Translation events in RNA granules in neurons are spatially clustered near individual granules, and translation output from individual granules is either sporadic or bursty. Translation of Venus-FMRP RNA is more bursty than translation of Venus-ARC RNA, and mGluR activation increases overall translation but decreases bursty translation.
Abstract
Dendritic RNAs are localized and translated in RNA granules. Here we use single-molecule imaging to count the number of RNA molecules in each granule and to record translation output from each granule using Venus fluorescent protein as a reporter. For RNAs encoding activity-regulated cytoskeletal-associated protein (ARC) or fragile X mental retardation protein (FMRP), translation events are spatially clustered near individual granules, and translational output from individual granules is either sporadic or bursty. The probability of bursty translation is greater for Venus-FMRP RNA than for Venus-ARC RNA and is increased in Fmr1-knockout neurons compared to wild-type neurons. Dihydroxyphenylglycine (DHPG) increases the rate of sporadic translation and decreases bursty translation for Venus-FMRP and Venus-ARC RNAs. Single-molecule imaging of translation in individual granules provides new insight into molecular, spatial, and temporal regulation of translation in granules.
INTRODUCTION
Local translation in dendrites provides a mechanism to convert ephemeral electrical signals at individual synapses into long-lasting structural and functional changes associated with learning and memory (Steward and Schuman, 2001). Dendritic RNAs are localized in granules containing multiple different RNA molecules, cognate RNA-binding proteins, and translational machinery (Gao et al., 2008). Conventional ensemble methods to measure translation lack sufficient molecular, spatial, and temporal resolution to determine where and when individual translation events occur in relation to individual granules. Here we describe a single-molecule imaging method using fluorescently tagged RNA to count RNA molecules in individual granules and Venus fluorescent protein (Nagai et al., 2002) as a reporter to record individual translation events. Each translation event produces a Venus protein molecule that undergoes rapid fluorophore maturation and subsequent photobleaching (Yu et al., 2006), resulting in a flash of fluorescence.
Venus is a variant of yellow fluorescent protein (YFP) that has more rapid fluorophore maturation than other fluorescent reporter proteins such as green fluorescent protein (GFP) because it contains multiple mutations (F64L/M153T/V163A/S175G/F46L) that increase tolerance to environmental changes, accelerate the rate of protein folding, and increase the rate of chromophore oxidation at 37°C. Time constants for Venus fluorophore maturation have been measured in vitro (t1/2 ∼ 2 min) and in bacterial cytoplasm (t1/2 ∼ 7 min; Xie et al., 2008). The time constant for Venus fluorophore maturation in hippocampal neurons has not been measured but, as shown here, is sufficiently rapid to detect increased translation rate within 1 min after synaptic stimulation in hippocampal neurons in culture.
After fluorophore maturation individual Venus fluorescent protein molecules undergo either photobleaching, in which the fluorophore undergoes a single-step transition to a stable low-emission state, or photoblinking, in which the fluorophore undergoes multiple single-step transitions between transient low-emission and high-emission states (McAnaney et al., 2005). If a newly synthesized Venus molecule undergoes fluorophore maturation followed by photobleaching, this provides a record of a single translation event. However, if a newly synthesized Venus molecule undergoes fluorophore maturation followed by multiple rounds of photoblinking, this could be erroneously interpreted as multiple independent translation events, resulting in overcounting. If the fluorescence signal from a single molecule is detected at the same location in sequential frames and if photoblinking events are relatively short (<1 s) and are preceded and followed by longer periods with no detectable signal, the short-duration photoblinking events can be ascribed to a single, newly synthesized molecule using an algorithm to identify short photoblinking events. However, longer-duration photoblinking events are difficult to distinguish from synthesis and photobleaching of new molecules. Because photoblinking is a probabilistic process, the frequency of long-duration photoblinking events is relatively low compared with the frequency of short-duration photoblinking events. Furthermore, as shown here, the rate of appearance of Venus molecules detected by single-molecule imaging in hippocampal neurons is affected by factors such as protein synthesis inhibitors, cis-acting sequences in different RNAs, trans-acting factors in different mutant neurons, antagonists of action potentials, and agonists of synaptic receptors that are known to affect translation rate but are not expected to affect photoblinking. These considerations suggest that single-molecule imaging of Venus molecules can be used to analyze translation events in hippocampal neurons, although the possibility that long-duration photoblinking events contribute a low level of background signal cannot be excluded. This approach was used to analyze times and locations of individual translation events for Venus–activity-regulated cytoskeletal-associated protein (ARC) and Venus–fragile X mental retardation protein (FMRP) RNA in individual granules in wild-type and Fmr1-knockout (KO) hippocampal neurons in culture treated with agonists and antagonists of synaptic activity.
ARC is an immediate early gene induced by synaptic stimulation (Lyford et al., 1995; Steward et al., 1998; Plath et al., 2006). ARC protein interacts with endophilin and dynamin to mediate endocytosis of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs; Chowdhury et al., 2006; Shepherd et al., 2006) in dendritic spines. Synaptic stimulation of group 1 metabotropic glutamate receptors (mGluRs) increases translation of ARC RNA in wild-type neurons (Park et al., 2008; Waung et al., 2008) but not in Fmr1 KO neurons (Castillo et al., 2008), suggesting that translation of ARC RNA is regulated by mGluR and FMRP. ARC RNA is localized in granules (Ostroff et al., 2002; Gao et al., 2008), but less than half of ARC RNA is associated with polysomes (Zalfa et al., 2003).
FMRP, encoded by the Fmr1 gene, is a translational repressor protein that regulates translation of several granule-associated RNAs, including ARC RNA and Fmr1 RNA itself (Bassell and Warren, 2008). In individuals with fragile X Syndrome (FXS) and in Fmr1 KO mice, lack of FMRP results in increased translation of FMRP-target RNAs (Park et al., 2008). Stimulation of group 1 mGluR results in transient activation of translation of FMRP target RNAs (Narayanan et al., 2007, 2008). FMRP RNA is localized in granules, and more than half of Fmr1 RNA is associated with polysomes (Zalfa et al., 2003).
RESULTS
Single-molecule imaging of translation in live cells
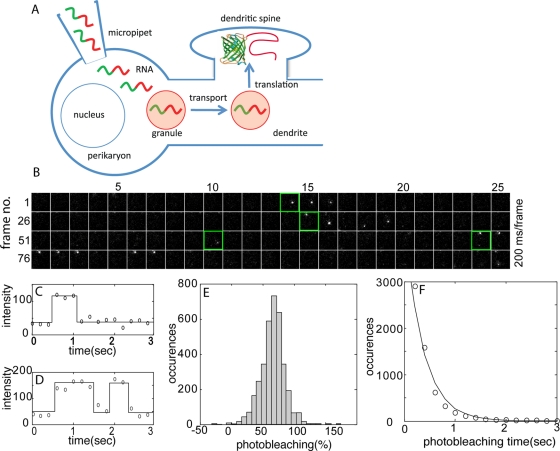
RNAs encoding Venus-ARC or Venus-FMRP were labeled by transcriptional incorporation of fluorescent ribonucleotide (Cy5 UTP) into the body of the RNA (∼10 fluorophores/RNA molecule). Labeled RNA was microinjected into the perikaryon of hippocampal neurons in culture. Fluorescent RNA was detected in granules and fluorescent Venus fusion protein was detected by single-molecule imaging (Figure 1A). After photobleaching of accumulated Venus-ARC protein, individual newly synthesized Venus fusion protein molecules were detected as discrete diffraction-limited fluorescence signals, which appear after fluorophore maturation and disappear after photobleaching (Figure 1B). Single molecules can be tracked in sequential frames because of the sparseness and infrequency of appearance of newly synthesized molecules and because most newly synthesized molecules remain relatively immobile during the time they are fluorescent. The properties of individual trajectories detected by single molecule imaging are consistent with single, newly synthesized Venus-ARC molecules. Signal intensities for individual trajectories are relatively constant over successive frames and are comparable to average signal intensities for single Venus molecules immobilized on a glass slide imaged under identical conditions. Individual trajectories are characterized by single-step appearance, occasional single-step "blinking” off and on, and single-step photobleaching (Figure 1, C and D), all of which are characteristic of single-molecule detection. Photobleaching or photoblinking result in a decrease (∼75% on average) in the fluorescence signal, which is consistent with a signal-to-background ratio of ∼3:1 for detection of individual trajectories (Figure 1E). Because photobleaching and photoblinking are probabilistic events, temporal trajectories for different molecules have different durations. A histogram of trajectory durations for newly synthesized Venus-fusion protein molecules (Figure 1F) reveals that most trajectories have durations <1 s, consistent with rapid photobleaching of single Venus molecules (Xie et al., 2008). The histogram of trajectory durations can be fit with a single exponential, which is characteristic of single-molecule photobleaching kinetics. Although the beginning and end of any individual trajectory could be due to either translation and photobleaching or photoblinking on and off, these observations suggest that most trajectories represent translation and photobleaching of single Venus molecules. The location and time of first detection for each newly synthesized Venus-ARC fusion protein molecule provide a record of where and when each molecule was translated. Because fluorophore maturation is a probabilistic process, there is a variable time lag between translation and detection for each molecule. Supplemental Movie S1 shows fluorescence signals corresponding to synthesis of individual Venus-ARC molecules in dendrites from a hippocampal neuron microinjected with Venus-ARC RNA. Supplemental Figure S1A shows a kymograph showing temporal appearance and disappearance (and photoblinking) of fluorescence signals in a segment of dendrite. In the kymograph photoblinking events appear as transient temporal interruptions in signal at a particular spatial location, whereas photobleaching events appear as long-term disappearance of signal at a particular spatial location.
FIGURE 1:
Single-molecule detection of newly synthesized Venus-ARC protein molecules in injected cells. (A) Experimental procedure. Fluorescently labeled RNA molecules (green/red squiggles) encoding Venus-ARC or Venus-FMRP were microinjected into the perikaryon of cultured hippocampal neurons. Microinjected RNA molecules assemble into RNA granules (light red circles) that are transported along dendrites to dendritic spines, where translation occurs. Newly synthesized Venus fusion protein molecules undergo fluorophore maturation and are detected by single-molecule imaging. (B) Time-lapse images (100 frames, 200 ms/frame, total time 20 s) of a single region of interest (ROI; 2.5 × 2.5 μm) within a dendrite. Four temporal trajectories corresponding to four separate translational events within the ROI are detected in the sequence shown. The initial frame of each trajectory is outlined in green. (C) Normalized fluorescence intensity trajectories for molecules corresponding to trajectories 1 and 4 in A illustrating single-step appearance, photobleaching, and blinking, consistent with single-molecule behavior. (D) Histogram of photobleaching coefficients (PBs) for single-molecule trajectories. (E) Distribution of photobleaching times for fluorescent molecules. The solid curve represents the fit to a single-exponential decay.
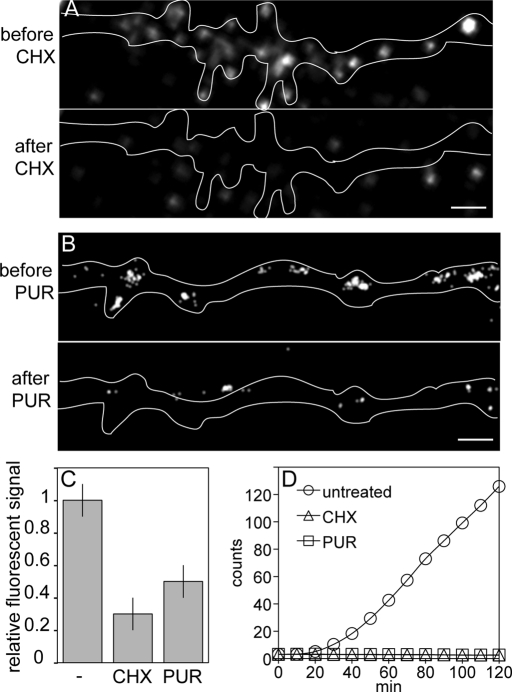
To determine whether fluorescence signal detected by single-molecule imaging in neurons microinjected with Venus-ARC RNA represents synthesis of new Venus fusion protein molecules, neurons were treated with either cycloheximide (CHX) to inhibit elongation or puromycin (PUR) to release nascent polypeptide chains from ribosomes (Darken, 1964; Kleiman et al., 1993; Schneider-Poetsch et al., 2010). Because residual translation of ARC RNA in neurons treated with CHX was reported previously (Bramham et al., 2008), we first determined whether translation of Venus-ARC RNA is sensitive to CHX or PUR in vitro. Venus-ARC RNA was incubated in wheat germ lysate in the presence and absence of CHX or PUR, and appearance of newly synthesized Venus-ARC protein molecules was measured by fluorescence correlation spectroscopy–in vitro translation (FCS-IVT; Figure 2D). In the absence of protein synthesis inhibitor, accumulation of newly synthesized Venus-ARC protein molecules is detected after a short lag. In the presence of CHX or PUR newly synthesized Venus-ARC protein molecules are not detected, indicating that translation of Venus-ARC RNA is completely inhibited by CHX and PUR in vitro. The lag time before newly synthesized Venus-ARC protein molecules is detected in the FCS-IVT assay may be due to a number of factors, including slow translation initiation of Venus-ARC RNA in vitro, time required for synthesis of new Venus-ARC protein molecules in vitro, and Venus fluorophore maturation time in vitro (t1/2 ∼ 2 min). In neurons injected with Venus-ARC RNA, fluorescence signal detected by single-molecule imaging is reduced by 60–70% after treatment with CHX or PUR compared with untreated neurons (Figure 2, A–C), indicating that most fluorescence signal detected in the absence of protein synthesis inhibitors represents newly synthesized protein molecules. Most of the residual fluorescence signal in the presence of CHX or PUR is located outside the cell, indicating that it does not represent newly synthesized Venus-ARC protein molecules but instead represents fluorescent debris on the coverslip that can be minimized but not completely eliminated by washing the coverslips extensively before plating the cells. There are several possible explanations for the small amount of residual fluorescence signal detected inside the cell in the presence of CHX or PUR. One possibility is that it represents Venus-ARC protein molecules that were synthesized outside the field of view (and therefore were not photobleached) before CHX or PUR was added that subsequently diffused into the field of view and were detected by single-molecule imaging. Another possibility is residual translation of RNA molecules in dendrites in the presence of protein synthesis inhibitors. In this regard, treatment of neurons with protein synthesis inhibitors increases transport of RNA molecules to distal dendrites (Kleiman et al., 1993), which may increase the potential for residual translation. A third possibility is photoblinking of Venus-ARC protein molecules that were synthesized and blinked off prior to addition of CHX or PUR and subsequently blinked on after addition of CHX or PUR. Such molecules would have to have photoblinking times longer than the time delay between addition of CHX or PUR and initiation of single-molecule imaging (several minutes), which may be relatively infrequent.
FIGURE 2:
Inhibition of protein synthesis with cycloheximide and puromycin. (A) Single-molecule imaging of Venus-ARC protein in a dendritic segment before and after treatment with CHX. Scale bar, 1 μm. (B) Single-molecule imaging of Venus-ARC protein in a dendritic segment before and after treatment with PUR. Scale bar, 1 μm. (C) Relative fluorescence signal in untreated and CHX- and PUR-treated dendritic segments shown in A and B. (D) FCS-IVT of Venus-ARC RNA in the presence and absence of CHX and PUR.
Spatial clustering of translation events near granules
Translation event maps were created by superimposing the locations of translation events on the locations of granules in the same cell (Figure 3). Granules were imaged first because extended illumination at the excitation wavelength used for detecting Venus fusion proteins causes photobleaching of fluorescent RNA. Coinjection of differentially labeled Venus-ARC RNA and Venus-FMRP RNA into hippocampal neurons revealed that both RNAs are colocalized in the same granules (Supplemental Figure S1). However, translation of both RNAs in the same granules cannot be analyzed simultaneously because Venus is the only available fluorescent protein with photophysical properties (rapid fluorophore maturation) suitable for monitoring translation by single-molecule imaging.
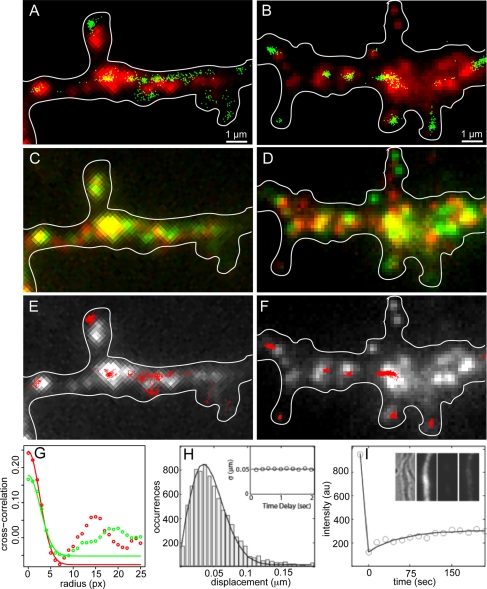
FIGURE 3:
Translation event maps for Venus-ARC and Venus-FMRP RNAs in wild-type hippocampal neurons. (A, B) Locations of individual Venus-ARC or Venus-FMRP granules (red) were determined by imaging microinjected fluorescent RNA in a dendritic segment. Centroid coordinates for newly synthesized Venus-ARC or Venus-FMRP protein molecules (green) determined by single-molecule imaging in the same dendritic segment are superimposed on the granule image. (C, D) Translocation of Venus-ARC and Venus-FMRP RNA granules over time. The first frame (t = 0 min; shown in red) was superimposed on the last frame (t = 5; min, shown in green) to evaluate how far each granule moved during the period immediately prior to imaging of translation events. (E, F) Spatial trajectories for individual newly synthesized Venus-ARC and Venus-FMRP protein molecules were calculated by plotting centroid coordinates for individual molecules in sequential frames. Single-molecule spatial trajectories for several individual protein molecules (red) are overlaid on granule images (white). (G) Image cross-correlation analysis of translation events and RNA granules for Venus-ARC and Venus-FMRP. Centroid coordinates for newly synthesized Venus-ARC and Venus-FMRP molecules, as shown in Figure 2, A and B, respectively, were mapped onto individual pixels to create translation intensity images that were cross-correlated with the corresponding RNA granule images, as shown in Figure 2, A and B, to generate the cross-correlation functions. In each case the initial amplitude of the cross-correlation function provides a measure of Pearson's coefficient of correlation between translation events and RNA granules. The decay of cross-correlation as a function of distance provides a measure of the spatial distributions of translation events in relation to RNA granules. The secondary peak in the cross-correlation function may indicate either correlation with secondary granules in the vicinity of the primary granule or granule translocation between the time granules were imaged and the time translation events were recorded. (H) Histogram of apparent single-molecule displacements between adjacent imaging frames. The displacement is defined as the distance between the centroid positions of the same fluorescence spot in two successive frames. The solid curve is fitted to the Gaussian random walk model: P(r) = r exp(-r2/2σ2). The inset shows the σ value vs. different time delays for calculating displacements. (I) FRAP results for Venus-ARC protein. Intensity data are plotted in open circles. Solid line is the exponential recovery fit. Dendrite ROI images in the inset are, from left to right, bright-field image, fluorescence image before photobleaching, fluorescence image after photobleaching, and fluorescence image after recovery for 3 min.
Locations of translation events were determined by calculating centroid coordinates for each newly synthesized Venus fusion protein molecule. The overall spatial relationship between translation events and granules was evaluated by cross-correlation analysis of the image of translation events with the image of granules in the same cell (Figure 3G). Centroid coordinates for translation events were mapped to individual pixels to generate a translation event density distribution image, which was cross-correlated with the corresponding fluorescent granule intensity distribution image. For both Venus-FMRP and Venus-ARC there is a primary peak of positive cross correlation values at 0 pixels, which decays over a distance of several pixels, indicating that density of translation events is highest in the vicinity of granules and declines with distance from granules. The amplitude of the cross-correlation function at 0 pixels provides a measure of Pearson's coefficient of correlation between translation events and granules (r = 0.17 for Venus-FMRP protein/RNA and r = 0.24 for Venus-ARC protein/RNA). For both Venus-ARC and Venus-FMRP a secondary peak of positive cross-correlation values at 15–20 pixels could represent correlation of translation events with nearby granule(s) located in the immediate vicinity of the primary granule, in which case the distance between the primary and secondary cross-correlation peaks would correspond to the average distance between granules. Alternatively, the secondary peak could be due to translocation of the primary granule between the time of granule imaging and the time of translation event recording, in which case the distance between the primary and secondary cross-correlation peaks would correspond to the average translocation distance for individual granules.
To examine translation output from individual granules, the number of translation events clustered in the vicinity of each granule was counted. Separate translation event maps for Venus-ARC RNA and Venus-FMRP RNA in wild-type neurons are shown in Figure 3, A and B, respectively. In Figure 3A, 12 distinct Venus-ARC RNA granules are resolved, with various intensities corresponding to various numbers of RNA molecules per granule. The locations of 521 translation events (recorded over a period of 5 min) are superimposed on the granule image. In Figure 3B, 15 distinct Venus-FMRP RNA granules are resolved, and the locations of 598 translation events are superimposed on the granule image. In both cases, translation events appear to be spatially clustered near individual granules. A cluster is defined as a restricted region with increased translation event density (>5 events/μm2), surrounded by regions with lower translation event densities (<2 events/μm2). In Figure 3A, 516/521 translation events can be assigned to 12 discrete clusters, all but 1 of which (11/12, 85%) are associated (located within <1 μm) with a proximate granule. In Figure 3B, 589/598 translation events can be assigned to 10 discrete clusters, all of which (10/10, 100%) are associated with proximate granules. The areas occupied by individual clusters sometimes appear smaller than the areas occupied by the associated granules because clusters are defined by precise centroid coordinates for individual translation events, whereas granules are defined by a diffuse cloud of fluorescence from multiple fluorophores in multiple RNA molecules in each granule. In some cases the location of a cluster of translation events appears shifted relative to the location of the proximate granule, which may mean that the granule moved slightly between the time it was imaged and the time the centroids of newly synthesized proteins in the cluster were recorded. Granules not associated with clusters of translational events may be translationally inactive or may have moved out of the frame before translational events were recorded. Clusters of translational events not associated with a proximate granule may represent translational output from a granule that moved into the frame after granule locations were recorded. Spatial clustering of translation events near individual granules suggests that each cluster represents translational output from the proximate granule.
Mapping translation events to individual granules requires that both the granule and the newly synthesized protein molecules remain relatively immobile during the experiment. If newly synthesized Venus-ARC or Venus-FMRP protein molecules diffuse away from their sites of synthesis before fluorophore maturation, they will be detected as isolated individual molecules not associated with granules. The fact that most newly synthesized Venus-ARC and Venus-FMRP molecules are found in clusters associated with proximate granules suggests that most translation events occur in granules, that most granules are relatively immobile, and that most newly synthesized molecules do not move far from their sites of synthesis during the lag time between translation and detection.
To measure movement of granules directly, sequential images of Venus-ARC or Venus-FMRP RNA granules separated by an interval of 5 min were superimposed in contrasting pseudocolors (red and green). In Figure 3C, 12 Venus-ARC RNA granules are detected in both the red and the green channels. In Figure 3D, 15 Venus-FMRP RNA granules are detected in both the red and the green images. In both cases none of the granules moved >1 μm during the time interval between the two images. This indicates that both Venus-ARC RNA and Venus-FMRP RNA granules are relatively immobile during the 5-min period immediately prior to single-molecule imaging of translation events.
To measure movement of newly synthesized Venus-ARC and Venus-FMRP protein molecules directly, individual molecules were tracked in sequential frames. Most single-molecule trajectories for both Venus-ARC (Figure 3E) and Venus-FMRP (Figure 3F) are constrained to the vicinity where the molecule first appears, indicating that newly synthesized protein molecules remain close to their sites of synthesis. The “apparent” displacements of centroid coordinates for individual molecules between adjacent frames (200-ms delay) was fitted with a random walk model, P(r) = r exp(-r2/2σ2) (Figure 3H). The value of σ = 0.0052 μm, obtained from the fitting, is well below the measurement noise level, and increasing the time delay did not increase the displacement, indicating that most of the “apparent” displacement can be attributed to deviation in centroid determination between frames rather than to actual displacement. Furthermore, because of rapid Venus photobleaching, it was usually not possible to track individual Venus fusion molecules for long enough to accurately measure very slow diffusion. We conclude that mobility of newly synthesized Venus-ARC and Venus-FMRP protein molecules is too slow to measure at the spatial and temporal resolution of single-molecule tracking measurements.
Mobility of Venus-ARC protein over longer spatial and temporal scales was measured by fluorescence redistribution after photobleaching (FRAP). FRAP measurements of Venus-ARC (Figure 3I) showed <25% fluorescence recovery over several minutes, indicating that >75% of Venus-ARC protein molecules are relatively immobile at this time scale. Similar FRAP studies with FMRP-GFP in neurons (Antar et al., 2005) also showed that most FMRP-GFP molecules are relatively immobile over a comparable time. These experiments indicate that most Venus-ARC and Venus-FMRP protein molecules are relatively immobile in hippocampal neurons.
To determine whether newly synthesized Venus-ARC molecules are translocated from the perikaryon to the dendrites, the dendrite was amputated from the perikaryon with a micropipette after microinjection but before single-molecule imaging, to prevent translocation of newly synthesized protein molecules from the perikaryon into the amputated dendrite during the imaging period. Newly synthesized Venus-ARC protein molecules still appeared spatially clustered near individual granules in the amputated dendrite (data not shown), indicating that clusters of Venus-ARC protein molecules associated with proximate granules were synthesized locally in the dendrite and did not originate in the perikaryon.
Specific translation activities for individual granules
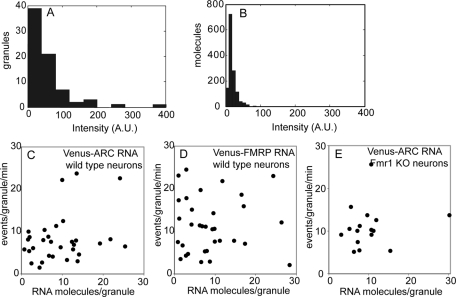
The digital nature of single-molecule imaging makes it possible to count the number of fluorescent RNA molecules in each granule and the number of newly synthesized Venus-fusion protein molecules produced by each granule per unit time. The number of RNA molecules in each granule was determined by comparing total fluorescence intensities for each RNA granule in microinjected cells (Figure 4A) with average intensities for single RNA molecules immobilized on a glass slide (Figure 4B). One caveat to this approach is that imaging conditions for granules in live cells are different from imaging conditions for single RNA molecules on a glass slide, which can affect the fluorescence quantum yield for individual RNA molecules. Imaging RNA granules in fixed cells would be more comparable to RNA molecules on a glass slide; however, to calculate specific translation activity for individual granules, it is necessary to image RNA granules in live cells prior to recording translation events because continuous illumination during translation event recording causes photobleaching of the RNA molecules, making it impossible to detect fluorescent RNA in fixed cells after the experiment. The number of labeled RNA molecules per granule determined using this approach was highly variable (1–30 RNA molecules per granule) for both Venus-ARC RNA and Venus-FMRP RNA. Translational output for each granule per unit time, determined by counting the number of centroids in the cluster associated with each granule, was also highly variable (1–30 events/granule per min) for both Venus-ARC RNA and Venus-FMRP RNA granules. Specific translation activity for each granule, defined as the number of newly synthesized Venus-fusion protein molecules produced per unit time divided by the number of fluorescent RNA molecules in the granule, was plotted with number of RNA molecules per granule on the x-axis and number of newly synthesized protein molecules per granule per unit time on the y-axis (Figure 4, C–E). The number of RNA molecules per granule is correlated with the number of newly synthesized protein molecules per granule per unit time for Venus-ARC RNA in wild-type neurons (r = 0.3) and in Fmr1 KO neurons (r = 0.15) but not for Venus-FMRP RNA in wild-type neurons (r = 0.0), indicating that translation output is proportional to number of RNA molecules in the granule for Venus-ARC RNA but not for Venus-FMRP RNA.
FIGURE 4:
Specific translation activities for Venus-ARC and Venus-FMRP RNA granules in wild-type and Fmr1 KO neurons. (A) Fluorescence intensities for individual well-resolved granules containing Venus-ARC RNA. (B) Fluorescence intensities for individual RNA molecules immobilized on glass coverslips, imaged under the same conditions as in A. (C–E) Specific translation activities for granules containing Venus-ARC RNA in wild-type neurons, Venus-FMRP RNA in wild-type neurons, and Venus-ARC in Fmr1 KO neurons are plotted with number of RNA molecules per granule (determined by comparing the total fluorescence intensity of the granule with the average intensity of individual RNA molecules) on the x-axis and number of events per granule per minute on the y-axis. The number of events per granule was determined by counting the number of centroids corresponding to newly synthesized Venus fusion proteins that appeared in the vicinity of each well-resolved granule over a period of 10 min.
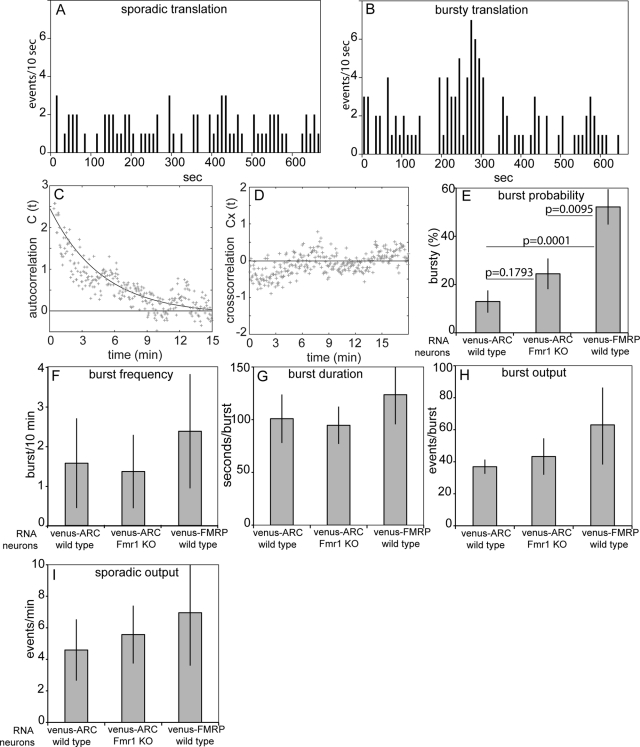
Translation output from individual granules is either sporadic or bursty
Translation event schedules for individual granules were generated by recording time of first detection for each newly synthesized Venus-fusion protein molecule in each cluster associated with a well-resolved granule. For both Venus-ARC and Venus-FMRP RNA granules two distinct modes of translation were observed—sporadic translation, consisting of isolated individual translation events occurring at apparently random intervals (Figure 5A), and bursty translation, consisting of multiple sequential translation events occurring over a relatively short time period interspersed with intervening periods of sporadic translation (Figure 5B). Autocorrelation analysis was used to measure parameters for bursty translation output from Venus-ARC RNA granules. If the duration of a single burst is less than the maturation time for Venus fluorophore and if sequential bursts from the same granule are uncorrelated independent events, then the theoretical autocorrelation function for bursty schedules follows the analytical form G(2)(τ) = (tb/tm)exp(—t/tm) where τm is the maturation time of the Venus fluorophore and τb is the average time interval between individual translation bursts (Yu et al., 2006; see Supplemental Material). Fitting the calculated autocorrelation function for Venus-ARC RNA granules into this form indicates that τm = 4.2 min and τb = 10.1 min (Figure 5C). The exact duration of each burst cannot be accurately determined from our data because burst duration time is convolved with fluorophore maturation time, which is unknown and varies probabilistically among different Venus molecules. However, from the autocorrelation analysis, it can be estimated that burst duration is shorter than fluorophore maturation time and may be as short as 1–2 min.
FIGURE 5:
Translation event schedules and translation output parameters for Venus-ARC and Venus-FMRP RNA granules in wild-type and Fmr1 KO neurons. Translation event schedules for individual granules were generated by recording times of first appearance for each newly synthesized molecule in a cluster of centroids associated with a well-resolved granule. (A, B) representative sporadic and bursty schedules for individual granules. (C) average autocorrelation function for bursty translation event schedules in granules containing Venus-ARC RNA in wild-type neurons. The solid line represents fitting of the autocorrelation data to a single-exponential decay function with a time constant of 4.2 min, corresponding to the average maturation time of Venus. (D) Average cross-correlation function for bursty translation event schedules in different granules. (E–I) Burst probabilities, burst frequencies, burst durations, burst outputs, and sporadic outputs for Venus-ARC and Venus-FMRP granules in wild-type and Fmr1 KO neurons. In each case average values and standard deviations were determined for schedules from > 40 different granules. Fisher's exact test was used to calculate p values for burst probabilities in E.
If translation in different granules were temporally regulated by a common upstream signal, then translation event schedules for different granules in the same dendrite would have a correlated component. If the regulatory signal were propagated along the dendritic process over time, translation event schedules for granules in the same dendrite would still be correlated but with a time delay. To test these possibilities, temporal cross-correlation functions were calculated for translation event schedules for different Venus-ARC RNA granules in the same dendrite (Figure 5D). Most of the cross correlation values are zero or negative, indicating that translation events in different granules are not correlated. This means that translation output from different granules is not globally regulated by overall regulatory mechanisms affecting whole dendrites but rather is locally regulated by spatially restricted mechanisms affecting individual RNA granules. Parenthetically, these results also indicate that bursty appearance of fluorescence signal is not due to focal drift, which would be expected to cause correlated fluctuations in multiple granules in the same dendrite.
Observation of sporadic and bursty translation event schedules from individual granules provides access to previously inaccessible quantitative parameters of translation. On the basis of examination of numerous translation event schedules, we defined the following operational criteria to quantify bursty and sporadic translation. A “burst” is defined as an uninterrupted period (1–2 min) of increased translational activity (≥3 events/10 s), preceded and followed by intervals (≥20 s) of sporadic translational activity (≤2 events/10 s). According to these criteria, burst probability (percentage of granules with bursty schedules; Figure 5E) is greater for Venus-FMRP RNA (52%) than for Venus-ARC RNA (12%), indicating that cis-acting sequences in the RNA determine burst probability. Burst probability for Venus-ARC RNA is increased in Fmr1 KO neurons (24%) compared with wild-type neurons (12%), indicating that trans-acting FMRP regulates burst probability. Other parameters of translation output, including burst frequency (1–2 bursts/10 min; Figure 5F), burst duration (∼100 s; Figure 5G), burst output (∼50 events/burst; Figure 5H), and sporadic output (∼5 events/min; Figure 5I), are comparable for Venus-ARC and Venus-FMRP RNAs in wild-type and Fmr1 KO neurons, indicating that these parameters reflect intrinsic properties of the granule that are not regulated by cis-acting sequences in the RNA or trans-acting FMRP in the cell.
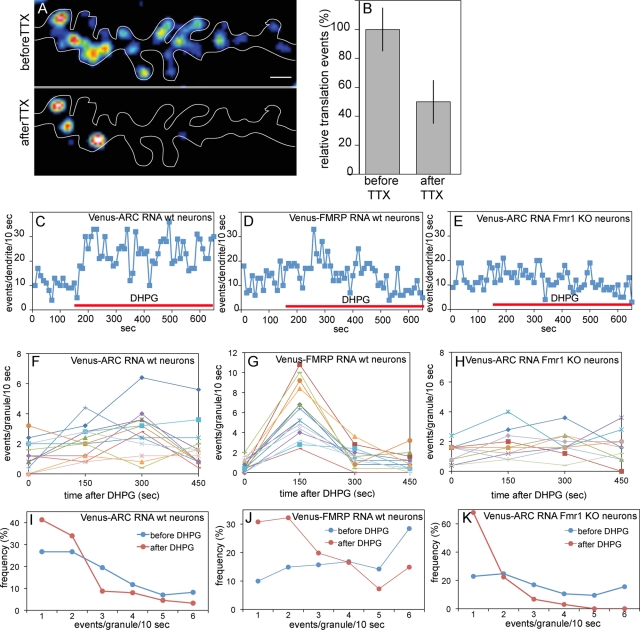
Synaptic activity regulates translation in granules
To determine whether synaptic activity affects translation output from individual granules, neurons were treated with either tetrodotoxin (TTX) to inhibit spontaneous action potentials or dihydroxyphenylglycine (DHPG) to stimulate group 1 mGluRs. TTX treatment decreased the overall number of translation events for Venus-ARC RNA (Figure 6, A and B), indicating that action potentials increase translation activity. DHPG increased the overall number of translation events per dendrite (Figure 6, C–E) and per granule (Figure 6, F–H) for both Venus-ARC RNA and Venus-FMRP RNA in wild-type neurons but not for Venus-ARC RNA in Fmr1 KO neurons, indicating that synaptic activity increases translation output from individual granules through an FMRP-dependent mechanism. In the case of Venus-ARC RNA, the increase in translational output is rapid and sustained, reaching a maximum ∼2 min after DHPG addition, and remaining elevated up to 6 min. The rapidity of the increase in translational output after DHPG addition indicates that Venus fluorophore maturation time must be <2 min in hippocampal neurons. In the case of Venus-FMRP RNA, the increase in translational output is delayed for ∼2 min and decays within 3 min. Frequency distribution analysis of translation events in individual granules (Figure 5, I–K) indicates that after DHPG addition the percentage of granules with a frequency of >3 events/10 s (characteristic of bursty translation) is reduced, with a compensatory increase in the percentage of granules with a frequency of <3 events/10 s (characteristic of sporadic translation), for Venus-ARC RNA and Venus-FMRP RNA in wild-type neurons and for Venus-ARC RNA in Fmr1 KO neurons. This indicates that DHPG either inhibits bursty translation, stimulates sporadic translation, or both.
FIGURE 6:
Synaptic activity affects translation output in granules containing Venus-ARC or Venus-FMRP RNA in wild type and Fmr1 KO neurons. (A, B) Translation activity in a dendritic segment before and after TTX treatment. (C–E) Translation activity in dendritic segments before and after DHPG treatment. (F–H) Translation activity in individual granules before and after DHPG treatment. The value at time 0 represents the average output during the 150 s prior to DHPG addition, the value at time 150 s represents the average output during the first 150 s after DHPG addition, the value at 300 s represents the average output between 150 and 300 s after DHPG addition, and the value at 450 s represents the average output between 300 and 450 s after DHPG addition. (I–K) Frequency distributions for translation events in granules before and after DHPG treatment.
DISCUSSION
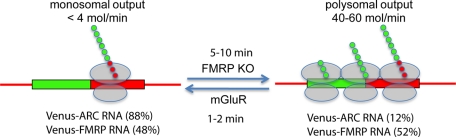
Sporadic translation (isolated translation events at random intervals) is consistent with translation by individual ribosomes (monosomes). Bursty translation (multiple sequential translation events within a short time period) is consistent with translation by polysomes (Figure 7). Burst probabilities (percentage of granules with bursty schedules) are also consistent with previous studies of percentage of RNA in polysomes (PMPs) in mouse brain (Zalfa et al., 2003). In wild-type mouse brain, PMP is lower for ARC RNA (∼42%) than for Fmr1 RNA (∼62%), and in wild-type neurons burst probability is also lower for Venus-ARC (12%) than for Venus-FMRP RNA (52%). In Fmr1 KO brain PMP for ARC RNA (∼ 60%) is increased compared with wild-type mouse brain (42%), and in Fmr1 KO neurons burst probability for Venus-ARC RNA (24%) is also increased compared with wild-type neurons (12%). Correlation between burst probabilities in neurons and PMP values in mouse brain supports the hypothesis that sporadic translation event schedules represent monosomal translation, whereas bursty schedules represent polysomal translation. The fact that PMP values and burst probabilities are not identical is not surprising given the differences in biological systems (mouse brain vs. microinjected neurons) and analytical methods (sedimentation velocity for PMP vs. translation event schedules for burst probability).
FIGURE 7:
Summary diagram for sporadic and bursty translation output in granules. RNA molecules are shown with Venus ORF as a green rectangle and ARC or FMRP ORF as a red rectangle and 5′ and 3′UTRs as red lines. Ribosomal subunits translating RNA molecules are shown as semitransparent ovals. Nascent polypeptide chains for Venus-ARC or Venus-FMRP fusion proteins are shown as linear arrays of small green and red circles. Left, sporadic output corresponding to monosomal translation. Right, bursty output corresponding to polysomal translation. mGluR increases the rate of monosomal translation. FMRP KO increases the probability of polysomal translation.
Parameters for monosomal and polysomal translation can be inferred from parameters for sporadic and bursty translation event schedules, respectively, as illustrated in Figure 7. Burst probability provides a measure of the probability of polysome assembly in individual granules. Burst probability is not correlated with the number of RNA molecules in the granule, suggesting that only a limited number of polysomes can be supported by each granule. Venus-ARC and Venus-FMRP RNAs, despite being colocalized in the same granules, have different burst probabilities, suggesting that cis-acting elements in the RNAs regulate polysome formation. Burst probability for Venus-ARC RNA is increased in Fmr1 KO neurons, suggesting that FMRP represses polysome formation for Venus-ARC RNA in wild-type neurons.
Burst frequency provides a measure of the average rate of polysome formation in each granule (∼1 polysome/5 min). Burst frequency is not correlated with number of RNA molecules per granule, suggesting that most granules only support polysome formation on one RNA molecule at a time. Multiple sequential bursts in granules containing one RNA molecule indicate that a single RNA molecule can undergo multiple rounds of polysome formation and runoff interspersed with periods of monosomal translation.
Burst duration provides a measure of polysome lifetime (∼100 s). The probabilistic lag time between translation and fluorophore maturation for each newly synthesized Venus molecule tends to delay and disperse detection of newly synthesized molecules, which means that actual polysome lifetimes may be somewhat shorter than burst durations. In granules that contain a single RNA molecule, the duration of a single burst may reflect the lifetime of a single polysome.
Burst output provides a measure of translation output per polysome (40–60 events/polysome). In granules that contain a single RNA molecule, burst output may represent the translation output from a single polysome, which means that an average polysome can generate 40–60 newly synthesized protein molecules during its lifetime (100 s). This is significantly more that the average number of ribosomes per polysome (<10), which means that individual ribosomes can undergo multiple cycles of initiation and runoff during the polysome lifetime.
Sporadic output provides a measure of monosomal translation rate (4–7 events/min). If initiation, elongation, and termination rates are comparable for monosomal and polysomal translation, the difference between monosomal output (4–7 events/min) and polysomal output (40–60 events/polysome) is consistent with an average polysome size of ∼10 ribosomes. This is comparable to the average size of polysomes visualized in dendrites by electron microscopy (Bourne et al., 2007).
Relative proportions of polysomes versus monosomes are determined by relative rates of initiation and elongation, respectively (Nelson and Winkler, 1987). Therefore, the ratio between burst probability and sporadic output reflects the balance between initiation and elongation rates in each granule. DHPG treatment decreases the ratio of burst probability to sporadic output, suggesting that stimulation of group 1 mGluR either increases elongation rate or decreases initiation rate (or both). Stimulation of group 1 mGluR is known to activate eEF2K, resulting in phosphorylation of EF2, which decreases elongation rates for most RNAs but increases translation of ARC RNA (Park et al., 2008; Waung and Huber, 2009). This is consistent with decreased ratio of burst probability and increased sporadic output for Venus-ARC RNA after DHPG treatment. Stimulation of group 1 mGluR is also known to cause transient inactivation of FMRP (Narayanan et al., 2007, 2008). Because FMRP represses translation of its own RNA, transient inactivation of FMRP after DHPG treatment is consistent with transient increase in sporadic output for Venus-FMRP RNA after DHPG treatment.
The physiological consequences of sporadic versus bursty translation in granules depend on the function(s) of the encoded proteins. ARC protein mediates endocytosis of AMPAR (Chowdhury et al., 2006) to modulate synaptic sensitivity in response to synaptic activity (Rial-Verde et al., 2006; Shepherd et al., 2006). Monosomal translation of ARC RNA would generate a steady-state level of ARC protein synthesis in dendritic spines, resulting in steady-state endocytosis of AMPAR from the synapse. Stimulation of monosomal translation of ARC RNA by activation of group 1 mGluR would increase the rate of ARC protein synthesis in the spine, resulting in increased endocytosis of AMPAR and decreased sensitivity of proximate synapses. Polysomal translation of ARC RNA in granules would generate localized bursts of ARC protein synthesis, with concomitant bursts of endocytosis of AMPAR from proximate synapses. Increased polysomal translation of ARC RNA in Fmr1 KO mice would increase fluctuations in endocytosis of AMPAR, with concomitant fluctuations in synaptic sensitivity. Because newly synthesized ARC protein molecules remain close to their sites of synthesis in granules, these effects would be restricted to proximate spines and would not spread to nearby regions of the dendrite. Polysomal translation of FMRP RNA would result in transient bursts of FMRP protein synthesis in specific granules, with concomitant repression of FMRP target RNAs such as ARC RNA (Park et al., 2008) that regulate synaptic sensitivity. Increased monosomal translation of FMRP RNA following group 1 mGluR activation would increase repression of FMRP target RNAs, decreasing synaptic sensitivity. Because newly synthesized FMRP protein molecules remain close to their sites of synthesis, these effects would be restricted to individual granules and would not spread to nearby granules, providing a mechanism for transient local regulation of translation in individual granules.
It is important to recognize that the experimental protocol outlined in Figure 1A is designed to assay the translational potential of specific exogenous RNA molecules microinjected directly into the cytoplasm and does not provide information about nuclear processes that can affect subsequent translation of the corresponding endogenous RNAs. For example, nuclear deposition of exon junction complex proteins on the 3′ untranslated region (UTR) of endogenous ARC RNA may affect its subsequent cytoplasmic translation and target it for nonsense-mediated decay (Giorgi et al., 2007), neither of which would be detected in the experiments reported here because the RNA is injected directly into the cytoplasm and does not pass through the nucleus. To assay nuclear phenomena of this type, exogenous RNA could be microinjected directly into the nucleus.
Sporadic and bursty translation output from different granules provides an explanation for the sparseness of polysomes detected in dendrites by electron microscopy (EM). Although the number of RNA molecules in dendrites is quite large (Eberwine et al., 2001; Poon et al., 2006; Suzuki et al., 2007), the number of polysomes detected by serial section EM is relatively small (on the order of one polyribosome per micrometer of dendritic length; Bourne et al., 2007). At any one point in time, many of the RNA molecules in dendrites may be engaged with single monosomes, which are difficult to detect by EM. Therefore, EM “snapshots” of dendrites might only detect granules undergoing bursty translation mediated by polysomes and fail to detect sporadic translation mediated by monosomes.
The method described here for single-molecule imaging of translation in individual granules represents a significant advance in molecular, spatial, and temporal resolution for analysis of translation in neurons. The inherently digital nature of the technique provides access to quantitative parameters of translation that have not been measured previously, revealing sporadic and bursty translation in granules in neurons for the first time. Differential regulation of sporadic and bursty translation in individual granules may mediate spatially restricted and temporally transient changes in synaptic function during learning and memory.
MATERIALS AND METHODS
RNA synthesis
Venus-ARC construct was derived from ARC cDNA clone (pBS II (SK+) r ARC, obtained from Paul Worley, Johns Hopkins University, Baltimore, MD). The ARC open reading frame (ORF) was inserted in-frame at the C-terminus of the Venus ORF in a Venus vector. The NotI-NdeI fragment from this clone, including the Venus ORF and the first 16 bases from ARC ORF, was excised and religated with the corresponding NdeI-NotI fragment of the original ARC cDNA construct. The final construct carries the complete ARC ORF and 3′-UTR but not the 5′-UTR. Venus-FMRP construct was derived from a GFP-FMRP clone obtained from Jennifer Darnell (Rockefeller University, New York, NY). The GFP ORF was replaced with the Venus ORF by restriction free cloning (Geiser et al., 2001). Venus DNA was obtained by PCR using the following primers: forward, 5′-CACCATGGTGAGCAAGGGCGA GGA-3′; reverse, 5′-CTTGTACAGCTCGTCCATGC-3′. The Venus PCR product was gel purified and used in a linear amplification reaction with GFP-FMRP plasmid as the template using a QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA). RNAs synthesized by in vitro transcription of linearized template DNA were capped and polyadenylated using mScript Kit (Epicentre Biotechnologies, Madison, WI) according to manufacturer's protocol. To make fluorescently labeled RNA, Cy5-conjugated UTP (Amersham, GE Healthcare Bio-Sciences, Piscataway, NJ) was added to the transcription reaction mixture to a final concentration of 250 μM. The resulting RNA was purified by precipitation with 5 M ammonium acetate and ethanol, dissolved in water, and filtered through 0.22-μm filters (Millipore, Billerica, MA) prior to microinjection. The concentration of unincorporated nucleotide, measured by fluorescence correlation spectroscopy, was <5% of the concentration of labeled RNA.
Cell culture
Hippocampal neuron cultures were prepared from E17 rat (Sprague Dawley) embryos or newborn mouse pups as described previously (Gao et al., 2008). Cells were seeded at low density on glass-bottom culture dishes (MatTek Corporation, Ashland, MA) that were cleaned by sonicating sequentially in ethanol (200 proof), 10% NaOH, and Millipore water and coated with polylysine (Sigma-Aldrich, St. Louis, MO) before use. Cell cultures were maintained for 14–19 days in vitro in Neurobasal medium (Life Technologies, Carlsbad, CA) supplemented with B27 before switching to Hibernate E medium (BrainBits, Springfield, IL) for several hours before microscopic analysis. To inhibit translation, cycloheximide (Sigma-Aldrich) or puromycin (Sigma-Aldrich) was added to the medium (final concentrations, 300 and 10 μM, respectively). To inhibit spontaneous action potentials in the culture, TTX was added to the medium (final concentration, 100 μM). To stimulate group 1 mGluR, DHPG was added to the medium (final concentration, 100 μM).
Microscopy and single-molecule imaging
Venus-ARC RNA or Venus-FMRP RNA were microinjected at 0.5–1 mg/ml concentration into the perikaryon of hippocampal neurons using an Eppendorf pressure injection system as described previously (Gao et al., 2008). In some cases dendrites were amputated near the cell body after microinjection by dragging a microinjection needle across the process. Injected cells were imaged with a modified Olympus IX81 microscope equipped with 60× objective (1.45 numerical aperture, total internal reflection fluorescence; Olympus, Tokyo, Japan) and an EM-charge-coupled device camera (PhotonMax, Princeton Instruments, Trenton, NJ). To image RNAs, Cy5-labeled RNA molecules were excited with the 635-nm laser line from a diode laser (57ICS; CVI Melles Griot, Albuquerque, NM). Single-molecule detection of fluorescent Venus protein was carried out with excitation from the 514-nm laser line from an argon ion laser (35LAP321; CVI Melles Griot) at 5-Hz frame rate and power density of 600 W/cm2 under a small-field aperture setting (25 μm × 25 μm). Images were acquired using the open-source microscopy software suite μmanager under the ImageJ platform (National Institutes of Health, Bethesda, MD).
Fluorescence correlation spectroscopy in vitro translation
Venus-ARC RNA (4 nM) was translated in wheat germ lysate (Promega, Madison, WI) precentrifuged (10,000 × g, 30 min) to remove fluorescent particles. Newly synthesized Venus-ARC protein molecules were detected by fluorescence correlation spectroscopy in a 96-well glass-bottom dish using a Confocor 3 system with a 40×, 1.2 numerical aperture, water immersion objective (Zeiss, Thornwood, NY). FCS measurements of total counts in each well were taken at 10-min intervals for 120 min.
Image analysis
Time-lapse images were analyzed by single-molecule tracking software developed in the lab. Centroid coordinates of individual molecules were determined using an algorithm based on the polynomial fit with Gaussian-weighted method (Rogers et al., 2007) and linked in time via a standard two-dimensional single-particle tracking algorithm to construct temporal trajectories. To account for fluorescence intensity “blinking” of single molecules, the particle-tracking algorithm allows for temporary loss of intensity for up to three image frames without losing tracking. This accounts for the majority of blinking events observed with Venus because further increasing the blinking time duration had little effect (<1%) on the total number of trajectories detected.
Image correlation analysis was performed as described previously (Gao et al., 2008). Centroid coordinates for newly synthesized Venus-fusion protein molecules (determined as described) were mapped onto individual pixels to create a translational intensity distribution image that was cross-correlated with a corresponding image showing fluorescence intensity distribution of individual granules in the same region of interest. The initial amplitude of the cross-correlation function provides a measure of Pearson's coefficient of correlation between the locations of translation and granules. Cross-correlation as a function of distance provides a measure of the spatial distribution of translation events and RNA molecules in granules.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants NS15190 and RR022232 to J.H.C., GM085301 to J.Y., and NS19943 to E.B. Richard Mains (University of Connecticut Health Center) provided critical comments on the manuscript.
Abbreviations used:
- AMPAR
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor
- ARC
activity-regulated cytoskeletal-associated protein
- CHX
cycloheximide
- DHPG
dihydroxyphenylglycine
- EM
electron microscopy
- FCS
fluorescence correlation spectroscopy
- FMRP
fragile X mental retardation protein
- FRAP
fluorescence redistribution after photobleaching
- FXS
fragile X syndrome
- GFP
green fluorescent protein
- IVT
in vitro translation
- mGluR
metabotropic glutamate receptor
- ORF
open reading frame
- PMP
percentage of mRNA in polysomes
- PUR
puromycin
- ROI
region of interest
- UTR
untranslated region
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-07-0622) on January 4, 2012.
REFERENCES
- Antar LN, Dictenberg JB, Plociniak M, Afroz R, Bassell GJ. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 2005;4:350–359. doi: 10.1111/j.1601-183X.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne JN, Sorra KE, Hurlburt J, Harris KM. Polyribosomes are increased in spines of CA1 dendrites 2 h after the induction of LTP in mature rat hippocampal slices. Hippocampus. 2007;17:1–4. doi: 10.1002/hipo.20238. [DOI] [PubMed] [Google Scholar]
- Bramham CR, Worley PF, Moore MJ, Guzowski JF. The immediate early gene arc/arg3.1: regulation, mechanisms, and function. J Neurosci. 2008;28:11760–11767. doi: 10.1523/JNEUROSCI.3864-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo PE, Francesconi A, Carroll RC. The ups and downs of translation-dependent plasticity. Neuron. 2008;59:1–3. doi: 10.1016/j.neuron.2008.06.016. [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52:445–459. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darken MA. Puromycin inhibition of protein synthesis. Pharmacol Rev. 1964;16:223–243. [PubMed] [Google Scholar]
- Eberwine J, Miyashiro K, Kacharmina JE, Job C. Local translation of classes of mRNAs that are targeted to neuronal dendrites. Proc Natl Acad Sci USA. 2001;98:7080–7085. doi: 10.1073/pnas.121146698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Tatavarty V, Korza G, Levin MK, Carson JH. Multiplexed dendritic targeting of {alpha} calcium calmodulin-dependent protein kinase II, neurogranin, and activity-regulated cytoskeleton-associated protein RNAs by the A2 pathway. Mol Biol Cell. 2008;19:2311–2327. doi: 10.1091/mbc.E07-09-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser M, Cèbe R, Drewello D, Schmitz R. Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. Biotechniques. 2001;31:88–90. doi: 10.2144/01311st05. [DOI] [PubMed] [Google Scholar]
- Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Kleiman R, Banker G, Steward O. Inhibition of protein synthesis alters the subcellular distribution of mRNA in neurons but does not prevent dendritic transport of RNA. Proc Natl Acad Sci USA. 1993;90:11192–11196. doi: 10.1073/pnas.90.23.11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- McAnaney TB, Zeng W, Doe CF, Bhanji N, Wakelin S, Pearson DS, Abbyad P, Shi X, Boxer SG, Bagshaw CR. Protonation, photobleaching, and photoactivation of yellow fluorescent protein (YFP 10C): a unifying mechanism. Biochemistry. 2005;44:5510–5524. doi: 10.1021/bi047581f. [DOI] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Narayanan U, Nalavadi V, Nakamoto M, Pallas DC, Ceman S, Bassell GJ, Warren ST. FMRP phosphorylation reveals an immediate-early signaling pathway triggered by group I mGluR and mediated by PP2A. J Neurosci. 2007;27:14349–14357. doi: 10.1523/JNEUROSCI.2969-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan U, Nalavadi V, Nakamoto M, Thomas G, Ceman S, Bassell GJ, Warren ST. S6K1 phosphorylates and regulates fragile X mental retardation protein (FMRP) with the neuronal protein synthesis-dependent mammalian target of rapamycin (mTOR) signaling cascade. J Biol Chem. 2008;283:18478–18482. doi: 10.1074/jbc.C800055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson EM, Winkler MM. Regulation of mRNA entry into polysomesParameters affecting polysome size and the fraction of mRNA in polysomes. J Biol Chem. 1987;262:11501–11506. [PubMed] [Google Scholar]
- Ostroff LE, Fiala JC, Allwardt B, Harris KM. Polyribosomes redistribute from dendritic shafts into spines with enlarged synapses during LTP in developing rat hippocampal slices. Neuron. 2002;35:535–545. doi: 10.1016/s0896-6273(02)00785-7. [DOI] [PubMed] [Google Scholar]
- Park S, et al. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath N, et al. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52:437–444. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J Neurosci. 2006;26:13390–13399. doi: 10.1523/JNEUROSCI.3432-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rial Verde EM, Lee-Osbourne J, Worley PF, Malinow R, Cline HT. Increased expression of the immediate-early gene arc/arg3.1 reduces AMPA receptor-mediated synaptic transmission. Neuron. 2006;52:461–474. doi: 10.1016/j.neuron.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SS, Waigh TA, Zhao X, Lu JR. Precise particle tracking against a complicated background: polynomial fitting with Gaussian weight. Phys Biol. 2007;4:220–227. doi: 10.1088/1478-3975/4/3/008. [DOI] [PubMed] [Google Scholar]
- Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52:475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Schuman EM. Protein synthesis at synaptic sites on dendrites. Annu Rev Neurosci. 2001;24:299–325. doi: 10.1146/annurev.neuro.24.1.299. [DOI] [PubMed] [Google Scholar]
- Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Tian QB, Kuromitsu J, Kawai T, Endo S. Characterization of mRNA species that are associated with postsynaptic density fraction by gene chip microarray analysis. Neurosci Res. 2007;57:61–85. doi: 10.1016/j.neures.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Waung MW, Huber KM. Protein translation in synaptic plasticity: mGluR-LTD, fragile X. Curr Opin Neurobiol. 2009;19:319–326. doi: 10.1016/j.conb.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waung MW, Pfeiffer BE, Nosyreva ED, Ronesi JA, Huber KM. Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron. 2008;59:84–97. doi: 10.1016/j.neuron.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XS, Choi PJ, Li GW, Lee NK, Lia G. Single-molecule approach to molecular biology in living bacterial cells. Annu Rev Biophys. 2008;37:417–444. doi: 10.1146/annurev.biophys.37.092607.174640. [DOI] [PubMed] [Google Scholar]
- Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.