expression vector and siRNA targeting EphA3 increased and decreased the laminin-binding glycan expression, respectively, in MCF-7 cells (Figure 8D). This result is consistent with the observation that either overall tyrosine phosphorylation or STAT3 phosphorylation was decreased following EphA3 transfection of MCF-7 cells (Figure 7). Of interest, siRNA-mediated down-regulation of CTTN in MCF-7 cells also increased levels of laminin-binding glycans (Figure 8D), indicating that both Fer and CTTN act as an inhibitor of laminin-binding glycan expression.
siRNA-mediated down-regulation of Fer decreases cell migration
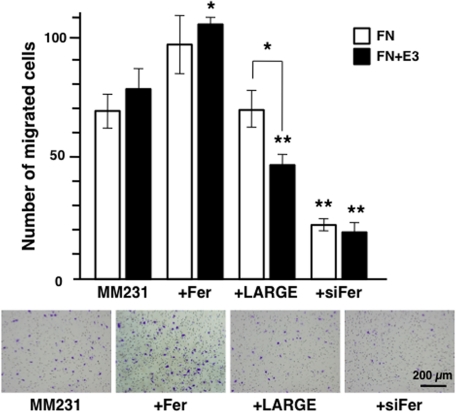
Previously we showed that DU-145 and T47D cells expressing moderate amount of laminin-binding glycans exhibited decreased migration on a laminin E3 fragment, which binds to the laminin-binding glycans (Bao et al., 2009). To analyze the effect of Fer expression on cell migration, we transfected MDA-MB-231 cells with expression vectors encoding Fer, LARGE, and siRNA-Fer and analyzed migration. Consistent with the changes in the amount of laminin-binding glycans, Fer increased migration, whereas siRNA-Fer decreased migration, of MDA-MB-231 cells (Figure 9). When the cell migration was assayed in the presence of E3, the cells migrated more for those expressing decreased amount of glycans (control and Fer), whereas the migration decreased after transfection of LARGE. Altogether, our results demonstrate a novel mechanism by which expression of antimigratory laminin-binding glycans on α-DG might be regulated by Fer/STAT3 signaling.
FIGURE 9:
siRNA-Fer or LARGE decreases breast cancer cell migration. MDA-MB-231 cells were transfected with Fer cDNA, LARGE cDNA, or siRNA-Fer, and migration through a Transwell chamber was measured. (Bottom) Typical results of MDA-MB-231 cell migration through the membrane coated with fibronectin alone (FN) and fibronectin and laminin E3 fragment (FN + E3) after staining with crystal violet. Experiments were repeated in triplicate. Columns and error bars represent average and SD, respectively, obtained after counting cell numbers migrated. LARGE-dependent laminin-binding glycan decreased cell migration specifically on FN + E3. siRNA-FER attenuated cell migration on both FN and FN + E3. Statistical significance was analyzed between original and transfected cells on the same substrates or between the same cells on different substrates (significance was observed only in LARGE-transfected cells). *p < 0.01, **p < 0.005.
DISCUSSION
Loss of the laminin-binding glycans in α-DG from epithelium-derived cancer cells is a critical step promoting cell detachment from the BM and consequent metastasis. Here we undertook an siRNA screen to identify pathways regulating synthesis of laminin-binding glycans and identified several candidate molecules, among them Fer kinase. Previously Fer was shown to associate with factors functioning in actin polymerization and with catenins, suggesting a role in cell migration and adhesion (Piedra et al., 2003; Xu et al., 2004). Fer kinase expressed in cancer cells is reportedly activated by interleukin 6 (IL-6) and IL-10 and functions to increase tumor cell growth (Hayun et al., 2007; Zoubeidi et al., 2009). Here, we identify a novel role of Fer kinase in regulating glycosylation of α-DG. Specifically, we show that Fer kinase inhibits laminin-binding glycan synthesis in breast and prostate cancer cells, suggesting that activity of this kinase is suppressed when epithelial cells interact with the BM.
Our screening also identified other factors associated with the cell cycle, proliferation, migration, and differentiation. Similar to the case with Fer, we found that siRNA targeting FGFR1 or 4 enhanced glycan expression. Both FGFR1 and 4 activate signaling relevant to phospholipase Cγ, phosphatidyl inositol 3-kinase, mitogen-activated protein kinase, and STAT pathways, and both have been linked to prostate cancer progression (Kwabi-Addo et al., 2004). The same screening also showed that EphA3 receptor upregulates the laminin-binding glycans. This finding is consistent with the previous report that EphA3 receptor gene is mutated in lung carcinoma, suggesting that EphA3 receptor functions as a tumor suppressor (Ding et al., 2008).
It is noteworthy that Fes, a kinase similar to Fer, was not identified in our screen. In separate experiments, transfection of siRNA-Fes moderately increased the laminin-binding glycan, and overexpression of Fes did not decrease the laminin-binding glycans. It was reported that N-terminal domain is important for Fer function (Priel-Halachmi et al., 2000). Fes and Fer differ most in that region, which might account for these functional differences.
It was reported that Fer activates STAT3 (Zoubeidi et al., 2009). Indeed, we found that STAT3 is phosphorylated in the presence of wild-type Fer. We also found that mutated Fer containing the SH2 domain but lacking kinase activity promoted increased glycan expression, suggesting that the SH2 domain mediates interaction of Fer with target molecules, such as STAT3. Overall, enhanced levels of laminin-binding glycans seen following Fer kinase knockdown suggest that Fer antagonizes glycan synthesis partly by activating STAT3. Down-regulation of Fer also down-regulates E-cadherin (Supplemental Figure S4; Xu et al., 2004). Because E-cadherin is critical for maintenance of an epithelial structure (Levayer and Lecuit, 2008), decreases in laminin-binding glycans may not necessarily promote an EMT. We also found that the amount of the laminin-binding glycan is inversely correlated well with the progression of prostate cancer as assessed by Gleason patterns on patients (Shimojo et al., 2011) but is not evidently correlated with the survival of patients assessed by Kaplan–Meier analysis. Thus a mechanism other than stimulation of EMT likely underlies malignancy caused by Fer activation.
The present work indicates that down-regulation of laminin-binding glycans by Fer is due to decreased transcription of mRNAs encoding glycosyltransferases, which are required for synthesis of laminin-binding glycans. Furthermore, STAT3 inactivation resulted in increased expression of laminin-binding glycans on cell surface. Because STAT3 itself only moderately down-regulates glycosyltransferases that form laminin-binding glycans, STAT3 might attenuate the transport of α-DG with laminin-binding glycans to the cell surface. This result is consistent with the absence of potential STA3-binding sites in the β3GnT1 gene. Recent studies show that STAT3 plays a critical role in initiation and progression of pancreatic cancer (Fukuda et al., 2011; Lesina et al., 2011; Li et al., 2011), an effect likely due to increased proliferation and decreased apoptosis seen following STAT3 activation. It was also suggested that STAT3 orchestrates tumor-associated inflammation by up-regulating chemokines, leading to recruitment of immune and inflammatory cells (Bromberg et al., 1999; Li et al., 2011). These results, combined with our findings, suggest that the Fer-STAT3 axis might represent a major pathway to link glycosylation to inflammatory response and tumor formation. Of interest, because Fer-deficient mice are viable (Craig et al., 2001), Fer might play a more critical role in cancer progression than it does in maintenance of normal tissues. Fer kinase might represent a good target for cancer therapy without major side effects.
MATERIALS AND METHODS
Cell lines
The DU145 prostate cancer cell line was obtained from the American Type Culture Collection (ATCC; Manassas, VA) and grown in RPMI-1640 with penicillin, streptomycin, and 10% fetal bovine serum (FBS) at 37°C with 5% CO2 as described (Bao et al., 2009). Breast cancer cell lines MCF-7, MDA-MB-231, and T47D were also purchased from ATCC and cultured in DME/high glucose supplemented with penicillin, streptomycin, and 10% FBS.
Antibodies
Laminin-binding glycans (IIH6; Santa Cruz Biotechnology, Santa Cruz, CA), Fer (Abcam, Cambridge, MA, and Cell Signaling, Beverly, MA), phosphorylated tyrosine (Santa Cruz Biotechnology), β-actin (Sigma-Aldrich, St. Louis, MO), E-cadherin (Invitrogen, Carlsbad, CA), and STAT3 and phospho-STAT3 (Cell Signaling) were used for fluorescence-activated cell sorting analysis, Western blotting, and immunoprecipitation. Goat anti–mouse immunoglobulin M (IgM) labeled with Alexa Fluor 488 (Invitrogen) and peroxidase-conjugated secondary antibodies for mouse IgG or rabbit IgG (GE Healthcare, Piscataway, NJ) were also used.
siRNA library screening
The 384-well, black, clear-bottom plates spotted with Silencer Human Kinase siRNAs targeting 704 genes (Applied Biosystems/Ambion, Austin, TX) were prepared at the Functional Genomics Facility of Sanford-Burnham Medical Research Institute (SBMRI; La Jolla, CA). Three different siRNAs per gene (a total of 2112 siRNAs) were analyzed in duplicate to avoid effects of nonspecific silencing. DU145 cells, which express laminin-binding glycans detectable by the IIH6 monoclonal antibody (Barresi and Campbell, 2006), were used for screening. DharmaFECT1 (0.1 μl; Thermo Fisher Scientific, Waltham, MA) in OPTI-MEM (10 μl; Invitrogen) was added to each well and incubated for 30 min, and then cells (1 × 104) suspended in 40 μl of complete medium were slowly added to wells for culturing for 3 d at 37°C. After fixation in 4% paraformaldehyde, cells were incubated with phosphate-buffered saline (PBS) containing 2% bovine serum albumin and 2% normal goat serum (2% BSA/NGS-PBS) for 1 h and then reacted with IIH6 antibody in 1% BSA/NGS-PBS for 1 h. Cells were washed three times with PBS containing 0.05% Tween 20 (PBST) and incubated with Alexa Fluor 488–conjugated anti–mouse IgM antibody in 1% BSA/NGS-PBS for 1 h. After four PBST washes, cells were incubated 1 h with 100 ng/ml 4′,6-diamidino-2-phenylindole (DAPI) in PBS.
Expression levels of laminin-binding glycans per cell were analyzed by an Eidaq 100 (Q3DM, San Diego, CA) at the SBMRI. Four images per well were collected for both DAPI and Alexa 488 fluorophores using an Eidaq 100 automated inverted fluorescence microscope with video and digital cameras. Cells were defined by DAPI staining, and captured images were analyzed using Cytoshop software (Beckman Coulter, Brea, CA). To compare the effect of each siRNA, total fluorescence intensity from each well was divided by the number of DAPI-positive cells.
siRNA
To analyze knockdown phenotypes, siRNAs were obtained from Applied Biosystems/Ambion, and control siRNAs were obtained from Qiagen (Valencia, CA). siRNAs were diluted to 20 μM using RNase-free distilled water and kept at −80°C until use. Cells were cultured in six-well plates and transfected with siRNAs at a final concentration of 25 nM using DharmaFECT1, according to the manufacturer's protocol.
RT-PCR
Total RNA was prepared from MCF-7, MDA-MB-231, T47D, and DU145 cells with or without siRNA transfection by using TRIzol as described previously (Angata et al., 2007). A total of 5 μg of RNA was used to synthesize cDNA by oligo(dT) and Superscript II (Invitrogen), followed by PCR using gene-specific primers. Primers for α-DG, POMT1, POMT2, POMGnT1, β3GnT1, LARGE, β-actin, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as described previously (Angata et al., 2007; Bao et al., 2009). For Fer, Fer-RT5; 5′-GACAGCCTGTCTACATCATTATGG-3′, and Fer-RT3; 5′-GGAAATATCCTCTGGACAGTGCTG-3′ were used. For Fes, Fes-RT5; 5′-GCCTCAGGGGGCCTCAGACA-3″ and Fes-RT3; 5’-TGCCAAAGCTCCACACGTCGC-3′ were used. For STAT3, STAT-RT5; 5′-CATCATGGGCTTTATCAGTAAGGA-3′ and STAT-RT3; 5′-GTCAATGGTATTGCTGCAGGTCGT-3′ were used. For LARGE2, LARGE2-RT5; 5′-GGGACCACAACCGCTCCGAC-3′ and LARGE2-RT3; 5′-CGGCTGGAGTTATGCCCCGC-3′ were used. For β-catenin, CTNB-RT5; 5′-CTAGCTCGGGATGTTCACAACCGA-3′, and CTNB-RT3; 5′-AATATCAAGTCCAAGATCAGCAGT-3′ were used. For p120 catenin, CTND-RT5; 5′-TCTGCCAGGAGGACAGCAGAACTC-3′, and CTND-RT3; 5′-CCGGTCAATGAGAGGGAGAGTACT-3z were used. For cortactin, CTTN-RT5; 5′-GATGCGGCTTCCTTCAAGGCAGAG-3′, and CTTN-RT3; 5′-ACCAGCCGTCGTCAATCATCTCGA-3′ were used. After amplified DNA was run in agarose gel, images were captured, and intensity of the DNA bands was analyzed by ImageJ (National Institutes of Health, Bethesda, MD) as described (Angata et al., 2007). Quantitative RT-PCR was conducted in the Gene Analysis Facility of Sanford-Burnham Medical Research Institute.
cDNA
Human Fer cDNA was amplified from DU145 RNA using primers and subcloned into pcDNA3.1-MycHis vector (Invitrogen). Mutated Fer, Fer(M) with D743R substitution (Cole et al., 1999), was generated by introducing the NruI site by PCR. PCR-amplified fragments using primers, Bgl-Fer5; 5′-GAAGATCTTCCTCAGGAATTGAAAATAAAA-3′ with Fer-Nru3; 5′-CATCGCGACTCTGAACTGTATCTCCCATAA-3′, and Nru-Fer5; 5′-CATCGCGAGTGTGGAGCTTTGGCATCCTTC-3′ with Fer-Xba3; 5′-CCTCTAGACTGTGTGAGTTTTCTCTTGATG-3′ were connected at the common NruI site (TCGCGA, underline for R743). BglII–XbaI fragment with D743R replaced Fer•MH cDNA at the same sites. Truncated Fer cDNAs were generated by using PCR primers, EV-Fer5; 5′-GGCACAGCTCCATCAGAATCAGTATTA-3′ as common 5′ primer for deletion mutants, Fer-Kpn3; 5′-AGTCTAGAATGGTACCAGTCCTGTTCTGCC-3′ for Fer(−S), and Fer-Bgl3; 5′-GGTCTAGAAAGATCTTCTTTACATGTTTTA-3′ for Fer(−K). Anti-Myc antibody was used to stain transfected cells and detect proteins by Western blotting. Rabbit antibody against Fer and protein A–agarose beads (Pierce, Thermo Fisher Scientific, Rockford, IL) were used for immunoprecipitation of wild-type and mutant Fer proteins.
Human STAT3 cDNA was purchased from Open Biosystems (Thermo Biosystems, Huntsville, AL) and DNA fragment amplified by PCR with primers, Hin-STAT3; 5′-TCAAGCTTTTAGCAGGATGGCCCAATG-3′ and STAT3-Nhe; 5′-TAGCTAGCCATGGGGGAGGTAGCGCACTCC-3′, was subcloned into the pcDNA3.1-MycHis vector. Constitutively activated STAT3 was generated based on previous work (Bromberg et al., 1999) using PCR primers, STAT3-Fsp; 5′-ATGCGCAATCCATGATCTTATAGCCCA-3′, Sca-STAT3; 5′-AAGTACTTGTATCCTGGTGTCTCCACTGGT-3′. STAT3 protein was immunoprecipitated with rabbit anti-STAT3 antibody and protein A–agarose beads to detect phosphorylated STAT3. Human EphA3 cDNA was a kind gift from Elena Pasquale of Sanford-Burnham Medical Research Institute.
FACS analysis
Single cells were prepared using enzyme-free cell dissociation solution (Millipore) and stained with IIH6 antibody and Alexa 488– labeled anti–mouse IgM antibody. Cells were then analyzed by FACSCalibur (BD Biosciences, San Diego, CA) and FlowJo (TreeStar, Ashland, OR; Bao et al., 2009).
Cell migration assay
MDA-MB-231 cells were cultured in serum-free DME/high glucose overnight and dissociated into single cells as described (Bao et al., 2009). Chambers with 8-μm pores (BD Biosciences) were placed into a 24-well plate, and the bottom of the chamber was treated with 5 μg/ml laminin or 10 μg/ml fibronectin for 2 h. Cells (2 × 104) suspended in serum-free DME/high glucose were plated into the chamber for another 3 h. For DU145 cells, 2 × 104 cells were plated and cultured for 24 h in serum-free DME. Cells migrating through the membrane were stained with crystal violet and counted in at least four different fields under the microscope. Statistical analysis was done using Student's t test.
Supplementary Material
Acknowledgments
We thank Elena Pasquale and Renate Koenig for useful suggestions and comments, Pedro Aza-Blanc for technical assistance in siRNA screening at the Functional Genomics Facility of Sanford-Burnham Medical Research Institute, and Giang Nguyen for technical assistance. We also thank Elise Lamar for critical reading of the manuscript and Aleli Morse, Misa Suzuki-Anekoji, and Motohiro Nonaka for help in preparing the manuscript. This work was supported by National Institutes of Health Grants P01CA071932 and R01CA048737 from the National Cancer Institute. The Functional Genomics Facility was supported by Cancer Center Grant P30 CA030199.
Abbreviations used:
- β1
2-N-acetylglucosaminyltransferase 1
- β3GnT1
β3-N-acetylglucosaminyltransferase 1
- BM
basement membrane
- EMT
epithelial–mesenchymal transition
- α-DG
α-dystroglycan
- POMGnT1
protein O-mannose
- POMT1
protein O-mannosyltransferase 1
- POMT2
protein O-mannosyltransferase 2
- LARGE
like-acetylglucosaminyltransferase
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-06-0517) on January 11, 2012.
REFERENCES
- Angata K, Huckaby V, Ranscht B, Terskikh A, Marth JD, Fukuda M. Polysialic acid-directed migration and differentiation of neural precursors are essential for mouse brain development. Mol Cell Biol. 2007;27:6659–6668. doi: 10.1128/MCB.00205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Kobayashi M, Hatakeyama S, Angata K, Gullberg D, Nakayama J, Fukuda MN, Fukuda M. Tumor suppressor function of laminin-binding alpha-dystroglycan requires a distinct beta3-N-acetylglucosaminyltransferase. Proc Natl Acad Sci USA. 2009;106:12109–12114. doi: 10.1073/pnas.0904515106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barresi R, Campbell KP. Dystroglycan: from biosynthesis to pathogenesis of human disease. J Cell Sci. 2006;119:199–207. doi: 10.1242/jcs.02814. [DOI] [PubMed] [Google Scholar]
- Barresi R, et al. LARGE can functionally bypass alpha-dystroglycan glycosylation defects in distinct congenital muscular dystrophies. Nat Med. 2004;10:696–703. doi: 10.1038/nm1059. [DOI] [PubMed] [Google Scholar]
- Brockington M, et al. Mutations in the fukutin-related protein gene (FKRP) cause a form of congenital muscular dystrophy with secondary laminin alpha2 deficiency and abnormal glycosylation of alpha-dystroglycan. Am J Hum Genet. 2001;69:1198–1209. doi: 10.1086/324412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Chiba A, Matsumura K, Yamada H, Inazu T, Shimizu T, Kusunoki S, Kanazawa I, Kobata A, Endo T. Structures of sialylated O-linked oligosaccharides of bovine peripheral nerve alpha-dystroglycan. The role of a novel O-mannosyl-type oligosaccharide in the binding of alpha-dystroglycan with laminin. J Biol Chem. 1997;272:2156–2162. doi: 10.1074/jbc.272.4.2156. [DOI] [PubMed] [Google Scholar]
- Cole LA, Zirngibl R, Craig AW, Jia Z, Greer P. Mutation of a highly conserved aspartate residue in subdomain IX abolishes Fer protein-tyrosine kinase activity. Protein Eng. 1999;12:155–162. doi: 10.1093/protein/12.2.155. [DOI] [PubMed] [Google Scholar]
- Craig AW, Zirngibl R, Williams K, Cole LA, Greer PA. Mice devoid of Fer protein-tyrosine kinase activity are viable and fertile but display reduced cortactin phosphorylation. Mol Cell Biol. 2001;21:603–613. doi: 10.1128/MCB.21.2.603-613.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bernabe DB, Inamori K, Yoshida-Moriguchi T, Weydert CJ, Harper HA, Willer T, Henry MD, Campbell KP. Loss of alpha-dystroglycan laminin binding in epithelium-derived cancers is caused by silencing of LARGE. J Biol Chem. 2009;284:11279–11284. doi: 10.1074/jbc.C900007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Sayegh TY, Arora PD, Fan L, Laschinger CA, Greer PA, McCulloch CA, Kapus A. Phosphorylation of N-cadherin-associated cortactin by Fer kinase regulates N-cadherin mobility and intercellular adhesion strength. Mol Biol Cell. 2005;16:5514–5527. doi: 10.1091/mbc.E05-05-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferletta M, Kikkawa Y, Yu H, Talts JF, Durbeej M, Sonnenberg A, Timpl R, Campbell KP, Ekblom P, Genersch E. Opposing roles of integrin alpha6Abeta1 and dystroglycan in laminin-mediated extracellular signal-regulated kinase activation. Mol Biol Cell. 2003;14:2088–2103. doi: 10.1091/mbc.E03-01-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda A, et al. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell. 2011;19:441–455. doi: 10.1016/j.ccr.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey C, et al. Refining genotype phenotype correlations in muscular dystrophies with defective glycosylation of dystroglycan. Brain. 2007;130:2725–2735. doi: 10.1093/brain/awm212. [DOI] [PubMed] [Google Scholar]
- Grady RM, Grange RW, Lau KS, Maimone MM, Nichol MC, Stull JT, Sanes JR. Role for alpha-dystrobrevin in the pathogenesis of dystrophin-dependent muscular dystrophies. Nat Cell Biol. 1999;1:215–220. doi: 10.1038/12034. [DOI] [PubMed] [Google Scholar]
- Greer P. Closing in on the biological functions of Fps/Fes and Fer. Nat Rev Mol Cell Biol. 2002;3:278–289. doi: 10.1038/nrm783. [DOI] [PubMed] [Google Scholar]
- Grewal PK, Holzfeind PJ, Bittner RE, Hewitt JE. Mutant glycosyltransferase and altered glycosylation of alpha-dystroglycan in the myodystrophy mouse. Nat Genet. 2001;28:151–154. doi: 10.1038/88865. [DOI] [PubMed] [Google Scholar]
- Hayun R, Shpungin S, Malovani H, Albeck M, Okun E, Nir U, Sredni B. Novel involvement of the immunomodulator AS101 in IL-10 signaling, via the tyrosine kinase Fer. Ann NY Acad Sci. 2007;1095:240–250. doi: 10.1196/annals.1397.028. [DOI] [PubMed] [Google Scholar]
- Jing J, Lien CF, Sharma S, Rice J, Brennan PA, Gorecki DC. Aberrant expression, processing and degradation of dystroglycan in squamous cell carcinomas. Eur J Cancer. 2004;40:2143–2151. doi: 10.1016/j.ejca.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Kanagawa M, et al. Molecular recognition by LARGE is essential for expression of functional dystroglycan. Cell. 2004;117:953–964. doi: 10.1016/j.cell.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, et al. An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature. 1998;394:388–392. doi: 10.1038/28653. [DOI] [PubMed] [Google Scholar]
- Konig R, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig R, et al. Human host factors required for influenza virus replication. Nature. 2010;463:813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwabi-Addo B, Ozen M, Ittmann M. The role of fibroblast growth factors and their receptors in prostate cancer. Endocr Relat Cancer. 2004;11:709–724. doi: 10.1677/erc.1.00535. [DOI] [PubMed] [Google Scholar]
- Larsen M, Artym VV, Green JA, Yamada KM. The matrix reorganized: extracellular matrix remodeling and integrin signaling. Curr Opin Cell Biol. 2006;18:463–471. doi: 10.1016/j.ceb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Lee SH, et al. Synapses are regulated by the cytoplasmic tyrosine kinase Fer in a pathway mediated by p120catenin, Fer, SHP-2, and beta-catenin. J Cell Biol. 2008;183:893–908. doi: 10.1083/jcb.200807188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesina M, et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19:456–469. doi: 10.1016/j.ccr.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Levayer R, Lecuit T. Breaking down EMT. Nat Cell Biol. 2008;10:757–759. doi: 10.1038/ncb0708-757. [DOI] [PubMed] [Google Scholar]
- Li N, Grivennikov SI, Karin M. The unholy trinity: inflammation, cytokines, and STAT3 shape the cancer microenvironment. Cancer Cell. 2011;19:429–431. doi: 10.1016/j.ccr.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman C, et al. Mutations in the human LARGE gene cause MDC1D, a novel form of congenital muscular dystrophy with severe mental retardation and abnormal glycosylation of alpha-dystroglycan. Hum Mol Genet. 2003;12:2853–2861. doi: 10.1093/hmg/ddg307. [DOI] [PubMed] [Google Scholar]
- Manya H, Chiba A, Yoshida A, Wang X, Chiba Y, Jigami Y, Margolis RU, Endo T. Demonstration of mammalian protein O-mannosyltransferase activity: coexpression of POMT1 and POMT2 required for enzymatic activity. Proc Natl Acad Sci USA. 2004;101:500–505. doi: 10.1073/pnas.0307228101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntoni F, et al. Muscular dystrophies due to defective glycosylation of dystroglycan. Acta Myol. 2007;26:129–135. [PMC free article] [PubMed] [Google Scholar]
- Muschler J, Levy D, Boudreau R, Henry M, Campbell K, Bissell MJ. A role for dystroglycan in epithelial polarization: loss of function in breast tumor cells. Cancer Res. 2002;62:7102–7109. [PubMed] [Google Scholar]
- Pasder O, Shpungin S, Salem Y, Makovsky A, Vilchick S, Michaeli S, Malovani H, Nir U. Downregulation of Fer induces PP1 activation and cell-cycle arrest in malignant cells. Oncogene. 2006;25:4194–4206. doi: 10.1038/sj.onc.1209695. [DOI] [PubMed] [Google Scholar]
- Patnaik SK, Stanley P. Mouse large can modify complex N- and mucin O-glycans on alpha-dystroglycan to induce laminin binding. J Biol Chem. 2005;280:20851–20859. doi: 10.1074/jbc.M500069200. [DOI] [PubMed] [Google Scholar]
- Peyrard M, et al. The human LARGE gene from 22q12.3-q13.1 is a new, distinct member of the glycosyltransferase gene family. Proc Natl Acad Sci USA. 1999;96:598–603. doi: 10.1073/pnas.96.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedra J, Miravet S, Castano J, Palmer HG, Heisterkamp N, Garcia de Herreros A, Dunach M. p120 Catenin-associated Fer and Fyn tyrosine kinases regulate beta-catenin Tyr-142 phosphorylation and beta-catenin-alpha-catenin Interaction. Mol Cell Biol. 2003;23:2287–2297. doi: 10.1128/MCB.23.7.2287-2297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- Priel-Halachmi S, Ben-Dor I, Shpungin S, Tennenbaum T, Molavani H, Bachrach M, Salzberg S, Nir U. FER kinase activation of Stat3 is determined by the N-terminal sequence. J Biol Chem. 2000;275:28902–28910. doi: 10.1074/jbc.M003402200. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Kurata-Miura K, Ujita M, Angata K, Nakagawa S, Sekine S, Nishi T, Fukuda M. Expression cloning of cDNA encoding a human beta-1,3-N-acetylglucosaminyltransferase that is essential for poly-N-acetyllactosamine synthesis. Proc Natl Acad Sci USA. 1997;94:14294–14299. doi: 10.1073/pnas.94.26.14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgambato A, De Paola B, Migaldi M, Di Salvatore M, Rettino A, Rossi G, Faraglia B, Boninsegna A, Maiorana A, Cittadini A. Dystroglycan expression is reduced during prostate tumorigenesis and is regulated by androgens in prostate cancer cells. J Cell Physiol. 2007;213:528–539. doi: 10.1002/jcp.21130. [DOI] [PubMed] [Google Scholar]
- Shimojo H, Kobayashi M, Kamigaito T, Shimojo Y, Fukuda M, Nakayama J. Reduced glycosylation of α-dystroglycans on carcinoma cells contributes to formation of highly infiltrative histological patterns in prostate cancer. Prostate. 2011;71:1151–1157. doi: 10.1002/pros.21330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence HJ, Dhillon AS, James M, Winder SJ. Dystroglycan, a scaffold for the ERK-MAP kinase cascade. EMBO Rep. 2004;5:484–489. doi: 10.1038/sj.embor.7400140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White DE, Kurpios NA, Zuo D, Hassell JA, Blaess S, Mueller U, Muller WJ. Targeted disruption of beta1-integrin in a transgenic mouse model of human breast cancer reveals an essential role in mammary tumor induction. Cancer Cell. 2004;6:159–170. doi: 10.1016/j.ccr.2004.06.025. [DOI] [PubMed] [Google Scholar]
- Xu G, Craig AW, Greer P, Miller M, Anastasiadis PZ, Lilien J, Balsamo J. Continuous association of cadherin with beta-catenin requires the non-receptor tyrosine-kinase Fer. J Cell Sci. 2004;117:3207–3219. doi: 10.1242/jcs.01174. [DOI] [PubMed] [Google Scholar]
- Yoshida A, et al. Muscular dystrophy and neuronal migration disorder caused by mutations in a glycosyltransferase, POMGnT1. Dev Cell. 2001;1:717–724. doi: 10.1016/s1534-5807(01)00070-3. [DOI] [PubMed] [Google Scholar]
- Yoshida-Moriguchi T, Yu L, Stalnaker SH, Davis S, Kunz S, Madson M, Oldstone MB, Schachter H, Wells L, Campbell KP. O-mannosyl phosphorylation of alpha-dystroglycan is required for laminin binding. Science. 2010;327:88–92. doi: 10.1126/science.1180512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoubeidi A, Rocha J, Zouanat FZ, Hamel L, Scarlata E, Aprikian AG, Chevalier S. The Fer tyrosine kinase cooperates with interleukin-6 to activate signal transducer and activator of transcription 3 and promote human prostate cancer cell growth. Mol Cancer Res. 2009;7:142–155. doi: 10.1158/1541-7786.MCR-08-0117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.