In vertebrates, pancreas and liver arise from bipotential progenitors located in the endoderm. At early stages, BMP and FGF are known to promote liver fate at the expense of pancreas. At later stages, bmp2a, fgf10, and fgf24 are essential for ventral pancreas specification, whereas they have an opposite effect on liver development.
Abstract
In vertebrates, pancreas and liver arise from bipotential progenitors located in the embryonic gut endoderm. Bone morphogenic protein (BMP) and fibroblast growth factor (FGF) signaling pathways have been shown to induce hepatic specification while repressing pancreatic fate. Here we show that BMP and FGF factors also play crucial function, at slightly later stages, in the specification of the ventral pancreas. By analyzing the pancreatic markers pdx1, ptf1a, and hlxb9la in different zebrafish models of BMP loss of function, we demonstrate that the BMP pathway is required between 20 and 24 h postfertilization to specify the ventral pancreatic bud. Knockdown experiments show that bmp2a, expressed in the lateral plate mesoderm at these stages, is essential for ventral pancreas specification. Bmp2a action is not restricted to the pancreatic domain and is also required for the proper expression of hepatic markers. By contrast, through the analysis of fgf10−/−; fgf24−/− embryos, we reveal the specific role of these two FGF ligands in the induction of the ventral pancreas and in the repression of the hepatic fate. These mutants display ventral pancreas agenesis and ectopic masses of hepatocytes. Overall, these data highlight the dynamic role of BMP and FGF in the patterning of the hepatopancreatic region.
INTRODUCTION
Pancreas is an endodermal organ composed of endocrine and exocrine tissues that respectively produce hormones and digestive enzymes. During embryogenesis, pancreas arises from two buds developing from the dorsal and ventral aspects of the gut epithelium. The ventral pancreatic bud appears adjacent to the liver bud, and previous studies on mouse embryonic explants and in zebrafish revealed the presence of bipotential endodermal progenitors that can give rise to both organs (Deutsch et al., 2001; Rossi et al., 2001; Chung et al., 2008). This cell fate decision is controlled by extrinsic factors released by the neighboring lateral plate mesoderm (LPM). Identification of such inducing stimuli is critical to the design of novel cellular therapies for pathologies affecting these organs.
In zebrafish, specification of the pancreatic region can be detected as early as 14 h postfertilization (hpf) through the activation of pdx1 expression in midtrunk endoderm (Biemar et al., 2001). The first pdx1-expressing cells, which are located near the medial line of the embryo just under the notochord, will delaminate to form the dorsal pancreatic bud by 24 hpf and will generate the first pancreatic endocrine cells. The first signs of hepatic development occur at 22 hpf with the activation of prox1 and hhex expression in a segment of the gut endoderm anterior to the dorsal pancreatic bud (Ober et al., 2006). These prox1+/hhex+ cells produce an outgrowth on the left side of the intestinal rod. The formation of this hepatic bud is quickly followed by the specification of the ventral pancreatic bud, which appears adjacent and posterior to the liver (Field et al., 2003a). Indeed, ptf1a and hlxb9la (also named mnr2a)—the two earliest markers of the ventral pancreas—are detected at 32 hpf between the liver and dorsal pancreatic buds (Wendik et al., 2004; Zecchin et al., 2004). The pdx1 homeobox gene, expressed in endocrine cells of the dorsal bud, is also detected in the adjacent segment of intestinal rod, which encompasses the prospective ventral pancreatic bud. In zebrafish, the ventral pancreatic bud (VPB) generates the whole exocrine tissue comprising the acinar and ductal cells. The ventral and dorsal pancreatic buds eventually merge by 52 hpf to form the pancreas (Field et al., 2003a, 2003b).
The ventral pancreatic and hepatic buds are induced by the adjacent mesodermal tissues through the release of signaling molecules such as fibroblast growth factors (FGFs) and bone morphogenic proteins (BMPs). However, conflicting data have been reported on the effect of these factors, revealing either an inducing or a repressing activity. For example, experiments on mouse embryonic explants showed that FGF from the cardiac mesoderm and BMP from the septum transversum are essential for the induction of liver markers and block pancreatic specification (Deutsch et al., 2001; Rossi et al., 2001). Similarly, FGFs and BMPs have also been shown in zebrafish to be essential for hepatic induction (Shin et al., 2007), and bmp2b can specify liver at the expense of pancreas (Chung et al., 2008). On the other hand, experiments with chicken embryonic explants indicate that BMP from the LPM is required for development of the ventral pancreas (Kumar et al., 2003), and we reported that FGF signaling is essential to specify the ventral pancreatic bud in zebrafish embryos (Manfroid et al., 2007). These contradictory results could be explained by a highly dynamic change in the inductive network. Indeed, Wandzioch and Zaret (2009) recently demonstrated that, whereas BMPs repress pancreatic specification at 3-4S in mouse embryos, they promote pancreatic fate a few hours later at 5-6S Another explanation could be that distinct members of the FGF and/or BMP ligand families have different activities. Thus a better understanding of liver and ventral pancreas development will require the identification of the BMP and FGF ligands expressed near the prospective hepatopancreatic region and their mutual relation.
In the present study, we show the crucial role of BMP pathway after 20 hpf for the specification of the ventral pancreatic bud in zebrafish embryos. We identify bmp2a as a crucial player in this induction and demonstrate its requirement for the activation of the first markers of the ventral pancreas (ptf1a and hlxb9la) as well as of the liver (prox1 and hhex). In contrast, by analyzing these pancreatic and hepatic markers in double fgf10−/−; fgf24−/− mutants, we find that both FGF10 and FGF24 ligands have completely opposed effect on the two organs, inducing all pancreatic markers while repressing hepatic markers. Thus our study provides new insights into the molecular mechanisms that initiate development of the liver and pancreas.
RESULTS
Requirement of BMP signaling in the specification of the ventral pancreatic bud
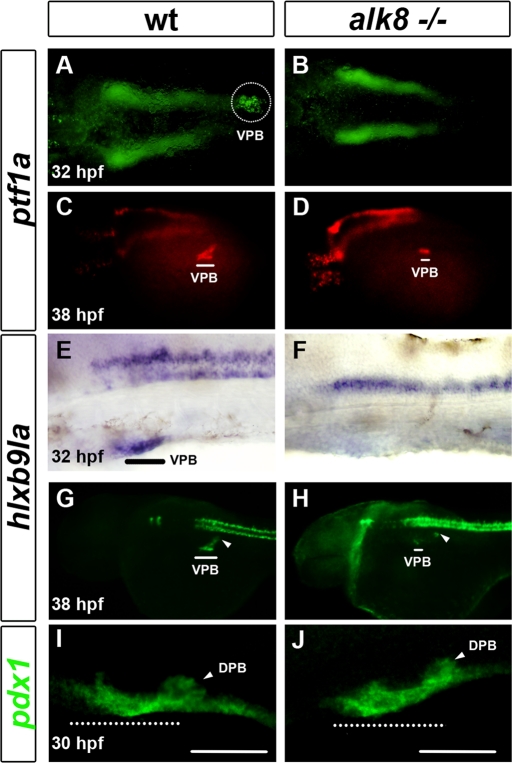
Involvement of BMP signaling in liver bud specification was previously investigated with laf/alk8 mutants (Chung et al., 2008). In addition, Chung et al. (2010) showed that VPB outgrowth was severely affected in alk8 mutants, and alk8 morphants have a hypoplasic ventral pancreas at 3 d postfertilization (dpf). However, it was unknown whether BMP signaling was necessary for the first steps of VPB development. Thus we evaluated VPB specification by analyzing the expression of ptf1a and hlxb9la (mnr2a), the two first VPB markers. At 32 hpf, ptf1a and hlxb9la were absent in the pancreatic region of alk8 mutants, whereas their expression in the neural tube remained normal (Figure 1, A, B, E, and F). However, expression of these two markers was detected a few hours later at around 38 hpf in the pancreatic region of alk8 mutants, although in very few cells and at a reduced level compared with wild-type siblings (Figure 1, C, D, G, and H). Although hlxb9la was strongly reduced in the prospective VPB of alk8 mutants at 38 hpf, its expression in the dorsal pancreatic bud was unchanged (see arrowhead in Figure 1H). In contrast to ptf1a and hlxb9la, the pan-pancreatic marker pdx1 was not reduced in the prospective ventral bud at 30 hpf (Figure 1, I and J). Taken together, these results indicate that BMP signaling is required for the proper activation of the two ventral pancreatic genes ptf1a and hlxb9la but not for the pan-pancreatic gene pdx1. These data suggest that the severe pancreas hypoplasia recently reported at 72 hpf in the alk8 mutant not only is due to an outgrowth defect of the VPB, but also results from a severe delay in its specification.
FIGURE 1:
Decrease in BMP signaling leads to a delay in VPB specification and to a reduction in ptf1a+ and hlxb9la+ VPB progenitors. (A, B) Ventral view of ptf1a expression in wild-type (wt) sibling compared with alk8 mutant (alk8−/−) at 32 hpf. (C, and D) Lateral view of ptf1a expression in wt and alk8 mutant at 36–38 hpf. (E–J) Lateral view of hlxb9la expression in wt and alk8 mutant at 32 hpf (E, and H) and 36–38 hpf. (I, J). Lateral view of pdx1 expression in wt and alk8 mutant. White and black lines and white-dotted circle indicate the ventral pancreatic bud (VPB); white-dotted lines indicate the prospective ventral pancreatic bud; and white arrowheads indicate the dorsal pancreatic bud (DPB).
BMP signaling is required at 20 hpf to induce ventral bud specification
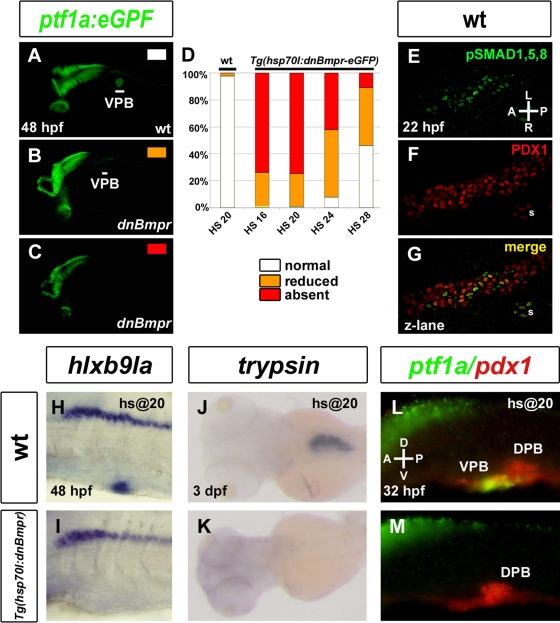
To determine more precisely the time window during which the BMP signaling is required to specify the VPB, we blocked this pathway using the transgenic fish Tg(hsp70l:dnBmpr-GFP) (Pyati et al., 2005) expressing a dominant-negative form of the BMP receptor fused to green fluorescent protein (GFP) under the control of the inducible heat shock promoter hsp70. These fish were crossed with Tg(ptf1a:eGFP) fish, and embryos from this cross were exposed to one heat shock at 16 hpf (n = 88), 20 hpf (n = 103), 24 hpf (n = 104), or 28 hpf (n = 100). Expression of the dominant-negative BMP receptor was verified after the heat shock by following the induced ubiquitous GFP expression, which was strong at 3 h and weak at 24 h after induction. Expression of ptf1a:eGFP in the ventral pancreatic region was analyzed at 48 hpf. Heat-shocked embryos could be sorted into three kinds of phenotype: unaffected (Figure 2A), strongly reduced (Figure 2B), and absent (Figure 2C) in the pancreatic region, whereas other expression domains such as eyes and brain remained unaltered. The strongest inhibition of ptf1a:eGFP in the VPB was observed in Tg(hsp701:dnBmpr-GFP) embryos heat shocked at 16 or 20 hpf, as 75% of transgenic embryos displayed no GFP in the VPB (Figure 2D). By contrast, heat shocking of embryos lacking the hsp70:dnBmpr-GFP transgene had no effect on the expression of ptf1a:GFP. Blocking BMP signaling from 24 hpf in the double-transgenic embryos still had an inhibitory effect on VPB specification, but the proportion of affected embryos was smaller compared with 16 and 20 hpf. This proportion was further reduced when the heat shock was performed at 28 hpf. These data indicate that BMP activity is required from 20 hpf onward for the specification of VPB. To determine whether, in wild-type embryos, the BMP pathway is activated in endodermal cells during this time window, we performed an immunostaining at 22 hpf with anti-phosphorylated Smad1/5/8 antibody and counterstained with anti-Pdx1 antibody to label pancreatic cells (Figure 2, E–G). Nuclear phospho-Smad 1/5/8 staining was indeed detected in some Pdx1+ ventral cells, indicating that BMP pathway is activated in pancreatic cells at 22 hpf.
FIGURE 2:
BMP signaling is essential at 20 hpf for VPB specification. (A–C, H–M) Embryos obtained from outcrossing a hemizygous Tg(hsp70l:dnBmpr-GFP) zebrafish with homozygous Tg(ptf1a:eGFP) were heat shocked at 20 hpf and harvested at 30 hpf (L, M) 48 hpf (A–C, H–I), and 3 dpf (J, K) and examined for ptf1a:eGFP fluorescence (A–C) and hlxb9la (H, I), trypsin (J, K), and ptf1a and pdx1 (L, M) expression. (D) Graph quantifying ptf1a:GFP expression in wild-type and Tg(hsp70l:dnBmpr-GFP) heat-shocked embryos at 16, 20, 24. and 28 hpf. Data are presented as the percentage of embryos displaying normal (white), reduced (orange), or absent (red) ptf1a:GFP fluorescence. (E–G) Confocal z-lane showing activation of the BMP pathway in pancreatic cells as revealed by immunostaining with anti-pSmad 1/5/8 (green) (E) and anti-Pdx1 (red) (H) antibodies. Double pSmad 1/5/8+ and Pdx1+ cells are observed on the merge picture (G). White lines indicate the VPB. DPB, dorsal pancreatic bud; s, somite.
As for alk8 mutants, we examined the expression of other pancreatic markers in embryos heat shocked at 20 hpf. Similar to the ptf1a gene, no expression of hlxb9la could be detected in the VPB when analyzed at 48hpf (Figure 2, H and I). Moreover, trypsin, a marker of the acinar pancreatic tissue—the major VPB derivative—is totally absent at 3 dpf (Figure 2, J and K). Despite the absence of ptf1a expression at 32 hpf after BMP inhibition, pdx1 expression was maintained, as observed in the alk8 mutants (Figure 2, L and M). Taken together, theses results indicate that activation of the BMP pathway in endodermal cells after 20 hpf is crucial for the induction of the early ventral pancreatic markers ptf1a and hlxb9la and for further acinar differentiation.
bmp2a is expressed in LPM adjacent to the ventral pancreatic bud and is involved in its specification
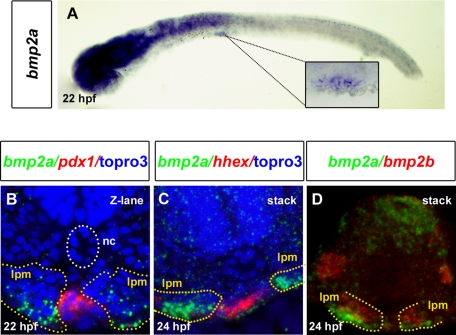
The LPM has been shown in vertebrates to be essential for proper specification of liver and pancreas (Manfroid et al., 2007; Chung et al., 2008). Zebrafish bmp2a and bmp2b were both reported to be expressed in LPM at 24 hpf (Chocron et al., 2007; Chung et al., 2008). bmp2b is expressed in LPM since the 10-somite stage and is required for hepatic specification at the expense of pancreatic fate, whereas bmp2a seems activated at later stages. Thus we investigated more closely the expression of bmp2a in the LPM adjacent to the prospective VPB when the BMP pathway is required for VPB specification. bmp2a expression starts to be detected in the LPM from 22 hpf at the level of the third somite, that is, in the hepatopancreatic region (Figure 3A), and its expression increases at 24 hpf (Figure 3C). To confirm that bmp2a was expressed adjacent to the pancreatic region, a double in situ hybridization with a pdx1 probe was performed. Bmp2a was expressed just next to and on both sides of the pdx1 expression domain (Figure 3B). Expression of bmp2a in LPM is also adjacent to the hepatic anlagen, as highlighted by hhex staining (shown at 24 hpf in Figure 3C). Furthermore, bmp2a is expressed more ventrally compared with bmp2b (Figure 3D) and more closely to the prospective VPB and the hepatic bud. Thus these results show that bmp2a is expressed at the correct time and place to be the BMP ligand responsible for proper ventral pancreatic bud specification.
FIGURE 3:
bmp2a is expressed in the LPM adjacent to the hepatopancreatic endoderm at 22 hpf. (A) Lateral view showing bmp2a expression in the LPM starting at 22 hpf. The square shows a close-up of the region indicated by the dotted lines. (B) bmp2a (green) and pdx1 (red) expression analyzed by fluorescence whole-mount in situ hybridization at 22 hpf. The transverse sections were analyzed by confocal microscopy. (C) bmp2a (green) and hhex (red) expression analyzed at 24 hpf. (D) bmp2a (green) and bmp2b (red) expression comparison at 24 hpf. The images of transverse sections are flat stacking of several consecutive optical sections. TO-PRO-3 (blue) labels nuclei. Yellow and white dots delineate the LPM and the notochord (nc), respectively.
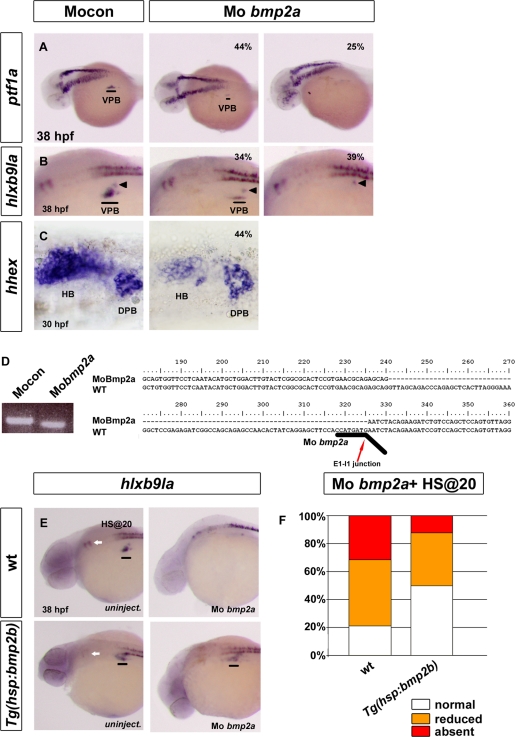
To test the role of bmp2a in VPB specification, we performed a knockdown of its expression using a morpholino targeting the splice donor site at the bmp2a exon 1–intron 1 junction. The efficiency of this knockdown was checked by performing reverse transcription (RT)-PCR on RNA extracted from injected embryos using primers annealing to exons 1 and 2 of bmp2a. The amplified cDNA was shorter in bmp2a morphants compared with control embryos, and sequencing of this fragment revealed that the bmp2a morpholino induces a deletion of 86 base pairs in exon 1 coding sequence (Figure 4D). This deletion induces a frameshift leading to a truncated protein of 101 instead of 386 amino acids. When the ventral pancreatic markers ptf1a and hlxb9la were analyzed at 36–38 hpf in bmp2a morphants, absence or strong reduction of expression was noted in 69 and 73% of bmp2a morphants for these two genes, respectively (n = 85 and 91; Figure 4, A and B). This effect was specific to the VPB, and expression of hlxb9la in dorsal pancreatic bud remained unaffected (see arrowheads in Figure 4B). Expression of insulin and pdx1 was not modified at 30 hpf (data not shown). As observed in alk8 mutants, expression of ptf1a and hlxb9la was detected at later stages (48 hpf) in almost all bmp2a morphants but was significantly reduced compared with control embryos (data not shown). Thus these data clearly demonstrate the involvement of bmp2a in VPB development, with its knockdown leading to a strong delay in VPB specification.
FIGURE 4:
Knockdown of bmp2a represses VPB specification. (A–C) Analysis of ventral pancreatic and hepatic markers in control morphants (left) and in bmp2a morphants (right) by in situ hybridization. (A) Lateral view of ptf1a expression at 36–38 hpf. (B) Laterial view of hlxb9la expression at 36–38 hpf. (C) Ventral view of hhex expression at 30 hpf. Percentage represents the proportion of morphants exhibiting a reduction (middle) or an absence (right) of gene expression. (D) Gel electrophoresis after RT-PCR (left) and sequencing result (right) illustrating the RNA splicing defects caused by the bmp2a morpholino in injected embryos. The sequence of the amplified bmp2a fragment was aligned with wt bmp2a cDNA and revealed a 86–base pair deletion in exon 1. Black line underlines the binding site of the bmp2a morpholino. Red arrow shows the junction between exon 1 and intron 1. (E, F) Lateral view of hlxb9la expression at 38 hpf in wt and Tg(hsp70l:bmp2b) in uninjected embryos (E) or in bmp2a morphants (F). Embryos were heat shocked at 20 hpf. (G) hlb9la expression in wt and Tg(hsp70l:bmp2b) embryos injected with MO bmp2a, heat shocked at 20 hpf, and fixed at 38 hpf. Data are presented as the percentage of embryos displaying normal (white), reduced (orange), or absent (red) expression of hlxb9la. Black lines indicate the VPB; black arrowheads indicate the dorsal pancreatic bud (DPB); white arrows indicate rhombomere 5 and 6 localization. HB, hepatic bud.
Because several studies showed a role of BMP in induction of the hepatic bud (Shin et al., 2007; Huang et al., 2008) and bmp2a is expressed adjacent to the hhex expression domain (Figure 3C), we also analyzed the hepatic markers hhex and prox1 in the bmp2a morphants. Although the expression of these two hepatic genes was still detected, their expression in the hepatic bud was clearly reduced in 44% of morphants (n = 153) for hhex (Figure 4C) and in 55% of morphants for prox1 (n = 64). These two genes were also expressed at normal level in the dorsal pancreatic bud. Thus, not only is bmp2a crucial for the specification of the VPB, but it is also involved in hepatic development, although it is less essential there than it is for the VPB.
To verify that the delay of the VPB specification caused by bmp2a morpholino is actually due to the loss of BMP2a activity, we next assessed whether induced ectopic BMP expression could rescue this defect. To activate BMP expression specifically at 20 hpf, we used the Tg(hsp70l:bmp2b) transgenic line allowing induction of bmp2b expression upon heat shock. bmp2a morpholinos were injected in eggs obtained from an outcross between Tg(hsp70l:bmp2b) and wild-type fish. Then injected embryos were heat shocked at 20 hpf, and hlxb9la expression was analyzed by in situ hybridization at 38 hpf. Identification of transgenic and nontransgenic embryos was performed after hlxb9la staining by genotyping. Tg(hsp70l:bmp2b) embryos also display loss of hlxb9la expression in rhombomeres 5 and 6 (see arrowheads in Figure 4, E and F). The rescue of hlxb9la expression in bmp2a morphants is shown in Figure 4G. The percentage of embryos exhibiting absence of hlxb9la expression in the pancreatic region was drastically reduced in the transgenic embryos (12%, n = 32) compared with the nontransgenic embryos (31%, n = 38). In addition, the percentage of embryos with normal hlxb9la expression in the VPB was significantly higher in the transgenic (50%) compared with nontransgenic (21%) embryos. Thus overexpression of bmp2b at 20 hpf can partially rescue the knockdown of bmp2a, and this indicates that the delay in VPB specification caused by the bmp2a morpholino is specific.
fgf10 and 24 are specifically involved in ventral pancreatic specification
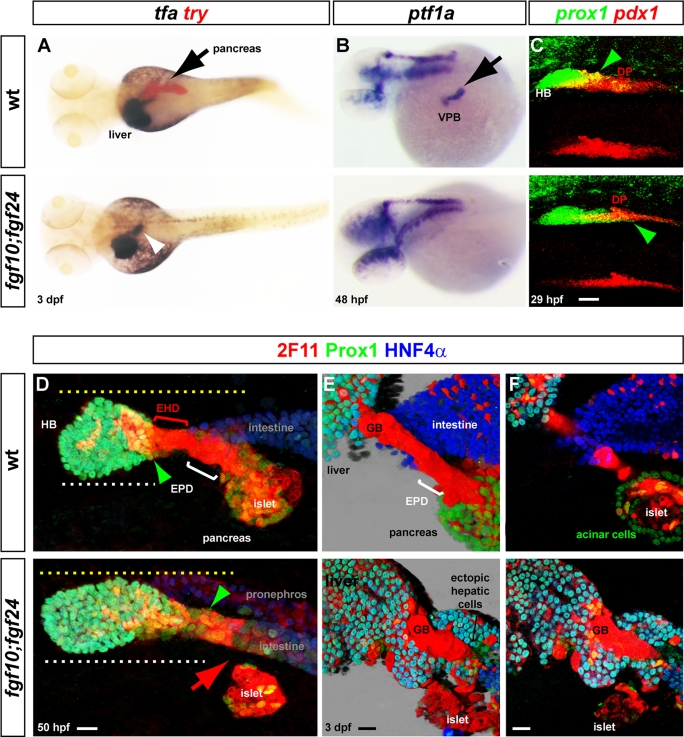
Because the specification of the ventral pancreas and the development of the neighboring hepatic bud both rely on bmp2a, other extrinsic factors must determine the choice between the pancreatic or hepatic fate. We recently reported by morpholino knockdown experiments that fgf10 acts redundantly with fgf24 to control the formation of the pancreatic exocrine tissue (Manfroid et al., 2007). However, the role of these two FGF ligands on hepatic specification has not been investigated. Therefore we generated fgf10; fgf24 compound mutants and examined in detail the development of the entire hepatopancreatic region. Analysis of trypsin and transferrin (tfa) expression at 3 dpf indicated a complete lack of the pancreatic exocrine tissue in the double mutants, whereas the liver was not reduced (Figure 5A), confirming our previous knockdown data. The absence of pancreatic exocrine tissue was due to a defect in VPB specification, as expression of ptf1a and hlxb9la was not detected in most double fgf10; fgf24 mutants at 48 hpf (Figure 5B and data not shown). Because the VPB also gives rise to the pancreatic ducts, their formation was next analyzed in the compound mutants. Immunolabeling with Prox1, HNF4α, and 2F11 antibodies revealed complete lack of the extrapancreatic duct (EPD) connecting the pancreas to the intestine in the mutants at 50 hpf (red arrowhead). Indeed, whereas the 2F11+ ductal cells delineating the extrahepatic duct (EHD) still make the junction between the hepatic bud and intestine in the mutants, extrapancreatic ductal cells could not be detected between the pancreatic islet, derived from the dorsal pancreatic bud (also 2F11+), and the intestine (Figure 5D). In addition, the hepatic domain appeared posteriorly expanded toward the pancreatic endocrine islet in all mutants examined (green arrowheads; n = 6 for wild-type embryos and n = 8 for mutant embryos). Indeed, whereas the distance from the anterior limit of the hepatic bud to the islet did not significantly change (125 μm in wild-type embryos and 127 μm in mutants; yellow dotted lines), the length of the hepatic domain increased (108 μm in mutants compared with 75 μm in wild-type embryos; white dotted lines). When the double fgf10; fgf24 mutants were analyzed at 3 dpf, no pancreatic acinar cell (Prox1+) could be detected around the islet nor at intra- and extrapancreatic ducts (Figure 5, E and F). In addition, masses of ectopic hepatocyte-like (Prox1+/HNF4α+/2F11−) cells and biliary-like cells (Prox1+/HNF4α−/2F11+ cells) were observed contiguous to the hepatopancreatic duct remnants and also in direct contact with the gut (Figure 5, E and F). Because ectopic tfa expression was also noticed in this region (see white arrowhead in Figure 5A), this supports ectopic hepatic differentiation occurring posterior to the hepatic domain. Thus all these data demonstrate a complete lack of pancreatic acinar and pancreatic ductal cells in the fgf10; fgf24 mutants, whereas hepatic cell differentiation seems increased. These results led us to examine whether, in the double mutants, specification of the hepatic bud was favored at the expense of the ventral pancreas. To that goal, we examined the expression of the hepatic marker prox1 and of the pancreatic marker pdx1 at 29 hpf, that is, just prior to the initiation of ventral pancreas specification (Figure 5C). Of interest, pdx1 labeling in the ventral aspect of the pancreatic domain was dramatically reduced in all the double mutants, whereas the prox1+ hepatic domain was expanded posteriorly and reached the dorsal pancreas (green arrowheads, n = 4). This observation suggests that, in the absence of FGF10/FGF24 signaling, hepatic progenitors are specified at the expense of the ventral pancreatic progenitors.
FIGURE 5:
Absence of VPB cells and ectopic hepatic cells in fgf10; fgf24 compound mutants. (A) Dorsal view of transferrin (tfa) and trypsin (try) expression, comparing wt and fgf10; fgf24 mutants at 3 dpf. (B) Dorsolateral view of ptf1a expression comparing wt and double mutants at 48 hpf. (C) Confocal projection of the pdx1 and prox1 expression domains at 29 hpf showing the severe decrease of pdx1 in the anterior part of the pancreatic region and the extension of prox1 in fgf10; fgf24 mutants (green arrowheads). (D–F) Immunolabeling of the hepatopancreatic region with 2F11, Prox1, and HNF4α in wild-type and fgf10; fgf24 double mutants. (D) At 50 hpf, the ventral pancreas and the EPD are absent (red arrow) in fgf10; fgf24 mutants (confocal projections). (E, F) Morphology and differentiation of the hepatopancreatic ducts connecting the pancreas and liver to the intestine in fgf10; fgf24 double mutants at 3 dpf. (E) Three-dimensional blend projection. (F). Z-planes). Black arrows indicate pancreas and VPB; white arrowhead indicates ectopic tfa expression. EHD, extrahepatic duct; EPD, extrapancreatic duct; GB, gall bladder; islet, endocrine islet. Scale bar, 20 μm.
Because bmp2a, fgf24, and fgf 10 are involved in specification of the VPB, we finally tested whether cross-regulation occurs between these extrinsic factors. Because fgf10 expression starts in the LPM ∼5 h after the onset of bmp2a expression (Manfroid et al., 2007), the activation of bmp2a cannot be controlled by fgf10. As previously reported, the fgf24 gene displays a dynamic expression pattern, being expressed in the gut endoderm before 24 hpf, and then endodermal expression progressively decreases at the level of the pancreas, whereas it appears in the adjacent pancreatic LPM. Expression of bmp2a was not modified in fgf24 mutants at 24 hpf (data not shown). Conversely, fgf24 and fgf10 expression was analyzed in bmp2a morphants. To accurately discern fgf24 expression in the endoderm and in the LPM, double in situ hybridization was performed with fgf24 and pdx1 probes. Comparison of wild-type and bmp2a morphant embryos at 32 hpf revealed that the expression of fgf24 is strongly decreased in the pancreatic LPM, whereas the level in the endoderm remains high (Supplemental Figure S1A). In contrast, fgf10 expression in the LPM was not affected in bmp2a morphants (Supplemental Figure S1B). All these data indicate that bmp2a acts not only on the endoderm, but also on the mesoderm, where it is involved in the activation of fgf24 expression.
DISCUSSION
Conflicting data have been reported on the effect of BMP and FGF signals on pancreas development, either showing a negative effect (Deutsch et al., 2001; Rossi et al., 2001; Shin et al., 2007; Chung et al., 2008; Spagnoli and Brivanlou, 2008; Tehrani and Lin, 2011) or a positive effect (Kumar et al., 2003; Manfroid et al., 2007; Wandzioch and Zaret, 2009). Studies on mouse embryonic explants first indicated that these two signaling pathways act on endodermal progenitors to favor hepatic differentiation while restricting formation of pancreatic cells (Rossi et al., 2001). Such role of the BMP pathway has also been confirmed in zebrafish; indeed, BMP2b secreted from the LPM around 14 hpf acts on endodermal cells to induce hepatic fate at the expense of pancreas (Chung et al., 2008). In the present study, we show that, a few hours later at around 20–24 hpf, the BMP pathway plays a different role and is actually essential for the specification of the ventral pancreatic bud. Furthermore, we identify BMP2a as a major BMP ligand responsible for this induction process. Similarly, FGF signaling has been shown to play a crucial role in development of liver in vertebrate (Deutsch et al., 2001) and notably in zebrafish embryos between 18 and 22 hpf, that is, just before hepatic specification (Shin et al., 2007). Here we demonstrate that fgf10 and fgf24, also expressed in the LPM around 26 hpf and required for ventral pancreas specification, impair hepatic development. Finally, we show that bmp2a positively regulates fgf24 expression in the LPM. In conclusion, our data underscore the importance of the developmental stage in the response to these extrinsic signals.
BMP signaling is crucial for ventral pancreatic specification
A recent analysis of the alk8 zebrafish mutants revealed an increase of endocrine pancreatic β cells but also a severe hypoplasy of the ventral pancreas (Chung et al., 2010). In this study, we focused our analyses on the first steps of ventral pancreatic bud development by analyzing three early pancreatic markers—pdx1, ptf1a, and hlxb9la. We show that these three pancreatic regulatory genes are differently controlled in response to BMP signaling. Indeed, whereas pdx1 was not significantly affected in the alk8 mutants, expression of ptf1a and hlxb9la was not detected at 32 hpf and was strongly reduced after 38 hpf in these mutants. A similar regulation was observed when BMP pathway was disrupted by two other means: first, by blocking BMP receptors after 20 hpf by using the Tg(hsp70l:dnbmpr-GFP) line, and second, by the knockdown of bmp2a.
The delay in the formation of the ptf1a+ cells was longer after induction of the dominant-negative receptor dnBMPR-GFP than in alk8 mutants and bmp2a morphants (a 16-h delay compared with 6 h). Such differences in phenotype severity are probably due to compensation by other BMP receptors I and other BMP ligands expressed at later stages. bmp2a expression in the LPM coincides in timing and space with the requirement of BMP for ventral pancreas specification. It is interesting to recall that its paralogue, bmp2b, expressed a few hours before, triggers an opposite action on pancreas specification, indeed favoring hepatic fate at the expense of pancreatic fate. This is not due to different biological activities of the two BMP2 proteins, as bmp2a morphant defects can be partially rescued by bmp2b overexpression, but rather results from a difference in their timing of expression. We can speculate that, after the genome duplication that occurred early during teleost evolution, each copy of the ancestral bmp2 gene retained distinct regulatory sequences driving expression in the LPM either at early (for bmp2b) or later (for bmp2a) developmental stages. As in zebrafish, the critical stages of liver specification precede ventral pancreas specification, and the role of the two bmp2 paralogues was split, with bmp2b acting on liver specification and bmp2a acting on ventral pancreas specification and on the maintenance of hepatic genes hhex and prox1.
fgf10 and -24 specify the VPB at the expense of the hepatic fate
One important finding of our study is the role fgf10 and fgf24 in the induction of ventral pancreas while they restrict hepatic competence. Activation of ptf1a and hlxb9la genes in endoderm is not detected at 32 hpf in the double fgf10; fgf24 mutants, and the pdx1+ domain becomes significantly reduced while the prox1+ hepatic domain extends posteriorly. Only a delay in ptf1a and hlxb9la endodermal expression was observed either in fgf24 homozygous mutants or in some double heterozygotes (data not shown), but expression of these two genes reappears later, around 44 hpf (data not shown). This indicates that a correct level of FGF is required for proper timing of ventral pancreas specification. At 3 dpf, the fgf10−/−; fgf24−/− larvae possess only the pancreatic endocrine islet, which is derived from the dorsal bud, and no pancreatic exocrine tissue. The liver bud in these mutant larvae is apparently normal, but ectopic masses of hepatocytes develop in the endoderm just posterior to the liver, near the remnant of the hepatopancreatic ducts and close to the junction with the intestine. The actual origin of these hepatocytes is unclear; they may derive from the posterior extension of the prox1+ hepatic domain observed at 30 hpf (Figure 3C). Two zebrafish studies recently described defects highly similar to the fgf10; fgf24 mutant phenotype, as they showed that the endoderm posterior to the hepatic domain, corresponding to the prospective intestinal bulb and pancreas, can give rise to ectopic hepatocytes upon Wnt overexpression (Poulain and Ober, 2011; Shin et al., 2011). Of interest, this hepatic competence can be induced by Wnt overexpression at 26 hpf, thus following liver specification in the hepatic progenitor domain at 22 hpf. Moreover, it is negatively regulated with time by FGF signaling (Shin et al., 2011), and fgf10a (here referred to as fgf10) was shown to be partially responsible of this inhibitory effect. The present data indicate that the combined loss of fgf10 and fgf24, both genes being expressed in the LPM after liver specification, is sufficient to trigger hepatic cell differentiation posterior to the normal hepatic domain without artificial Wnt overexpression. Whether Wnt signaling is activated in the fgf10; fgf24 compound mutants remains to be elucidated.
Our data showing the ability of FGF signaling to induce pancreatic fate versus hepatic fate contrast drastically with previous data on mouse embryonic explants demonstrating the opposite effect of FGF (Deutsch et al., 2001). This difference does not seem to be due to intrinsic biological activities of FGF10 and FGF24 protein ligands, as the phenotype of the fgf10; fgf24 mutant can be almost recapitulated in wild-type zebrafish embryos treated with FGF inhibitor SU5402 between 24 and 48 hpf (data not shown; Manfroid et al., 2007). In contrast, if FGF signaling is blocked at earlier stage (i.e., from 18 hpf onward), liver specification is disrupted (Shin et al., 2007). This again underscores the importance of developmental stage in the response to extrinsic factors.
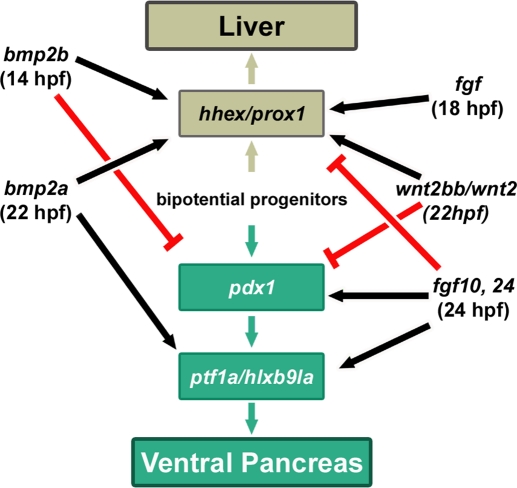
From data of this and other zebrafish studies, we can build a model in which hepatic versus pancreatic commitment is controlled by the sequential action of distinct extrinsic factors (Figure 6). First, bmp2b expressed in the LPM from 14 hpf promotes hepatic fate at the expense of pancreatic fate, notably through a down-regulation of pdx1 in gut endoderm (Shin et al., 2007; Chung et al., 2008). Then, around 18–22 hpf, a FGF signal, the exact identity of which is still unknown, is required to activate hhex and prox1 genes in hepatoblasts (Shin et al., 2007). Activation of these two hepatic genes also necessitates Wnt2bb expression from the LPM from 22 hpf (Ober et al., 2006). BMP2A is also secreted from that stage to allow activation of ptf1a and hlxb9la genes in the VPB and to maintain high expression level of hhex and prox1 in hepatoblasts. Secretion of FGF10 and FGF24 from around 24–26 hpf is essential for the onset of ptf1a and hlxb9la expression determining ventral pancreatic bud. In the absence of these two FGF ligands, the prox1/hhex+ hepatic anlagen expands posteriorly at the expense of pdx1+ cells. Finally, our results also reveal that bmp2a stimulates fgf24 expression in the pancreatic LPM. Because fgf24 expression in the LPM, reflecting proper pancreatic LPM patterning (Manfroid et al., 2007), is important for VPB specification, this thereby demonstrates that cross-talks between BMP and FGF pathways contribute to coordinated liver and pancreas development.
FIGURE 6:
Model for the role of BMP and FGF pathways in hepatopancreatic patterning. Bipotential endodermal progenitors, shown in the center of the scheme, can give rise to liver and pancreatic cells through the consecutive action of extrinsic signals secreted from the lateral mesoderm. Hepatic specification is determined by hhex and prox1 expression, whereas pdx1, ptf1a and hlxb9la are markers of pancreatic cell commitment.
MATERIALS AND METHODS
Embryos
Zebrafish (Danio rerio) were raised and cared for according to standard protocols (Westerfield, 1995). Wild-type embryos from the AB strain were used and staged according to Kimmel et al. (1995). We used the following mutant and transgenic lines: laftm110b (Mintzer et al., 2001), Tg(hsp70l:dnBmpr-GFP) (Pyati et al., 2005), Tg(ptf1a:eGFP), (Godinho et al., 2007) and Tg(hsp70l:bmp2b) (Chocron et al., 2007).
Heat shock conditions
Embryos were heat shocked at various stages by transferring them into a prewarmed Falcon tube containing egg water in a water bath. Tg(hsp70l:dnBmpr-GFP) embryos were heat shocked for 30 min at 40°C; Tg(hsp70l:bmp2b) embryos for 30 min at 37°C. After heat shock, embryos were transferred at 28°C and then were harvested between 30 and 72 hpf. Hemizygous Tg(hsp70l:dnBmpr-GFP) embryos were sorted 3 h after heat shock based on GFP-positive expression. Hemizygous Tg(hsp70l:bmp2b) embryos were tested for presence of the hsp70l:bmp2b transgene by PCR using genomic DNA after in situ hybridization, using the primers previously used Shin et al. (2007). In our hands, a nested PCR was needed, and the following primers were used: 5′-GCAAAAGGCCAGGAACCGTAA-3′ and 5′- GCACACAGCCCAGCTTGGAGC-3′.
Whole-mount in situ hybridization and immunochemistry
Visible in situ hybridization was performed as described (Hauptmann and Gerster, 1994). Fluorescence labeling was performed as described (Mavropoulos et al., 2005).
The riboprobes used were ptf1a (Zecchin et al., 2004), hlxb9la (Wendik et al., 2004), trypsin (Biemar et al., 2001), pdx1 (Milewski et al., 1998), bmp2a (Thisse and Thisse, 2005), tfa (Mudumana et al., 2004), prox1 (Glasgow and Tomarev, 1998), and hhex (Ho et al., 1999).
Whole-mount immunohistochemistry was described in Dong et al. (2007). We used the following antibodies: polyclonal rabbit anti-Prox1 (1:1000; Chemicon, Temecula, CA), polyclonal rabbit anti–phospho-Smad (1:200; Cell Signaling Technology, Beverly, MA), polyclonal guinea pig against zebrafish Pdx1 (1:200; a gift from C. Wright, Vanderbilt University, Nashville, TN), polyclonal goat anti-HNF4α (1:100; Santa Cruz Biotechnology, Santa Cruz, CA), monoclonal mouse 2F11 (Crosnier et al., 2005; 1:1000; a gift from J. Lewis, London Research Institute), and fluorescently conjugated Alexa antibodies (Molecular Probes, Invitrogen, Carlsbad, CA).
Fluorescence images were acquired with a Leica SP2 (Wetzlar, Germany) or Olympus FluoView FV1000 (Tokyo, Japan) confocal microscope, and three-dimensional blend projections were performed with the Imaris software (Bitplane, Zurich, Switzerland).
Injection of morpholino antisense oligonucleotides
Morpholino oligonucleotides (MOs) were purchased from Gene Tools (Philomath, OR). Each MO was resuspended in Danieau's solution at the stock concentration of 8 μg/μl. For injections, they were further diluted in Danieau's solution at 4 ng/nl with 0.5% rhodamine dextran to check injection efficiency. The splice inhibition MO bmp2a (AGTAAACACTTGCTTACCATCATGG) targets the exon 1–intron 1 boundary.
Control of the morpholino efficiency
Zebrafish embryos were injected at one-cell stage with Mo Bmp2a (4 ng/embryo), and mRNAs were extracted at 30 hpf. RT-PCR was performed on 1-μg mRNAs. The primers used for PCR amplification were bmp2a exon 1 (upstream primer; 5′-GGCTCCAGTGGACTCGTTCCTCA-3′) and bmp2a exon 2 (downstream primer, 5′-CTCCTGAAGAGAACCGGACGGCCT-3z), and PCR cycles were performed as follows: 40 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C, followed by a final 7-min extension at 72°C. Amplified cDNAs were analyzed by gel electrophoresis and sequencing.
Genotyping
Genotyping was performed on genomic DNA extracted from adult tails or tails obtained from embryos processed through in situ hybridization or immunohistochemistry. The ikarus mutation in the fgf24 locus generates a restriction site for the AccI endonuclease. The PCR fragment obtained with forward 5′-CTGTCAGTCCCACAGCAGTGGACCA-3′ and reverse 5′-CCATGTAGATTTTATTACATGTAGGT-3′ primers (615 base pairs) digested by AccI produces two fragments (185 and 430 base pairs) in the mutant. The daedalus mutation in fgf10 generates a single-nucleotide polymorphism that was identified by the TaqMan SNP Genotyping Assays (Applied Biosystems, Foster City, CA). The region encompassing the mutation was amplified with the forward primer dae-SNp1F 5′-CCGAGCTCCAGGACAATGTG-3′ and reverse primer dae-SNp1R 5′-GCAGGACAGACGGAACCA-3′. Allelic discrimination was performed by dae-SNp1V2 VIC primer 5′-CCCTTAGTCACTTTCCATTT-3′ (wild-type allele) and dae-SNp1M2 FAM primer 5′-CCTTAGTCACTTACCATTT-3′ (mutant allele) according to the manufacturer.
Supplementary Material
Acknowledgments
We thank S. Ormenese, G. Moraes, and the GIGA-Cell Imaging Facility, W. Coppieters and the GIGA-GenoTranscriptomics Facility, and M. Winandy and the GIGA-Zebrafish Facility (Liège, Belgium). I.M. was supported by the Fonds de la Recherche Scientifique (F.R.S.-FNRS) and by the Action de Recherches Concertées (University of Liège). B.P. and M.L.V. are Chercheurs qualifiés of the F.R.S.-FNRS. N.D. and V.V.B. are funded by Waleo (Région Wallonne). F.N. has a postdoctoral fellowship from the University of Liège. This work was funded by the Belgian State Program on Interuniversity Poles of Attraction (SSTC, PAI) and by the Sixth European Union Framework Program (BetaCellTherapy Integrated Project).
Abbreviations used:
- BMP
bone morphogenic protein
- EHD
extrahepatic duct
- EPD
extrapancreatic duct
- FGF
fibroblasts growth factor
- VPB
ventral pancreatic bud
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-08-0664) on January 4, 2012.
REFERENCES
- Biemar F, Argenton F, Schmidtke R, Epperlein S, Peers B, Driever W. Pancreas development in zebrafish: early dispersed appearance of endocrine hormone expressing cells and their convergence to form the definitive islet. Dev Biol. 2001;230:189–203. doi: 10.1006/dbio.2000.0103. [DOI] [PubMed] [Google Scholar]
- Chocron S, Verhoeven MC, Rentzsch F, Hammerschmidt M, Bakkers J. Zebrafish Bmp4 regulates left-right asymmetry at two distinct developmental time points. Dev Biol. 2007;305:577–588. doi: 10.1016/j.ydbio.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Chung WS, Andersson O, Row R, Kimelman D, Stainier DY. Suppression of Alk8-mediated Bmp signaling cell-autonomously induces pancreatic beta-cells in zebrafish. Proc Natl Acad Sci USA. 2010;107:1142–1147. doi: 10.1073/pnas.0910205107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WS, Shin CH, Stainier DY. Bmp2 signaling regulates the hepatic versus pancreatic fate decision. Dev Cell. 2008;15:738–748. doi: 10.1016/j.devcel.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosnier C, Vargesson N, Gschmeissner S, Ariza-McNaughton L, Morrison A, Lewis J. Delta-Notch signalling controls commitment to a secretory fate in the zebrafish intestine. Development. 2005;132:1093–1104. doi: 10.1242/dev.01644. [DOI] [PubMed] [Google Scholar]
- Deutsch G, Jung J, Zheng M, Lora J, Zaret KS. A bipotential precursor population for pancreas and liver within the embryonic endoderm. Development. 2001;128:871–881. doi: 10.1242/dev.128.6.871. [DOI] [PubMed] [Google Scholar]
- Dong PD, Munson CA, Norton W, Crosnier C, Pan X, Gong Z, Neumann CJ, Stainier DY. Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat Genet. 2007;39:397–402. doi: 10.1038/ng1961. [DOI] [PubMed] [Google Scholar]
- Field HA, Dong PD, Beis D, Stainier DY. Formation of the digestive system in zebrafish. II. Pancreas morphogenesis. Dev Biol. 2003a;261:197–208. doi: 10.1016/s0012-1606(03)00308-7. [DOI] [PubMed] [Google Scholar]
- Field HA, Ober EA, Roeser T, Stainier DY. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev Biol. 2003b;253:279–290. doi: 10.1016/s0012-1606(02)00017-9. [DOI] [PubMed] [Google Scholar]
- Glasgow E, Tomarev SI. Restricted expression of the homeobox gene prox 1 in developing zebrafish. Mech Dev. 1998;76:175–178. doi: 10.1016/s0925-4773(98)00121-x. [DOI] [PubMed] [Google Scholar]
- Godinho L, Williams PR, Claassen Y, Provost E, Leach SD, Kamermans M, Wong RO. Nonapical symmetric divisions underlie horizontal cell layer formation in the developing retina in vivo. Neuron. 2007;56:597–603. doi: 10.1016/j.neuron.2007.09.036. [DOI] [PubMed] [Google Scholar]
- Hauptmann G, Gerster T. Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 1994;10:266. doi: 10.1016/0168-9525(90)90008-t. [DOI] [PubMed] [Google Scholar]
- Ho CY, Houart C, Wilson SW, Stainier DY. A role for the extraembryonic yolk syncytial layer in patterning the zebrafish embryo suggested by properties of the hex gene. Curr Biol. 1999;9:1131–1134. doi: 10.1016/s0960-9822(99)80485-0. [DOI] [PubMed] [Google Scholar]
- Huang H, et al. Mypt1-mediated spatial positioning of Bmp2-producing cells is essential for liver organogenesis. Development. 2008;135:3209–3218. doi: 10.1242/dev.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kumar M, Jordan N, Melton D, Grapin-Botton A. Signals from lateral plate mesoderm instruct endoderm toward a pancreatic fate. Dev Biol. 2003;259:109–122. doi: 10.1016/s0012-1606(03)00183-0. [DOI] [PubMed] [Google Scholar]
- Manfroid I, Delporte F, Baudhuin A, Motte P, Neumann CJ, Voz ML, Martial JA, Peers B. Reciprocal endoderm-mesoderm interactions mediated by fgf24 and fgf10 govern pancreas development. Development. 2007;134:4011–4021. doi: 10.1242/dev.007823. [DOI] [PubMed] [Google Scholar]
- Mavropoulos A, Devos N, Biemar F, Zecchin E, Argenton F, Edlund H, Motte P, Martial JA, Peers B. sox4b is a key player of pancreatic alpha cell differentiation in zebrafish. Dev Biol. 2005;285:211–223. doi: 10.1016/j.ydbio.2005.06.024. [DOI] [PubMed] [Google Scholar]
- Milewski WM, Duguay SJ, Chan SJ, Steiner DF. Conservation of PDX-1 structure, function, and expression in zebrafish. Endocrinology. 1998;139:1440–1449. doi: 10.1210/endo.139.3.5768. [DOI] [PubMed] [Google Scholar]
- Mintzer KA, Lee MA, Runke G, Trout J, Whitman M, Mullins MC. Lost-a-fin encodes a type I BMP receptor, Alk8, acting maternally and zygotically in dorsoventral pattern formation. Development. 2001;128:859–869. doi: 10.1242/dev.128.6.859. [DOI] [PubMed] [Google Scholar]
- Mudumana SP, Wan H, Singh M, Korzh V, Gong Z. Expression analyses of zebrafish transferrin, ifabp, and elastaseB mRNAs as differentiation markers for the three major endodermal organs: liver, intestine, and exocrine pancreas. Dev Dyn. 2004;230:165–173. doi: 10.1002/dvdy.20032. [DOI] [PubMed] [Google Scholar]
- Ober EA, Verkade H, Field HA, Stainier DY. Mesodermal Wnt2b signalling positively regulates liver specification. Nature. 2006;442:688–691. doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]
- Poulain M, Ober EA. Interplay between Wnt2 and Wnt2bb controls multiple steps of early foregut-derived organ development. Development. 2011;138:3557–3568. doi: 10.1242/dev.055921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyati UJ, Webb AE, Kimelman D. Transgenic zebrafish reveal stage-specific roles for Bmp signaling in ventral and posterior mesoderm development. Development. 2005;132:2333–2343. doi: 10.1242/dev.01806. [DOI] [PubMed] [Google Scholar]
- Rossi JM, Dunn NR, Hogan BL, Zaret KS. Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes Dev. 2001;15:1998–2009. doi: 10.1101/gad.904601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Lee Y, Poss KD, Stainier DY. Restriction of hepatic competence by Fgf signaling. Development. 2011;138:1339–1348. doi: 10.1242/dev.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Shin CH, Tucker J, Ober EA, Rentzsch F, Poss KD, Hammerschmidt M, Mullins MC, Stainier DY. Bmp and Fgf signaling are essential for liver specification in zebrafish. Development. 2007;134:2041–2050. doi: 10.1242/dev.000281. [DOI] [PubMed] [Google Scholar]
- Spagnoli FM, Brivanlou AH. The Gata5 target, TGIF2, defines the pancreatic region by modulating BMP signals within the endoderm. Development. 2008;135:451–461. doi: 10.1242/dev.008458. [DOI] [PubMed] [Google Scholar]
- Tehrani Z, Lin S. Antagonistic interactions of hedgehog, Bmp and retinoic acid signals control zebrafish endocrine pancreas development. Development. 2011;138:631–640. doi: 10.1242/dev.050450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High throughput expression analysis of ZF-models consortium clones. ZFIN. 2005. Direct Data Submission ZDB-PUB-051025-1.
- Wandzioch E, Zaret KS. Dynamic signaling network for the specification of embryonic pancreas and liver progenitors. Science. 2009;324:1707–1710. doi: 10.1126/science.1174497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendik B, Maier E, Meyer D. Zebrafish mnx genes in endocrine and exocrine pancreas formation. Dev Biol. 2004;268:372–383. doi: 10.1016/j.ydbio.2003.12.026. [DOI] [PubMed] [Google Scholar]
- Westerfield M. A Guide for the Laboratory Use of Zebrafish (Danio rerio) 3rd. Eugene: University of Oregon Press; 1995. The Zebrafish Book. [Google Scholar]
- Zecchin E, Mavropoulos A, Devos N, Filippi A, Tiso N, Meyer D, Peers B, Bortolussi M, Argenton F. Evolutionary conserved role of ptf1a in the specification of exocrine pancreatic fates. Dev Biol. 2004;268:174–184. doi: 10.1016/j.ydbio.2003.12.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.