Autophagy is an intracellular degradation process that is mediated by autophagosomes. Mammalian Atg2 proteins Atg2A and Atg2B are identified and characterized as essential for autophagy. They are also present on lipid droplets and are involved in regulation of lipid droplet volume and distribution.
Abstract
Macroautophagy is an intracellular degradation system by which cytoplasmic materials are enclosed by the autophagosome and delivered to the lysosome. Autophagosome formation is considered to take place on the endoplasmic reticulum and involves functions of autophagy-related (Atg) proteins. Here, we report the identification and characterization of mammalian Atg2 homologues Atg2A and Atg2B. Simultaneous silencing of Atg2A and Atg2B causes a block in autophagic flux and accumulation of unclosed autophagic structures containing most Atg proteins. Atg2A localizes on the autophagic membrane, as well as on the surface of lipid droplets. The Atg2A region containing amino acids 1723–1829, which shows relatively high conservation among species, is required for localization to both the autophagic membrane and lipid droplet and is also essential for autophagy. Depletion of both Atg2A and Atg2B causes clustering of enlarged lipid droplets in an autophagy-independent manner. These data suggest that mammalian Atg2 proteins function both in autophagosome formation and regulation of lipid droplet morphology and dispersion.
INTRODUCTION
Macroautophagy, hereafter referred to simply as autophagy, is an intracellular degradation process accompanied by unique membrane dynamics. An isolation membrane extends to enclose the cytoplasmic contents, resulting in formation of a double-membrane autophagosome. The autophagosome fuses with acidic compartments, endosomes and lysosomes, to degrade the materials inside the autophagosome. Autophagy is important for a wide range of physiological processes such as adaptation to starvation, quality control of intracellular proteins and organelles, embryonic development, elimination of intracellular microbes, and prevention of neurodegeneration and tumor formation (Cecconi and Levine, 2008; Deretic and Levine, 2009; Mizushima and Levine, 2010; Levine et al., 2011; Mizushima and Komatsu, 2011; White et al., 2010).
Thirty-five autophagy-related (ATG) genes have been identified in yeast, and among them, ATG1–10, 12–14, 16–18, 29, and 31 are essential for autophagosome formation (Nakatogawa et al., 2009). The products of these genes are classified into six functional groups: the starvation-responsive Atg1 kinase complex (Atg1–Atg13–Atg17–Atg29–Atg31), the class III phosphatidylinositol 3 (PtdIns3)-kinase complex (Atg6–Atg14–Vps15–Vps34), the Atg12—Atg5–Atg16 complex (— indicates covalent attachment), the Atg8—phosphatidylethanolamine (PE) conjugate, the multimembrane-spanning protein Atg9, and the Atg2–Atg18 complex (Suzuki et al., 2007; Nakatogawa et al., 2009; Mizushima et al., 2011). These Atg products and their functional groups are highly conserved in other species, including mammals.
It has been suggested that the Atg1 complex, the PtdIns3-kinase complex, and Atg9 appear to be important for nucleation of the isolation membrane and that the Atg12 and Atg8 conjugation systems are important for membrane elongation and/or membrane closure. However, the function of Atg2 has not been well characterized. Yeast Atg2, a ∼200-kDa protein, localizes to a perivacuolar punctate structure termed the preautophagosomal structure (PAS), where most Atg proteins gather and autophagosome formation is initiated (Shintani et al., 2001; Stromhaug et al., 2001; Suzuki et al., 2001, 2007; Wang et al., 2001). Recruitment of Atg2 to the PAS requires the Atg1 complex, the PtdIns3-kinase complex, and Atg9. Atg2 interacts with Atg18, a PtdIns(3)P-binding protein (Suzuki et al., 2001; Dove et al., 2004; Obara et al., 2008), and Atg9 (Wang et al., 2001). Atg2 and Atg18 are required for retrograde transport of Atg9 from the PAS (Reggiori et al., 2004). However, Atg2 is not essential for Atg8—PE conjugate and PAS localization of most of Atg proteins (besides Atg18), suggesting that Atg2 is genetically the most downstream factor in the autophagy pathway (Suzuki et al., 2007).
In addition to identification of autophagy-related molecules, the membrane dynamics of autophagosome formation has also been extensively analyzed. Elongating isolation membranes are frequently observed to closely attach to the endoplasmic reticulum (ER; Kovács et al., 2007), and single or multiple connections between the isolation membrane and the ER have been detected (Hayashi-Nishino et al., 2009; Yla-Anttila et al., 2009). Consistent with these observations, autophagosomes appear to be generated at a subdomain of the ER termed the omegasome, which is labeled with another PtdIns(3)P-binding protein, double FYVE-containing protein 1 (DFCP1; Axe et al., 2008). Furthermore, many mammalian Atg proteins are also observed on or close to the ER membrane (Itakura and Mizushima, 2010; Matsunaga et al., 2010). All these findings suggest that the ER plays an important role in autophagosome formation as a membrane source or a platform.
The ER is a central organelle in many cellular functions, such as protein biogenesis/degradation, membrane trafficking, biogenesis of organelles and lipid droplets, lipid metabolism, calcium storage/release, stress sensing, and regulation of cell death (Holthuis and Levine, 2005; Levine and Rabouille, 2005; English et al., 2009; Fagone and Jackowski, 2009; Hotamisligil, 2010; Ma et al., 2011). It is therefore reasonable to assume that autophagosome formation may share some machinery with the other ER-associated functions. However, such cross-talk has not been analyzed at the molecular level.
Here we report the identification and characterization of human Atg2 homologues Atg2A and Atg2B. These Atg2 proteins are essential for autophagosome formation, presumably at a late stage. Atg2A is present on not only autophagic membranes but also lipid droplets. Silencing of both Atg2A and Atg2B increases the size and number of lipid droplets and causes their clustering. These changes are not observed in Atg5-depleted cells. These data suggest that mammalian Atg2A and Atg2B function both in autophagosome formation and regulation of lipid droplet volume and distribution.
RESULTS
Mammalian Atg2A and Atg2B are required for autophagy
Humans have two Atg2 homologues, Atg2A (KIAA0404) and Atg2B (FLJ10242). Atg2A and Atg2B are similar to each other (44.5% of Atg2A residues are identical to those of Atg2B), and Saccharomyces cerevisiae Atg2 shows 15.5 and 15.8% identity to human Atg2A and Atg2B, respectively.
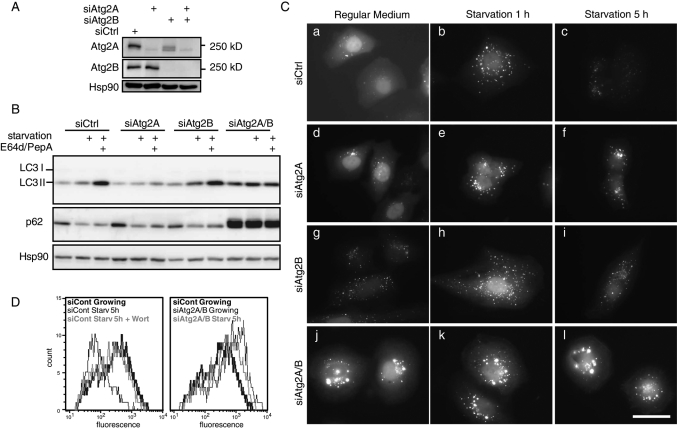
We first tested whether human Atg2 homologues are essential for autophagy. In cells treated with small interfering RNA (siRNA) directed against Atg2A, Atg2B, or both (Atg2A/B), target proteins were efficiently depleted (Figure 1A). These results also confirmed that these antibodies recognize and distinguish the Atg2 isoforms. We used four different methods to measure autophagic activity. First, we performed the microtubule-associated protein light chain 3 (LC3) turnover assay to monitor autophagy flux. In control siRNA-treated cells, the amount of LC3-II (LC3—PE) increased during starvation, which further increased as a result of treatment with lysosomal protease inhibitors, indicating that LC3 was degraded by autophagy during starvation (Figure 1B). Although siRNA against Atg2B (siAtg2B) alone did not affect autophagic flux, it was partially inhibited by siRNA against Atg2A (siAtg2A; Figure 1B). Combination of both siRNAs (siAtg2A/B) completely abrogated the increase in LC3-II caused by starvation and lysosomal inhibition. In these cells, LC3-II accumulated even without starvation; this accumulation is often observed following abrupt depletion of autophagy factors such as Atg14, Vps34 (Itakura et al., 2008), and vacuole membrane protein 1 (VMP1; Itakura and Mizushima, 2010).
FIGURE 1:
Atg2A and Atg2B are essential for autophagy in mammalian cells. (A) HeLa cells stably expressing GFP-LC3 were transfected with the indicated siRNAs twice. Cell lysates were analyzed by immunoblotting with anti-Atg2A, anti-Atg2B, and anti-Hsp90 (internal control) antibodies. (B) HeLa cells stably expressing GFP-LC3 were treated with siRNA directed against Atg2A and/or Atg2B twice for 5 d and then cultured in regular DMEM or starvation medium in the presence or absence of protease inhibitors (E64d and pepstatin A) for 3 h. Cell lysates were analyzed by immunoblotting using the indicated antibodies. (C) HeLa cells stably expressing GFP-LC3 were treated with control siRNA (a–c) and siRNAs against Atg2A (d–f), Atg2B (g–i), and both Atg2A and Atg2B (j–l) for 5 d and then cultured in regular DMEM (a, d, g, and j) or starvation medium for 1 h (b, e, h, and k) or 5 h (c, f, i, and l). GFP-LC3 signals were analyzed by fluorescence microscopy. Scale bar, 50 μm. (D) HeLa cells stably expressing GFP-LC3 were treated with control siRNA or siRNA against Atg2A and Atg2B and cultured as in C in the presence or absence of 0.2 μM wortmannin. Total cellular GFP-LC3 signals were analyzed by flow cytometry.
Second, starvation-induced degradation of p62, a selective autophagy substrate, was suppressed in cells treated with siAtg2A/B (Figure 1B). Accordingly, p62 markedly accumulated in these cells, suggesting that autophagy flux is blocked in siAtg2A/B-treated cells.
Third, we investigated the distribution of green fluorescent protein (GFP)–tagged LC3 (GFP-LC3), a marker for the autophagosomal membrane. HeLa cells stably expressing GFP-LC3 demonstrated diffuse GFP signals in the cytoplasm and nucleus, as well as a small number of tiny punctate structures, which likely represent autophagosomes (Figure 1C, a). The number of GFP-LC3 puncta increased following starvation for 1 h (Figure 1C, b). Prolonged starvation treatment (5 h) destroyed the GFP-LC3 signals because GFP-LC3 was degraded as an autophagic substrate in the lysosome (Figure 1C, c). By contrast, in siAtg2A/B-treated cells, large GFP-LC3 structures accumulated even before starvation treatment (Figure 1C, j and k). Of note, these larger structures remained after prolonged starvation, suggesting that they represent abnormal structures, which were not delivered to the lysosome (Figure 1C, l). Cells treated with either siAtg2A or siAtg2B were only slightly or not affected (Figure 1C, d–i). These data suggest that normal autophagosome formation was perturbed in siAtg2A/B-treated cells.
Finally, we quantified the total GFP-LC3 level by flow cytometry, as it is a good measure of autophagic degradation (Shvets et al., 2008). In control siRNA-treated HeLa cells expressing GFP-LC3, GFP fluorescence was reduced following starvation for 5 h (Figure 1D, left). The fluorescence level was completely restored by addition of wortmannin, a PtdIns 3-kinase inhibitor, to suppress autophagosome formation, confirming that the reduction in the GFP-LC3 signal depends on autophagy. By contrast, such a reduction was not observed in siAtg2A/B-treated cells; a higher level of GFP fluorescence was maintained even after starvation for 5 h (Figure 1D, right).
Taken together these results suggest that both Atg2A and Atg2B are required for autophagy and that they have redundant and overlapping functions, although Atg2A may have a dominant role in HeLa cells.
Accumulation of unclosed autophagosome-related membranes in Atg2-depleted cells
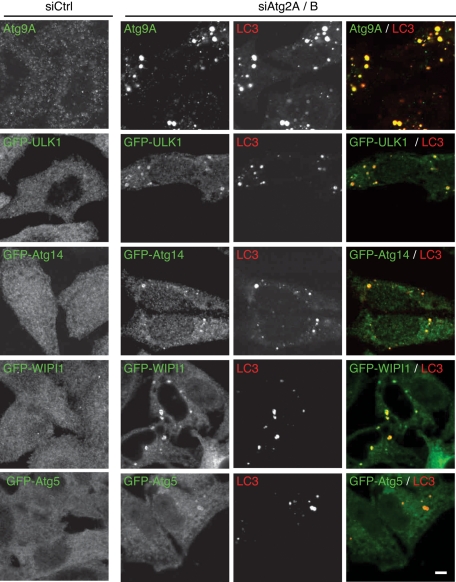
Although Atg2A/B-depleted cells showed defective autophagy, PE conjugation of LC3 (LC3-II formation) and formation of large GFP-LC3 puncta were observed (Figure 1, B and C). In these cells, LC3 might be included in aberrant autophagic structures, or LC3 protein might simply be aggregated. We therefore sought to determine whether other Atg proteins also accumulated in these structures. Punctate structures of endogenous Atg9A, GFP-ULK1 (an Atg1 homologue), GFP-Atg14, GFP–WD-repeat protein interacting with phosphoinoside (WIPI; an Atg18 homologue), and GFP-Atg5 were rarely observed in control siRNA-treated cells under nutrient-rich conditions (Figure 2). However, all these Atg proteins formed several punctate structures in siAtg2A/B cells without starvation treatment, and they colocalized with LC3 (Figure 2). Because these accumulated Atg proteins are representatives of major Atg functional complexes, these results suggest that Atg2A/B play an essential role, probably at a late step of autophagosome formation.
FIGURE 2:
Autophagy-related proteins accumulate in Atg2A/B-depleted cells. HeLa cells or HeLa cells stably expressing the indicated GFP-fused proteins were treated with control siRNA or siRNA against Atg2A and Atg2B. Cells cultured in regular medium were fixed and stained with the anti-Atg9A, anti-LC3 (CTB-LC3-2-1C; Cosmo Bio), and anti-GFP (A6455; Invitrogen) antibodies. Immunofluorescence images were obtained using a confocal microscope. Signal color is indicated by color of typeface. Scale bar, 5 μm.
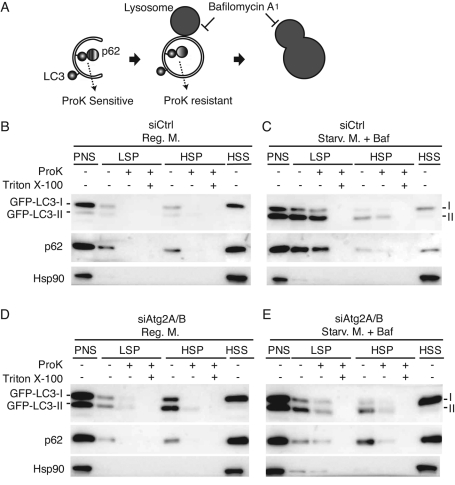
Next we analyzed the autophagosome-related membranes in siAtg2A/B cells by differential centrifugation. HeLa cells stably expressing GFP-LC3 were homogenized and the postnuclear supernatant (PNS) was fractionated by differential centrifugation. We used GFP-LC3 as a marker of the autophagosome-related membrane and p62 as a representative autophagy substrate (Figure 3A). In control siRNA-treated HeLa cells, under nutrient-rich conditions GFP-LC3 was present mostly in an LC3-I form (cytosolic form), which was recovered in a high-speed supernatant (HSS) fraction (Figure 3B and Supplemental Figure S1). To accumulate autophagosomes, we starved cells in the presence of bafilomycin A1, an inhibitor of V-type ATPase, which inhibits intralysosomal degradation and/or autophagosome–lysosome fusion (Klionsky et al., 2008; Figure 3, A and C). In these cells, GFP-LC3-II (membrane-bound PE-conjugated form) accumulated and was recovered in the low-speed pellet (LSP) fraction together with p62 (Figure 3C). GFP-LC3-II and p62 in the LSP fraction were resistant to proteinase K, but they became sensitive in the presence of Triton X-100 (Figure 3, A and C). LAMP1, a lysosomal marker, was recovered in both LSP and HSP fractions under nutrient-rich conditions but mainly in the LSP fraction following starvation treatment (Supplemental Figure S1). These data suggest that GFP-LC3-II and p62 collected in the LSP fraction were enclosed by autophagosomes/autolysosomes.
FIGURE 3:
Unsealed autophagosomes accumulate in Atg2A/B-depleted cells. (A) Schematic drawing of autophagosome formation and protease protection assay. (B–E) HeLa cells expressing GFP-LC3 were treated with control siRNA (B, C) or a mixture of siRNAs against Atg2A and Atg2B (D, E), and cultured in regular DMEM (B, D) or starvation medium containing 0.2 μM bafilomycin A1 (Baf) for 2 h (C, E). The PNS was separated into LSP, HSP, and HSS fractions and then analyzed by SDS–PAGE and immunoblotting using anti-GFP, anti-p62, and anti-Hsp90 antibodies. The subfractions were treated with proteinase K (ProK) with or without Triton X-100.
In siAtg2A/B-treated cells, GFP-LC3-II and p62 were collected mainly in the HSP fraction under nutrient-rich conditions (Figure 3D) and both in the LSP and HSP fractions under conditions of starvation with and without bafilomycin A1 (Figure 3E and Supplemental Figure S1). These data suggest that LC3-II–positive structures accumulate in the absence of Atg2A/B. However, GFP-LC3-II and p62 collected in the LSP and HSP fractions were mostly sensitive to proteinase K, suggesting that complete closure of the autophagosome was impaired. Starvation-induced fraction shift of LAMP1 was less apparent in Atg2-depleted cells (Supplemental Figure S1). GFP-LC3-II in the LSP and HSP fractions in siAtg2A/B-treated cells was membrane bound, not an insoluble protein aggregate, because it was solubilized by Triton X-100 (Supplemental Figure S1).
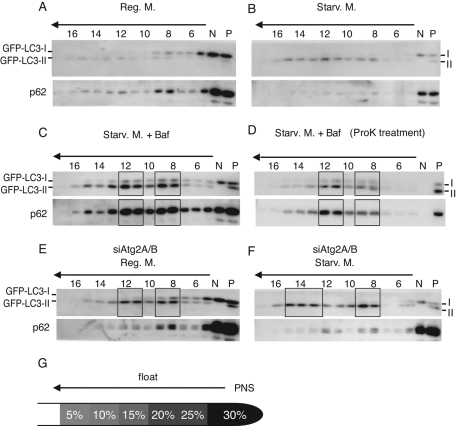
We further characterized the autophagy-related membrane structures generated in siAtg2A/B-treated cells by membrane floatation assay on OptiPrep density gradients (Figure 4). PNS fractions were subjected to equilibrium density gradient centrifugation, and membrane fractions were floated up to the top fractions. When the control cells were cultured in starvation medium containing bafilomycin A1, GFP-LC3-II and p62 were floated up to two peaks: fractions 8 and 9 and fractions 11 and 12 (Figure 4C). GFP-LC3-II and p62 in fractions 11 and 12 were highly resistant to proteinase K, whereas they were more sensitive to proteinase K in fractions 8 and 9 (Figure 4D). These results suggest that complete autophagosomes were mostly recovered in fractions 11 and 12 and partially in fractions 8 and 9. On the other hand, only small amounts of GFP-LC3 and p62 were floated up in control cells cultured under nutrient-rich and starvation conditions (Figure 4, A and B). In Atg2A/B-depleted cells, GFP-LC3-II was floated up to the two-peak fractions (8 and 9; 11 and 12) even under nutrient-rich conditions (Figure 4E). Following starvation, the peak around fractions 11 and 12 appeared to shift to a new peak at fractions 13–15, suggesting that aberrant autophagosome-related membranes were induced by starvation (Figure 4F). The amounts of p62 floated up with GFP-LC3 were less than those in control cells, suggesting that p62 cannot remain on these aberrant autophagic structures (Figure 4, C–F). All these morphological and biochemical experiments suggest that unclosed autophagosome-related membranes accumulate in Atg2A/B-depleted cells.
FIGURE 4:
Aberrant membrane structures accumulate in Atg2A/B-depleted cells. HeLa cells expressing GFP-LC3 were treated with control siRNA (A–D) or mixture of siRNAs against Atg2A and Atg2B (E–F) and cultured in regular DMEM (A, E), starvation medium (B, F), or starvation medium with bafilomycin A1 (C, D). PNS fractions were layered at the bottom of the OptiPrep density gradient and fractionated by centrifugation as illustrated in G. Fractions in C were treated with proteinase K after fractionation (D). The fractions were analyzed by SDS–PAGE and immunoblotting using anti-GFP and anti-p62 antibodies. N and P, nonfloat and PNS fractions, respectively.
Atg2A localizes to the isolation membrane and lipid droplet
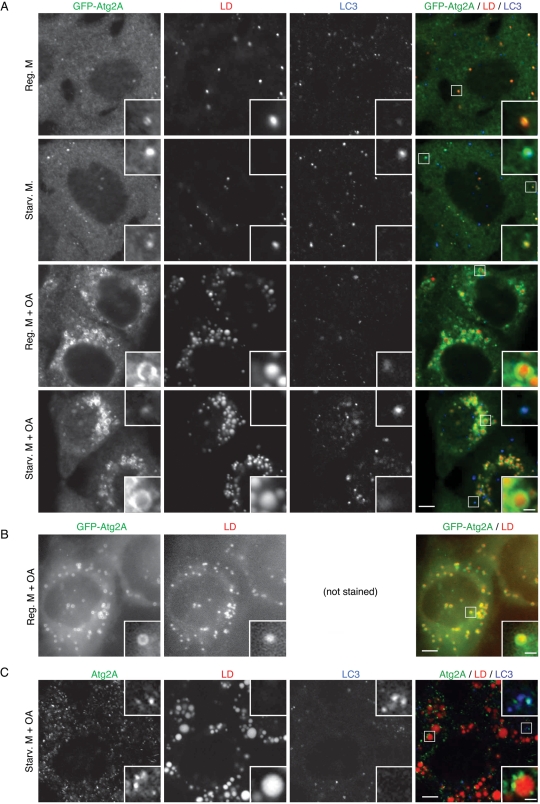
Having demonstrated that Atg2 is essential for autophagy, we next determined the intracellular localization of Atg2A in HeLa cells. Exogenously expressed GFP-Atg2A forms many small punctate structures even under nutrient-rich conditions, with the number increasing only slightly during starvation (Figure 5A). Staining of endogenous LC3 showed that only a small population of GFP-Atg2A puncta colocalized with LC3, and majority of GFP-Atg2A structures were negative for LC3. We therefore tested various organelle markers and realized that most GFP-Atg2A puncta colocalized with lipid droplets. When we incubated cells with oleic acid, lipid droplets, which were stained with labeled fatty acids (BODIPY 558/568-C12), increased both in size and number, and GFP-Atg2A was observed on the surface of these lipid droplets both under nutrient-rich and starvation condition (Figure 5A). The shape of Atg2A signals was irregular, but it seemed to be affected during fixation and antibody staining; when we took live images of these cells without fixation and antibody staining, GFP-Atg2A smoothly surrounded the surface of lipid droplets (Figure 5B). Endogenous Atg2A was also detected on the surface of lipid droplets as small patches (Figure 5C). Similar to GFP-Atg2A, endogenous Atg2A partially colocalized with LC3 (Figure 5C). Costaining of Atg2A with LC3 or lipid droplets was specific because siAtg2A abolished these costaining signals (we still observed many small puncta in siAtg2A-treated cells, indicating that these are nonspecific reactions; Supplemental Figure S2).
FIGURE 5:
Atg2A is present on both autophagic structures and lipid droplets. (A, B) HeLa cells stably expressing GFP-Atg2A were cultured in the presence or absence of oleic acid-BSA (OA) for 16 h and then incubated in regular DMEM or starvation medium without oleic acid for 2 h. Cells were incubated with BODIPY 558/568-C12 during the last 1 h of culture. Cells were fixed, permeabilized, stained with anti-GFP and anti-LC3 antibodies, and examined by confocal microscopy (A). Cells were also directly observed by fluorescence microscopy without fixation and antibody staining (B). (C) HeLa cells were cultured in the presence of oleic acid-BSA for 24 h and then starved for 2 h. Cells were stained with BODIPY 558/568-C12 as in A. Endogenous Atg2A and LC3 were detected with anti-Atg2A and anti-LC3 antibodies. Signal color is indicated by color of typeface. Scale bars, 5 μm, and 1 μm in inset (A–C).
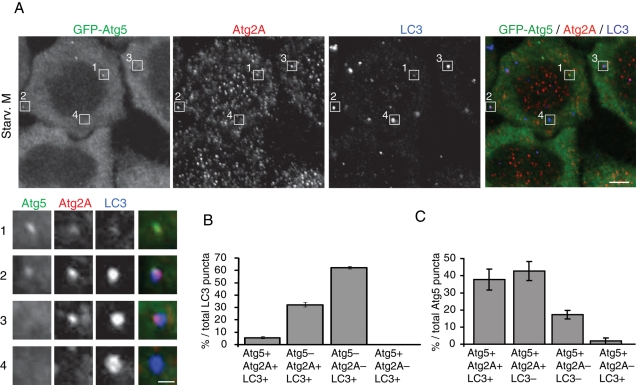
Next we determined the stage of autophagosome formation at which Atg2A is recruited to the autophagic structures. Approximately 40% of LC3 puncta were positive for Atg2A, indicating that Atg2A is not always present together with LC3 (Figure 6, A and B). About 14% of the Atg2A+LC3+ structures were positive for Atg5, a typical isolation membrane marker (Mizushima et al., 2001), suggesting that Atg2A remains on the isolation membrane or autophagosome even after Atg5 dissociates from the structures (Figure 6, A and B). By contrast, we observed that ∼20% of the Atg5-positive puncta were negative for both Atg2 and LC3, and ∼50% of the Atg5- and Atg2-double positive puncta were negative for LC3 (Figure 6, A and C), suggesting that Atg2 is recruited to the autophagosome formation site later than Atg5 but before LC3. Autophagosome formation seems not to be directly related to lipid droplets because Atg2A–LC3–lipid droplet triple colocalization was rarely observed (Figure 5C). Taken together these data show that Atg2A is a unique Atg protein that localizes to both isolation membranes (possibly also to early autophagosomes) and lipid droplets.
FIGURE 6:
Atg2 is recruited to the autophagosome formation site later than Atg5 but before LC3. HeLa cells stably expressing GFP-Atg5 were cultured in starvation medium for 2 h. Cells were stained with anti-GFP, anti-Atg2A, and anti-LC3 antibodies (A). The ratios (%) of Atg2−Atg5−, Atg2+Atg5−, Atg2−Atg5+, and Atg2+Atg5+ populations to the total LC3-positive structures (B) and of Atg2−LC3−, Atg2+LC3−, Atg2−LC3+, and Atg2+LC3+ populations to the total Atg5-positive structures (C) were calculated from 10 cells. Data represent mean ± SE of three independent cultures. Signal color is indicated by color of typeface. Scale bars, 5 μm, and 1 μm in inset.
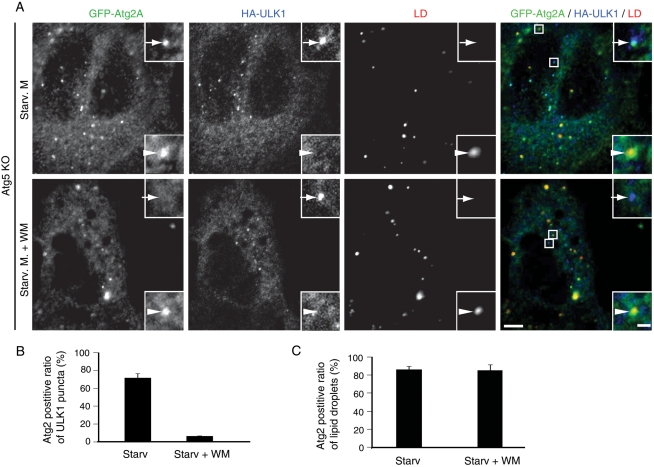
We previously reported the genetic hierarchy of mammalian Atg proteins in terms of recruitment to the autophagosome formation site (Itakura and Mizushima, 2010). We performed similar analysis for Atg2A. Atg2A colocalized with ULK1, an upstream Atg protein, in Atg5-knockout (KO) mouse embryonic fibroblasts (MEFs) under starvation conditions (Figure 7A). As we previously reported, inhibition of PtdIns 3-kinase with wortmannin did not affect ULK1 puncta formation, but it inhibited recruitment of Atg2A to the ULK1 structures (Figure 7, A and B). Thus recruitment of Atg2A to the autophagosome formation depends on PtdIns 3-kinase but not on Atg5. This hierarchical position of Atg2A is exactly the same as that of WIPI1 in mammals (Itakura and Mizushima, 2010) and Atg18 in yeast (Suzuki et al., 2007), suggesting that the WIPI proteins could be important for Atg2 recruitment.
FIGURE 7:
Targeting of Atg2A to autophagic structures depends on PI3K activity but not on Atg5, and that to lipid droplets depends on neither PI3K activity nor Atg5. Atg5 KO MEFs stably expressing GFP-Atg2A and HA-ULK1 were cultured in starvation medium containing BODIPY 558/568-C12 with or without 0.2 μM wortmannin for 1 h. Cells were subjected to immunofluorescence microscopy using anti-GFP and anti-HA antibodies. Arrows indicate ULK1-positive structures, and arrowheads indicate lipid droplets. Signal color is indicated by color of typeface (A). Atg2A-positive ratio of total ULK1 structures (B) and that of total lipid droplets (C) were quantified from 10 cells. Data represent mean ± SE of three independent experiments. Scale bars, 5 μm and 1 μm in inset.
In contrast, localization of Atg2A to lipid droplets was not affected by 1-h and overnight wortmannin treatment (Figure 7, A and B, and data not shown) and did not depend on other Atg proteins such as focal adhesion kinase interacting protein of 200 kDa (FIP200) and Atg5 (Supplemental Figure S3).
Amino acids 1723–1829 of Atg2A are essential for localization to both lipid droplets and autophagosomes and are required for autophagy
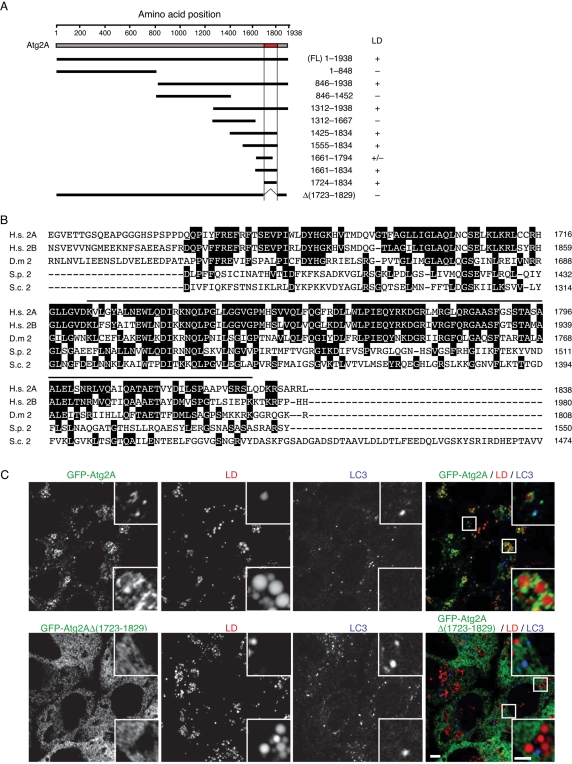
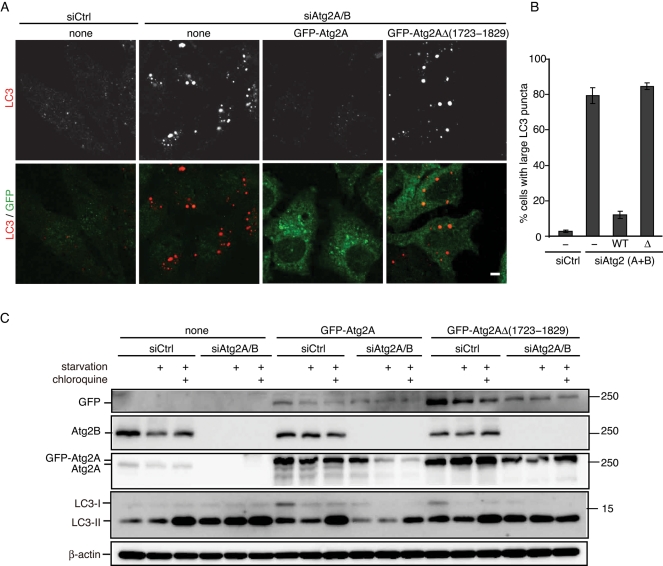
We determined which region of Atg2A is required for targeting lipid droplets using Atg2-deletion mutants. A short fragment containing amino acids 1723–1829 was both required and sufficient for association with lipid droplets (Figure 8, A and C, and Supplemental Figure S4). This region has no sequence similarity with known lipid droplet–binding proteins but is highly conserved in Atg2B (Figure 8B). Consistently, the corresponding region of Atg2B (1863–1982) also has affinity to lipid droplets (Supplemental Figure S4). This region also shows relatively high similarity to Atg2 homologues in other organisms (Figure 8B), suggesting that this region should have some evolutionarily conserved function. In fact, this region is also important for localization to the autophagic membrane; an Atg2A mutant lacking this region—GFP-Atg2AΔ(1723–1829)—was able to colocalize with neither lipid droplets nor LC3 structures (Figure 8C).
FIGURE 8:
Amino acids 1723–1829 of Atg2A are essential for localization to both lipid droplets and autophagosomes. (A) GFP-tagged Atg2A fragments were expressed in HeLa cells, and their colocalization with lipid droplets was examined. Representative images are shown in Supplemental Figure S4. (B) Amino acid alignment of human Atg2A (NP_055919) with human Atg2B (NP_060506), Drosophila melanogaster Atg2 (NP_647748), Schizosaccharomyces pombe Atg2 (XP_001713120), and S. cerevisiae Atg2 (NP_014157). The alignment was generated using CLUSTAL W. Identical residues are indicated with filled boxes. The black line above the human Atg2A sequence shows the minimal region required for lipid droplet binding (1723–1829). (C) HeLa cells stably expressing wild-type GFP-Atg2A or its mutant (GFP-Atg2A Δ1723–1829) were cultured in regular medium containing oleic acid-BSA for 24 h and then in starvation medium containing BODIPY 558/568-C12) for 1 h. Cells were fixed, stained with anti-GFP and anti-LC3 antibodies, and subjected to confocal microscopy. Signal color is indicated by color of typeface. Scale bars 5 μm, and 2 μm in inset.
We therefore determined whether the lipid droplet–targeting domain of Atg2 is essential for autophagy. As shown in Figures 1C and 2, silencing of both Atg2A and Atg2B caused accumulation of large LC3 aggregates, and it was suppressed by reexpression of GFP-Atg2A (Figure 9, A and B). However, reexpression of GFP-Atg2AΔ(1723–1829) showed no effect. Similarly, reexpression of GFP-Atg2A, but not of GFP-Atg2AΔ(1723–1829), restored autophagic flux in siAtg2A/B-treated cells (Figure 9C). Thus the Atg2A region containing amino acids 1723–1829 is important not only for targeting to lipid droplets, but also for autophagy.
FIGURE 9:
Amino acids 1723–1829 of Atg2A are required for autophagy. (A) HeLa cells stably expressing either siRNA-resistant wild-type GFP-Atg2A or GFP-Atg2A Δ1723–1829 were treated with control siRNA or a mixture of siRNAs against Atg2A and Atg2B. Cells grown in regular medium were fixed and subjected to immunofluorescence microscopy using anti-LC3 and anti-GFP antibodies. Scale bar, 5 μm. (B) Ratio of cells containing large LC3 punctate structures was quantified. Data represent mean ± SE of three independent treatments with siRNA. (C) HeLa cells used in A were cultured in regular DMEM or starvation medium in the presence or absence of 20 μM of chloroquine for 2 h. Cell lysates were analyzed by immunoblotting using the indicated antibodies.
Depletion of Atg2A/B causes aggregation of enlarged lipid droplets
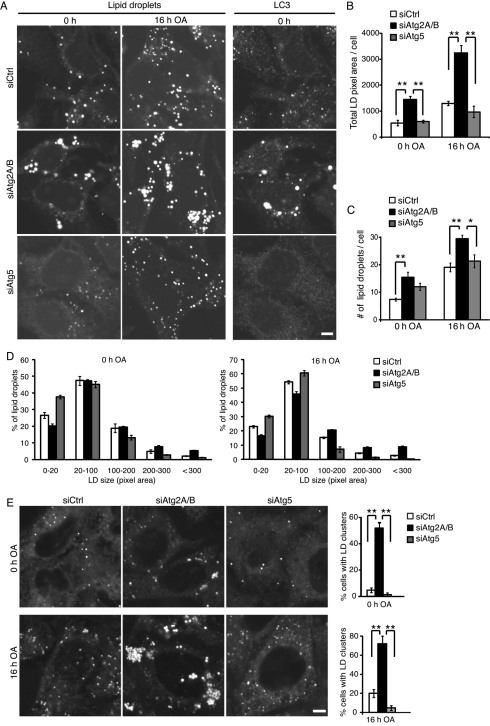
Because mammalian Atg2 was detected on lipid droplets, we analyzed the role of Atg2 in lipid droplet formation and turnover. In control siRNA-treated cells, small lipid droplets were observed, which increased both in number and size following 16 h of treatment with oleic acid (Figure 10, A–C). On the other hand, in siAtg2A/B-treated cells, the size and number of lipid droplets increased even before oleic acid treatment (total pixel area of lipid droplets was approximately twofold higher than that of control cells; Figure 10, A–C). Some lipid droplets were markedly enlarged (Figure 10, A and D). Both total volume and number of lipid droplets further increased following oleic acid treatment (Figure 10, A–C). Of note, the distribution of lipid droplets was also altered. Lipid droplets are usually dispersed in control cells, but they tended to stick to each other and formed clusters (Figure 10, A and E, and Supplemental Movies 1 and 2). Successful knockdown of Atg2A/B was confirmed by immunoblotting (Supplemental Figure S5) and accumulation of large LC3 structures (Figure 10A).
FIGURE 10:
Silencing of Atg2A/B causes aggregation of enlarged lipid droplets. (A–D) HeLa cells were treated with the indicated siRNAs for 5 d and cultured without (0 h) or with oleic acid (16 h). Cells were fixed, stained with Sudan III (2 mg/ml), and immediately analyzed by fluorescence microscopy (B–D), or stained with anti-LC3 antibody and Sudan III and analyzed by confocal microscopy (A). Total lipid droplet pixel area (B) and the number of lipid droplets (C) were quantified in 20 randomly selected cells. Histogram analysis of size variation of 1000 lipid droplets in each group is shown (D). Data represent mean ± SE of three independent treatments with siRNA and oleic acid. Statistical difference determined by one-way analysis of variance (ANOVA) with Bonferroni–Dunn posttest (*p < 0.05, **p < 0.01). Scale bar, 5 μm (A). (E) Quantification of cluster formation of lipid droplets. Cells used in A–C were cultured without (0 h) or with oleic acid (16 h). After additional 1 h incubation with BODIPY 558/568-C12, the BODIPY signals were directly captured by confocal microscopy without fixation to avoid artificial disruption of lipid droplet clusters during the fixation processes. Ratio of cells having lipid droplet clusters was quantified from 50 randomly selected cells as described in Materials and Methods. Data represent mean ± SE of three independent treatments with siRNA and oleic acid. Statistical difference determined by one-way ANOVA with Bonferroni–Dunn posttest (**p < 0.01). Scale bar, 5 μm.
Because autophagy was found to be involved in degradation of lipid droplets in hepatocytes (known as lipophagy; Singh et al., 2009), we investigated whether the increased volume of lipid droplets in siAtg2A/B-treated HeLa cells was simply due to a defect of lipophagy or was a specific phenotype for Atg2 depletion. We performed a similar analysis using siRNA directed against Atg5, another essential factor for autophagy. As Atg5 is essential for LC3—PE conjugation, formation of LC3 puncta (Figure 10A) and LC3-II (Supplemental Figure S5) was suppressed in siAtg5-treated cells, which confirmed successful depletion of Atg5. However, the total volume of lipid droplets was not affected by Atg5 knockdown either before or after oleic acid treatment, although small lipid droplets might be slightly increased in number (Figure 10, A–D). Thus lipophagy in HeLa cells may not be as active as that reported in hepatocytes and MEFs (Singh et al., 2009). Furthermore, lipid droplets were normally dispersed in Atg5-silencing cells (Figure 10, A and E). These data suggest that the effect of Atg2A/B on regulation of lipid droplet is independent of autophagic degradation, and Atg2A/B have a more direct role on lipid droplet morphology and distribution.
DISCUSSION
We showed that mammalian Atg2A and Atg2B have redundant functions and both are required for autophagosome formation. Aberrant or intermediate autophagic structures, which are not completely closed, accumulate in Atg2A/B-silencing cells under growing conditions (Figures 3 and 4). Atg9A, ULK1, Atg14, WIPI1, Atg5, and LC3 localize to these structures (Figure 2). These data are generally consistent with results in yeast cells (Suzuki et al., 2007).
Accumulation of most Atg proteins to punctate structures has also been observed in VMP1-knockdown cells, in which ULK1, WIPI1, DFCP1, Atg16L1, and LC3 form puncta under nutrient-rich conditions (Itakura and Mizushima, 2010; Tian et al., 2010). Phenotypic similarity between Atg2 and VMP1 mutants is also observed in Caenorhabditis elegans; accumulation of large puncta of LGG-1 (an Atg8/LC3 homologue) is observed in both epg-3/VMP1 and atg-2 mutants (Tian et al., 2010). Thus Atg2 and VMP1 may function at a similar step—for example, assembly of different membrane sources, maturation of the omegasome, or completion of isolation membrane closure, but not at initial steps to recruit other Atg proteins.
Our hierarchical analysis shows that localization of mammalian Atg2 depends on PtdIns 3-kinase activity but not on Atg5 (Figure 7). This hierarchical position is exactly the same as that of WIPI1 (Itakura and Mizushima, 2010), confirming the previous reports that Atg2 interacts with the Atg18/WIPI-family proteins in S. cerevisiae (Suzuki et al., 2007), C. elegans (Lu et al., 2011), and mammals (Behrends et al., 2010). Recent studies showed that Atg18/WIPIs are required for omegasome maturation both in mammals and C. elegans (Polson et al., 2010; Lu et al., 2011). However, their functions appear to differ among the isoforms. C. elegans mutants of apg-18 and epg-6 show accumulation of LGG-1 puncta, but Atg-18 (a WIPI1/2 homologue) appears to function upstream of Epg-6 (a WIPI4 homologue; Lu et al., 2011). In addition, in mammalian cells, silencing of WIPI2—a major WIPI form in HEK293A cells—suppresses LC3 puncta formation (Polson et al., 2010), whereas silencing of WIPI4 in NRK cells leads to accumulation of LC3 puncta (Lu et al., 2011). It would be important to dissect the function of mammalian WIPI family proteins in more depth and determine whether Atg2 proteins are required for all functions.
One novel aspect of the present study is the finding that mammalian Atg2 proteins are present on lipid droplets, which are lipid storage organelles present in essentially all cell types (Martin and Parton, 2006; Walther and Farese, 2009; Beller et al., 2010; Digel et al., 2010). Lipid droplets consist of a hydrophobic core containing neutral lipids (mainly triglyceride and cholesterol ester) and a surface monolayer of phospholipids and cholesterol. Although it is generally believed that they are generated on the ER, their biogenesis and turnover involve complicated processes, which are not yet completely understood. Although several proteins, such as PAT proteins (e.g., perilipin, adipose differentiation-related protein, and TIP47) and Rab18 localize on the surface of lipid droplets, no autophagy-related proteins have been identified on the surface. We identified the Atg2 region required for lipid droplet targeting (Figure 8), but it does not show any homology with known lipid droplet proteins. Despite the clear association of Atg2 with lipid droplet, we could not detect endogenous Atg2A/B in the lipid droplet fractions obtained by membrane floatation method (Nishimura and Mizushima, unpublished data). This suggests that Atg2 proteins only weakly associate with lipid droplets and dissociate during the purification procedure or that Atg2 associates with some structures close to lipid droplets. Further experiments will be required to test whether Atg2 proteins directly or indirectly associate with lipid droplets. In contrast to targeting to the autophagic structures, Atg2 localization to lipid droplets seems to be independent of other Atg proteins (Figure 7 and Supplemental Figure S3). We also found that the WIPI4-binding region of Atg2A is distinct from the lipid droplet–binding region (Oita and Mizushima, unpublished data), suggesting that association of Atg2 with lipid droplets is not mediated by WIPIs. The Atg-independent targeting of Atg2 to lipid droplets might also explain why exogenously expressed Atg2 associates with lipid droplets better than with autophagic structures (Figure 5).
Although we found clear change in lipid droplet morphology and dispersion in Atg2-silencing cells, the precise function of Atg2 in this process is unknown, particularly in relationship to autophagosome formation. Because knockdown of Atg5 (Figure 10) does not cause clustering of enlarged lipid droplet under the same conditions, it is not simply due to a defect in lipophagy. Lipophagy activity may be different between cell types and tissues, or prolonged suppression may cause more obvious changes as observed in vivo. Apart from the degradation step, the lipid droplet phenotype observed in Atg2-silencing cells could be explained at the formation step. A genome-wide screen in Drosophila S2 cells identified many proteins involved in regulation of size and dispersion of lipid droplets, which were classified into five groups (Guo et al., 2008). Mutant cells defective in phospholipid metabolism and COPI vesicle formation exhibit larger lipid droplets. Although these do not have exactly the same phenotype as observed in Atg2-deficient cells, the data suggest that defects in the ER and/or Golgi apparatus could affect lipid droplet morphology. In particular, phosphatidylcholine appears to be one of the major regulators (Krahmer et al., 2011; Walker et al., 2011). Considering that the ER is involved in formation of both the autophagosome and lipid droplet, it is reasonable to hypothesize that a common Atg2 function is shared by the two pathways. It would be important to test whether other Atg proteins play roles in formation of lipid droplets, as well as in other processes occurring in the ER.
MATERIALS AND METHODS
Plasmids, antibodies, and reagents
cDNA encoding human Atg2A (KIAA0404) was obtained from Kazusa DNA Research Institute (Kisarazu, Japan). The full-length or fragments of Atg2A open reading frame were amplified by PCR and subcloned into pEGFP-C1 (Clontech, Mountain View, CA). To generate antibodies against human Atg2A protein, a cDNA fragment of Atg2A (amino acid residues 700–1400) was subcloned into pET28a vector (Novagen, EMD4Biosciences, Gibbstown, NJ). The bacterially expressed protein was recovered into insoluble inclusions, extracted from SDS–PAGE gel, and used to immunize rabbits. Antisera were affinity purified. Anti–human Atg2B antibody (HPA 019665) was purchased from Sigma-Aldrich (St. Louis, MO). A siRNA-resistant Atg2A silent mutant was created by substituting four nucleotides in the Atg2A siRNA-targeting region (G3267A, T3268C, G3270T, and G3273A).
Rabbit polyclonal anti-Atg16L1 (Mizushima et al., 2003), anti-Atg5 (Mizushima et al., 2001), anti-Atg9A (Itakura et al., 2012), anti-GFP (A6455; Invitrogen, Carlsbad, CA), anti-LC3 (Hosokawa et al., 2006; L7543; Sigma-Aldrich), anti-p62 (PM045; MBL, Woburn, MA), anti-Tom40 (Suzuki et al., 2000), and anti-LAMP1 (Niwa et al., 2003) antibodies, guinea pig polyclonal anti-p62 antibody (GP62-C; Progen, Heidelberg, Germany), mouse monoclonal anti-LC3 (CTB-LC3-2-1C, clone 1703; Cosmo Bio Co., Tokyo, Japan), anti-Atg16L1 (IF12; MBL), anti-Hsp90 (BD Biosciences, San Diego, CA), anti–β actin (clone AC74; Sigma-Aldrich), and anti-HA (16B12; Covance, Berkeley, CA) antibodies, and rat monoclonal anti-GFP antibody (Nacalai Tesque, GF090R) were used. For immunostaining, Alexa Fluor 488–conjugated anti–rat immunoglobulin G (IgG) and anti–rabbit IgG, Alexa Fluor 564–conjugated anti–mouse IgG, and Alexa Fluor 660–conjugated anti–mouse IgG secondary antibodies (Molecular Probes, Invitrogen) were used.
E64d and pepstatin A were purchased from Peptide Institute (Osaka, Japan), proteinase K was purchased from Merck (Darmstadt, Germany), bafilomycin A1 was purchased from Wako Chemicals (Osaka, Japan), and chloroquine and wortmannin were purchased from Sigma-Aldrich.
Cell culture and transfection
HeLa cells expressing GFP-LC3 (Hosokawa et al., 2009) have been published. HeLa cells stably expressing GFP-ULK1, GFP-Atg14, GFP-WIPI1, and GFP-Atg5 were generated using mCAT-expressing HeLa cells (a gift from S. Hatakeyama [Hokkaido University]; Bohgaki et al., 2008) and retroviral transfection systems (Kitamura et al., 2003) using pMXs-IP-GFP-ULK1 (Hara et al., 2008), pMXs-IP-GFP-Atg14 (Itakura et al., 2008), pMXs-puro-GFP-WIPI1 (Itakura and Mizushima, 2010), and pMXs-IP-GFP-Atg5 (Hara et al., 2008), respectively. HeLa cells stably expressing GFP-Atg2A and Atg2AΔ1723–1829 were generated as follows: HEK293T cells were transiently transfected using Lipofectamine 2000 (Invitrogen) reagent with pMXs-IP-GFP-Atg2A or pMXs-IP-GFP-Atg2AΔ1723–1829 together with pCG-VSV-G and pCG-gag-pol to render retroviruses infectious to human cells. Wild-type MEFs, FIP200 KO MEFs (Gan et al., 2006), and Atg5 KO MEFs (Kuma et al., 2004) were also transfected using pMXs-IP-GFP-Atg2A–derived retrovirus. Cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) in a 5% CO2 incubator. For starvation, cells were washed with phosphate-buffered saline (PBS) and incubated in amino acid-free DMEM without FBS (starvation medium).
RNA interference
Stealth RNA interference (RNAi) oligonucleotides were used for RNAi experiments (Invitrogen). The target sequences were as follows: human Atg2A siRNA (target position: 3255–3279) 5′-gcauucccaguuguuggaguuccua-3′, human Atg2B siRNA (target position: 4655–4679) 5′-aggucucucuugucuggcaucuuua-3′, and human Atg5 siRNA (target position: 35–59) 5′-gguuuggacgaauuccaacuuguuu-3′. For the negative control, medium GC duplex of Stealth RNAi (Invitrogen) or siRNA against luciferase (5′-cgcggucgguaaaguuguuccauuu-3′) was used. The Stealth RNAi oligonucleotides were transfected into cells using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's protocols. After 2 d, the cells were again transfected with the same siRNA and cultured for an additional 3 d before analysis.
Flow cytometry
Cells were harvested with 0.05% trypsin-EDTA (Gibco, Life Technologies, Carlsbad, CA) and diluted with PBS. The samples were analyzed using a FACSCalibur HG flow cytometer (BD Biosciences). Data were analyzed using BD CellQuest Pro software (BD Biosciences).
Fluorescence microscopy
For fluorescence microscopy, HeLa cells expressing GFP-fused protein were cultured on a glass-bottomed dish and directly observed. For examination by immunofluorescence microscopy, cells grown on gelatin-coated coverslips were fixed with 4% paraformaldehyde, permeabilized using 50 or 100 μg/ml digitonin, and then stained with specific antibodies as previously described (Itakura et al., 2008). These cells were observed with a fluorescence microscope (IX81; Olympus, Tokyo, Japan) equipped with a charged-coupled device camera (ORCA ER; Hamamatsu Photonics, Hamamatsu, Japan). A 60× PlanApo oil immersion lens (1.42 numerical aperture; Olympus) was used. Images were acquired using MetaMorph image analysis software, and the size and number of lipid droplets were quantified using the Top Hat program (Molecular Devices, Sunnyvale, CA). For confocal microscopy, cells were examined under a confocal laser microscope (FV1000D IX81; Olympus), using a 60× PlanApoN oil immersion lens (1.42 numerical aperture; Olympus). For live-cell imaging, the culture dish was mounted in a chamber (Tokai Hit, Shizuoka, Japan) to maintain incubation conditions at 37°C.
Subcellular fractionation and proteinase K treatment
HeLa cells were suspended in homogenization buffer (10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES]-KOH, pH 7.4, 0.22 M mannitol, 0.07 M sucrose, and protease inhibitors) and homogenized with 10 strokes using a syringe with 27-gauge needle. The PNS was recovered by centrifugation at 300 × g for 5 min. The lysate was spun at 7700 × g for 5 min to separate the LSP, and the supernatant was centrifuged again at 100,000 × g for 30 min to generate an HSP and HSS. The LSP and HSP were resuspended in the same buffer and washed. To analyze detergent solubility, each sample was incubated with 1% Triton X-100 on ice for 30 min and then centrifuged at 100,000 × g for 30 min. To examine proteinase K sensitivity, each fraction was treated with 100 μg/ml proteinase K on ice for 20 min with or without 0.5% Triton X-100. The samples were precipitated with 10% trichloroacetic acid, washed once with ice-cold acetone, resuspended in SDS–PAGE sample buffer, immediately boiled, and analyzed by SDS–PAGE.
For density-gradient centrifugation, OptiPrep solutions (Axis-Shield PoC, Oslo, Norway) were prepared in 20 mM HEPES-KOH, pH 7.4, supplemented with 1 mM EDTA and protease inhibitors. The PNS fractions containing 30% OptiPrep (3 ml) were layered at the bottom of a 12-ml centrifuge tube (Beckman, Brea, CA), and then density gradients were prepared as follows: 1.5 ml of 25%, 2 ml of 20%, 2 ml of 15%, 1.5 ml of 10%, and 1 ml of 5%. The gradients were centrifuged at 107,680 × g for 12 h at 4°C using a swing rotor SW40 in a Beckman L90 centrifuge with slow acceleration and slow brake. The centrifuged solution was separated for each 0.5-ml fraction and subjected to immunoblotting.
Preparation of the oleic acid–albumin solution
Oleic acid was conjugated to albumin as previously described (Spector and Hoak, 1969). Oleic acid (Nacalai Tesque, Kyoto, Japan) was dissolved in hexane and added to Celite Hyflo Super-Cel (Nacalai Tesque), and the solvent was evaporated under nitrogen. Oleic acid–coated Celite was incubated with 6.7% fatty acid–free bovine serum albumin (BSA; Nacalai Tesque) in PBS for 30 min at room temperature. The suspension was stirred slowly with a magnetic bar. Celite was removed by centrifugation at 4°C at 9900 × g for 20 min. The supernatant solution was passed through a Millipore filter of pore size 0.22 μm to ensure complete removal of the Celite. To promote lipid droplet formation, cells were cultured with 100–1000 μM oleic acid.
Lipid droplet staining and quantification of clustering
For costaining experiments, live cells were cultured with either 1 μg/ml (in regular medium containing 10% serum) or 0.01 μg/ml (in starvation medium) BODIPY 558/568-C12 (Molecular Probes) for 1 h. To stain lipid droplets in fixed samples, Sudan III Cerasin Red (Sigma-Aldrich) prepared in 70% ethanol was directly added to paraformaldehyde-fixed cells at 2 mg/ml for 20 min. For quantification of lipid droplets, live cells stained with BODIPY 558/568-C12 were directly subjected to confocal microscopy and analyzed. Lipid droplet cluster was defined as 1) accumulation of >3 lipid droplets in a circular region 2 μm in diameter in the absence of exogenous oleic acid or 2) accumulation of >10 lipid droplets in a circular region 5 μm in diameter following 16-h- oleic acid treatment. Cells having these lipid droplet clusters were counted.
Statistical analysis
Differences were statistically analyzed using analysis of variance with Bonferroni–Dunn posttest.
Supplementary Material
Acknowledgments
We thank Jun-Lin Guan for providing FIP200 KO MEFs, Shigetsugu Hatakeyama and Kentaro Hanada for the mCAT-1 plasmid and the mCAT-1–expressing HeLa cell line, Teruhito Yasui for the pCG-VSV-G and pCG-gag-pol plasmids, Yoshitaka Tanaka for anti-Lamp1 antibody, Hisashi Ichikawa for his help in cloning of human Atg2A, Taichi Hara and Eisuke Itakura for preparing HeLa stable transformants, and M. Ohashi for preparation of oleic acid solution. This work was supported in part by the Funding Program for Next Generation World-Leading Researchers and Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to N.I. and N.M.) and by the Naito Foundation (to T.N.).
Abbreviations used:
- Atg
autophagy related
- DFCP1
double FYVE-containing protein 1
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- LC3
microtubule-associated protein light chain 3
- MEF
mouse embryonic fibroblast
- PAS
preautophagosomal structure
- PE
phosphatidylethanolamine
- PtdIns 3-kinase
phosphatidylinositol 3-kinase
- VMP1
vacuole membrane protein 1
- WIPI
WD-repeat protein interacting with phosphoinoside
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-09-0785) on January 4, 2012.
REFERENCES
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beller M, Thiel K, Thul PJ, Jackle H. Lipid droplets: a dynamic organelle moves into focus. FEBS Lett. 2010;584:2176–2182. doi: 10.1016/j.febslet.2010.03.022. [DOI] [PubMed] [Google Scholar]
- Bohgaki M, Tsukiyama T, Nakajima A, Maruyama S, Watanabe M, Koike T, Hatakeyama S. Involvement of Ymer in suppression of NF-kappaB activation by regulated interaction with lysine-63-linked polyubiquitin chain. Biochim Biophys Acta. 2008;1783:826–837. doi: 10.1016/j.bbamcr.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell. 2008;15:344–357. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digel M, Ehehalt R, Fullekrug J. Lipid droplets lighting up: insights from live microscopy. FEBS Lett. 2010;584:2168–2175. doi: 10.1016/j.febslet.2010.03.035. [DOI] [PubMed] [Google Scholar]
- Dove SK, et al. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 2004;23:1922–1933. doi: 10.1038/sj.emboj.7600203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AR, Zurek N, Voeltz GK. Peripheral ER structure and function. Curr Opin Cell Biol. 2009;21:596–602. doi: 10.1016/j.ceb.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagone P, Jackowski S. Membrane phospholipid synthesis and endoplasmic reticulum function. J Lipid Res. 2009;50:S311–S316. doi: 10.1194/jlr.R800049-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B, Peng X, Nagy T, Alcaraz A, Gu H, Guan JL. Role of FIP200 in cardiac and liver development and its regulation of TNFα and TSC–mTOR signaling pathways. J Cell Biol. 2006;175:121–133. doi: 10.1083/jcb.200604129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453:657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- Holthuis JC, Levine TP. Lipid traffic: floppy drives and a superhighway. Nat Rev Mol Cell Biol. 2005;6:209–220. doi: 10.1038/nrm1591. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Hara Y, Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 2006;580:2623–2629. doi: 10.1016/j.febslet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009;5:973–979. doi: 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C, Koyama-Honda I, Mizushima N. Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. J Cell Sci. 2012 doi: 10.1242/jcs.094110. (in press) [DOI] [PubMed] [Google Scholar]
- Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Koshino Y, Shibata F, Oki T, Nakajima H, Nosaka T, Kumagai H. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol. 2003;31:1007–1014. [PubMed] [Google Scholar]
- Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy. 2008;4:849–950. doi: 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- Kovács AL, Pálfia Z, Réz G, Vellai T, Kávacs J. Sequestration revisited: integrating traditional electron microscopy, de novo assembly and new results. Autophagy. 2007;3:655–662. doi: 10.4161/auto.4590. [DOI] [PubMed] [Google Scholar]
- Krahmer N, et al. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 2011;14:504–515. doi: 10.1016/j.cmet.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine T, Rabouille C. Endoplasmic reticulum: one continuous network compartmentalized by extrinsic cues. Curr Opin Cell Biol. 2005;17:362–368. doi: 10.1016/j.ceb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Lu Q, Yang P, Huang X, Hu W, Guo B, Wu F, Lin L, Kovacs AL, Yu L, Zhang H. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev Cell. 2011;21:343–357. doi: 10.1016/j.devcel.2011.06.024. [DOI] [PubMed] [Google Scholar]
- Ma C, Agrawal G, Subramani S. Peroxisome assembly: matrix and membrane protein biogenesis. J Cell Biol. 2011;193:7–16. doi: 10.1083/jcb.201010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, Noda T, Yoshimori T. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190:511–521. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–667. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Niwa K, Tanaka R, Murase H, Ishikawa T, Fujita H, Himeno M, Tanaka Y. Two lysosomal membrane proteins, LGP85 and LGP107, are delivered to late endosomes/lysosomes through different intracellular routes after exiting from the trans-Golgi network. Biochem Biophys Res Commun. 2003;301:833–840. doi: 10.1016/s0006-291x(03)00046-9. [DOI] [PubMed] [Google Scholar]
- Obara K, Sekito T, Niimi K, Ohsumi Y. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J Biol Chem. 2008;283:23972–23980. doi: 10.1074/jbc.M803180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, Tooze SA. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506–522. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Shintani T, Suzuki K, Kamada Y, Noda T, Ohsumi Y. Apg2p functions in autophagosome formation on the perivacuolar structure. J Biol Chem. 2001;276:30452–30460. doi: 10.1074/jbc.M102346200. [DOI] [PubMed] [Google Scholar]
- Shvets E, Fass E, Elazar Z. Utilizing flow cytometry to monitor autophagy in living mammalian cells. Autophagy. 2008;4:621–628. doi: 10.4161/auto.5939. [DOI] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector AA, Hoak JC. An improved method for the addition of long-chain free fatty acid to protein solutions. Anal Biochem. 1969;32:297–302. doi: 10.1016/0003-2697(69)90089-x. [DOI] [PubMed] [Google Scholar]
- Stromhaug PE, Bevan A, Dunn WA., Jr GSA11 encodes a unique 208-kDa protein required for pexophagy and autophagy in Pichia pastoris. J Biol Chem. 2001;276:42422–42435. doi: 10.1074/jbc.M104087200. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Okazawa Y, Komiya T, Saeki K, Mekada E, Kitada S, Ito A, Mihara K. Characterization of rat TOM40, a central component of the preprotein translocase of the mitochondrial outer membrane. J Biol Chem. 2000;275:37930–37936. doi: 10.1074/jbc.M006558200. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Tian Y, et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 2010;141:1042–1055. doi: 10.1016/j.cell.2010.04.034. [DOI] [PubMed] [Google Scholar]
- Walker AK, et al. A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell. 2011;147:840–852. doi: 10.1016/j.cell.2011.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Farese RV., Jr The life of lipid droplets. Biochim Biophys Acta. 2009;1791:459–466. doi: 10.1016/j.bbalip.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CW, Kim J, Huang WP, Abeliovich H, Stromhaug PE, Dunn WA, Jr, Klionsky DJ. Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. J Biol Chem. 2001;276:30442–30451. doi: 10.1074/jbc.M102342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E, Karp C, Strohecker AM, Guo Y, Mathew R. Role of autophagy in suppression of inflammation and cancer. Curr Opin Cell Biol. 2010;22:212–217. doi: 10.1016/j.ceb.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.