Matrix rigidity regulates a switch between TGF-β1–induced cell functions in two epithelial cell lines. On compliant polyacrylamide gels, TGF-β1 induced apoptosis, whereas on rigid gels, cells underwent an epithelial–mesenchymal transition (EMT). Compliant gels reduced PI3K/Akt activity, which was essential for cell survival and EMT on rigid gels.
Abstract
The transforming growth factor-β (TGF-β) signaling pathway is often misregulated during cancer progression. In early stages of tumorigenesis, TGF-β acts as a tumor suppressor by inhibiting proliferation and inducing apoptosis. However, as the disease progresses, TGF-β switches to promote tumorigenic cell functions, such as epithelial–mesenchymal transition (EMT) and increased cell motility. Dramatic changes in the cellular microenvironment are also correlated with tumor progression, including an increase in tissue stiffness. However, it is unknown whether these changes in tissue stiffness can regulate the effects of TGF-β. To this end, we examined normal murine mammary gland cells and Madin–Darby canine kidney epithelial cells cultured on polyacrylamide gels with varying rigidity and treated with TGF-β1. Varying matrix rigidity switched the functional response to TGF-β1. Decreasing rigidity increased TGF-β1–induced apoptosis, whereas increasing rigidity resulted in EMT. Matrix rigidity did not change Smad signaling, but instead regulated the PI3K/Akt signaling pathway. Direct genetic and pharmacologic manipulations further demonstrated a role for PI3K/Akt signaling in the apoptotic and EMT responses. These findings demonstrate that matrix rigidity regulates a previously undescribed switch in TGF-β–induced cell functions and provide insight into how changes in tissue mechanics during disease might contribute to the cellular response to TGF-β.
INTRODUCTION
Transforming growth factor-β (TGF-β) is a pleiotropic cytokine essential for many physiological processes, including embryonic development, immune function, and wound healing (Wu and Hill, 2009). Misregulation of TGF-β signaling can contribute to the progression of disease states such as organ fibrosis and cancer, and a key to treating these diseases will be a better understanding of the TGF-β signal transduction machinery (Massague, 2008). However, due to its widespread effects, the role of TGF-β is not well understood. This is perhaps best illustrated in the context of tumor progression, although analogous situations can be found in other settings. During early stages of tumorigenesis, TGF-β acts as a tumor suppressor. TGF-β induces growth arrest and apoptosis in most normal epithelial cells in vitro (Pietenpol et al., 1990; Hannon and Beach, 1994; Siegel and Massague, 2003). Mice in which the TGFB1 or SMAD genes are disrupted are prone to the development of cancer (Zhu et al., 1998; Engle et al., 1999; Go et al., 1999). Retrospective studies of various human tumor types have also found frequent down-regulation or mutations inactivating the TGF-β signaling pathway (Kaklamani et al., 2005; Stuelten et al., 2006; Bacman et al., 2007). In later stages of cancer progression, however, TGF-β is believed to switch roles and promote tumor progression and metastasis (Derynck et al., 2001; Wakefield and Roberts, 2002; Tang et al., 2003). Within the tumor, TGF-β enhances migration, invasion, survival, and epithelial–mesenchymal transition (EMT) (Massague, 2008). High levels of TGF-β in clinical settings are associated with a poor prognosis (Friess et al., 1993; Wikstrom et al., 1998; Fukai et al., 2003), and treatment with TGF-β in animal models results in larger, more metastatic tumors (Wikstrom et al., 1998; Fukai et al., 2003; Muraoka et al., 2003). TGF-β also plays an active role in remodeling of the tumor microenvironment, promoting activation of fibroblasts, increasing angiogenesis, and suppressing immune surveillance (Bierie and Moses, 2006). Although the switch in TGF-β from a tumor suppressor to promoter during disease progression is well documented, it is still unclear how this switch occurs. One possibility is that changes in the cellular microenvironmental context guide the cellular response to TGF-β.
Although many aspects of the cellular microenvironment change during disease, including soluble factors, cell–cell interactions, and cell–extracellular matrix (ECM) adhesion, changes in the mechanical properties of the microenvironment may also modulate the response to the TGF-β. The mechanical stiffness of tissue microenvironments varies widely, as adipose tissue is less rigid than muscle, which is less rigid than bone, and tissue stiffness can also change within the same type of tissue during disease states (Butcher et al., 2009). In the context of cancer progression, as well as tissue fibrosis, increased tissue stiffness is well documented and is due to a number of factors, including extracellular matrix remodeling, deposition, and cross-linking (Ebihara et al., 2000; Levental et al., 2009). Several recent studies have shown that such changes in matrix rigidity can regulate many cellular functions, including focal adhesion maturation, cell spreading, actin stress fiber formation, and cell motility (Pelham and Wang, 1997; Lo et al., 2000; Yeung et al., 2005). Several cell types cultured on compliant substrates decrease proliferation and increase apoptosis as compared with cells on rigid substrates (Wang et al., 2000; Klein et al., 2009; Tilghman et al., 2010; Mih et al., 2011). Differentiation of many cell types can also be regulated by matrix rigidity, including human mesenchymal stem cells, portal fibroblasts, mammary epithelial cells, and endothelial cells (Vailhe et al., 1997; Paszek et al., 2005; Engler et al., 2006; Li et al., 2007; Alcaraz et al., 2008). Because matrix rigidity can regulate a number of cell functions also regulated by TGF-β, such as proliferation, apoptosis, and differentiation, and tissues become stiffer during disease progression, we hypothesized that changes in matrix rigidity could regulate TGF-β–induced cellular functions.
In this study, we examined whether matrix rigidity regulates TGF-β–induced cell function. We examined two cell functions—apoptosis and EMT—as representative responses to TGF-β classically associated with tumor suppression or promotion, respectively (Massague, 2008). In most nontransformed epithelial cells, TGF-β induces programmed cell death, or apoptosis; this is one way TGF-β suppresses tumorigenesis during early stages of the disease (Rahimi and Leof, 2007). In contrast, EMT is a key step during metastasis, which occurs during later stages of disease, and is characterized by dissolution of epithelial cell–cell junctions, remodeling of cell–matrix adhesion, and increased motility (Lee et al., 2006). In studies presented here, we found a novel PI3K/Akt-mediated switch in which substrate rigidity controlled TGF-β1–induced cell functions—epithelial cells cultured on compliant substrates underwent apoptosis when treated with TGF-β1, whereas on more rigid substrates, TGF-β1 induced EMT. These findings suggest that matrix mechanics plays a key role in regulating the opposing functional effects of TGF-β1.
RESULTS
Matrix rigidity regulates TGF-β1–induced cell fate
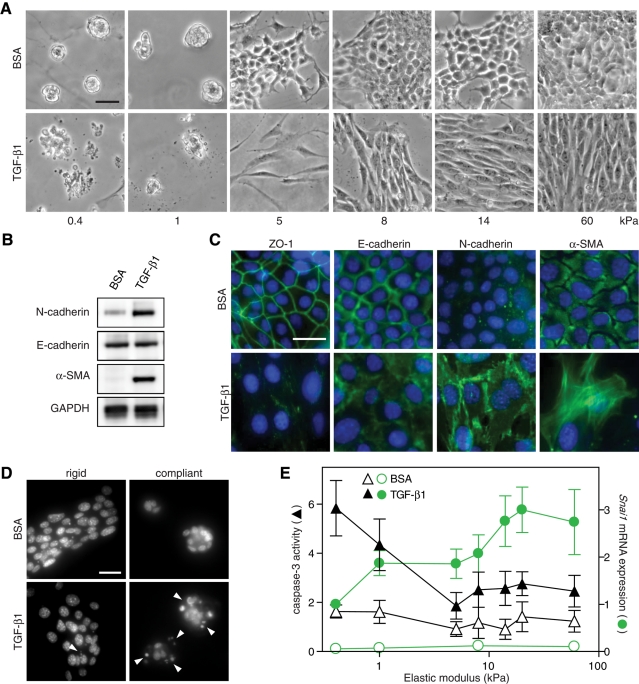
To explore whether matrix rigidity influences cellular responses to TGF-β1 in a noncancerous genetic background, we used normal murine mammary gland epithelial cells (NMuMG) and Madin–Darby canine kidney epithelial cells (MDCK), both well established in vitro model systems of EMT (Miettinen et al., 1994). We first examined NMuMG cells that were cultured on fibronectin-conjugated polyacrylamide (PA) gels with a range of elastic modulus (E) from 0.4 to 60 kPa and then treated with TGF-β1. NMuMG cells cultured on PA gels exhibited differences in morphology as a function of substrate compliance (Figure 1A). Cells on the most rigid gels (E > 14 kPa) appeared cuboidal and formed a monolayer on the surface identical to cells on tissue culture plastic. In contrast, cells on compliant gels (E < 1 kPa) were rounded and formed spherical clusters. On rigid PA gels (E > 5 kPa) or on tissue culture plastic, TGF- β1 treatment induced an elongated morphology and scattering of cells, characteristic of an EMT (Figure 1A). Examination of known EMT markers confirmed this response, as evidenced by delocalization of the epithelial junctional markers zonula occludens-1 (ZO-1) and E-cadherin and increased expression of mesenchymal markers N-cadherin, α-smooth muscle actin (α-SMA), and the EMT-associated transcription factor Snail (Figure 1, B, C, and E). Although E-cadherin was displaced from adherens junctions, no significant decrease was observed in E-cadherin protein expression, similar to observations from other groups using the NMuMG cell line (Figure 1B; Bakin et al., 2000; Shintani et al., 2006; Bailey and Liu, 2008). Although TGF-β1 did not appear to induce EMT on compliant gels, as indicated by decreased N-cadherin, α-SMA, and Snail expression as compared with rigid gels, phase and immunofluorescence imaging revealed a dramatic increase in TGF-β1–induced apoptosis on compliant gels (Figure 1, A and D, and Supplemental Figure S1B). Apoptosis was confirmed by nuclear fragmentation and caspase activity (Figure 1, D and E). On compliant gels, ∼28% of cells were positive for cleaved caspase-3 by immunofluorescence after 24 h of TGF-β1 treatment, whereas 13% of cells were positive on rigid gels (Supplemental Figure S1C). Increased apoptosis on compliant gels was observed across three orders of magnitude of TGF-β1 concentration, from 0.1 to 10 ng/ml (Supplemental Figure S1D). TGF-β1 treatment was necessary for the observed increase in apoptosis, as basal levels of apoptosis were similar across substrates of different stiffness (Figure 1, D and E, and Supplemental Figure S1C). On substrates with modulus ranging from 1 to 8 kPa, there was a decrease in apoptosis, as well as a concomitant increase in Snail expression (Figure 1E); this range of modulus is also associated with measurements of excised tumor tissue (Paszek et al., 2005). Snail, a transcription factor induced by TGF-β and responsible for the down-regulation of E-cadherin, is a critical factor regulating EMT and was used here as an early marker of EMT, as the rapid onset of apoptosis on compliant gels prevented reliable measurement of later-stage markers (Supplemental Figure S1, A and B; Cano et al., 2000; Peinado et al., 2003; Cho et al., 2007). These data suggest that matrix rigidity regulates a functional switch between apoptosis and EMT in response to TGF-β1.
FIGURE 1:
Matrix rigidity regulates TGF-β1–induced EMT and apoptosis in NMuMG cells. (A) Phase contrast images of cells cultured on PA gels with elastic modulus ranging from 0.4 to 60 kPa and treated with TGF-β1 or BSA control. (B) Western blot of N-cadherin (135 kDa), E-cadherin (120 kDa), α-SMA (42 kDa), and GAPDH control (38 kDa) in cells cultured on rigid (8 kPa) PA gels. (C) Immunofluorescence images of cells cultured on rigid PA gels. (D) Hoechst-stained nuclei of cells cultured on rigid (8 kPa) and compliant (0.4 kPa) gels. Fragmented nuclei indicated by white triangles. (E) Caspase-3 activity (▲, ∆) and Snai1 mRNA expression (•, ○) in cells cultured on PA gels. n = 5 ± SEM. Bars, 50 μm.
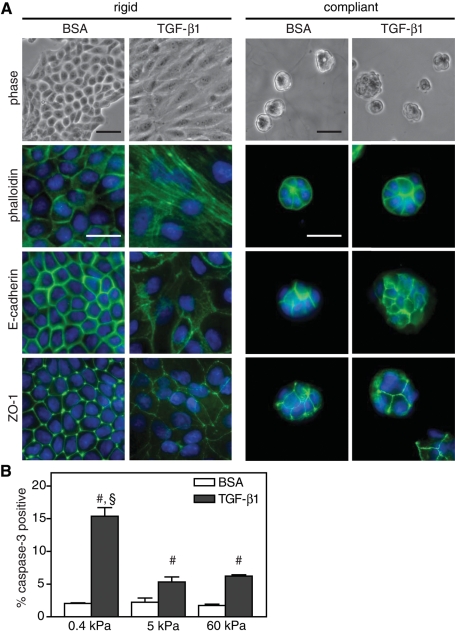
A similar switch in cell fate between apoptosis and EMT was also observed in MDCK epithelial cells, suggesting that this control mechanism is not restricted to mammary epithelia. MDCK cells cultured on rigid gels underwent a classic EMT with TGF-β1 treatment, as indicated by an elongated morphology, actin stress fibers, and delocalization of the epithelial cell–cell adhesion markers E-cadherin and ZO-1 (Figure 2A). Compliant substrates, conversely, inhibited this transition, and MDCK cells retained a rounded morphology, cortical actin, and epithelial adherens and tight junctions with TGF-β1 treatment. Similar to the NMuMGs, increased apoptosis was also observed in MDCK cells cultured on compliant substrates (Figure 2B).
FIGURE 2:
TGF-β1–induced EMT and apoptosis in MDCK cells cultured on polyacrylamide gels. (A) Immunostaining on rigid (5 kPa) and compliant (0.4 kPa) PA gels in cells treated with TGF-β1 or BSA control. (B) Graph of percentage of cells positive for cleaved caspase-3 immunofluorescence. n = 3 ± SEM. #p < 0.01 as compared with BSA conditions; §p < 0.01 as compared with 5- and 60-kPa TGF-β1 conditions. Bars, 50 μm.
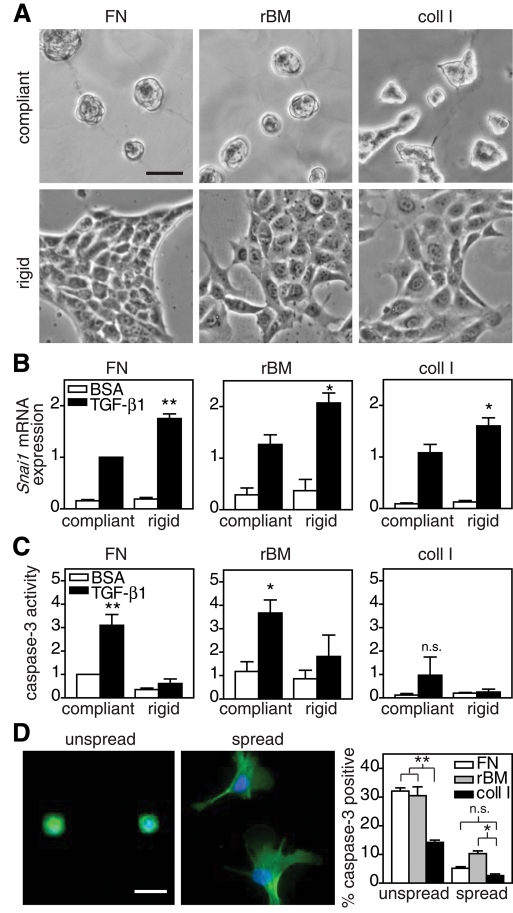
ECM proteins promote cell adhesion but also provide biochemical and biophysical cues that can regulate cell function and signaling (Hynes, 2009). Thus it was not clear whether the rigidity-regulated switch between EMT and apoptosis was specific to the fibronectin (FN) conjugated to the PA gels or occurred irrespective of the ECM protein to which cells attached. To address this question, NMuMG cells were cultured on polyacrylamide gels conjugated with FN, reconstituted basement membrane (rBM; commercially known as Matrigel), or collagen I (coll I) and treated with TGF-β1. NMuMGs cultured on substrates conjugated with FN or rBM were morphologically similar; however on coll I substrates, cell spreading was increased (Figure 3A). Substrate compliance inhibited TGF-β1–induced EMT regardless of ECM type, as indicated by decreased Snail expression (Figure 3B). Compliant substrates conjugated with FN or rBM also increased TGF-β1–induced apoptosis (Figure 3C). However, cells cultured on coll I substrates had dramatically reduced levels of caspase-3 activity in all conditions, although increased compliance still enhanced levels of apoptosis. Because cell spreading can regulate apoptosis and coll I increased cell spreading on compliant substrates, it was unclear whether the decrease in apoptosis was due to increased cell spreading or more specifically to coll I signaling (Chen et al., 1997). To address this question, each ECM was microcontact printed onto polydimethylsiloxane (PDMS)–coated coverslips to restrict cell spreading (area of 289 μm2) or to allow cells to fully spread (Figure 3D). Restricting cell spreading, similar to compliant substrates, increased TGF-β1–induced apoptosis compared with fully spread cells for all ECM types (Figure 3D). Whereas increased apoptosis was observed in unspread cells on coll I substrates as compared with fully spread cells, apoptosis on coll I substrates was significantly less than for cells cultured on FN or rBM, suggesting that coll I specifically inhibits apoptosis on compliant substrates in addition to regulating cell spreading.
FIGURE 3:
TGF-β1–induced EMT and apoptosis in NMuMG cells cultured on polyacrylamide gels conjugated with ECM. (A) Phase contrast images of cells cultured on FN, rBM, or coll I conjugated PA gels. (B) Snai1 mRNA expression in cells on compliant and rigid gels treated with TGF-β1. (C) Caspase-3 activity in cells on compliant and rigid gels treated with TGF-β1. (D) Phalloidin (green)- and Hoechst (blue)-stained cells on 289 μm2 islands (unspread) or large areas (spread) of microcontact-printed FN. Graph of percentage of cells treated with TGF-β1 positive for cleaved caspase-3 immunofluorescence. n = 3 ± SEM. *p < 0.05; **p < 0.01. Bars, 50 μm.
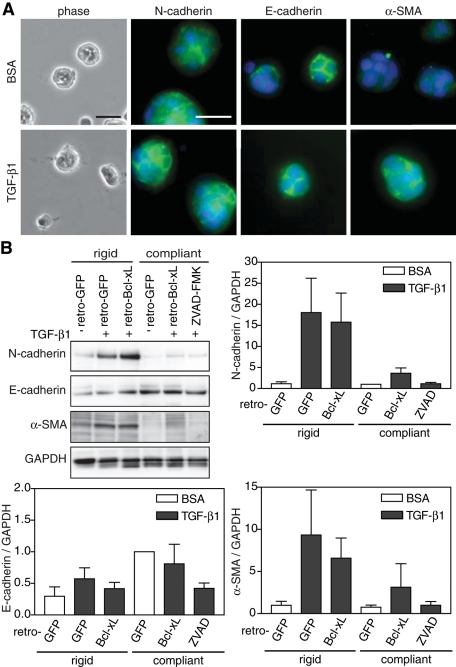
Given that the apoptotic response occurred within hours, whereas a full EMT required at least 48 h of TGF-β1 treatment, it was not clear whether the decreased EMT on compliant gels was a result of TGF-β1–induced cell death or compliance directly regulated EMT independent of its effects on cell survival. To address this, we blocked the apoptotic response by either overexpressing the survival factor, Bcl-xL, or treating with a pan-caspase inhibitor, ZVAD-FMK, and observed whether EMT on compliant gels would be rescued (Boise et al., 1993). As a control, both reagents decreased caspase-3 activity and prevented nuclear fragmentation (Supplemental Figure S2). When apoptosis was inhibited, NMuMGs cultured on compliant gels still failed to undergo EMT. E-cadherin remained localized to junctions, N-cadherin and α-SMA failed to express, and cells did not transition to an elongated phenotype (Figure 4, A and B). Together these data suggest that substrate stiffness regulates a switch in the response of cells to TGF-β1 between EMT and apoptosis and that these two responses are independently regulated.
FIGURE 4:
Decreased matrix rigidity inhibits EMT independent of apoptosis. (A) Phase contrast and immunostaining for N-cadherin, E-cadherin, α-SMA, and nuclei of NMuMG cells infected with retro-Bcl-xL on compliant gels. (B) Western blot and quantification of N-cadherin, E-cadherin, α-SMA, and GAPDH in NMuMG cells infected with retro-GFP or retro-Bcl-xL or treated with 400 μM ZVAD-FMK, plated on rigid and compliant gels, and treated with TGF-β1. n = 4 ± SEM. Bars, 50 μm.
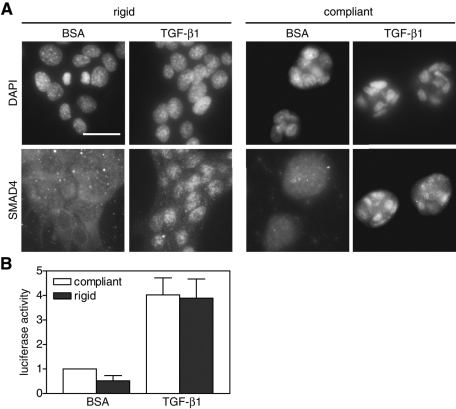
Previous studies showed that cell density can regulate TGF-β–induced cell functions and that cells grown to confluence do not undergo EMT (Petridou et al., 2000; Nelson et al., 2008). Specifically, less TGF-β bound to TGF-β receptors and Smad translocation was reduced in confluent cells (Petridou et al., 2000). To investigate whether matrix rigidity regulates TGF-β signaling through a similar mechanism, NMuMGs were plated at 60% confluence (1 × 105 cells/cm2) and treated with TGF-β1. As early as 2 h after TGF-β1 treatment, Smad4 translocated to the nucleus in NMuMGs to similar degrees on both rigid and compliant substrates (Figure 5A). Furthermore, use of a Smad-responsive 3TP-luciferase reporter plasmid also showed no difference in Smad transcriptional activity on rigid versus compliant substrates (Figure 5B; Wrana et al., 1992; Yingling et al., 1997). Together these results suggest that TGF-β receptor/Smad signaling functions at similar levels on compliant and rigid substrates and is not responsible for the matrix rigidity–induced switch in TGF-β function.
FIGURE 5:
Smad signaling in NMuMG cells on compliant and rigid gels. (A) Immunostaining for Smad4 and nuclei in NMuMG cells cultured on rigid and compliant PA gels treated with TGF-β1. (B) Luciferase activity in cells transfected with 3TP-luciferase reporter plasmid on compliant and rigid gels. n = 4 ± SEM. Bars, 50 μm.
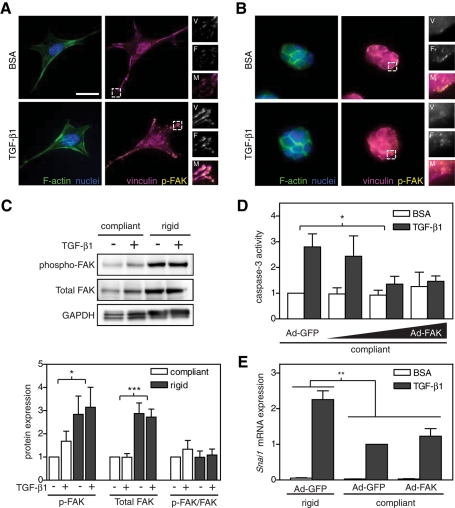
Matrix rigidity could modulate TGF-β1 signaling at numerous levels in addition to the Smad signaling pathway. Previous work showed that increased matrix rigidity and treatment with TGF-β1 each can promote actin stress fiber and focal adhesion formation (Pelham and Wang, 1997; Yeung et al., 2005). Similar stress fiber and focal adhesion responses are seen upon treatment with TGF-β1 (Miettinen et al., 1994; Edlund et al., 2002). In addition, focal adhesion kinase (FAK), one of the main signaling components within focal adhesions, can also be regulated by matrix rigidity and TGF-β1 and is associated with cell survival and EMT (Ilic et al., 1998; Wang et al., 2004; Paszek et al., 2005; Cicchini et al., 2008; Zouq et al., 2009). To investigate whether FAK may be involved in this system, we first examined whether both matrix compliance and TGF-β1 modulated focal adhesion formation and FAK phosphorylation. Prominent focal adhesions, as indicated by punctate immunofluorescence staining for vinculin, and actin stress fibers were observed in NMuMGs cultured on rigid substrates (Figure 6A). On compliant substrates, focal adhesion markers were diffuse, and cortical actin was observed (Figure 6B). Treatment with TGF-β1 qualitatively increased focal adhesion size on rigid substrates, but no effect was observed on compliant gels. We also observed increased phospho-FAK localization to focal adhesions in a manner that directly correlated with vinculin localization (Figure 6A). Western blot analysis confirmed this observation, showing increased levels of phospho-FAK (Figure 6C). However, specific activity of FAK (phospho-FAK normalized to total FAK) showed no significant difference between compliant and rigid gels, as FAK protein levels were greatly decreased on compliant gels. In a recent study, FAK protein expression was found to be critical for the mesenchymal phenotype and Snail expression in mouse embryonic fibroblasts, so we hypothesized that overexpressing FAK in cells on compliant gels may rescue Snail expression and EMT (Li et al., 2011). To test this possibility, we overexpressed FAK using an adenoviral vector (Supplemental Figure S3A). Overexpression of FAK did not rescue Snail mRNA expression on compliant gels; however a decrease in apoptosis was observed (Figure 6, D and E). Previous reports showed decreased FAK expression on collagen gels due to FAK degradation by calpain (Wang et al., 2003). In this system, however, we did not observe lower–molecular weight bands associated with FAK degradation by Western blot, and treatment with a calpain inhibitor, ALLN, did not increase FAK expression or inhibit apoptosis on compliant gels (data not shown).
FIGURE 6:
Effect of matrix rigidity and TGF-β1 on the actin cytoskeleton and focal adhesion formation in NMuMG cells. (A,B) Immunofluorescence images of F-actin (green), nuclei (blue), vinculin (magenta), and phospho-FAK (yellow) on rigid (8 kPa) (A) and compliant (0.4 kPa) (B) gels. Inset shows magnification of vinculin (V), phospho-FAK (F), and merged (M) images. (C) Western blot and quantification of phospho-FAK (125 kDa), total FAK (125 kDa), and GAPDH (38 kDa). Caspase-3 activity (D) and Snai1 mRNA expression (E) in NMuMG cells infected with Ad-GFP or Ad-FAK cultured on rigid and compliant gels treated with TGF-β1. n = 4 ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001. Bars, 25 μm.
Whereas overexpression of wild-type FAK rescued cell survival on compliant gels, expression of CD2-FAK, an activated FAK allele (Frisch et al., 1996), failed to inhibit apoptosis on compliant gels (Supplemental Figure S3E). Further supporting these data, pharmacological inhibition of FAK activity with the small-molecule inhibitor PF 573228 reduced Y397 FAK phosphorylation but did not affect EMT or apoptosis (Supplemental Figure S3, B and C). Expression of the dominant-negative FRNK and the phosphorylation mutant FAK Y397F, both at physiological levels and highly overexpressed, did not reduce FAK phosphorylation at Y397 and did not affect EMT or apoptosis (Supplemental Figure S3E). These data suggest that matrix rigidity is regulating FAK signaling by modulating FAK protein levels and that FAK levels in turn regulate compliance-induced apoptosis but not EMT. Other important regulators of stress fiber and focal adhesion formation, the RhoGTPases, were also investigated using the pharmacological inhibitors Y27632—a ROCK (Rho kinase) inhibitor—and NSC 23766—a Rac1 inhibitor. Neither inhibitor significantly affected apoptosis or EMT on compliant or rigid gels (Supplemental Figure S4, A and B).
Matrix rigidity regulates apoptosis and EMT through PI3K and Akt
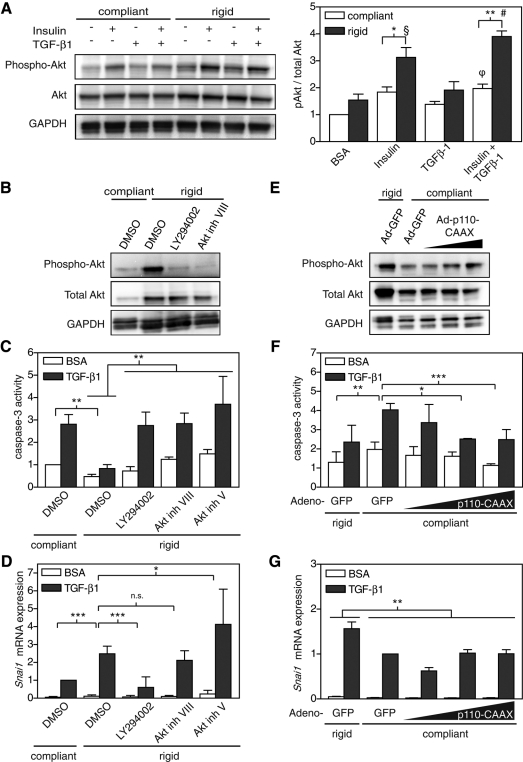
Similar to FAK, the PI3K/Akt pathway has been shown to regulate both EMT and survival in a variety of settings and can be regulated by matrix stiffness (Chen et al., 1998; Bakin et al., 2000; Levental et al., 2009). To investigate whether substrate rigidity regulates the PI3K/Akt signaling pathway, we first measured Akt phosphorylation at serine 473. Because insulin is an essential component of the growth media of NMuMGs and insulin is known to stimulate Akt activity, exposure to insulin was included as a background control (Burgering and Coffer, 1995). In all cases, NMuMGs cultured on compliant gels showed decreased Akt activation compared with cells on rigid gels (Figure 7A). Inhibition of PI3K or Akt activity with pharmacological inhibitors LY294002 and two Akt inhibitors increased TGF-β1–induced apoptosis (Figure 7, B and C). Inhibition of PI3K decreased Snail mRNA expression on rigid gels; however, inhibition of Akt did not (Figure 7D). Although these studies suggest that PI3K is necessary for survival and EMT following TGF-β1 treatment, it was not clear whether it was also sufficient. We increased PI3K activity by adenoviral expression of p110-CAAX, a membrane-localized subunit of PI3K, and observed suppression of apoptosis on compliant gels to similar levels observed on rigid gels (Figure 7, E and F). p110-CAAX expression, however, did not rescue Snail mRNA expression (Figure 7G). Together these data demonstrate a role for PI3K and Akt in transducing substrate compliance and regulating the compliance-induced switch in cellular response to TGF-β1.
FIGURE 7:
Regulation of Akt activity by matrix rigidity in NMuMG cells. (A) Western blot and quantification of phospho-Akt (60 kDa), total Akt (60 kDa), and GAPDH (38 kDa) in cells plated on compliant and rigid polyacrylamide gels. (B–D) Caspase-3 activity and Snai1 mRNA expression in cells treated with DMSO control, 10 μM LY294002, 1 μM Akt inhibitor VIII, or 10 μM Akt inhibitor V. (E–G) Caspase-3 activity and Snai1 mRNA expression in cells infected with Ad-GFP or Ad-p110-CAAX. n = 3 ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001; §p < 0.01 rigid + insulin compared with BSA and TGF-β1 conditions; #p < 0.01 rigid + insulin/TGF-β1 compared with all conditions; φp < 0.01 compliant + insulin/TGF-β1 compared with compliant BSA condition.
DISCUSSION
We find that decreasing matrix rigidity inhibits PI3K/Akt activity and through this action impinges on both survival and EMT. Numerous previous studies demonstrated the importance of the PI3K/Akt signaling pathways for cell survival (Dudek et al., 1997; Khwaja et al., 1997) and EMT (Bakin et al., 2000; Kattla et al., 2008). Although we found inhibition of Akt activity by two pharmacological inhibitors increased apoptosis on rigid substrates, EMT was unaffected. This could be explained by demonstration of distinct regulatory roles for the Akt isoforms (Irie et al., 2005), and here pharmacological inhibition would not differentiate between the isoforms. It is perhaps not surprising that up-regulation of PI3K failed to rescue EMT on low-rigidity substrates. Given the many disparate processes that are necessary to drive EMT, it is likely that additional points of regulation are affected by rigidity (Cannito et al., 2010). In mesenchymal cells (as opposed to epithelial cells), one component of EMT has been reported to exhibit remnants of this control mechanism. TGF-β–induced smooth muscle actin expression in fibroblasts and trabecular meshwork cells (associated with myofibroblast differentiation) appears to be suppressed with decreased matrix rigidity (Arora et al., 1999; Li et al., 2007; Chen et al., 2011; Han et al., 2011). Moreover, inhibitors of PI3K/Akt signaling can block α-SMA expression (Han et al., 2011). Although it would be inappropriate to suggest that mechanical regulation of EMT is equivalent to α-SMA expression, since EMT involves many additional regulatory steps, including loss of epithelial markers, cell–cell adhesions, and polarity, these studies do suggest some conserved mechanisms. More starkly, the stiffness-induced switch between apoptosis and EMT that we report here in two epithelial cell systems is absent in fibroblastic cells, suggesting a new function of matrix rigidity to regulate a switch between TGF-β–induced functions. Further elucidation of these mechanisms is likely forthcoming, as recent studies are beginning to uncover the wide array of signaling pathways affected by rigidity, including integrin activation, focal adhesion assembly, and numerous signaling pathways, including Rho GTPases, mitogen-activated protein kinases, FAK, and phosphoinositide kinase-3 (Fringer and Grinnell, 2001; Wozniak et al., 2003; Paszek et al., 2005; Friedland et al., 2009; Klein et al., 2009; Levental et al., 2009).
The results presented here also highlight the complex interplay among matrix rigidity, cell spreading, and ECM subtypes. Here we found that TGF-β–induced EMT is inhibited on compliant substrates independent of ECM subtypes, but that the compliance-induced apoptosis was more dramatic when cells engaged FN or rBM as compared with coll I. Of interest, we observed that cell spreading was enhanced on coll I, and it was previously reported that cell spreading can antagonize apoptosis (Chen et al., 1997). Indeed, the coll I–induced reduction in apoptosis appears to be due in part to the increased cell spreading, since controlling cell area through microcontact printing partially accounted for this difference. However, coll I was still partially able to inhibit cell death even when cell spreading was restricted. Thus binding this ECM appears to have an additional benefit, possibly through specific collagen receptors such as α2β1 integrin or the discoidin domain receptors 1/2 (Ongusaha et al., 2003). In other cell types, such as fibroblasts and endothelial cells, adhesion to coll I reduced cell spreading (Yeung et al., 2005), and in a melanoma cell line coll I did not affect cell spreading but increased cell stiffness and adhesion strength (Byfield et al., 2009). Thus, although there is a widely demonstrated link between substrate stiffness and cell spreading (Pelham and Wang, 1997; Yeung et al., 2005; Fu et al., 2010), how specific ECMs can impact this response may depend on the cell type. These results highlight that regulation of cell function by matrix rigidity can be affected by other cell–matrix adhesion inputs, such as cell spreading and ECM subtype.
TGF-β regulates a diverse array of cellular functions, including proliferation, motility, and differentiation, and these effects are distinct in many cell types. How TGF-β regulates often divergent functions even in a single cell type, particularly in disease contexts such as tumorigenesis, is not well understood. It has been reported that some cell types spontaneously undergo apoptosis on compliant substrates (Wang et al., 2000, 2007), but in the work presented here TGF-β was a required trigger for death. Here we found that on compliant substrates, with a modulus similar to that of native breast tissue, TGF-β induces apoptosis, whereas on rigid substrates, with a modulus similar to that of tumor or fibrotic tissue, TGF-β induces EMT. These results provide a possible explanation for the switch in TGF-β's action from tumor suppressor to promoter during tumorigenesis. Furthermore, these studies highlight the central role for matrix mechanics in regulating cell signaling and fate.
MATERIALS AND METHODS
Cell culture and reagents
NMuMG and MDCK cells were obtained from the American Type Culture Collection (Manassas, VA) and cultured according to their recommendations. Reagents were obtained as follows. Monoclonal antibodies: α-smooth muscle actin (1A4), Smad4 (DCS-46), and vinculin (hVIN-1) (Sigma-Aldrich, St. Louis, MO); glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 6C5; Applied Biosystems/Ambion, Austin, TX); and E-cadherin (36), N-cadherin (32), and FAK (77; BD Biosciences, San Diego, CA). Polyclonal antibodies: ZO-1 (Zymed Laboratories, San Francisco, CA); pY397 FAK (Invitrogen, Carlsbad, CA); and pAkt, Akt, Bcl-xL, cleaved caspase-3, and FAK (Cell Signaling Technology, Beverly, MA). ECMs (FN, coll I, and rBM), all from BD Biosciences.
Cells were plated at a density of 0.1 × 106 cells/cm2 on FN-functionalized polyacrylamide gels for 16 h in growth medium. Compliant gels referred to in the text indicate gels with an elastic modulus of 0.4 kPa, and rigid gels have E > 5 kPa. The cells were rinsed in sterile phosphate-buffered saline (PBS) and then growth factor starved in high-glucose DMEM for 2 h. Cells were treated with 10 μg/ml insulin (Sigma-Aldrich) and 2 ng/ml TGF-β1 (R&D Systems, Minneapolis, MN) for 2 h (RNA isolation, FAK, and Akt Western blotting), 4 h (caspase activity, focal adhesion staining, luciferase assays), 24 h (nuclei fragmentation, cleaved caspase-3 staining), or 48 h (for EMT staining and Western blotting). For inhibitor studies, cells were treated 1 h prior to TGF-β treatment with ZVAD-FMK (400 μM; Enzo Life Sciences, Plymouth, PA), Y-276322 (10 μM) and PF 573228 (1 μM; Tocris Biosciences, Ellisville, MO), and NSC 23766 (10 μM), LY294002 (10 μM), Akt Inhibitor VIII (1 μM), and Akt Inhibitor V (10 μM) (Calbiochem, La Jolla, CA).
Polyacrylamide gel preparation
Polyacrylamide gels were prepared as previously described (Winer et al., 2009). Mechanical properties of the polyacrylamide gels were controlled by varying the percentage of acrylamide and bis-acrylamide as follows: elastic modulus (% acrylamide; % bis-acrylamide), 0.4 kPa (3; 0.05), 1 kPa (3; 0.1), 5 kPa (5.5; 0.15), 8 kPa (5; 0.3), 14.5 kPa (7.5; 0.15), 20 kPa (8; 0.264), and 60 kPa (10; 0.5). Gels were functionalized with 20 μg/ml FN, 20 μg/ml coll I, or 140 μg/ml rBM in 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 8, for 1 h at room temperature (RT; 2 h on ice for rBM), rinsed in double-distilled H2O (ddH2O), incubated with 1% (vol/vol) ethanolamine in 50 mM HEPES, pH 8, for 30 min, and rinsed with ddH2O. The gels were sterilized in 5% (vol/vol) isopropanol in PBS for 1 h at RT and rinsed two times with sterile PBS before plating with cells.
Preparation of micropatterned substrates
Micropatterned substrates were prepared as described (Pirone et al., 2006). Briefly, micropatterned stamps were fabricated by casting PDMS (Sylgard 184; Dow Corning, Midland, MI) on a photolithographically generated master. Stamps were immersed for 1 h in 20 μg/ml fibronectin or 20 μg/ml collagen I or 2 h on ice in 140 μg/ml rBM, washed two times in water, and thoroughly dried with nitrogen. Protein was transferred to surface-oxidized PDMS-coated glass coverslips. Stamped coverslips were immersed in 0.2% Pluronic F127 (BASF, Florham Park, NJ) in PBS for 1 h and rinsed in PBS before cell seeding.
Adenovirus production
FAK, FRNK, FAK-Y397F, and green fluorescent protein (GFP) recombinant adenoviruses were constructed as described previously (Pirone et al., 2006) using the AdEasy XL system (Stratagene, Santa Clara, CA) according to manufacturer's instructions. The CD2-FAK adenovirus was generated by C. Henke (University of Minnesota, Minneapolis, MN) and p110-CAAX by L. Romer (Johns Hopkins University, Baltimore, MD). Expression was optimized and verified by Western blot.
Retrovirus production
Retrovirus was produced as described (Ory et al., 1996). Bcl-xL plasmid was obtained from Addgene (Cambridge, MA; plasmid 8790; Cheng et al., 2001).
Caspase-3 activity assays
Caspase-3 activity was determined by EnzChek Caspase-3 Assay Kit #1 (Invitrogen) according to the manufacturer's instructions. Caspase activity was normalized to total DNA content as determined by CyQUANT Cell Proliferation Assay (Invitrogen).
Western blotting
Cells were rinsed in PBS, lysed in ice-cold modified RIPA buffer (25 mM HEPES, 75 mM NaCl, 1% NP-40, 0.25% deoxycholate, 1 mM EDTA, 1 mM NaF, 1× Halt protease and phosphatase inhibitor cocktail [Thermo Scientific, Waltham, MA]), and centrifuged at 14,000 RPM for 10 min at 4°C. Protein concentration was determined by Precision Red Advanced Protein Assay (Cytoskeleton, Denver, CO). A 25-μg amount of protein was separated by denaturing SDS–PAGE, electroblotted onto polyvinylidene fluoride blocked with 5% bovine serum albumin (BSA) or milk in 0.3% Tween-20 in Tris-buffered saline (TBS), immunoblotted with specific antibodies (1:1000), and detected using horseradish peroxidase–conjugated secondary antibodies (1:5000; Jackson ImmunoResearch Laboratories, West Grove, PA) and SuperSignal West Dura (Pierce, Thermo Fisher Scientific, Rockford, IL) as a chemiluminescent substrate. Densitometric analysis was performed using a VersaDoc imaging system with QuantityOne software (Bio-Rad Laboratories, Hercules, CA).
Microscopy, immunofluorescence, and image acquisition
Samples were rinsed in PBS and fixed in 4% paraformaldehyde at RT for 10 min, or, for E-cadherin and ZO-1 staining, cells were fixed in 1:1 acetone/methanol on ice for 20 min. After fixation, samples were permeabilized with 0.5% Triton-X, blocked in 10% goat serum for 1 h at RT, incubated with primary antibodies (1:200) for 1 h at RT, rinsed with PBS, then incubated with Alexa Fluor 488, 555, or 647 secondary antibodies (1:200), Alexa Fluor 488 phalloidin (1:200; Invitrogen), and Hoechst 33342 (1:1000; Invitrogen) for 1 h at RT. Samples were rinsed in PBS, then mounted with Fluormount-G (Electron Microscopy Sciences, Hatfield, PA). Images were acquired at RT using an epifluorescence microscope (model TE200; Nikon, Melville, NY) equipped with Plan Fluor 10×, 0.3 numerical aperture (NA), and Plan Apo 60×, 1.4 NA, oil immersion lenses, Spot camera, and software (Diagnostic Instruments, Sterling Heights, MI). Some image levels were adjusted using Photoshop (Adobe, San Jose, CA).
For pY397 FAK and vinculin immunofluorescence samples were rinsed with ice-cold cytoskeleton extraction buffer (10 mM 1,4-piperazinediethanesulfonic acid, 50 mM NaCl, 150 mM sucrose, 3 mM MgCl2, 1× Halt protease, and phosphatase inhibitor cocktail) for 1 min on ice, followed by two 30-s incubations with cytoskeleton buffer plus 0.5% Triton, one rinse with cytoskeleton buffer, and fixation with 4% paraformaldehyde for 10 min at RT. Staining was completed as described. Images were acquired at RT using an epifluorescence microscope (Axiovert 200M; Carl Zeiss MicroImaging, Jena, Germany) equipped with 63× Plan-Apochromat, 1.4 NA, oil immersion objective, an AxioCam camera, and AxioVision software.
Real-time RT-PCR
Total RNA was isolated using an RNeasy Mini or Micro Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. cDNA was transcribed with a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA) with 0.5 μg of total RNA per reaction. Quantitative PCR was performed in an ABI 7300 system (Applied BioSystems) using TaqMan gene expression assays according to the manufacturer's instructions. Results were analyzed using the relative quantitation method, and all mRNA expression data were normalized to 18S expression in the corresponding sample and then to the control sample. TaqMan gene expression assays used were as follows: Snai1 (Mm00441533_g1), 18S (Hs99999901_s1).
Luciferase assays
Cells were transfected with p3TP-lux (plasmid 11767; Addgene; Wrana et al., 1992) using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions 24 h before plating. Transfected cells were treated with TGF-β1 for 6 h and then lysed and analyzed using the dual-luciferase reporter assay (Promega, Madison, WI). Luminescence was measured with GloMax 20/20 Luminometer (Promega). Luciferase values were normalized to DNA content as described for caspase-3 activity assays.
Statistical analysis
Data were analyzed using GraphPad Prism software (GraphPad Software, La Jolla, CA) to perform two-way analysis of variance with Bonferroni posttests to test for significance (p < 0.05) between conditions.
Supplementary Material
Acknowledgments
We thank C. Henke, D. Pirone, and L. Romer for generously providing reagents and R. Assoian, R. Wells, and C. Sarkar for helpful discussions. This work was supported in part by grants from the National Institutes of Health (EB00262, HL73305, GM74048) and the Center for Engineering Cells and Regeneration of the University of Pennsylvania. J.L.L. was supported by the National Science Foundation and M.A.W. by a Ruth L. Kirschstein National Research Service Award (F32 AR054219-01).
Abbreviations used:
- α-SMA
α-smooth muscle actin
- coll I
collagen type I
- EMT
epithelial–mesenchymal transition
- FN
fibronectin
- MDCK
Madin–Darby canine kidney cells
- NMuMG
normal murine mammary gland cells
- PA
polyacrylamide
- rBM
reconstituted basement membrane
- TGF-β
transforming growth factor-β
- ZO-1
zonula occludens-1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-06-0537) on January 11, 2012.
REFERENCES
- Alcaraz J, Xu R, Mori H, Nelson CM, Mroue R, Spencer VA, Brownfield D, Radisky DC, Bustamante C, Bissell MJ. Laminin and biomimetic extracellular elasticity enhance functional differentiation in mammary epithelia. EMBO J. 2008;27:2829–2838. doi: 10.1038/emboj.2008.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora PD, Narani N, McCulloch CA. The compliance of collagen gels regulates transforming growth factor-beta induction of alpha-smooth muscle actin in fibroblasts. Am J Pathol. 1999;154:871–882. doi: 10.1016/s0002-9440(10)65334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacman D, Merkel S, Croner R, Papadopoulos T, Brueckl W, Dimmler A. TGF-beta receptor 2 downregulation in tumour-associated stroma worsens prognosis and high-grade tumours show more tumour-associated macrophages and lower TGF-beta1 expression in colon carcinoma: a retrospective study. BMC Cancer. 2007;7:156. doi: 10.1186/1471-2407-7-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey KM, Liu J. Caveolin-1 up-regulation during epithelial to mesenchymal transition is mediated by focal adhesion kinase. J Biol Chem. 2008;283:13714–13724. doi: 10.1074/jbc.M709329200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakin AV, Tomlinson AK, Bhowmick NA, Moses HL, Arteaga CL. Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. J Biol Chem. 2000;275:36803–36810. doi: 10.1074/jbc.M005912200. [DOI] [PubMed] [Google Scholar]
- Bierie B, Moses HL. Tumour microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6:506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- Boise LH, Gonzalez-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nunez G, Thompson CB. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- Burgering BM, Coffer PJ. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature. 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- Butcher DT, Alliston T, Weaver VM. A tense situation: forcing tumour progression. Nat Rev Cancer. 2009;9:108–122. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byfield FJ, Wen Q, Levental I, Nordstrom K, Arratia PE, Miller RT, Janmey PA. Absence of filamin A prevents cells from responding to stiffness gradients on gels coated with collagen but not fibronectin. Biophys J. 2009;96:5095–5102. doi: 10.1016/j.bpj.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannito S, Novo E, di Bonzo LV, Busletta C, Colombatto S, Parola M. Epithelial-mesenchymal transition: from molecular mechanisms, redox regulation to implications in human health and disease. Antioxid Redox Signal. 2010;12:1383–1430. doi: 10.1089/ars.2009.2737. [DOI] [PubMed] [Google Scholar]
- Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- Chen JH, Chen WL, Sider KL, Yip CY, Simmons CA. beta-Catenin mediates mechanically regulated, transforming growth factor-beta1-induced myofibroblast differentiation of aortic valve interstitial cells. Arterioscler Thromb Vasc Biol. 2011;31:590–597. doi: 10.1161/ATVBAHA.110.220061. [DOI] [PubMed] [Google Scholar]
- Chen RH, Su YH, Chuang RL, Chang TY. Suppression of transforming growth factor-beta-induced apoptosis through a phosphatidylinositol 3-kinase/Akt-dependent pathway. Oncogene. 1998;17:1959–1968. doi: 10.1038/sj.onc.1202111. [DOI] [PubMed] [Google Scholar]
- Cheng EH, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T, Korsmeyer SJ. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol Cell. 2001;8:705–711. doi: 10.1016/s1097-2765(01)00320-3. [DOI] [PubMed] [Google Scholar]
- Cho HJ, Baek KE, Saika S, Jeong MJ, Yoo J. Snail is required for transforming growth factor-beta-induced epithelial-mesenchymal transition by activating PI3 kinase/Akt signal pathway. Biochem Biophys Res Commun. 2007;353:337–343. doi: 10.1016/j.bbrc.2006.12.035. [DOI] [PubMed] [Google Scholar]
- Cicchini C, Laudadio I, Citarella F, Corazzari M, Steindler C, Conigliaro A, Fantoni A, Amicone L, Tripodi M. TGFbeta-induced EMT requires focal adhesion kinase (FAK) signaling. Exp Cell Res. 2008;314:143–152. doi: 10.1016/j.yexcr.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- Dudek H, Datta SR, Franke TF, Birnbaum MJ, Yao R, Cooper GM, Segal RA, Kaplan DR, Greenberg ME. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science. 1997;275:661–665. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- Ebihara T, Venkatesan N, Tanaka R, Ludwig MS. Changes in extracellular matrix and tissue viscoelasticity in bleomycin-induced lung fibrosisTemporal aspects. Am J Respir Crit Care Med. 2000;162:1569–1576. doi: 10.1164/ajrccm.162.4.9912011. [DOI] [PubMed] [Google Scholar]
- Edlund S, Landstrom M, Heldin CH, Aspenstrom P. Transforming growth factor-beta-induced mobilization of actin cytoskeleton requires signaling by small GTPases Cdc42 and RhoA. Mol Biol Cell. 2002;13:902–914. doi: 10.1091/mbc.01-08-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engle SJ, Hoying JB, Boivin GP, Ormsby I, Gartside PS, Doetschman T. Transforming growth factor beta1 suppresses nonmetastatic colon cancer at an early stage of tumorigenesis. Cancer Res. 1999;59:3379–3386. [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Friedland JC, Lee MH, Boettiger D. Mechanically activated integrin switch controls alpha5beta1 function. Science. 2009;323:642–644. doi: 10.1126/science.1168441. [DOI] [PubMed] [Google Scholar]
- Friess H, Yamanaka Y, Buchler M, Ebert M, Beger HG, Gold LI, Korc M. Enhanced expression of transforming growth factor beta isoforms in pancreatic cancer correlates with decreased survival. Gastroenterology. 1993;105:1846–1856. doi: 10.1016/0016-5085(93)91084-u. [DOI] [PubMed] [Google Scholar]
- Fringer J, Grinnell F. Fibroblast quiescence in floating or released collagen matrices: contribution of the ERK signaling pathway and actin cytoskeletal organization. J Biol Chem. 2001;276:31047–31052. doi: 10.1074/jbc.M101898200. [DOI] [PubMed] [Google Scholar]
- Frisch SM, Vuori K, Ruoslahti E, Chan-Hui PY. Control of adhesion-dependent cell survival by focal adhesion kinase. J Cell Biol. 1996;134:793–799. doi: 10.1083/jcb.134.3.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Wang YK, Yang MT, Desai RA, Yu X, Liu Z, Chen CS. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukai Y, Fukuchi M, Masuda N, Osawa H, Kato H, Nakajima T, Kuwano H. Reduced expression of transforming growth factor-beta receptors is an unfavorable prognostic factor in human esophageal squamous cell carcinoma. Int J Cancer. 2003;104:161–166. doi: 10.1002/ijc.10929. [DOI] [PubMed] [Google Scholar]
- Go C, Li P, Wang XJ. Blocking transforming growth factor beta signaling in transgenic epidermis accelerates chemical carcinogenesis: a mechanism associated with increased angiogenesis. Cancer Res. 1999;59:2861–2868. [PubMed] [Google Scholar]
- Han H, Wecker T, Grehn F, Schlunck G. Elasticity-dependent modulation of TGF-beta responses in human trabecular meshwork cells. Invest Ophthalmol Vis Sci. 2011;52:2889–2896. doi: 10.1167/iovs.10-6640. [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Beach D. p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- Hynes RO. The extracellular matrix: not just pretty fibrils. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic D, Almeida EA, Schlaepfer DD, Dazin P, Aizawa S, Damsky CH. Extracellular matrix survival signals transduced by focal adhesion kinase suppress p53-mediated apoptosis. J Cell Biol. 1998;143:547–560. doi: 10.1083/jcb.143.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie HY, Pearline RV, Grueneberg D, Hsia M, Ravichandran P, Kothari N, Natesan S, Brugge JS. Distinct roles of Akt1 and Akt2 in regulating cell migration and epithelial-mesenchymal transition. J Cell Biol. 2005;171:1023–1034. doi: 10.1083/jcb.200505087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaklamani VG, et al. Combined genetic assessment of transforming growth factor-beta signaling pathway variants may predict breast cancer risk. Cancer Res. 2005;65:3454–3461. doi: 10.1158/0008-5472.CAN-04-2961. [DOI] [PubMed] [Google Scholar]
- Kattla JJ, Carew RM, Heljic M, Godson C, Brazil DP. Protein kinase B/Akt activity is involved in renal TGF-beta1-driven epithelial-mesenchymal transition in vitro and in vivo. Am J Physiol Renal Physiol. 2008;295:F215–F225. doi: 10.1152/ajprenal.00548.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khwaja A, Rodriguez-Viciana P, Wennstrom S, Warne PH, Downward J. Matrix adhesion and Ras transformation both activate a phosphoinositide 3-OH kinase and protein kinase B/Akt cellular survival pathway. EMBO J. 1997;16:2783–2793. doi: 10.1093/emboj/16.10.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein EA, Yin L, Kothapalli D, Castagnino P, Byfield FJ, Xu T, Levental I, Hawthorne E, Janmey PA, Assoian RK. Cell-cycle control by physiological matrix elasticity and in vivo tissue stiffening. Curr Biol. 2009;19:1511–1518. doi: 10.1016/j.cub.2009.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental KR, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-Y, Zhou X, Rowe RG, Hu Y, Schlaepfer DD, Ilić D, Dressler G, Park A, Guan J-L, Weiss SJ. Snail1 controls epithelial–mesenchymal lineage commitment in focal adhesion kinase–null embryonic cells. J Cell Biol. 2011;195:729–738. doi: 10.1083/jcb.201105103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Dranoff JA, Chan EP, Uemura M, Sevigny J, Wells RG. Transforming growth factor-beta and substrate stiffness regulate portal fibroblast activation in culture. Hepatology. 2007;46:1246–1256. doi: 10.1002/hep.21792. [DOI] [PubMed] [Google Scholar]
- Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. TGFbeta in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen PJ, Ebner R, Lopez AR, Derynck R. TGF-beta induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J Cell Biol. 1994;127:2021–2036. doi: 10.1083/jcb.127.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mih JD, Sharif AS, Liu F, Marinkovic A, Symer MM, Tschumperlin DJ. A multiwell platform for studying stiffness-dependent cell biology. PLoS One. 2011;6:e19929. doi: 10.1371/journal.pone.0019929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka RS, Koh Y, Roebuck LR, Sanders ME, Brantley-Sieders D, Gorska AE, Moses HL, Arteaga CL. Increased malignancy of Neu-induced mammary tumors overexpressing active transforming growth factor beta1. Mol Cell Biol. 2003;23:8691–8703. doi: 10.1128/MCB.23.23.8691-8703.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Khauv D, Bissell MJ, Radisky DC. Change in cell shape is required for matrix metalloproteinase-induced epithelial-mesenchymal transition of mammary epithelial cells. J Cell Biochem. 2008;105:25–33. doi: 10.1002/jcb.21821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongusaha PP, Kim JI, Fang L, Wong TW, Yancopoulos GD, Aaronson SA, Lee SW. p53 induction and activation of DDR1 kinase counteract p53-mediated apoptosis and influence p53 regulation through a positive feedback loop. EMBO J. 2003;22:1289–1301. doi: 10.1093/emboj/cdg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek MJ, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Peinado H, Quintanilla M, Cano A. Transforming growth factor beta-1 induces snail transcription factor in epithelial cell lines: mechanisms for epithelial mesenchymal transitions. J Biol Chem. 2003;278:21113–21123. doi: 10.1074/jbc.M211304200. [DOI] [PubMed] [Google Scholar]
- Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petridou S, Maltseva O, Spanakis S, Masur SK. TGF-beta receptor expression and smad2 localization are cell density dependent in fibroblasts. Invest Ophthalmol Vis Sci. 2000;41:89–95. [PubMed] [Google Scholar]
- Pietenpol JA, Stein RW, Moran E, Yaciuk P, Schlegel R, Lyons RM, Pittelkow MR, Munger K, Howley PM, Moses HL. TGF-beta 1 inhibition of c-myc transcription and growth in keratinocytes is abrogated by viral transforming proteins with pRB binding domains. Cell. 1990;61:777–785. doi: 10.1016/0092-8674(90)90188-k. [DOI] [PubMed] [Google Scholar]
- Pirone DM, Liu WF, Ruiz SA, Gao L, Raghavan S, Lemmon CA, Romer LH, Chen CS. An inhibitory role for FAK in regulating proliferation: a link between limited adhesion and RhoA-ROCK signaling. J Cell Biol. 2006;174:277–288. doi: 10.1083/jcb.200510062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi RA, Leof EB. TGF-beta signaling: a tale of two responses. J Cell Biochem. 2007;102:593–608. doi: 10.1002/jcb.21501. [DOI] [PubMed] [Google Scholar]
- Shintani Y, Wheelock MJ, Johnson KR. Phosphoinositide-3 kinase-Rac1-c-Jun NH2-terminal kinase signaling mediates collagen I-induced cell scattering and up-regulation of N-cadherin expression in mouse mammary epithelial cells. Mol Biol Cell. 2006;17:2963–2975. doi: 10.1091/mbc.E05-12-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- Stuelten CH, Buck MB, Dippon J, Roberts AB, Fritz P, Knabbe C. Smad4-expression is decreased in breast cancer tissues: a retrospective study. BMC Cancer. 2006;6:25. doi: 10.1186/1471-2407-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B, Vu M, Booker T, Santner SJ, Miller FR, Anver MR, Wakefield LM. TGF-beta switches from tumor suppressor to prometastatic factor in a model of breast cancer progression. J Clin Invest. 2003;112:1116–1124. doi: 10.1172/JCI18899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilghman RW, Cowan CR, Mih JD, Koryakina Y, Gioeli D, Slack-Davis JK, Blackman BR, Tschumperlin DJ, Parsons JT. Matrix rigidity regulates cancer cell growth and cellular phenotype. PLoS One. 2010;5:e12905. doi: 10.1371/journal.pone.0012905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vailhe B, Ronot X, Tracqui P, Usson Y, Tranqui L. In vitro angiogenesis is modulated by the mechanical properties of fibrin gels and is related to alpha(v)beta3 integrin localization. In Vitro Cell Dev Biol Anim. 1997;33:763–773. doi: 10.1007/s11626-997-0155-6. [DOI] [PubMed] [Google Scholar]
- Wakefield LM, Roberts AB. TGF-beta signaling: positive and negative effects on tumorigenesis. Curr Opin Genet Dev. 2002;12:22–29. doi: 10.1016/s0959-437x(01)00259-3. [DOI] [PubMed] [Google Scholar]
- Wang H, Radjendirane V, Wary KK, Chakrabarty S. Transforming growth factor beta regulates cell-cell adhesion through extracellular matrix remodeling and activation of focal adhesion kinase in human colon carcinoma Moser cells. Oncogene. 2004;23:5558–5561. doi: 10.1038/sj.onc.1207701. [DOI] [PubMed] [Google Scholar]
- Wang HB, Dembo M, Wang YL. Substrate flexibility regulates growth and apoptosis of normal but not transformed cells. Am J Physiol Cell Physiol. 2000;279:C1345–C1350. doi: 10.1152/ajpcell.2000.279.5.C1345. [DOI] [PubMed] [Google Scholar]
- Wang YH, Chiu WT, Wang YK, Wu CC, Chen TL, Teng CF, Chang WT, Chang HC, Tang MJ. Deregulation of AP-1 proteins in collagen gel-induced epithelial cell apoptosis mediated by low substratum rigidity. J Biol Chem. 2007;282:752–763. doi: 10.1074/jbc.M604801200. [DOI] [PubMed] [Google Scholar]
- Wang YK, Wang YH, Wang CZ, Sung JM, Chiu WT, Lin SH, Chang YH, Tang MJ. Rigidity of collagen fibrils controls collagen gel-induced down-regulation of focal adhesion complex proteins mediated by alpha2beta1 integrin. J Biol Chem. 2003;278:21886–21892. doi: 10.1074/jbc.M300092200. [DOI] [PubMed] [Google Scholar]
- Wikstrom P, Stattin P, Franck-Lissbrant I, Damber JE, Bergh A. Transforming growth factor beta1 is associated with angiogenesis, metastasis, and poor clinical outcome in prostate cancer. Prostate. 1998;37:19–29. doi: 10.1002/(sici)1097-0045(19980915)37:1<19::aid-pros4>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Winer JP, Janmey PA, McCormick ME, Funaki M. Bone marrow-derived human mesenchymal stem cells become quiescent on soft substrates but remain responsive to chemical or mechanical stimuli. Tissue Eng Part A. 2009;15:147–154. doi: 10.1089/ten.tea.2007.0388. [DOI] [PubMed] [Google Scholar]
- Wozniak MA, Desai R, Solski PA, Der CJ, Keely PJ. ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. J Cell Biol. 2003;163:583–595. doi: 10.1083/jcb.200305010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrana JL, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang XF, Massague J. TGF beta signals through a heteromeric protein kinase receptor complex. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- Wu MY, Hill CS. Tgf-beta superfamily signaling in embryonic development and homeostasis. Dev Cell. 2009;16:329–343. doi: 10.1016/j.devcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming W, Weaver V, Janmey PA. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- Yingling JM, Datto MB, Wong C, Frederick JP, Liberati NT, Wang XF. Tumor suppressor Smad4 is a transforming growth factor beta-inducible DNA binding protein. Mol Cell Biol. 1997;17:7019–7028. doi: 10.1128/mcb.17.12.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Richardson JA, Parada LF, Graff JM. Smad3 mutant mice develop metastatic colorectal cancer. Cell. 1998;94:703–714. doi: 10.1016/s0092-8674(00)81730-4. [DOI] [PubMed] [Google Scholar]
- Zouq NK, Keeble JA, Lindsay J, Valentijn AJ, Zhang L, Mills D, Turner CE, Streuli CH, Gilmore AP. FAK engages multiple pathways to maintain survival of fibroblasts and epithelia: differential roles for paxillin and p130Cas. J Cell Sci. 2009;122:357–367. doi: 10.1242/jcs.030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.