The unfolded protein response regulates lipid metabolism, but the functional benefit of this regulation to ER function is not clear. This work shows that inhibition of fatty acid oxidation raises cellular oxidation potential, facilitates ER oxidative folding, and protects hepatocytes from ER stress.
Abstract
The unfolded protein response (UPR) signals protein misfolding in the endoplasmic reticulum (ER) to effect gene expression changes and restore ER homeostasis. Although many UPR-regulated genes encode ER protein processing factors, others, such as those encoding lipid catabolism enzymes, seem unrelated to ER function. It is not known whether UPR-mediated inhibition of fatty acid oxidation influences ER function or, if so, by what mechanism. Here we demonstrate that pharmacological or genetic inhibition of fatty acid oxidation renders liver cells partially resistant to ER stress–induced UPR activation both in vitro and in vivo. Reduced stress sensitivity appeared to be a consequence of increased cellular redox potential as judged by an elevated ratio of oxidized to reduced glutathione and enhanced oxidative folding in the ER. Accordingly, the ER folding benefit of inhibiting fatty acid (FA) oxidation could be phenocopied by manipulating glutathione recycling during ER stress. Conversely, preventing cellular hyperoxidation with N-acetyl cysteine partially negated the stress resistance provided by blocking FA oxidation. Our results suggest that ER stress can be ameliorated through alteration of the oxidizing environment within the ER lumen, and they provide a potential logic for the transient regulation of metabolic pathways by the UPR during stress.
INTRODUCTION
Although it is best known as the site of folding of secretory proteins and resident proteins of the endomembrane system, the endoplasmic reticulum (ER) is intimately connected, both functionally and physically, with essentially every other compartment of the cell and the processes that occur therein. This extensive interconnectivity suggests that conditions in the ER will affect, and be affected by, conditions elsewhere in the cell (Rutkowski and Hegde, 2010). Accordingly, it is reasonable to expect that signaling mechanisms have evolved to sense ER disruption and to influence the activity of other cellular pathways that affect ER function either directly or indirectly.
The unfolded protein response (UPR) is the best-understood signaling mechanism linking conditions in the ER to the regulation of gene expression. Its activation is driven by ER stress—an imbalance between the need for nascent proteins to be properly folded, modified, assembled, and sent onward in the ER (collectively, “ER protein processing”) versus the capacity of the ER to match that need. Disruption to ER protein processing in vertebrates is sensed by three ER-resident transmembrane proteins—PERK, IRE1α, and ATF6 (α and β isoforms)—each of which initiates a unique signaling cascade that culminates in alterations in transcriptional regulation (Ron and Walter, 2007). For PERK, this effect is mediated by the eIF2α-dependent transcription factor ATF4; IRE1α's effect occurs through its unconventional splicing of mRNA encoding the XBP1 transcription factor; and ATF6 is itself a transcription factor that is liberated by intramembrane proteolysis. Genes encoding ER chaperones, ER-associated degradation factors, and other proteins participating in processes with obvious relevance to the protein-processing environment in the secretory pathway are all up-regulated by the UPR independent of cell type. Yet the UPR also regulates the expression of genes with less obvious connections to ER protein folding; during ER stress in the liver, these include a host of genes encoding metabolic regulators, the majority of which are down-regulated by the UPR (Rutkowski et al., 2008; Yamamoto et al., 2010). Because the role of the UPR is to protect ER function, these findings suggest that metabolism is functionally connected to ER homeostasis.
The best-characterized example of an interconnection between metabolism and ER function is the feedback loop that accompanies feeding. Insulin signaling upon nutrient availability activates mTOR, which promotes protein synthesis and presumably increases the protein-folding burden on the ER (Hietakangas and Cohen, 2009). The ensuing UPR activation promotes JNK-mediated dephosphorylation and inactivation of the insulin receptor substrate in a pathway of negative feedback that would then suppress further activation of mTOR (Özcan et al., 2008). Unfortunately, chronic activation of this regulatory loop by the modern problem of overnutrition leads to insulin resistance and various downstream problems (Hotamisligil, 2010). Nevertheless, this regulatory pathway illustrates how the UPR acts on a metabolic pathway (in this case, by nontranscriptional means) to protect ER function. More recently, the UPR has been shown to exert both transcriptional and translational control over metabolic pathways, including lipogenesis (Lee et al., 2008; Oyadomari et al., 2008), gluconeogenesis (Wang et al., 2009), lipoprotein production (Rutkowski et al., 2008; Yamamoto et al., 2010), and lipid catabolism (Rutkowski et al., 2008). It is thus possible that suppression of these processes might benefit the ER; however, it is not known whether flux through these metabolic pathways influences ER function or, if so, by what mechanism.
In previous work, we demonstrated that ER stress in the liver leads to suppression of genes encoding transcriptional master regulators of metabolism, including C/EBPα, SREBP1, SREBP2, PPARα, PGC1α, and others, as well as the downstream targets of these master regulators (Rutkowski et al., 2008). Some of the metabolic pathways controlled by these master regulators have intuitive connections to ER function, such as SREBP-mediated synthesis of very low density lipoprotein particles, which takes place in the ER lumen and heavily taxes the ER quality control machinery (Shelness and Sellers, 2001). However, fatty acid (FA) oxidation, controlled by PPARα, as well as C/EBPα, PGC1α, and other regulators, takes place predominantly in mitochondria, and so its potential connection to ER homeostasis is unclear. Here we test the hypothesis that suppression of FA oxidation improves ER function.
RESULTS
Inhibition of fatty acid oxidation attenuates UPR activation in vivo
The observation that the UPR suppresses the expression of mRNAs encoding key components of the FA oxidation machinery—including both transcriptional regulators of the process such as PPARα and PGC1α and rate-limiting FA oxidation enzymes such as CPT1A and ACOX1 (Rutkowski et al., 2008)—led us to speculate that suppression of FA oxidation might improve ER function during stress. The most sensitive readout for the protein-folding capacity of the ER is UPR activation; when ER protein folding is compromised, a clear effect can often be seen in the extent of UPR activation even when changes in the protein-folding environment per se are subtle (Rutkowski et al., 2006; Wu et al., 2007). Thus, using UPR activation as a proxy for ER functionality, we tested the hypothesis that inhibition of FA oxidation improves ER protein processing during stress.
Intraperitoneal challenge of animals with the ER stress–inducing agent tunicamycin (TM), which blocks N-linked glycosylation, leads to UPR activation in the liver, suppression of a wide range of metabolic genes, and dramatic accumulation of microvesicular lipid droplets in the organ (Rutkowski et al., 2008). Etomoxir (ET) is an irreversible inhibitor of CPT1A, which carries out the rate-limiting step in FA β-oxidation (Weis et al., 1994). If inhibition of FA oxidation improves ER function, then ER stress in animals treated with TM and ET should result in attenuated UPR activation but also in substantially fattier livers compared with mice challenged with TM alone.
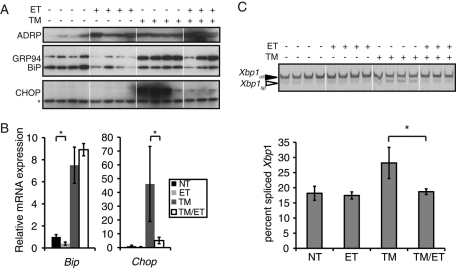
Whereas both TM and ET alone led to an increase in hepatic lipid accumulation, treatment of animals with both agents led to much more dramatic fatty liver, shown here by up-regulation of the lipid droplet marker protein ADRP (Figure 1A). However, ET almost completely blocked the up-regulation of the PERK-dependent UPR target gene CHOP caused by TM at both the protein and mRNA levels (Figure 1, A and B). ET also attenuated IRE1α-dependent splicing of Xbp1 mRNA (Figure 1C). Although ET did not appear to block TM-induced up-regulation of the ER chaperone BiP, ET treatment alone suppressed Bip mRNA expression (Figure 1, A and B). (Bip mRNA expression is also reduced in the livers of Atf6α-/- animals [D.T.R. lab, unpublished results], suggesting that, at least in the liver, the UPR contributes to basal Bip expression). These results show that the UPR is at least in part attenuated in the liver when FA oxidation is concomitantly inhibited. They raise the possibility that inhibition of FA oxidation influences ER functionality during stress.
FIGURE 1:
Inhibition of FA oxidation reduces UPR activation in vivo. (A) Wild-type mice were injected with 0.25 mg/kg TM or 20 mg/kg ET alone or in combination. Animals were killed 14 h after injection and liver homogenates prepared for immunoblotting against the ER chaperones BiP and GRP94, the PERK-dependent UPR target gene CHOP, or the lipid droplet marker protein ADRP. Asterisk denotes a nonspecific background band that shows equal protein loading. (B, C) In parallel, expression of Bip and Chop mRNAs taken from the same livers as in A were analyzed by qRT-PCR (B), or Xbp1 mRNA splicing was analyzed by conventional RT-PCR, and splicing was quantitated and is shown below the gel image (C). Xbp1 image is shown in black-to-white inverted form for visual clarity.
Inhibition of FA oxidation protects cultured hepatocytes from ER stress
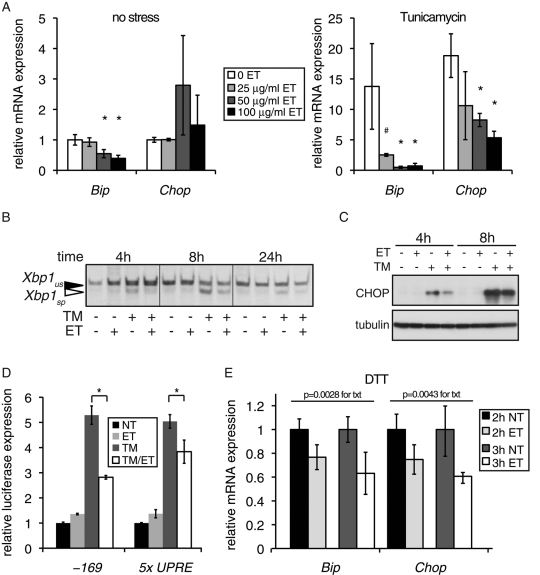
To better understand the potential mechanistic connection between inhibition of FA oxidation and ER function in a more experimentally tractable system, we turned to cultured rat FaO hepatoma cells. We previously showed that these cells suppress expression of key metabolic genes during UPR activation in a manner similar to liver cells in vivo (Rutkowski et al., 2008). Thus important in vivo functional connections between FA oxidation and ER homeostasis might be retained in these cells. As in vivo, we treated cells with TM in the presence or absence of ET, after first confirming that ET does not block the pharmacological activity of TM (Supplemental Figure S1). We monitored the expression of the UPR target genes Bip and Chop, which are sentinels for UPR activation. In the absence of ER stress, there was a dose-dependent decrease in Bip mRNA expression (Figure 2A, left), as in vivo. When cells were treated with TM, ET led to a substantial attenuation of the stress-induced up-regulation of both Bip and Chop (Figure 2A, right).
FIGURE 2:
Inhibition of FA oxidation by etomoxir attenuates UPR activation. (A) FaO hepatoma cells were treated for 6 h with the indicated doses of ET in triplicate, in the absence (left) or presence (right) of 250 ng/ml TM. Expression of Bip and Chop mRNA was quantitated by qRT-PCR. Expression is given relative to cells treated with neither chemical (note the change in scale). In this and all subsequent figures, error bars denote SDs taken from three independent treatments, unless indicated otherwise. *p < 0.05; #p < 0.1. (B) Cells were treated with 250 ng/ml TM and/or 20 μg/ml ET for the indicated times, followed by RT-PCR to detect spliced (sp) and unspliced (us) Xbp1 mRNA. (C) Cells were treated for 4 or 8 h with TM in the presence or absence of 50 μg/ml ET, and expression of CHOP was detected by immunoblot. (D) Cells were transfected with a control β-galactosidase expression plasmid and a construct containing the firefly luciferase gene controlled by either a minimal Bip promoter, encompassing the 169 proximal nucleotides and containing three ER stress response elements (–169; Luo and Lee, 2002), or by five tandem copies of a UPR element (5x UPRE; Wang et al., 2000). Cells were then treated in triplicate for 14 h with 250 ng/ml TM and/or 100 μg/ml ET, followed by quantitation of luciferase activity from cell lysates, which was normalized against β-galactosidase activity. (E) Cells were pretreated for 3 h with 50 μg/ml ET or vehicle, and then 1 mM DTT was added for a further 2 or 3 h to all samples. Relative expression of Bip and Chop mRNAs was quantitated by qRT-PCR and is given for cells pretreated with ET relative to cells that were not treated with ET. Statistical significance was determined by two-factor analysis of variance (treatment and time), with the p values for ET treatment shown.
ET treatment attenuated UPR activation by other independent measures as well. At several time points, the IRE1-dependent splicing of Xbp1 mRNA was reduced in TM-treated cells that were exposed to ET (Figure 2B). Similarly, ET suppressed the up-regulation of CHOP protein, which occurs largely as a consequence of PERK activation (Figure 2C), as well as up-regulation of UPR-dependent luciferase reporters (Figure 2D). Pretreatment with ET also reduced up-regulation of Bip and Chop mRNA and the splicing of Xbp1 mRNA in cells treated with the ER stress-inducing reductant dithiothreitol (DTT), which elicits ER stress by a mechanism distinct from that of TM (Figures 2E and S2). Taken together, these results indicate that ET broadly diminishes global UPR activation during ER stress.
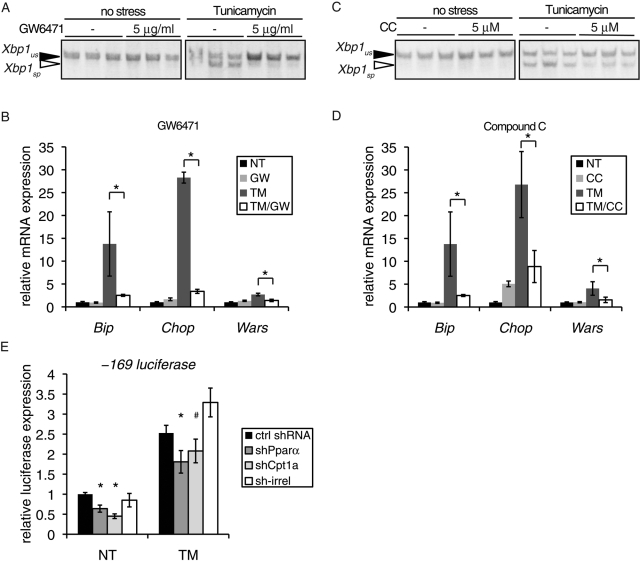
To determine whether inhibition of FA oxidation by other means could also lead to suppression of UPR activation, we treated cells with TM in the presence or absence of either GW6471 or Compound C. GW6471 antagonizes the transcription factor PPARα, which transcriptionally activates the FA oxidation gene program (Xu et al., 2002). Compound C is a competitive inhibitor of AMPK, a kinase that stimulates FA oxidation through phosphorylation-dependent inhibition of acetyl-CoA carboxylase (Zhou et al., 2001). Both chemicals substantially reduced splicing of Xbp1 and up-regulation of the UPR target genes Bip, Chop, and Wars (Wars encodes the ATF4-dependent tryptophanyl-tRNA synthetase) upon TM treatment (Figure 3, A–D). Suppression of UPR transcriptional activity could also be achieved by short hairpin RNA (shRNA; despite incomplete knockdown; Supplemental Figure S3) against either Pparα or Cpt1a compared with a control shRNA; an shRNA against an irrelevant target had no such effect (Figure 3E). Collectively, the data in Figures 1–3 demonstrate that inhibition of FA oxidation, by any of multiple means, suppresses UPR activation.
FIGURE 3:
Inhibition of FA oxidation by several means attenuates UPR activation. (A, B) Cells were treated in triplicate with 250 ng/ml TM or vehicle in the presence or absence of GW6471 and RNA was analyzed for Xbp1 splicing (A) or for expression of the UPR target genes Bip, Chop, and Wars by qRT-PCR (B). (C, D) Cells were treated as in A and B, but with Compound C (CC) instead of GW6471. (E) Cells were cotransfected with the –169 luciferase construct, a β-galactosidase transfection control, and a plasmid containing either a control shRNA or an shRNA targeting Pparα, Cpt1a, or an irrelevant gene. Luciferase expression was quantitated 8 h after TM treatment from three independent plates per condition.
Inhibition of FA oxidation enhances oxidative folding status in the ER
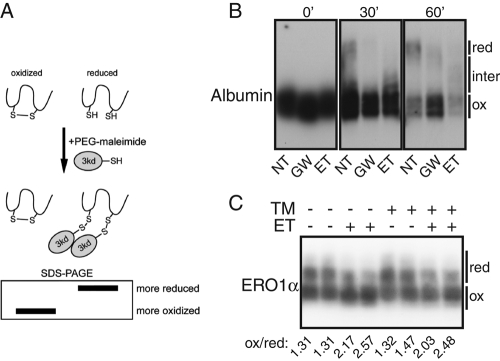
Inhibition of FA oxidation might suppress UPR activation during ER stress either by improving the ER folding environment or simply by directly inhibiting the activation of the ER-resident stress sensors (ATF6, IRE1, and PERK) during stress. These models can in principle be distinguished based on whether ER protein folding improves or worsens as a consequence of inhibition of FA oxidation. However, the ability of the ER to efficiently fold and process proteins for export is difficult to assess, particularly in mammalian cells, for which there are few reliable probes for ER protein folding (Lai et al., 2010). Indeed, even a fairly significant perturbation of this capacity, in the form of deletion of ATF6α, results in only subtle changes in the efficiency with which proteins progress through the secretory pathway (Wu et al., 2007). Consistent with this limitation, treatment of cells with ET had no appreciable effect on the passage of ER client proteins through ER quality control or their secretion, which can serve as indirect measures of ER protein processing (Supplemental Figure S4). However, one of the essential functions of the ER as a folding environment is the oxidation of client proteins, catalyzed by resident oxidases and thiol isomerases; this need accounts for why reducing agents such as DTT elicit ER stress. To assess this aspect of ER function, we monitored the oxidative folding status of the ER by assessing the resistance of an ER client protein—albumin, which forms 17 disulfide bonds—to reduction by DTT. To discriminate oxidized from reduced albumin, we modified lysate proteins with polyethylene glycol (PEG)–maleimide, which adds an ∼3-kDa PEG moiety to reduced, but not oxidized, cysteine residues and can readily be detected as a mobility shift upon SDS–PAGE (Figure 4A). Both ET and GW6471 rendered albumin partially resistant to the reducing effects of DTT treatment (Figure 4B), suggesting a relatively hyperoxidizing ER lumen. Even absent DTT, we also found a preponderance of the ER-resident oxidoreductase ERO1α to exist in a more highly oxidized state in the presence of ET than in its absence (Figure 4C). Although other effects of ET and GW6471 on ER functionality cannot be ruled out, these results demonstrate a measurable effect of inhibition of FA oxidation on ER protein folding, which could conceivably improve the folding environment during stress.
FIGURE 4:
Inhibition of FA oxidation leads to a hyperoxidizing ER lumen. (A) PEG-maleimide reacts with reduced cysteines to form 3-kDa adducts, leading to mobility retardation on SDS–PAGE. (B) Cells were pretreated for 2 h with 100 μg/ml ET or 15 μg/ml GW6471, and then 2 mM DTT, which leads to partial reduction of intracellular disulfide bonds, was added for the indicated times. Lysates were collected and treated with PEG-maleimide, and endogenous albumin was detected by immunoblot. Maximally reduced, maximally oxidized, and intermediate forms are indicated. (C) Cells were treated in duplicate with TM and/or 50 μg/ml ET for 8 h, followed by reaction of lysates with PEG-maleimide. Endogenous ERO1α was then detected by immunoblot. The ratio of more-oxidized to more-reduced forms of ERO1α was quantitated by densitometry and is shown below the image.
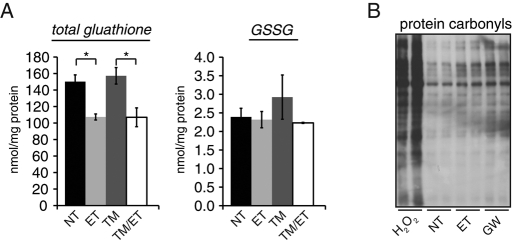
ET was previously reported to decrease the level of intracellular reduced glutathione in both human hepatoma and glioblastoma cells (Vickers et al., 2006; Pike et al., 2010). We likewise observed decreases in total cellular glutathione levels without decreases in oxidized glutathione (glutathione disulfide [GSSG]) upon treatment with ET (Figure 5A), indicative of an increased cellular GSSG/reduced glutathione (GSH) ratio. GSSG likely does not act in nascent protein oxidation directly, but rather acts as a redox buffer to maintain the oxidative potential of the ER lumen (Bulleid and Ellgaard, 2011). Thus an increased GSSG/GSH ratio would be expected to create a hyperoxidizing ER lumen. In addition, the primary role of reduced glutathione is to neutralize reactive oxygen species (ROS; Yuan and Kaplowitz, 2009). Consistent with the decrease in reduced glutathione, inhibition of FA oxidation led to an increase in oxidative damage to proteins, detected by increased protein carbonylation in the presence of ET or GW6471 (Figure 5B). This result is consistent with previous reports linking ET to ROS accumulation (Merrill et al., 2002; Pike et al., 2010). Taken together, the results in Figures 4 and 5 suggest that inhibition of FA oxidation leads to a hyperoxidizing cellular environment, both in the ER, seen in protein oxidative folding, and in the cytoplasm, seen in decreased antioxidative potential and increased ROS-mediated damage to proteins.
FIGURE 5:
Inhibition of FA oxidation impairs cellular antioxidant defense. (A) Total glutathione (left) and GSSG (right) were measured in lysates from cells treated with or without TM and 50 μg/ml ET for 4 h. Both species were measured as described in Materials and Methods. n = 3 independent plates. (B) Cells were treated for 8 h with vehicle, 100 μg/ml ET, or 5 μg/ml GW6471, or for 1 h with 1 mM H2O2. Protein carbonylation was detected in lysates by reaction with 2,4-dinitrophenylhydrazine (DNP) and detection by an anti-DNP antibody.
Protection from ER stress can be phenocopied by direct modulation of glutathione
Our data demonstrate a correlation among inhibition of FA oxidation, increased cellular oxidizing potential, and attenuated UPR activation. We hypothesize that inhibiting FA oxidation protects ER function during stress by influencing glutathione recycling. If this model is correct, then enhancing the production of GSSG should phenocopy inhibition of FA oxidation and protect against ER stress. Conversely, suppressing the production of GSSG should block the ability of inhibition of FA oxidation to protect against ER stress.
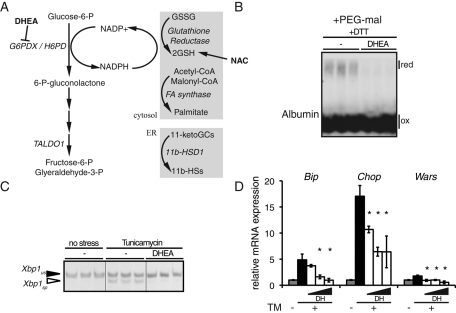
A primary source of reducing equivalents from the recycling of GSSG to GSH is the pentose phosphate pathway (PPP)—an alternate pathway of glucose metabolism that is particularly active in certain tissues, including liver, and is used to generate both NADPH and 5-carbon sugars (Wamelink et al., 2008). NADPH generated by PPP is used in the reductive biosynthesis of FAs, as well as in reduction of GSSG to GSH by glutathione reductase (Figure 6A). Inhibition of PPP in general, or glutathione reductase in particular, increases the cellular GSSG/GSH ratio (Sies et al., 1983; Zhao et al., 2009). Thus we reasoned that direct inhibition of PPP should mimic the effects of FA oxidation inhibitors. To test this hypothesis, we treated cells with dehydroepiandrosterone (DHEA), which is an uncompetitive inhibitor of glucose-6-phosphate dehydrogenase (G6PDX; Gordon et al., 1995; Seefeldt et al., 2009). Similar to treatment with ET (Figure 4B), DHEA treatment led to a resistance of albumin to reduction by DTT (Figure 6B). DHEA reduced splicing of Xbp1 mRNA and up-regulation of UPR target genes in a dose-dependent manner upon treatment with TM (Figure 6, C and D).
FIGURE 6:
Inhibition of the pentose phosphate pathway phenocopies inhibition of FA oxidation. (A) Schematic representation of PPP, with targets of DHEA and NAC indicated. NADPH is used in production of reduced glutathione, reductive fatty acid synthesis, and production of 11-β-hydroxysteroids from 11-ketoglucocorticoids. (B) Cells were treated with 150 μM DHEA for 4 h, and then 2 mM DTT was added for 1 h. Reduced and oxidized forms of albumin were detected by immunoblot following modification of lysates with PEG-maleimide as in Figure 4B. (C) Cells were treated in triplicate with 150 μM DHEA in the presence or absence of TM. Xbp1 splicing was detected by RT-PCR. (D) Cells were treated with TM in the presence of increasing concentrations of DHEA (50, 150, 500 μM), and the expression of Bip, Chop, and Wars mRNA was quantitated by qRT-PCR.
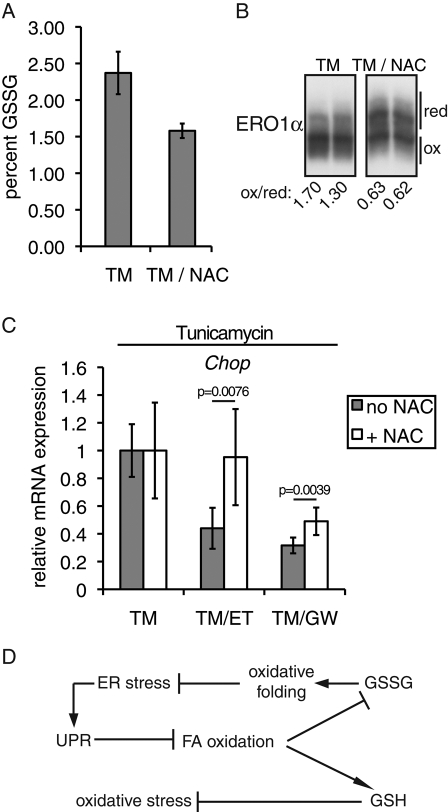
To test whether the protective effects of FA oxidation inhibition required elevation of the GSSG/GSH ratio, we challenged cells with TM in the presence or absence of both an FA oxidation inhibitor (ET or GW6471) and N-acetyl cysteine (NAC), which stimulates GSH production at the expense of GSSG (Figure 7A; Simons et al., 2007). NAC treatment led to a greater percentage of ERO1α being found in a more reduced form (Figure 7B), which is in contrast to the effects of ET and GW6471 (Figure 4C). (By comparison, the bona fide reducing agent DTT leaves no ERO1α fully oxidized, which likely explains why DTT induces ER stress but NAC appears not to.) We found that NAC blocked the ability of ET to suppress Chop mRNA expression during TM treatment, and it also modestly lessened the ability of GW6471 to do so (Figure 7C). (Results using other readouts—e.g., Xbp1 splicing or Bip expression—were more ambiguous due to large error terms, which likely were a consequence of the combination of several experimental manipulations.) These results suggest that at least part of the protective effects of inhibition of FA oxidation arise through modulation of glutathione oxidation status and its attendant consequences on cellular redox potential.
FIGURE 7:
Diminishing GSSG ablates protection from ER stress by inhibition of FA oxidation. (A) Cellular levels of total glutathione and GSSG were determined as in Figure 5A in cells treated for 4 h with 250 ng/ml TM alone or TM plus 20 mM NAC. The percentage of glutathione as GSSG, shown here, was calculated from those two measurements. (B) The relative oxidation state of ERO1α was determined in cells treated in duplicate as in A. The densitometrically quantitated ratio of oxidized to reduced ERO1α is given below the image. (C) Cells were pretreated for 1 h with 20 mM NAC. Then 250 μg/ml TM was added to the media, along with vehicle, 50 μg/ml ET, or 5 μg/ml GW6471. After 4 h, Chop mRNA expression was assessed by qRT-PCR and is expressed here relative to cells treated with or without NAC, in the absence of an inhibitor of FA oxidation. Thus ET and GW6471 reduce Chop expression during TM treatment when NAC is absent but not (or not as much) when it is present. n = 6 samples for each condition from two separate experiments. (D) Working model to describe the relationship among fatty acid oxidation, glutathione, oxidative folding, and ER stress. See Discussion for details.
DISCUSSION
In this study, we showed that inhibiting FA oxidation suppresses UPR activation during ER stress. Inhibition of FA oxidation promotes a more-oxidizing cellular environment, both for glutathione and for ER-resident and client proteins, at the apparent expense of antioxidant defense. Inhibition of PPP-dependent glutathione reduction also alleviated ER stress, underscoring the impact of changes in cellular redox potential on the ER folding environment. Our work suggests that flux through at least one metabolic pathway nominally unconnected to the ER can have surprising consequences for ER function during stress.
Our data support a model in which inhibition of FA oxidation protects the ER environment by promoting oxidative folding during stress and so reduces the effects of that stress (Figure 7D). This model is consistent with the observation that ER stress, induced either pharmacologically or by overexpression of misfolded proteins, creates a hypo-oxidizing environment in the ER lumen (Merksamer et al., 2008). The resistance of an ER client protein to reduction (Figure 4B) suggests that inhibition of FA oxidation leads to a bona fide alteration in the ER folding environment, presumably for the better. We believe this model best explains our data when taken together, although we cannot exclude the possibility that ET, GW6471, Compound C, and DHEA alleviate ER stress by some other shared mechanism. Conceivably, these agents might improve ER function by simply inhibiting protein synthesis, although we have not consistently found evidence to support this view; although ET and GW6471 appear to inhibit protein synthesis to a small extent immediately after addition, they do not do so at later time points (Supplemental Figure S4). The observation that knockdown of CPT1A or PPARα also suppresses UPR signaling supports the notion that the effects of inhibiting FA oxidation are biological rather than pharmacological. We also cannot formally exclude the possibility that inhibition of FA oxidation simply suppresses activation of the UPR sensors; indeed, at least ATF6 requires disulfide bond reduction for its activation (Nadanaka et al., 2007), and so a hyperoxidizing ER could suppress the transit of ATF6 to the Golgi, where it is cleaved. We would expect that direct impairment of the UPR absent an improvement in ER folding would actually lead to an exacerbated protein folding problem. However, we found no evidence that ET impairs ER protein folding or secretion (Supplemental Figure S4). The resolution of methods probing the ER folding environment in mammalian cells limits the conclusions that can be drawn at present.
Because suppression of FA oxidation protects against ER stress, an obvious question is whether the suppression of metabolic genes by the UPR, including those involved in FA oxidation (Pparα, Cpt1a, etc.), is carried out for this purpose. If this is true, then maintenance of expression of genes important in FA oxidation should exacerbate ER stress. This prediction is the subject of ongoing investigation. Similarly, we are still studying whether stimulation of FA oxidation alone is sufficient to induce ER stress absent some additional challenge, as might be expected, given that suppression of FA oxidation appears to alleviate ER stress. However, consistent with this idea, two recent articles demonstrate that sustained FA oxidation reduces hepatic lipid accumulation but induces ER stress (Huang et al., 2011a, 2011b).
A growing body of evidence suggests that activation of the UPR is accompanied by impaired defense against oxidative stress (Marciniak et al., 2004; Malhotra et al., 2008; Li et al., 2009). In this work as well, UPR activation appears to exacerbate ROS formation. In fact, the concomitant increase in oxidative stress that accompanies improvement in ER functionality upon inhibition of FA oxidation suggests that protecting or enhancing the environment in one compartment sacrifices it in another. Choosing between priorities presumably reflects a triaging mechanism on the part of the cell. Although treatment with TM is an obviously superphysiological stressor, there is reason to believe that such triaging decisions are relevant to physiological stresses as well. For example, the UPR is activated in the liver by feeding after a fast and then ebbs in the postprandial state (Oyadomari et al., 2008; Pfaffenbach et al., 2010). Accordingly, during such stresses antioxidant defense could be temporarily compromised while the ER adapts to stress caused by increased mTOR-dependent protein load and then restored once insulin signaling is suppressed.
Finally, the observation that inhibition of PPP phenocopies the effects of inhibiting FA oxidation raises the question of whether FA oxidation is directly connected to PPP. Of interest, Pparα−/− mice show reduced PPP activity, whereas dietary stimulation of PPARα increases PPP activity, suggesting that inhibition of FA oxidation might diminish levels of reduced glutathione via inhibition of PPP (Xu et al., 2004; Oosterveer et al., 2009). The oxidative phase of PPP is activated by the mTOR pathway, which also is required for eliciting ER stress during feeding (Oyadomari et al., 2008; Duvel et al., 2010; Pfaffenbach et al., 2010). Thus inhibition of PPP via suppression of FA oxidation could in principle represent a mechanism for attenuating mTOR-induced ER stress. Furthermore, genetic manipulation of PPP in both yeast and mammals alters sensitivity to ER stress, underscoring an emerging and complex functional connection between the two processes (Lavery et al., 2008; Tan et al., 2009). Together, these results suggest a nexus among ER stress, FA oxidation, and PPP, and it will be interesting to determine whether these processes are indeed coordinately regulated.
Our findings support the idea that flux through metabolic pathways influences the maintenance of homeostasis in the ER during stress, even when these pathways take place elsewhere in the cell. To the extent that inhibition of FA oxidation results in lipid accumulation and oxidative stress, they suggest that the efforts of the UPR to protect ER homeostasis might in some cases occur even at the expense of cellular well-being.
MATERIALS AND METHODS
Materials
Materials were purchased from the following sources: TM, EMD Biosciences, San Diego, CA; ET, GW6471, Compound C, DHEA, and NAC, Sigma-Aldrich, St. Louis, MO; and PEG–maleimide (TMM(PEG)12), Thermo Scientific, Waltham, MA. TM, GW6471, Compound C, and DHEA were prepared and aliquoted as stock solutions in dimethyl sulfoxide (DMSO). ET was prepared in 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, pH 7.9), and NAC was prepared fresh as a 1 M solution in sodium bicarbonate. Some lot-to-lot variability was observed in the strength of protection against ER stress conferred by ET. Antibody sources were as follows: KDEL (SPA-827; Enzo Life Sciences, Plymouth, PA); CHOP (sc-793; Santa Cruz Biotechnology, Santa Cruz, CA); β-tubulin (TUB 2.1, Sigma-Aldrich); alpha–1-antitrypsin (A0012; Dako, Carpinteria, CA); and anti–albumin-horseradish peroxidase (PA1-29267; Thermo Scientific. Anti-ERO1α was a kind gift of Roberto Sitia (Fondazione San Raffaele, Milan, Italy). Oxyblot kit for detecting protein carbonyls was from Millipore (Billerica, MA).
Animal experiments
The University of Iowa Institutional Animal Care and Use Committee approved all animal procedures. Animals were from the C57BL/6J strain, housed in the University of Iowa pathogen-free facility on a 12-h light/dark cycle. Mice were injected intraperitoneally with TM and/or ET dissolved in 10 mM HEPES, pH 7.9, or vehicle control. Animals were killed by isofluorane overdose, and liver pieces were flash frozen in liquid nitrogen. Tissue samples were homogenized in either TRIzol (Invitrogen, Carlsbad, CA) for RNA analysis or in RIPA buffer for protein analysis as described (Rutkowski et al., 2008).
Cell culture experiments
Procedures for molecular cloning, TM injections, cell culture, and molecular analysis (including immunoblotting, RT-PCR, quantitative RT-PCR (qRT-PCR), endoglycosidase H digestion, and luciferase assays) used methods that have been described exhaustively elsewhere (Rutkowski et al., 2006, 2008; Wu et al., 2007). Full details are given in the Supplemental Material, including sequences of oligonucleotides not previously published (Supplemental Table S1). All bar graphs show means ± SD. Statistical significances were determined by Student's two-tailed t test, except, where indicated, by analysis of variance. All statistically significant values (p < 0.05) are represented by asterisks; in some cases, exact p values are given. #, 0.1 > p > 0.05. For PEG-maleimide treatment, cells were lysed in 2% SDS 100 mM Tris (final pH 6.8) and heated to 100°C for 15 min. Equal aliquots were treated with either 4 mM PEG-maleimide or DMSO for 30 min at 37°C, followed by quenching with 100 mM DTT. Total and oxidized glutathione levels were measured using a spectrophotometric recycling method based on measuring the rate of reduction of Ellman's reagent in the presence of glutathione reductase and NADPH (Simons et al., 2007). Increasing quantities of genuine GSH were used to construct a standard curve, and results from samples were expressed per milligram of protein determined using the Lowry protein assay. Preincubation with 2-vinylpyridine was used to measure GSSG in the presence of GSH.
Supplementary Material
Acknowledgments
We thank R. Sitia for the ERO1α antibody, A. S. Lee for the –169 luciferase construct, R. Prywes for the 5x UPRE (also known as 5x ATF6) construct, M. L. McCormick for technical assistance, and C. Blaumueller, R. Hegde, E. Snapp, C. Thorpe, and J. Wu for comments and discussions. This work was funded by grants to D.T.R. from the National Institute of Diabetes and Digestive and Kidney Diseases (R01 DK084058) and the Carver Medical Research Trust Initiative and to D.R.S. from the National Cancer Institute (P30 CA086862). D.T.R. conceived and designed the experiments and wrote the article. D.T.R. and H.T. performed the experiments. D.T.R. and D.R.S. analyzed the data.
Abbreviations used:
- DTT
dithiothreitol
- ET
etomoxir
- FA
fatty acid
- GSH
reduced glutathione
- GSSG
glutathione disulfide
- NAC
N-acetyl cysteine
- PEG
polyethylene glycol
- PPP
pentose phosphate pathway
- TM
tunicamycin
- UPR
unfolded protein response
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-12-1011) on January 19, 2012.
REFERENCES
- Bulleid NJ, Ellgaard L. Multiple ways to make disulfides. Trends Biochem Sci. 2011;36:485–492. doi: 10.1016/j.tibs.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Duvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon G, Mackow MC, Levy HR. On the mechanism of interaction of steroids with human glucose 6-phosphate dehydrogenase. Arch Biochem Biophys. 1995;318:25–29. doi: 10.1006/abbi.1995.1199. [DOI] [PubMed] [Google Scholar]
- Hietakangas V, Cohen SM. Regulation of tissue growth through nutrient sensing. Annu Rev Genet. 2009;43:389–410. doi: 10.1146/annurev-genet-102108-134815. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Jia Y, Fu T, Viswakarma N, Bai L, Rao MS, Zhu Y, Borensztajn J, Reddy JK. Sustained activation of PPAR{alpha} by endogenous ligands increases hepatic fatty acid oxidation and prevents obesity in ob/ob mice. FASEB J. 2011a doi: 10.1096/fj.11-194019. Oct 18, doi: 10.1096/fj.11-194019 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, et al. Progressive endoplasmic reticulum stress contributes to hepatocarcinogenesis in fatty acyl-CoA oxidase 1-deficient mice. Am J Pathol. 2011b;179:703–713. doi: 10.1016/j.ajpath.2011.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CW, Aronson DE, Snapp EL. BiP availability distinguishes states of homeostasis and stress in the endoplasmic reticulum of living cells. Mol Biol Cell. 2010;21:1909–1921. doi: 10.1091/mbc.E09-12-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery GG, et al. Deletion of hexose-6-phosphate dehydrogenase activates the unfolded protein response pathway and induces skeletal myopathy. J Biol Chem. 2008;283:8453–8461. doi: 10.1074/jbc.M710067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Mongillo M, Chin KT, Harding H, Ron D, Marks AR, Tabas I. Role of ERO1-alpha-mediated stimulation of inositol 1,4,5-triphosphate receptor activity in endoplasmic reticulum stress-induced apoptosis. J Cell Biol. 2009;186:783–792. doi: 10.1083/jcb.200904060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Lee AS. Requirement of the p38 mitogen-activated protein kinase signalling pathway for the induction of the 78 kDa glucose-regulated protein/immunoglobulin heavy-chain binding protein by azetidine stress: activating transcription factor 6 as a target for stress-induced phosphorylation. Biochem J. 2002;366:787–795. doi: 10.1042/BJ20011802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra JD, Miao H, Zhang K, Wolfson A, Pennathur S, Pipe SW, Kaufman RJ. Antioxidants reduce endoplasmic reticulum stress and improve protein secretion. Proc Natl Acad Sci USA. 2008;105:18525–18530. doi: 10.1073/pnas.0809677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merksamer PI, Trusina A, Papa FR. Real-time redox measurements during endoplasmic reticulum stress reveal interlinked protein folding functions. Cell. 2008;135:933–947. doi: 10.1016/j.cell.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill CL, et al. Etomoxir-induced oxidative stress in HepG2 cells detected by differential gene expression is confirmed biochemically. Toxicol Sci. 2002;68:93–101. doi: 10.1093/toxsci/68.1.93. [DOI] [PubMed] [Google Scholar]
- Nadanaka S, Okada T, Yoshida H, Mori K. Role of disulfide bridges formed in the luminal domain of ATF6 in sensing endoplasmic reticulum stress. Mol Cell Biol. 2007;27:1027–1043. doi: 10.1128/MCB.00408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterveer MH, Grefhorst A, van Dijk TH, Havinga R, Staels B, Kuipers F, Groen AK, Reijngoud DJ. Fenofibrate simultaneously induces hepatic fatty acid oxidation, synthesis, and elongation in mice. J Biol Chem. 2009;284:34036–34044. doi: 10.1074/jbc.M109.051052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyadomari S, Harding HP, Zhang Y, Oyadomari M, Ron D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008;7:520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan U, Özcan L, Yilmaz E, Duvel K, Sahin M, Manning BD, Hotamisligil GS. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol Cell. 2008;29:541–551. doi: 10.1016/j.molcel.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffenbach KT, Nivala AM, Reese L, Ellis F, Wang D, Wei Y, Pagliassotti MJ. Rapamycin inhibits postprandial-mediated X-box-binding protein-1 splicing in rat liver. J Nutr. 2010;140:879–884. doi: 10.3945/jn.109.119883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike LS, Smift AL, Croteau NJ, Ferrick DA, Wu M. Inhibition of fatty acid oxidation by etomoxir impairs NADPH production and increases reactive oxygen species resulting in ATP depletion and cell death in human glioblastoma cells. Biochim Biophys Acta. 2010;1807:726–734. doi: 10.1016/j.bbabio.2010.10.022. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Arnold SM, Miller CN, Wu J, Li J, Gunnison KM, Mori K, Sadighi Akha AA, Raden D, Kaufman RJ. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006;4:e374. doi: 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, Hegde RS. Regulation of basal cellular physiology by the homeostatic unfolded protein response. J Cell Biol. 2010;189:783–794. doi: 10.1083/jcb.201003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, et al. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15:829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seefeldt T, Zhao Y, Chen W, Raza AS, Carlson L, Herman J, Stoebner A, Hanson S, Foll R, Guan X. Characterization of a novel dithiocarbamate glutathione reductase inhibitor and its use as a tool to modulate intracellular glutathione. J Biol Chem. 2009;284:2729–2737. doi: 10.1074/jbc.M802683200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelness GS, Sellers JA. Very-low-density lipoprotein assembly and secretion. Curr Opin Lipidol. 2001;12:151–157. doi: 10.1097/00041433-200104000-00008. [DOI] [PubMed] [Google Scholar]
- Sies H, Brigelius R, Wefers H, Muller A, Cadenas E. Cellular redox changes and response to drugs and toxic agents. Fundam Appl Toxicol. 1983;3:200–208. doi: 10.1016/s0272-0590(83)80126-2. [DOI] [PubMed] [Google Scholar]
- Simons AL, Ahmad IM, Mattson DM, Dornfeld KJ, Spitz DR. 2-Deoxy-d-glucose combined with cisplatin enhances cytotoxicity via metabolic oxidative stress in human head and neck cancer cells. Cancer Res. 2007;67:3364–3370. doi: 10.1158/0008-5472.CAN-06-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SX, Teo M, Lam YT, Dawes IW, Perrone GG. Cu, Zn superoxide dismutase and NADP(H) homeostasis are required for tolerance of endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol Biol Cell. 2009;20:1493–1508. doi: 10.1091/mbc.E08-07-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers AE, Bentley P, Fisher RL. Consequences of mitochondrial injury induced by pharmaceutical fatty acid oxidation inhibitors is characterized in human and rat liver slices. Toxicol In Vitro. 2006;20:1173–1182. doi: 10.1016/j.tiv.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Wamelink MM, Struys EA, Jakobs C. The biochemistry, metabolism and inherited defects of the pentose phosphate pathway: a review. J Inherit Metab Dis. 2008;31:703–717. doi: 10.1007/s10545-008-1015-6. [DOI] [PubMed] [Google Scholar]
- Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufma RJ, Prywes R. Activation of ATF6 and an ATF6 binding site by the endoplasmic reticulum stress response. J Biol Chem. 2000;275:27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Vera L, Fischer WH, Montminy M. The CREB coactivator CRTC2 links hepatic ER stress and fasting gluconeogenesis. Nature. 2009;460:534–537. doi: 10.1038/nature08111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis BC, Cowan AT, Brown N, Foster DW, McGarry JD. Use of a selective inhibitor of liver carnitine palmitoyltransferase I (CPT I) allows quantification of its contribution to total CPT I activity in rat heart. Evidence that the dominant cardiac CPT I isoform is identical to the skeletal muscle enzyme. J Biol Chem. 1994;269:26443–26448. [PubMed] [Google Scholar]
- Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Xu HE, et al. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature. 2002;415:813–817. doi: 10.1038/415813a. [DOI] [PubMed] [Google Scholar]
- Xu J, Chang V, Joseph SB, Trujillo C, Bassilian S, Saad MF, Lee WN, Kurland IJ. Peroxisomal proliferator-activated receptor alpha deficiency diminishes insulin-responsiveness of gluconeogenic/glycolytic/pentose gene expression and substrate cycle flux. Endocrinology. 2004;145:1087–1095. doi: 10.1210/en.2003-1173. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Takahara K, Oyadomari S, Okada T, Sato T, Harada A, Mori K. Induction of liver steatosis and lipid droplet formation in ATF6alpha-knockout mice burdened with pharmacological endoplasmic reticulum stress. Mol Biol Cell. 2010;21:2975–2986. doi: 10.1091/mbc.E09-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Kaplowitz N. Glutathione in liver diseases and hepatotoxicity. Mol Aspects Med. 2009;30:29–41. doi: 10.1016/j.mam.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Seefeldt T, Chen W, Wang X, Matthees D, Hu Y, Guan X. Effects of glutathione reductase inhibition on cellular thiol redox state and related systems. Arch Biochem Biophys. 2009;485:56–62. doi: 10.1016/j.abb.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108:1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.