Apolipoprotein B-100 after lipidation is dislocated from the ER lumen to the cytoplasmic surface of lipid droplets for proteasomal degradation. UBXD8 in lipid droplets and Derlin-1 in the ER membrane interact with each other and with ApoB and are engaged in the pre- and postdislocation steps, respectively.
Abstract
Apolipoprotein B-100 (ApoB) is the principal component of very low density lipoprotein. Poorly lipidated nascent ApoB is extracted from the Sec61 translocon and degraded by proteasomes. ApoB lipidated in the endoplasmic reticulum (ER) lumen is also subjected to proteasomal degradation, but where and how it dislocates to the cytoplasm remain unknown. In the present study, we demonstrate that ApoB after lipidation is dislocated to the cytoplasmic surface of lipid droplets (LDs) and accumulates as ubiquitinated ApoB in Huh7 cells. Depletion of UBXD8, which is almost confined to LDs in this cell type, decreases recruitment of p97 to LDs and causes an increase of both ubiquitinated ApoB on the LD surface and lipidated ApoB in the ER lumen. In contrast, abrogation of Derlin-1 function induces an accumulation of lipidated ApoB in the ER lumen but does not increase ubiquitinated ApoB on the LD surface. UBXD8 and Derlin-1 bind with each other and with lipidated ApoB and show colocalization around LDs. These results indicate that ApoB after lipidation is dislocated from the ER lumen to the LD surface for proteasomal degradation and that Derlin-1 and UBXD8 are engaged in the predislocation and postdislocation steps, respectively.
INTRODUCTION
Apolipoprotein B-100 (ApoB) is a large glycoprotein (>500 kDa) and the principal component of very low density lipoprotein (VLDL) secreted by hepatocytes. Given the physiological importance of ApoB in lipoprotein transport, its secretion is regulated at several intracellular steps (Brodsky and Fisher, 2008). ApoB is lipidated cotranslationally, where the microsomal triglyceride transfer protein (MTP) plays a critical role (Hussain et al., 2003). When lipidation is perturbed, nascent ApoB is subject to endoplasmic reticulum–associated degradation (ERAD): poorly lipidated ApoB is ubiquitinated, extracted from the Sec61 translocon, and degraded by the proteasome. An E3 ubiquitin ligase, gp78, as well as hsp70, hsp90, p97, p58IPK, and BiP, were shown to be involved in ERAD of the nascent ApoB at the translocon (Fisher et al., 1997, 2008; Gusarova et al., 2001; Liang et al., 2003; Oyadomari et al., 2006; Rutledge et al., 2009).
After cotranslational lipidation, ApoB acquires more lipids to make a mature VLDL in the secretory compartment. Under certain conditions, however, lipidated ApoB is degraded in the ER and/or post-ER compartments, but this posttranslational degradation mechanism is less well delineated than ERAD at the translocon. A post-ER mechanism involving macroautophagy has been reported (Pan et al., 2004, 2008), but lipidated ApoB may also be degraded by other mechanisms (Liao et al., 1998; Fisher et al., 2001; Olofsson and Boren, 2005).
We previously reported that ubiquitinated ApoB accumulated at lipid droplets (LDs) when proteasomes or autophagy was suppressed and inferred that the LD might function as a platform for ApoB degradation (Ohsaki et al., 2006). Another study showed that ApoB accumulating at LDs is in the lipidated form (Ohsaki et al., 2008). Because lipidation occurs only in the ER lumen, the result suggested that ApoB after lipidation is dislocated to the cytoplasmic side in the vicinity of LDs. The study also demonstrated that the accumulation of lipidated ApoB induces formation of a unique ER–LD amalgamation structure called an ApoB-crescent (Supplemental Figure S1; Ohsaki et al., 2008). The LD-resident ApoB degradation mechanism was clearly distinguished from ERAD of poorly lipidated ApoB at the Sec61 translocon because MTP inhibition virtually abolished the former while enhancing the latter (Zhou et al., 1998).
In the present study, we showed that lipidated ApoB is dislocated to the cytoplasmic surface of LDs and explored the mechanism. The results indicated that Derlin-1, a putative dislocon component in the ER membrane (Lee et al., 2008; Mueller et al., 2008; Claessen et al., 2010; Phan et al., 2010), and UBXD8, an UBX domain–containing protein, in the LD (Schuberth and Buchberger, 2008) interact with each other and with ApoB and play crucial roles in degrading lipidated ApoB. We also found that Derlin-1 is needed for the dislocation of lipidated ApoB, whereas UBXD8 is engaged in the postdislocation process by recruiting p97, an AAA-ATPase, to LDs. The results demonstrated that the LD serves as a platform of dislocation, ubiquitination, and proteasomal degradation of lipidated ApoB.
RESULTS
UBXD8 and other ERAD-related proteins localize in LDs
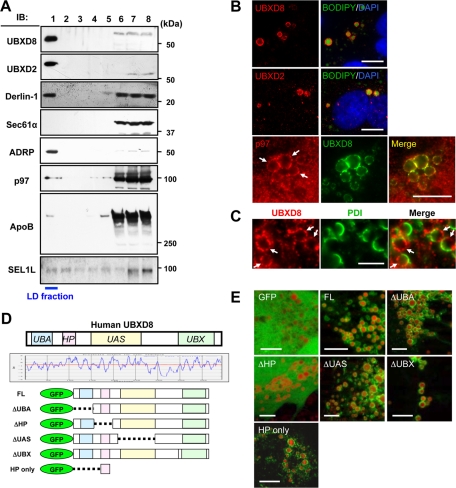
On the basis of the assumption that the ApoB-crescent should harbor molecules engaged in the dislocation and proteasomal degradation of lipidated ApoB (Ohsaki et al., 2008), we searched for candidates by using LDs purified from a human liver cell line, Huh7 (Higashi et al., 2003). Through proteomic analysis, two UBX domain–containing proteins, UBXD8 and UBXD2 (or erasin), and p97 were identified, along with authentic LD proteins, including adipocyte differentiation–related protein (ADRP) and tail-interacting protein of 47 kDa. UBXD8, UBXD2, and p97 were also reported in other proteomic studies on LDs (Fujimoto et al., 2004; Liu et al., 2004; Sato et al., 2006; Bartz et al., 2007). Western blotting confirmed the proteomics results and showed that a majority of both UBXD8 and UBXD2 was recovered in the LD fraction (Figure 1A and Supplemental Figure S2, A and B). The LD fraction also contained Derlin-1 and SEL1L but not other ER proteins such as Sec61α (Figure 1A). Derlin-1 and SEL1L were not detected proteomically but were identified by Western blotting, probably because appropriate peptides were not obtained from transmembrane segments by in-gel digestion (Washburn et al., 2001).
FIGURE 1:
UBXD8 and other ERAD-related proteins were present in LDs of Huh7 cells. The cells were maintained in the normal culture medium without any fatty acid supplement in this and subsequent experiments unless otherwise stated. (A) Fractions obtained by sucrose density-gradient ultracentrifugation were analyzed by Western blotting. LDs were recovered in the fraction of the lowest density (fraction 1), which was verified by enrichment of ADRP. In addition to UBXD8, UBXD2, and p97, which were identified by mass spectrometric analysis, Derlin-1 and SEL1L were also found in the LD fraction. In contrast, Sec61α was found only in the bottom fractions (fractions 6–8), in which most membrane and soluble proteins were contained. SEL1L was immunoprecipitated before SDS–PAGE for efficient detection. (B) Immunofluorescence microscopy confirmed that UBXD8, UBXD2, and p97 distributed in LDs. LDs and nuclei were labeled with BODIPY493/503 (green) and 4′,6-diamidino-2-phenylindole (blue), respectively. p97 distributed throughout the cytoplasm and nucleoplasm, but a conspicuous concentration around LDs was observed. Bars, 10 μm. (C) Double immunofluorescence labeling revealed that UBXD8 (red) and an ER luminal protein, PDI (green), were distributed on the opposite hemisphere of LDs. Bar, 5 μm. (D) Domain structure and hydropathy plot of UBXD8. A domain showing high hydrophobicity was named the HP domain. Full-length UBXD8 and mutants lacking one of the domains (ΔUBA, ΔHP, ΔUAS, or ΔUBX) were conjugated to the carboxy terminus of GFP and expressed in Huh7 cells. (E) GFP-UBXD8(ΔUBA), GFP-UBXD8(ΔUAS), GFP-UBXD8(ΔUBX), and GFP-HP showed concentration around LDs like GFP-UBXD8(FL), but GFP alone and GFP-UBXD8(ΔHP) distributed diffusely in the cytoplasm, indicating that the HP domain is necessary and sufficient for UBXD8 to localize in the LD. LDs were stained red by BODIPY558/568-C12. Bars, 10 μm.
The presence of UBXD8 and UBXD2 in LDs in situ was confirmed by immunofluorescence microscopy (Figure 1B). p97 showed prominent concentration around LDs, although it was distributed diffusely in both the cytoplasm and the nucleoplasm (Figure 1B). In ApoB-crescents, UBXD8 and an ER luminal protein, protein disulfide isomerase (PDI), exhibited a complementary distribution, indicating that UBXD8 exists on the cytoplasmic surface of LDs (Figure 1C and Supplemental Figure S1).
UBXD8 and UBXD2 were also observed in LDs of other cells that have fewer LDs than Huh7 (Supplemental Figure S3, A and B). In those cells, even after LD induction by incubation with oleic acid (OA), the proportion of UBXD2 in the LD fraction was lower than that of UBXD8 (Supplemental Figure S3C). The results indicated that UBXD8 and UBXD2 distribute to LDs irrespective of the ApoB expression, but the propensity to localize in LDs differs between them.
Because UBXD2 turned out to be involved less in the ApoB degradation than UBXD8 (see further results), we focused on UBXD8 in the present study. UBXD8 was shown to be related to the proteasomal degradation of several proteins (Lee et al., 2008; Mueller et al., 2008; Claessen et al., 2010; Phan et al., 2010), but details of its molecular function at LDs are yet to be clarified. The amino acid sequence shows that UBXD8 has UBA, UAS, and UBX domains, as well as a hydrophobic segment, which is tentatively named the HP domain (Figure 1D). To specify the domain(s) critical for LD localization, UBXD8 mutants lacking each of the four domains were analyzed. Mutants devoid of the UBA, UAS, or UBX domain (ΔUBA, ΔUAS, and ΔUBX, respectively) were still targeted to LDs, whereas the mutant lacking the HP domain (ΔHP) distributed diffusely in the cytoplasm. Furthermore, the HP domain alone (amino acids 80–130) was targeted to LDs (Figure 1E). These results indicated that the HP domain is necessary and sufficient for LD localization of UBXD8, which is consistent with previous reports (Mueller et al., 2008; Zehmer et al., 2009; Lee et al., 2010). For LD distribution of UBXD2, the UBA-like, UAS, and UBX domains of UBXD2 were dispensable, whereas the HP domain was necessary (data not shown).
UBXD8 recruits p97 to LDs by the UBX domain
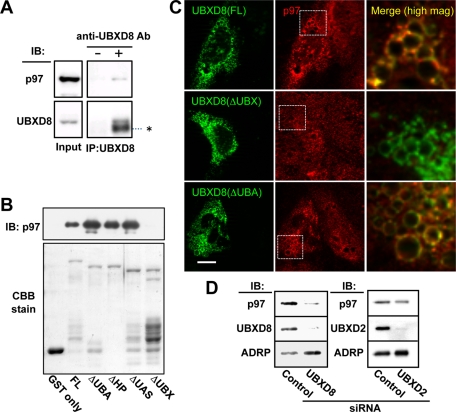
As expected from properties of other UBX domain–containing proteins (Schuberth and Buchberger, 2008), immunoprecipitation of UBXD8 from total Huh7 cell lysate caused cosedimentation of p97 (Figure 2A). To examine which domain of UBXD8 is responsible for binding to p97, in vitro glutathione S-transferase (GST) pull-down assays were performed using recombinant deletion mutants. GST-UBXD8(ΔUBX) did not bind to recombinant GST-free p97 (Figure 2B), whereas the other GST-UBXD8 proteins did, indicating that the UBX domain is responsible for the p97 binding.
FIGURE 2:
UBXD8 recruited p97 to LDs by the UBX domain. (A) Immunoprecipitation of endogenous UBXD8 from Huh7 cell lysates caused cosedimentation of endogenous p97. The input lane was loaded with 2% of the cell lysate. The IgG heavy chain is indicated by an asterisk. (B) In vitro pull-down assay between recombinant p97 and recombinant GST-UBXD8 proteins. An equal amount of GST-free p97 was incubated with full-length and mutant GST-UBXD8 proteins, and the bound fraction was examined by Western blotting. All but GST and GST-UBXD8(ΔUBX) pulled down p97. The GST proteins in the input are shown by Coomassie brilliant blue staining. (C) Endogenous UBXD8 in Huh7 cells was knocked down by siRNA, and siRNA-resistant cDNA of GFP-tagged UBXD8(FL), UBXD8(ΔUBX), or UBXD8(ΔUBA) was transfected. In cells expressing the GFP-tagged proteins (green), LDs that harbored GFP-UBXD8(FL) and UBXD8(ΔUBA) showed an intense accumulation of endogenous p97 (red), but those with UBXD8(ΔUBX) did not. Merged pictures show a high magnification of the rectangular areas. Bar, 10 μm. (D) Huh7 cells were depleted of UBXD8 or UBXD2 by RNAi and the amount of p97 in the LD fraction was compared. p97 in the LD fraction was reduced significantly after UBXD8 knockdown but less so by UBXD2 knockdown. ADRP is shown as a loading control of LDs.
Next we examined whether the UBX domain mediates the p97 recruitment to LDs in the cellular context. To avoid interference of endogenous UBXD8, Huh7 cells were subjected to UBXD8 RNA interference (RNAi) and then transfected with small interfering RNA (siRNA)–resistant green fluorescent protein (GFP)–UBXD8 cDNA constructs. GFP-UBXD8(full length [FL]) and GFP-UBXD8(ΔUBA) induced conspicuous concentration of endogenous p97 in LDs, whereas GFP-UBXD8(ΔUBX) did not (Figure 2C). The result demonstrated that the UBX domain of UBXD8 recruits p97 to LDs in vivo.
The recruitment of p97 to LDs in Huh7 cells was believed to be mediated by UBXD8 for the most part, because p97 in the LD fraction decreased significantly by knockdown of UBXD8 (Figure 2D). In contrast, the effect of UBXD2 knockdown on p97 was negligible. The difference between the two UBX proteins appears peculiar, considering that UBXD2 can also recruit p97 (Liang et al., 2006). In fact, the overexpression of GFP-UBXD2 increased p97 in LDs (Supplemental Figure S4). We speculate that the amount of UBXD8 available in LDs is larger than that of UBXD2 in Huh7 cells.
UBXD8 binds to lipidated ApoB and is involved in its degradation at LDs
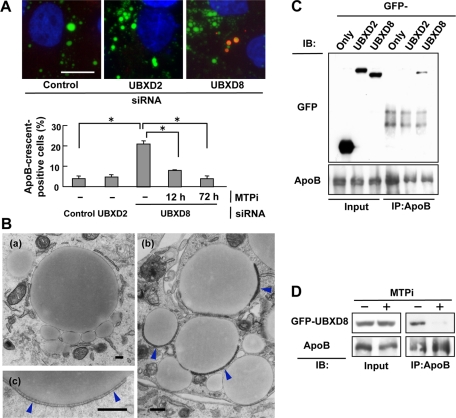
UBX domain–containing proteins have been shown to facilitate the proteasomal degradation of ubiquitinated proteins in both mammals and yeasts (Schuberth and Buchberger, 2008). We thus speculated that either or both of UBXD8 and UBXD2 may be involved in the degradation of ApoB. To test this hypothesis, the effect of their knockdown on the number of ApoB-crescents, which is indicative of the accumulation of lipidated ApoB (Ohsaki et al., 2008), was examined. As a result, the number of ApoB-crescents increased significantly by knockdown of UBXD8 but not by knockdown of UBXD2 (Figure 3A). The increase of ApoB-crescents after UBXD8 knockdown was suppressed by MTP inhibitor (MTPi), suggesting that it was indeed caused by the accumulation of lipidated ApoB (Figure 3A). The ultrastructure of ApoB-crescents in the UBXD8-knockdown cells showed the same LD–ER amalgamation structure as those induced by proteasomal inhibition (Figure 3B; Ohsaki et al., 2008). The results indicated that knockdown of UBXD8 caused the accumulation of lipidated ApoB.
FIGURE 3:
Knockdown of UBXD8 increased ApoB-crescents. (A) Huh7 cells were depleted of either UBXD8 or UBXD2, and the proportion of cells bearing ApoB-crescents was determined after labeling ApoB (red), LDs (green), and nuclei (blue). ApoB in the bulk ER lumen was not labeled when cells were permeabilized with digitonin (Ohsaki et al., 2008). Knockdown of UBXD8, but not that of UBXD2, significantly increased the number of ApoB-crescents in Huh7 cells (Student's t test; *p < 0.01). The increase of ApoB-crescents in cells depleted of UBXD8 was suppressed by treating cells with 100 nM BAY13-9952 (MTPi) for 12 or 72 h before fixation. The result is representative of three independent experiments. Bar, 10 μm. (B) Huh7 cells transfected with control siRNA (a) or UBXD8 siRNA (b, c) were observed by electron microscopy. ApoB-crescents made of an LD and a thin cistern fusing to it (arrowheads) were observed frequently in cells depleted of UBXD8. Bars, 500 nm. (C) Huh7 cells expressing GFP-UBXD8 or GFP-UBXD2 were treated with 10 μM acetyl-leucinyl-leucinyl-norleucinal (ALLN) for 12 h, lysed, and immunoprecipitated with anti-ApoB antibody. GFP-UBXD8, but not GFP-UBXD2, coprecipitated with ApoB. The ALLN treatment was carried out to increase ubiquitinated ApoB, but essentially the same result was obtained without the treatment. (D) Huh7 cells were treated with 10 μM ALLN alone or with 10 μM ALLN and 100 nM BAY13-9952 for 12 h before lysis and immunoprecipitation. GFP-UBXD8 showed coimmunoprecipitation with ApoB, but its amount was reduced drastically when MTP was inhibited.
UBXD8 showed coimmunoprecipitation with ApoB, whereas UBXD2 did not (Figure 3C). ApoB that bound with UBXD8 was believed to be lipidated because the coimmunoprecipitation decreased significantly when cells were pretreated with an MTPi (Figure 3D). These results implied that UBXD8 is engaged in the process of lipidated ApoB degradation.
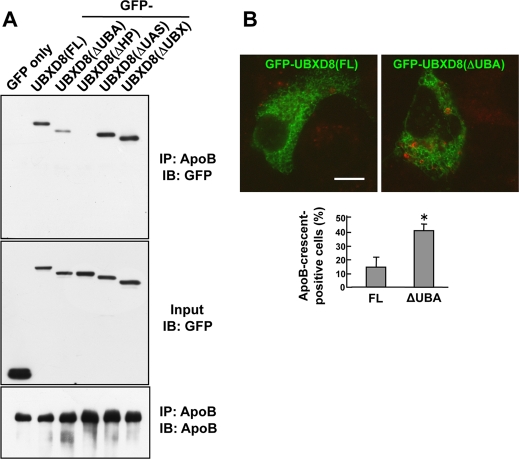
To determine which portion of UBXD8 is related to the ApoB binding, coimmunoprecipitation of ApoB- and UBXD8-deletion mutants was examined. Among the mutants, GFP-UBXD8(ΔUBX) and GFP-UBXD8(ΔUAS) cosedimented with ApoB to the same degree as GFP-UBXD8(FL), but GFP-UBXD8(ΔUBA) and GFP-UBXD8(ΔHP) showed little or no coprecipitation with ApoB, respectively (Figure 4A). The lack of coprecipitation of GFP-UBXD8(ΔHP) is likely to be caused by its absence in LDs (Figure 1E), whereas the weak interaction of GFP-UBXD8(ΔUBA) suggested that the UBA domain, which is known to recognize ubiquitinated proteins (Wilkinson et al., 2001; Hurley et al., 2006), is involved in binding to ApoB at least partially.
FIGURE 4:
The UBA domain of UBXD8 is important for degradation of lipidated ApoB. (A) Huh7 cells expressing GFP-tagged UBXD8 proteins were treated with 10 μM ALLN for 12 h, lysed, and immunoprecipitated with anti-ApoB antibody. GFP was used as a negative control. GFP-UBXD8(ΔUBA) and GFP-UBXD8(ΔHP) showed significantly less coprecipitation with ApoB than the others. (B) Huh7 cells were transfected with cDNAs of GFP-UBXD8(FL) or GFP-UBXD8(ΔUBA) and labeled for ApoB (red). The proportion of cells bearing ApoB-crescents was considerably higher in cells expressing GFP-UBXD8(ΔUBA) than in those expressing GFP-UBXD8(FL). The result shown in the bar graph is representative of three independent experiments. Bar, 10 μm.
To study the functionality of the UBA domain of UBXD8 in the cellular context, the effect of GFP-UBXD8(ΔUBA) overexpression in Huh7 cells was examined. Cells expressing GFP-UBXD8(ΔUBA) showed a significant increase of ApoB-crescents, whereas those expressing GFP-UBXD8(FL) did not (Figure 4B). The result implied that GFP-UBXD8(ΔUBA) acted in a dominant-negative manner and suppressed the normal degradation process of lipidated ApoB.
UBXD8 is engaged in a process after cytoplasmic dislocation of lipidated ApoB
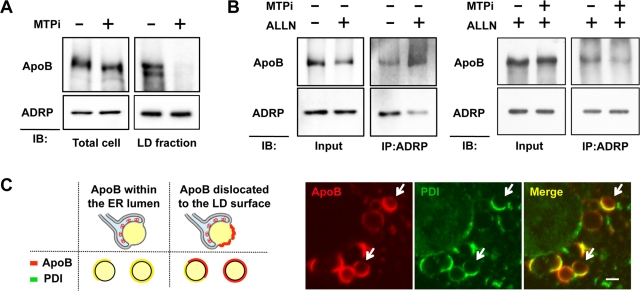
ApoB recovered in the LD fraction by subcellular fractionation showed increased ubiquitination upon proteasomal inhibition (Ohsaki et al., 2006). Because ApoB in this fraction decreased drastically when cells were treated with MTPi, most of it was believed to be derived from the lipidated population (Figure 5A). Because lipidation of ApoB is mediated by MTP in the ER lumen (Hussain et al., 2003), the results indicated that ApoB should traverse the ER membrane to reach the LD surface. Consistently, ApoB was found to be cross-linked to ADRP, a protein that distributes exclusively in the cytoplasmic surface of LDs, and the ApoB–ADRP cross-linking was increased by proteasomal inhibition but was suppressed by MTPi (Figure 5B).
FIGURE 5:
ApoB after lipidation was detected on the cytoplasmic surface of LDs. (A) Huh7 cells were treated with or without 100 nM BAY13-9952 for 48 h, and ApoB in the total cell lysate and the LD fraction was examined. ApoB in the LD fraction was virtually eliminated by the MTPi treatment, whereas the total ApoB in the cell was reduced to only a minor degree. ADRP is shown as a loading control of the LD fraction. (B) Huh7 cells were treated with 10 μM ALLN alone or with 10 μM ALLN and 100 nM BAY13-9952 for 12 h, and then reacted with 1 mM DSP, a membrane-permeable cross-linker, for 30 min on ice before lysis. ApoB cross-linkable with ADRP increased by proteasomal inhibition (left), but the increase was suppressed when MTP was inhibited simultaneously (right). (C) Huh7 cells were treated with 10 μM ALLN for 12 h and labeled for ApoB (red) and PDI (green). The diagram shows the rationale that dislocated ApoB should give rise to ApoB-positive, PDI-negative labeling on the entire or partial LD surface. Confocal microscopy showed that ApoB is present on the surface devoid of PDI in some ApoB-crescents (arrows). Bar, 1 μm.
Dislocation of ApoB to the cytoplasmic surface of LDs was also confirmed by confocal immunofluorescence microscopy by taking advantage of the easily recognizable topology of ApoB-crescents (Supplemental Figure S1). ApoB on the cytoplasmic surface of the LD was segregated from ER luminal proteins, giving rise to ApoB-positive, PDI-negative labeling (Figure 5C). That is, whereas ApoB in most ApoB-crescents was confined to the ER lumen and showed a complete overlap with PDI, some ApoB-crescents showed ApoB-positive, PDI-negative labeling (Figure 5C).
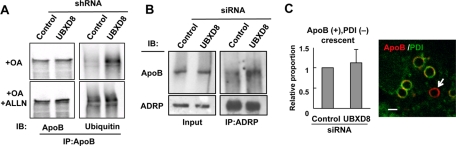
Next we explored whether and how UBXD8 depletion influences the process by which lipidated ApoB is dislocated to the LD surface. First, when UBXD8 was knocked down, ubiquitinated ApoB in the LD fraction showed a significant increase (Figure 6A). Second, ApoB cross-linked with ADRP also increased by UBXD8 knockdown (Figure 6B). Finally, immunofluorescence microscopy showed that ApoB on the cytoplasmic side of ApoB-crescents persisted after UBXD8 knockdown (Figure 6C). These results indicated that UBXD8 depletion did not block dislocation of ApoB, but rather increased the amount of dislocated ApoB on the LD surface. We inferred that UBXD8 is dispensable for the dislocation of ApoB and is instead involved in a subsequent process in which ubiquitinated ApoB is sent to proteasomes for degradation.
FIGURE 6:
Knockdown of UBXD8 increased ApoB on the cytoplasmic surface of LDs. (A) Huh7 cells stably harboring control or UBXD8 shRNA were incubated with or without 10 μM ALLN for 12 h, and the LD fractions were solubilized and immunoprecipitated with anti-ApoB antibody. OA (0.4 mM) was added to the culture medium to increase LDs. ApoB in the LD fraction after UBXD8 knockdown showed increased ubiquitination, and the intensity was comparable to that induced by ALLN. (B) Huh7 cells transfected with control or UBXD8 siRNA were treated with 1 mM DSP for cross-linking. ApoB cross-linkable to ADRP was increased significantly by UBXD8 depletion. (C) The dislocation assay by immunofluorescence microscopy. Huh7 cells were transfected with control or UBXD8 siRNA, treated with 10 μM ALLN for 12 h, and labeled for ApoB (red) and PDI (green). For unambiguous quantification, the proportion of entirely red labels among ApoB-crescents was compared (arrow). The result in the bar graph is the average of three independent experiments. The ApoB-positive, PDI-negative labeling persisted after UBXD8 knockdown, indicating that UBXD8 is dispensable for the ApoB dislocation. Bar, 1 μm.
When UBXD8 was depleted, increased ubiquitination was not limited to ApoB but was observed in a wide range of proteins in the LD fraction (Supplemental Figure S5). The result suggested that UBXD8 might also be engaged in the degradation of proteins other than ApoB.
Derlin-1 is necessary for the cytoplasmic dislocation of lipidated ApoB
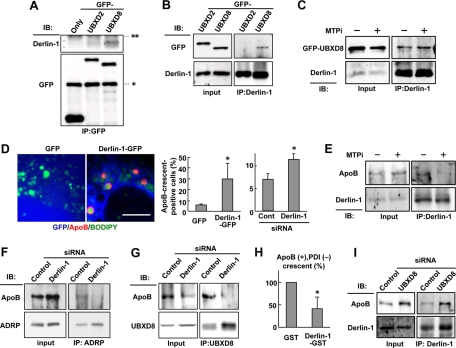
The foregoing results suggested that molecules other than UBXD8 should mediate the dislocation of lipidated ApoB. In this respect, it was notable that Derlin-1, a putative dislocon component (Lilley and Ploegh, 2004; Ye et al., 2004), was identified in the LD fraction (Figure 1A). Of interest, GFP-UBXD8, but not GFP-UBXD2, showed significant coimmunoprecipitation with Derlin-1 (Figure 7, A and B). This coimmunoprecipitation of Derlin-1 and GFP-UBXD8 was not affected by MTPi (Figure 7C), suggesting that Derlin-1 and UBXD8 interact with each other even in the absence of lipidated ApoB.
FIGURE 7:
Inhibition of Derlin-1 function suppressed the cytoplasmic dislocation of ApoB. (A) Huh7 cells expressing GFP, GFP-UBXD2, or GFP-UBXD8 were lysed and immunoprecipitated by anti-GFP antibody. Endogenous Derlin-1 coprecipitated with GFP-UBXD8 significantly, whereas it bound with no GFP or little GFP-UBXD2. The IgG heavy and light chains are indicated by asterisk and double asterisks, respectively. (B) Huh7 cells expressing GFP-UBXD2 or GFP-UBXD8 were lysed and immunoprecipitated by anti–Derlin-1 antibody. GFP-UBXD8 coimmunoprecipitated significantly with endogenous Derlin-1, whereas little GFP-UBXD2 did so. (C) Huh7 cells treated with 10 μM ALLN alone or with 10 μM ALLN and 100 nM BAY13-9952 for 12 h were lysed, and endogenous Derlin-1 was immunoprecipitated. Comparable amounts of GFP-UBXD8 coprecipitated with Derlin-1 irrespective of the MTPi treatment. (D) Huh7 cells expressing GFP or dominant-negative Derlin-1 (Derlin-1–GFP; green) were labeled for ApoB (red) and LDs (blue). Bar, 10 μm. The frequency of ApoB-crescent–positive cells was significantly higher in cells expressing Derlin-1–GFP than in those expressing GFP. ApoB-crescents also increased by Derlin-1 knockdown. The average of results from three independent experiments is shown (Student's t test; *p < 0.05). (E) Huh7 cells were treated as in (C). ApoB coprecipitating with Derlin-1 was reduced significantly by the MTPi treatment for 12 h. (F) Huh7 cells transfected with control or Derlin-1 siRNA were incubated with 10 μM ALLN for 12 h. ApoB cross-linkable to ADRP with 1 mM DSP was reduced by Derlin-1 knockdown. (G) Huh7 cells were treated as in (F). ApoB coimmunoprecipitating with UBXD8 was reduced by Derlin-1 knockdown. (H) The dislocation assay by immunofluorescence microscopy. Huh7 cells were transfected with cDNA of either GST alone or Derlin-1–GST, treated with 10 μM ALLN for 12 h, and labeled for ApoB (red), PDI (green), and GST (blue). Derlin-1–GST was used instead of Derlin-1–GFP to employ the same fluorophore combination for ApoB and PDI as in Figures 5C and 6C. The proportion of ApoB+, PDI− spheres was significantly lower in Derlin-1-GST–positive cells than that in the control, indicating that the dominant-negative Derlin-1 abrogated the cytoplasmic dislocation of ApoB. The average of three independent experiments is shown. (I) Huh7 cells were transfected with control or UBXD8 siRNA. UBXD8 knockdown did not influence the amount of ApoB coimmunoprecipitating with Derlin-1.
When the Derlin-1 function was suppressed either by overexpression of a dominant-negative mutant, Derlin-1-GFP (Ye et al., 2004), or by Derlin-1 knockdown, the frequency of ApoB-crescents increased significantly (Figure 7D). Expression of Derlin-2-GFP did not change the frequency of ApoB-crescents (data not shown). Moreover, Derlin-1 and ApoB showed coimmunoprecipitation, and this was reduced significantly when cells were treated with MTPi (Figure 7E). These results suggested the engagement of Derlin-1 in the process of lipidated ApoB degradation.
Next we examined where in the ApoB degradation process Derlin-1 is involved. When Derlin-1 was knocked down, the amount of ApoB cross-linked with ADRP decreased (Figure 7F). The amount of ApoB that coimmunoprecipitated with UBXD8 was also reduced significantly upon Derlin-1 knockdown (Figure 7G). Furthermore, immunofluorescence labeling revealed that ApoB dislocated to the cytoplasmic LD surface decreased significantly when the dominant-negative Derlin-1 was expressed (Figure 7H). The results corroborated that Derlin-1 is engaged in a step before ApoB dislocates and interacts with UBXD8 on the LD surface. Consistent with this, knockdown of UBXD8 did not affect coimmunoprecipitation of ApoB and Derlin-1 (Figure 7I).
Derlin-1 is a transmembrane protein of the ER and is unlikely to exist in the phospholipid monolayer of the LD surface. To probe where Derlin-1 interacts with UBXD8, the in situ proximity ligation assay (PLA) was performed. This assay gives positive fluorescence signals only when antibodies binding to two different proteins exist within 40 nm of each other (Soderberg et al., 2006). Through this assay, a significant proportion of Derlin-1 and UBXD8 approximation was found to occur at LDs (Figure 8A). In a control, many positive signals were observed between two translocon subunits—Sec61α and Sec61β—but few of them were associated with LDs (Figure 8A).
FIGURE 8:
Derlin-1 and UBXD8 were distributed in proximity around LDs. (A) In situ PLA to identify the location where two endogenous proteins exist in proximity. The approximation signal (red), LDs (green), and nuclei (blue) are shown. The number of the approximation signal was much higher for the combination of Sec61α and Sec61β (30.7/cell) than for the combination of Derlin-1 and UBXD8 (3.4/cell), but the proportion of the approximation signal associated with LDs was significantly higher for Derlin-1–UBXD8 than for Sec61α-Sec61β (Student's t test; *p < 0.05). The relative LD area was equivalent in the two samples (Derlin-1–UBXD8, 2.8%; Sec61α-Sec61β, 2.6%). The average of results from three independent experiments is shown. Negative control using the combination of anti–Derlin-1 antibody and nonimmune goat IgG did not give any approximation signal. Bar, 10 μm. (B) Schematic diagram of UBXD8, Derlin-1, and p97 interactions at the LD intercalated in the ER membrane. Only lipidated ApoB in the ER lumen is depicted here, but the present data indicated that some ApoB after lipidation is dislocated and present on the cytoplasmic surface of the LD.
DISCUSSION
UBXD8 in LDs is engaged in a process after ApoB dislocation
We previously found that ubiquitinated ApoB is present in LDs and that it increases by proteasomal inhibition (Ohsaki et al., 2006). The present study confirmed that ApoB exists on the cytoplasmic surface of LDs by using cross-linking with ADRP and immunofluorescence microscopy. ApoB on the LD surface was believed to be largely derived from lipidated ApoB because MTP inhibition virtually eliminated it.
Knockdown of UBXD8 recapitulated most of the results observed after proteasomal inhibition. That is, it induced formation of ApoB-crescents and increased ApoB existing on the cytoplasmic surface of LDs. UBXD8 in Huh7 cells was present almost exclusively in LDs and was found to play a major role in recruiting p97 to LDs. On the basis of the result, we inferred that UBXD8 is dispensable for dislocation of lipidated ApoB to the LD surface but is engaged in a p97-dependent process that sends the dislocated and ubiquitinated ApoB to proteasomes. This UBXD8 functionality appears different from that shown in previous studies, in which the dysfunction of UBXD8 suppressed the dislocation of transmembrane proteins but did not affect their ubiquitination (Lee et al., 2008; Mueller et al., 2008). This may not be surprising, however, because dislocation, ubiquitination, and proteasomal degradation are correlated in different ways for different ERAD substrates (Vembar and Brodsky, 2008; Bagola et al., 2011).
The engagement of UBXD8 in a postdislocation process is consistent with the presence of UBXD8 not only in LDs forming the ApoB-crescent but also in LDs that are not apparently associated with the ER. That is, as indicated by the general increase of ubiquitination in the LD fraction after knockdown, UBXD8 is likely to be related to the degradation of proteins other than ApoB. In fact, several ERAD-related proteins were shown to participate in the degradation of cytosolic proteins (Ghislain et al., 1996; Dai and Li, 2001; Metzger et al., 2008). It would be interesting to study what kinds of proteins are ubiquitinated and degraded in LDs and how the process is regulated.
UBXD2 had a domain structure similar to that UBXD8 and is also distributed in LDs, but the present results indicated that UBXD2 does not play a major role in the disposal of lipidated ApoB at LDs. As shown for other UBX domain–containing proteins, the difference between UBXD8 and UBXD2 indicates that they are involved in the degradation of different sets of substrates (Alexandru et al., 2008; Mueller et al., 2008) and probably form distinct dislocation complexes for ERAD (Ernst et al., 2009). It remains to be studied whether the presence of UBXD2 in LDs has any relevance to its ERAD-related functions.
Functional implications of the dual UBXD8 distribution
The hairpin-like configuration of UBXD8 is shared by several proteins, enabling them to distribute in both the ER and the LD (Fujimoto et al., 2001; Liang et al., 2006; Turro et al., 2006; Zehmer et al., 2009). UBXD8 was almost confined to LDs in Huh7 cells that have many abundant LDs. However, in other types of cells (e.g., HeLa cells) that contain a few small LDs, most UBXD8 was found in the ER, and the concentration in LDs became obvious only after fatty acid loading (Supplemental Figure S3; Zehmer et al., 2009; Jacquier et al., 2011). If we consider that the LD surface can be a direct extension of the ER membrane as in the ApoB-crescent (Supplemental Figure S1), the redistribution of UBXD8 is likely to occur by the translational movement from the bulk ER membrane to the LD. An LD-associated ER domain in which ERAD of 3-hydroxy-3-methyglutaryl CoA reductase was assumed to occur (Hartman et al., 2010) may also correspond to the LD continuous to the ER membrane.
In this context, it is intriguing that unsaturated fatty acids promoted the dissociation of UBXD8 from ubiquitinated Insig-1, which led to an increase of Insig-1 in the ER and down-regulation of the SREBP pathway (Yang et al., 2002; Lee et al., 2008). This dissociation may be occur because UBXD8 was translocated to the increased LD domain, whereas polytopic Insig-1 was not. That is, only UBXD8 in the bulk ER membrane may be able to bind Insig-1. On the other hand, the present result indicated that UBXD8 in the LD, and not that in the bulk ER, facilitates degradation of lipidated ApoB. These results suggest that UBXD8 modulates lipid metabolism by changing its lateral distribution according to the lipid content of the ER membrane.
A recent study showed that free unsaturated fatty acids directly bind and inactivate UBXD8 in vitro (Lee et al., 2010). The concentration of fatty acids used in the experiment was much higher than that in living cells (Kampf and Kleinfeld, 2004; Carley and Kleinfeld, 2011), but the shown property might underlie an alternative and nonexclusive mechanism to link the cellular lipid content and the UBXD8 function.
Derlin-1 and the ER–LD juncture
Derlin-1 is a putative dislocon component that is believed to span the ER membrane four times (Lilley and Ploegh, 2004; Ye et al., 2004, 2005; Katiyar et al., 2005; Sun et al., 2006; Wahlman et al., 2007; Bernardi et al., 2008; Schwieger et al., 2008), but its precise role in ERAD has not been determined (Hebert et al., 2010; Greenblatt et al., 2011). A previous study showed that Derlin-1 coimmunoprecipitates with ApoB, but the interaction was believed to be related to ERAD of nascent ApoB at the Sec61 translocon (Rutledge et al., 2009). The present study demonstrated that Derlin-1 plays an additional ApoB-related role. On the basis of the result that the abrogation of Derlin-1 reduced ApoB–UBXD8 coimmunoprecipitation, ApoB–ADRP cross-linking, and ApoB detected on the cytoplasmic LD surface, Derlin-1 was believed to be related to the dislocation of lipidated ApoB in a step upstream of UBXD8.
Because of the transmembrane configuration, Derlin-1 is not likely to exist in the phospholipid monolayer of LDs, but the in situ PLA assay suggested that Derlin-1 interacted with UBXD8 near LDs. The Derlin-1–UBXD8 interaction could occur either in-cis or in-trans, but the recovery of Derlin-1 in the LD fraction, from which other transmembrane ER proteins were excluded, suggested that Derlin-1 distributed in the ER membrane directly continuous to the LD domain. That is, Derlin-1 in the ER membrane and UBXD8 in the LD are likely to interact in-cis at the ER–LD juncture (Figure 8B).
It has been generally assumed that ERAD substrates are dislocated via protein-based mechanisms, and Sec61, Derlins, and E3 ligases have been proposed to support the transport (Rapoport, 2007; Nakatsukasa and Brodsky, 2008; Hebert et al., 2010). However, how large, misfolded proteins are transported across the ER membrane has remained enigmatic (Ploegh, 2007; Schliebs et al., 2010). We speculate that ApoB might be dislocated at the ER–LD juncture, a unique structure where the phospholipid bilayer abuts the monolayer vertically (Supplemental Figure S1; Figure 3 of Ohsaki et al., 2008). Here the Derlin-1–UBXD8 interaction might be important to form an ERAD machinery for ApoB. LDs were shown to be dispensable for ERAD of some proteins in yeast (Olzmann and Kopito, 2011), but mammalian ERAD, especially with regard to lipidated ApoB, might be different. Manipulation of the Derlin-1–UBXD8 interaction should be used to analyze the functionality of the ER–LD juncture.
Proteasomes are distributed in the vicinity of LDs (Ohsaki et al., 2006). AUP1, which binds to an E2 ubiquitin conjugase Ube2g2 (Spandl et al., 2011), SPG20, which binds to an E3 ligase WWP1 (Eastman et al., 2009), and an E3 ligase AMFR/gp78 (Hartman et al., 2010) were found in LDs. In conjunction with the present result, the observations indicated that the LD is a platform of protein dislocation, ubiquitination, and proteasomal degradation. Further clarification of LD molecules and dissection of the ER–LD juncture structure should illuminate the LD function in this regard.
MATERIALS AND METHODS
Cells
Huh7, HeLa, and Cos7 cells were obtained from the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan). They were cultured in DME supplemented with 10% fetal calf serum and antibiotics at 37°C in a humidified atmosphere containing 5% CO2. In some experiments as specified, OA complexed with fatty acid–free bovine serum albumen (Wako, Osaka, Japan) at a molar ratio of 6:1 (Brasaemle et al., 1997) was applied at a final fatty acid concentration of 0.4 mM. An MTPi, BAY13-9952, was kindly provided by Bayer Healthcare (Leverkusen, Germany).
Antibodies
Mouse monoclonal anti–human ApoB-100 (clone 6H12; Intracel, Frederick, MD), goat polyclonal anti–human ApoB-100 (Rockland Immunochemicals, Gilbertsville, PA), mouse anti-ADRP (Progen Biotechnik, Heidelberg, Germany), rabbit anti-Sec61a (Stressgen, Enzo Life Sciences, San Diego, CA), goat anti-ETEA/UBXD8, goat anti-Sec61b (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti–Derlin-1 (Sigma-Aldrich, St. Louis, MO), mouse anti-PDI (Affinity Bioreagents, Golden, CO), mouse anti-p97/VCP (GeneTex, Irvine, CA), mouse anti-GFP (GeneTex, Santa Cruz Biotechnology), rabbit anti-GST (Bethyl Laboratories, Montgomery, TX), mouse anti-ubiquitin (FK2; Nippon Bio-Test, Kokubunji, Japan), mouse anti-SEL1L (LifeSpan Biosciences, Seattle, WA), rabbit anti-V5 (Chemicon, Temecula, CA) antibodies, and secondary antibodies conjugated to fluorochromes (Jackson ImmunoResearch Laboratories, West Grove, PA; Invitrogen, Carlsbad, CA) were obtained from the respective suppliers. Rabbit anti-ADRP and rabbit anti-SEL1L antibodies used for immunoprecipitation were kindly provided by Tom Keenan (Virginia Polytechnic Institute, Blacksburg, VA) and Tatsuya Moriyama (Kinki University, Osaka, Japan), respectively.
Rabbit anti-human UBXD2 antibody was raised to a peptide (GCDDGEDENNTWNGNSTQQM) corresponding to the carboxy terminus of the protein and affinity purified with the peptide. The specificity of the antibody was confirmed by Western blotting and loss of the positive band after RNAi.
Subcellular fractionation and Western blotting
Cells were fractionated as previously described (Ohsaki et al., 2006). Briefly, after nitrogen cavitation, the cell homogenate was centrifuged at 1500 rpm for 10 min in a low-speed centrifuge, and the postnuclear supernatant adjusted to 0.54 M sucrose was overlaid with 0.27 M sucrose, 0.135 M sucrose, and a buffer alone and centrifuged in a Beckman SW41Ti rotor at 30,000 rpm for 60 min. Eight fractions were taken from the top, dissolved in an SDS-containing sample buffer, and analyzed by Western blotting. The Western blot signal was detected by chemiluminescence and captured either by Hyperfilm (GE Healthcare, Piscataway, NJ) or by a Light-Capture II imager (ATTO, Tokyo, Japan).
Proteomic analysis
The LD fraction purified from Huh7 cells was subjected to proteomic analysis. The sample was electrophoresed in SDS–PAGE gels, and the gel was stained with Coomassie brilliant blue, destained, and subjected to in-gel digestion with trypsin after reduction by dithiothreitol and alkylation by iodoacetamide (Kikuchi et al., 2004). The resulting peptides were extracted and subjected to liquid chromatography–tandem mass spectrometry (LC/MS) and data-dependent tandem LC-MS/MS analyses using a hybrid Fourier-transform ion cyclotron resonance mass spectrometer (Thermo Finnigan [San Jose, CA] LTQ-FT) interfaced online to nano nigh-performance liquid chromatography (LC Packings UltiMate 3000). The data were processed using a Mascot Distiller and subjected to database search (Mascot, version 2.1; Matrix Science, Boston, MA).
Transfection
Lipofectamine 2000 (Invitrogen) was used for all transfection experiments according to the manufacturer's instruction. Cells were used 1 or 2 d after transfection of cDNA and 3 d after transfection of siRNA. The targeted mRNA sequences for siRNA were 5′-GAAGUUAUUUCACUAAUAA-3′ (UBXD8), 5′-GAUCAGUACCCGUAUUUGA-3′ (UBXD2), and 5′-AACGAUUUAAGGCCUGCUA-3′ (Derlin-1).
siRNA-resistant GFP-UBXD8 cDNAs were prepared by substituting three nucleotides near the 3′-terminus of the siRNA target sequence of UBXD8. The cDNAs were transfected 2 d after RNAi, and the result was examined 24–36 h after the cDNA transfection.
Stable Huh7 cell lines harboring control, UBXD2, or UBXD8 sort hairpin RNA were also prepared by transfecting pRS plasmid carrying shRNA (Origene, Rockville, MD). The short hairpin sequences of shRNA were 5′-CTCTAACAGTAATCCACGACTTCTTATTC-3′ (UBXD8) and 5′-AGCAGGAAGACCAACTGCATCCATTGTAC-3′ (UBXD2). The cells were selected by G418 and used in experiments that required a large number of cells.
Immunoprecipitation
Cells or isolated LDs were solubilized at 4°C by a Nonidet P-40 solution (1% Nonidet P-40, 25 mM Tris-HCl, 150 mM NaCl, pH 7.5) for 30–60 min or by a Triton-deoxycholate buffer (1% Triton X-100, 1% sodium deoxycholate, 25 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH 7.5) for 16 h. The Triton-deoxycholate mixture was used to solubilize ApoB. For cross-linking experiments, cells were treated with a membrane-permeable cross-linker, 1 mM dithiobis(succinimidyl propionate) (DSP; Thermo Scientific, Waltham, MA) for 30 min on ice and then solubilized. Proteins that were bound to the antibodies were captured by protein A–agarose (Santa Cruz Biotechnology), washed extensively by solubilizing solution, dissolved in an SDS buffer, and analyzed by Western blotting.
GST-pull down
Escherichia coli was transformed with pGEX-6P vectors (GE Healthcare), and recombinant GST-tagged proteins were generated by induction with isopropyl β-d-1-thiogalactopyranoside and purified by a glutathione–agarose resin (Sigma-Aldrich). Some proteins were digested with a ProScission protease (GE Healthcare) to obtain the GST-free form. A GST-tagged protein bound to the glutathione–agarose resin was mixed with a GST-free protein in phosphate-buffered saline (PBS) containing 1% Triton X-100 and 10 mM dithiothreitol for 90 min at 4°C. After extensive rinsing, the interacting proteins were eluted from the resin by heating in an SDS buffer and examined by Western blotting.
Immunofluorescence microscopy and data analysis
Cells were fixed with 3% formaldehyde with or without 0.025% glutaraldehyde in 0.1 M phosphate buffer for 15 min and permeabilized with either 0.01% digitonin in PBS for 30 min or with 0.1% Triton X-100 in PBS for 5 min before blocking and incubation with antibodies. LDs were stained with BODIPY493/503 (Invitrogen) after fixation or with BODIPY 558/568-C12 (Invitrogen) added to the culture medium before fixation.
Images were captured using an Axiovert 200M fluorescence microscope (Carl Zeiss, Jena, Germany), with an Apochromat 63× lens with a 1.40 numerical aperture. Some images were obtained using the Apotome processing system. The labeling intensity was analyzed quantitatively using ImageJ software (National Institutes of Health, Bethesda, MD). The color, brightness, and contrast of the presented images were adjusted using Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA) for presentation.
For statistical analyses, more than 10 random fields, each containing three to eight cells, were taken for each sample, so that more than 50 cells were examined. The results obtained in three independent experiments were subject to Student's t test for statistical analysis.
In situ PLA
Cells were fixed, permeablized, and incubated with primary antibodies as in conventional immunofluorescence microscopy. Specimens were incubated with PLA PLUS and MINUS antibodies, and hybridization, ligation, and amplification were done according to the manufacturer's instruction to generate approximation signals (Olink Bioscience, Uppsala, Sweden). The combination of anti–Derlin-1 antibody and nonimmune goat immunoglobulin G (IgG) was used as negative control. The proportion of fluorescence signals associated with BODIPY493/503-positive LDs was determined.
Electron microscopy
For conventional electron microscopy, cells cultured on coverslips were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) and postfixed in a mixture of 1% osmium tetroxide and 0.1% potassium ferrocyanide in the same buffer (White et al., 1979). After ethanol dehydration, samples were embedded in a Quetol 812 resin. Ultrathin sections were observed using a JEOL (Peabody, MA) 1400EX electron microscope operated at 100 kV.
Supplementary Material
Acknowledgments
We thank Akira Kakizuka (Kyoto University, Kyoto, Japan), Tom Keenan (Virginia Polytechnic Institute), and Tatsuya Moriyama (Kinki University) for the gifts of the p97 vector, anti-ADRP antibody, and anti-SEL1L antibody, respectively, and Tsuyako Tatematsu and Razia Sultana for their technical assistance. This work was supported by Grants-in-Aid for Scientific Research and the Global Center of Excellence Program “Integrated Molecular Medicine for Neuronal and Neoplastic Disorders” of the Ministry of Education, Culture, Sports, Science and Technology of Japan (to T.F., M.S.) and by grants from the Kazato Research Foundation and Takeda Science Foundation (to Y.O.).
Abbreviations used:
- ADRP
adipocyte differentiation-related protein
- ALLN
acetyl-leucinyl-leucinyl-norleucinal
- ApoB
apolipoprotein B-100
- ERAD
endoplasmic reticulum–associated degradation
- FL
full length
- LD
lipid droplet
- MTP
microsomal triglyceride transfer protein
- MTPi
MTP inhibitor
- OA
oleic acid
- PDI
protein disulfide isomerase
- PLA
proximity ligation assay
- VLDL
very low density lipoprotein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-11-0950) on January 11, 2012.
REFERENCES
- Alexandru G, Graumann J, Smith GT, Kolawa NJ, Fang R, Deshaies RJ. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134:804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagola K, Mehnert M, Jarosch E, Sommer T. Protein dislocation from the ER. Biochim Biophys Acta. 2011;1808:925–936. doi: 10.1016/j.bbamem.2010.06.025. [DOI] [PubMed] [Google Scholar]
- Bartz R, Zehmer JK, Zhu M, Chen Y, Serrero G, Zhao Y, Liu P. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J Proteome Res. 2007;6:3256–3265. doi: 10.1021/pr070158j. [DOI] [PubMed] [Google Scholar]
- Bernardi KM, Forster ML, Lencer WI, Tsai B. Derlin-1 facilitates the retro-translocation of cholera toxin. Mol Biol Cell. 2008;19:877–884. doi: 10.1091/mbc.E07-08-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasaemle DL, Barber T, Kimmel AR, Londos C. Post-translational regulation of perilipin expression. Stabilization by stored intracellular neutral lipids. J Biol Chem. 1997;272:9378–9387. doi: 10.1074/jbc.272.14.9378. [DOI] [PubMed] [Google Scholar]
- Brodsky JL, Fisher EA. The many intersecting pathways underlying apolipoprotein B secretion and degradation. Trends Endocrinol Metab. 2008;19:254–259. doi: 10.1016/j.tem.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carley AN, Kleinfeld AM. Fatty acid (FFA) transport in cardiomyocytes revealed by imaging unbound FFA is mediated by an FFA pump modulated by the CD36 protein. J Biol Chem. 2011;286:4589–4597. doi: 10.1074/jbc.M110.182162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen JH, Mueller B, Spooner E, Pivorunas VL, Ploegh HL. The transmembrane segment of a tail-anchored protein determines its degradative fate through dislocation from the endoplasmic reticulum. J Biol Chem. 2010;285:20732–20739. doi: 10.1074/jbc.M110.120766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai RM, Li CC. Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat Cell Biol. 2001;3:740–744. doi: 10.1038/35087056. [DOI] [PubMed] [Google Scholar]
- Eastman SW, Yassaee M, Bieniasz PD. A role for ubiquitin ligases and Spartin/SPG20 in lipid droplet turnover. J Cell Biol. 2009;184:881–894. doi: 10.1083/jcb.200808041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst R, Mueller B, Ploegh HL, Schlieker C. The otubain YOD1 is a deubiquitinating enzyme that associates with p97 to facilitate protein dislocation from the ER. Mol Cell. 2009;36:28–38. doi: 10.1016/j.molcel.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher EA, Lapierre LR, Junkins RD, McLeod RS. The AAA-ATPase p97 facilitates degradation of apolipoprotein B by the ubiquitin-proteasome pathway. J Lipid Res. 2008;49:2149–2160. doi: 10.1194/jlr.M800108-JLR200. [DOI] [PubMed] [Google Scholar]
- Fisher EA, Pan M, Chen X, Wu X, Wang H, Jamil H, Sparks JD, Williams KJ. The triple threat to nascent apolipoprotein BEvidence for multiple, distinct degradative pathways. J Biol Chem. 2001;276:27855–27863. doi: 10.1074/jbc.M008885200. [DOI] [PubMed] [Google Scholar]
- Fisher EA, Zhou M, Mitchell DM, Wu X, Omura S, Wang H, Goldberg AL, Ginsberg HN. The degradation of apolipoprotein B100 is mediated by the ubiquitin-proteasome pathway and involves heat shock protein 70. J Biol Chem. 1997;272:20427–20434. doi: 10.1074/jbc.272.33.20427. [DOI] [PubMed] [Google Scholar]
- Fujimoto T, Kogo H, Ishiguro K, Tauchi K, Nomura R. Caveolin-2 is targeted to lipid droplets, a new “membrane domain” in the cell. J Cell Biol. 2001;152:1079–1085. doi: 10.1083/jcb.152.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto Y, Itabe H, Sakai J, Makita M, Noda J, Mori M, Higashi Y, Kojima S, Takano T. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim Biophys Acta. 2004;1644:47–59. doi: 10.1016/j.bbamcr.2003.10.018. [DOI] [PubMed] [Google Scholar]
- Ghislain M, Dohmen RJ, Levy F, Varshavsky A. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 1996;15:4884–4899. [PMC free article] [PubMed] [Google Scholar]
- Greenblatt EJ, Olzmann JA, Kopito RR. Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant alpha-1 antitrypsin from the endoplasmic reticulum. Nat Struct Mol Biol. 2011;18:1147–1152. doi: 10.1038/nsmb.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarova V, Caplan AJ, Brodsky JL, Fisher EA. Apoprotein B degradation is promoted by the molecular chaperones hsp90 and hsp70. J Biol Chem. 2001;276:24891–24900. doi: 10.1074/jbc.M100633200. [DOI] [PubMed] [Google Scholar]
- Hartman IZ, Liu P, Zehmer JK, Luby-Phelps K, Jo Y, Anderson RG, DeBose-Boyd RA. Sterol-induced dislocation of 3-hydroxy-3-methylglutaryl coenzyme A reductase from endoplasmic reticulum membranes into the cytosol through a subcellular compartment resembling lipid droplets. J Biol Chem. 2010;285:19288–19298. doi: 10.1074/jbc.M110.134213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert DN, Bernasconi R, Molinari M. ERAD substrates: which way out? Semin Cell Dev Biol. 2010;21:526–532. doi: 10.1016/j.semcdb.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Higashi Y, Itabe H, Fukase H, Mori M, Fujimoto Y, Takano T. Transmembrane lipid transfer is crucial for providing neutral lipids during very low density lipoprotein assembly in endoplasmic reticulum. J Biol Chem. 2003;278:21450–21458. doi: 10.1074/jbc.M301376200. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain MM, Shi J, Dreizen P. Microsomal triglyceride transfer protein and its role in apoB-lipoprotein assembly. J Lipid Res. 2003;44:22–32. doi: 10.1194/jlr.r200014-jlr200. [DOI] [PubMed] [Google Scholar]
- Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, Schneiter R. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Sci. 124:2424–2437. doi: 10.1242/jcs.076836. [DOI] [PubMed] [Google Scholar]
- Kampf JP, Kleinfeld AM. Fatty acid transport in adipocytes monitored by imaging intracellular free fatty acid levels. J Biol Chem. 2004;279:35775–35780. doi: 10.1074/jbc.M403630200. [DOI] [PubMed] [Google Scholar]
- Katiyar S, Joshi S, Lennarz WJ. The retrotranslocation protein Derlin-1 binds peptide:N-glycanase to the endoplasmic reticulum. Mol Biol Cell. 2005;16:4584–4594. doi: 10.1091/mbc.E05-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi M, Hatano N, Yokota S, Shimozawa N, Imanaka T, Taniguchi H. Proteomic analysis of rat liver peroxisome: presence of peroxisome-specific isozyme of Lon protease. J Biol Chem. 2004;279:421–428. doi: 10.1074/jbc.M305623200. [DOI] [PubMed] [Google Scholar]
- Lee JN, Kim H, Yao H, Chen Y, Weng K, Ye J. Identification of Ubxd8 protein as a sensor for unsaturated fatty acids and regulator of triglyceride synthesis. Proc Natl Acad Sci USA. 2010;107:21424–21429. doi: 10.1073/pnas.1011859107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JN, Zhang X, Feramisco JD, Gong Y, Ye J. Unsaturated fatty acids inhibit proteasomal degradation of Insig-1 at a postubiquitination step. J Biol Chem. 2008;283:33772–33783. doi: 10.1074/jbc.M806108200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Yin C, Doong H, Fang S, Peterhoff C, Nixon RA, Monteiro MJ. Characterization of erasin (UBXD2): a new ER protein that promotes ER-associated protein degradation. J Cell Sci. 2006;119:4011–4024. doi: 10.1242/jcs.03163. [DOI] [PubMed] [Google Scholar]
- Liang JS, Kim T, Fang S, Yamaguchi J, Weissman AM, Fisher EA, Ginsberg HN. Overexpression of the tumor autocrine motility factor receptor Gp78, a ubiquitin protein ligase, results in increased ubiquitinylation and decreased secretion of apolipoprotein B100 in HepG2 cells. J Biol Chem. 2003;278:23984–23988. doi: 10.1074/jbc.M302683200. [DOI] [PubMed] [Google Scholar]
- Liao W, Yeung SC, Chan L. Proteasome-mediated degradation of apolipoprotein B targets both nascent peptides cotranslationally before translocation and full-length apolipoprotein B after translocation into the endoplasmic reticulum. J Biol Chem. 1998;273:27225–27230. doi: 10.1074/jbc.273.42.27225. [DOI] [PubMed] [Google Scholar]
- Lilley BN, Ploegh HL. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, Anderson RG. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J Biol Chem. 2004;279:3787–3792. doi: 10.1074/jbc.M311945200. [DOI] [PubMed] [Google Scholar]
- Metzger MB, Maurer MJ, Dancy BM, Michaelis S. Degradation of a cytosolic protein requires endoplasmic reticulum-associated degradation machinery. J Biol Chem. 2008;283:32302–32316. doi: 10.1074/jbc.M806424200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B, Klemm EJ, Spooner E, Claessen JH, Ploegh HL. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc Natl Acad Sci USA. 2008;105:12325–12330. doi: 10.1073/pnas.0805371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K, Brodsky JL. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic. 2008;9:861–870. doi: 10.1111/j.1600-0854.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki Y, Cheng J, Fujita A, Tokumoto T, Fujimoto T. Cytoplasmic lipid droplets are sites of convergence of proteasomal and autophagic degradation of apolipoprotein B. Mol Biol Cell. 2006;17:2674–2683. doi: 10.1091/mbc.E05-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki Y, Cheng J, Suzuki M, Fujita A, Fujimoto T. Lipid droplets are arrested in the ER membrane by tight binding of lipidated apolipoprotein B-100. J Cell Sci. 2008;121:2415–2422. doi: 10.1242/jcs.025452. [DOI] [PubMed] [Google Scholar]
- Olofsson SO, Boren J. Apolipoprotein B: a clinically important apolipoprotein which assembles atherogenic lipoproteins and promotes the development of atherosclerosis. J Intern Med. 2005;258:395–410. doi: 10.1111/j.1365-2796.2005.01556.x. [DOI] [PubMed] [Google Scholar]
- Olzmann JA, Kopito RR. Lipid droplet formation is dispensable for endoplasmic reticulum-associated degradation. J Biol Chem. 2011;286:27872–27874. doi: 10.1074/jbc.C111.266452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyadomari S, et al. Cotranslocational degradation protects the stressed endoplasmic reticulum from protein overload. Cell. 2006;126:727–739. doi: 10.1016/j.cell.2006.06.051. [DOI] [PubMed] [Google Scholar]
- Pan M, Cederbaum AI, Zhang YL, Ginsberg HN, Williams KJ, Fisher EA. Lipid peroxidation and oxidant stress regulate hepatic apolipoprotein B degradation and VLDL production. J Clin Invest. 2004;113:1277–1287. doi: 10.1172/JCI19197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M, Maitin V, Parathath S, Andreo U, Lin SX, St Germain C, Yao Z, Maxfield FR, Williams KJ, Fisher EA. Presecretory oxidation, aggregation, and autophagic destruction of apoprotein-B: a pathway for late-stage quality control. Proc Natl Acad Sci USA. 2008;105:5862–5867. doi: 10.1073/pnas.0707460104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan VT, Ding VW, Li F, Chalkley RJ, Burlingame A, McCormick F. The RasGAP proteins Ira2 and neurofibromin are negatively regulated by Gpb1 in yeast and ETEA in humans. Mol Cell Biol. 2010;30:2264–2279. doi: 10.1128/MCB.01450-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploegh HL. A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature. 2007;448:435–438. doi: 10.1038/nature06004. [DOI] [PubMed] [Google Scholar]
- Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- Rutledge AC, Qiu W, Zhang R, Kohen-Avramoglu R, Nemat-Gorgani N, Adeli K. Mechanisms targeting apolipoprotein B100 to proteasomal degradation: evidence that degradation is initiated by BiP binding at the N terminus and the formation of a p97 complex at the C terminus. Arterioscler Thromb Vasc Biol. 2009;29:579–585. doi: 10.1161/ATVBAHA.108.181859. [DOI] [PubMed] [Google Scholar]
- Sato S, Fukasawa M, Yamakawa Y, Natsume T, Suzuki T, Shoji I, Aizaki H, Miyamura T, Nishijima M. Proteomic profiling of lipid droplet proteins in hepatoma cell lines expressing hepatitis C virus core protein. J Biochem. 2006;139:921–930. doi: 10.1093/jb/mvj104. [DOI] [PubMed] [Google Scholar]
- Schliebs W, Girzalsky W, Erdmann R. Peroxisomal protein import and ERAD: variations on a common theme. Nat Rev Mol Cell Biol. 2010;11:885–890. doi: 10.1038/nrm3008. [DOI] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci. 2008;65:2360–2371. doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieger I, Lautz K, Krause E, Rosenthal W, Wiesner B, Hermosilla R. Derlin-1 and p97/valosin-containing protein mediate the endoplasmic reticulum-associated degradation of human V2 vasopressin receptors. Mol Pharmacol. 2008;73:697–708. doi: 10.1124/mol.107.040931. [DOI] [PubMed] [Google Scholar]
- Soderberg O, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- Spandl J, Lohmann D, Kuerschner L, Moessinger C, Thiele C. Ancient ubiquitous protein 1 (AUP1) localizes to lipid droplets and binds the E2 ubiquitin conjugase G2 (Ube2g2) via its G2 binding region. J Biol Chem. 2011;286:5599–5606. doi: 10.1074/jbc.M110.190785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Zhang R, Gong X, Geng X, Drain PF, Frizzell RA. Derlin-1 promotes the efficient degradation of the cystic fibrosis transmembrane conductance regulator (CFTR) and CFTR folding mutants. J Biol Chem. 2006;281:36856–36863. doi: 10.1074/jbc.M607085200. [DOI] [PubMed] [Google Scholar]
- Turro S, Ingelmo-Torres M, Estanyol JM, Tebar F, Fernandez MA, Albor CV, Gaus K, Grewal T, Enrich C, Pol A. Identification and characterization of associated with lipid droplet protein 1: a novel membrane-associated protein that resides on hepatic lipid droplets. Traffic. 2006;7:1254–1269. doi: 10.1111/j.1600-0854.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlman J, DeMartino GN, Skach WR, Bulleid NJ, Brodsky JL, Johnson AE. Real-time fluorescence detection of ERAD substrate retrotranslocation in a mammalian in vitro system. Cell. 2007;129:943–955. doi: 10.1016/j.cell.2007.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- White DL, Mazurkiewicz JE, Barrnett RJ. A chemical mechanism for tissue staining by osmium tetroxide-ferrocyanide mixtures. J Histochem Cytochem. 1979;27:1084–1091. doi: 10.1177/27.7.89155. [DOI] [PubMed] [Google Scholar]
- Wilkinson CR, Seeger M, Hartmann-Petersen R, Stone M, Wallace M, Semple C, Gordon C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, Goldstein JL, Brown MS. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Kikkert M, van Voorden S, Wiertz E, Rapoport TA. Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc Natl Acad Sci USA. 2005;102:14132–14138. doi: 10.1073/pnas.0505006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- Zehmer JK, Bartz R, Bisel B, Liu P, Seemann J, Anderson RG. Targeting sequences of UBXD8 and AAM-B reveal that the ER has a direct role in the emergence and regression of lipid droplets. J Cell Sci. 2009;122:3694–3702. doi: 10.1242/jcs.054700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Fisher EA, Ginsberg HN. Regulated co-translational ubiquitination of apolipoprotein B100. A new paradigm for proteasomal degradation of a secretory protein. J Biol Chem. 1998;273:24649–24653. doi: 10.1074/jbc.273.38.24649. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.