ABSTRACT
The N-ethyl-N-nitrosourea-induced repro42 mutation, identified by a forward genetics strategy, causes both male and female infertility, with no other apparent phenotypes. Positional cloning led to the discovery of a nonsense mutation in Spata22, a hitherto uncharacterized gene conserved among bony vertebrates. Expression of both transcript and protein is restricted predominantly to germ cells of both sexes. Germ cells of repro42 mutant mice express Spata22 transcript, but not SPATA22 protein. Gametogenesis is profoundly affected by the mutation, and germ cells in repro42 mutant mice do not progress beyond early meiotic prophase, with subsequent germ cell loss in both males and females. The Spata22 gene is essential for one or more key events of early meiotic prophase, as homologous chromosomes of mutant germ cells do not achieve normal synapsis or repair meiotic DNA double-strand breaks. The repro42 mutation thus identifies a novel mammalian germ cell-specific gene required for meiotic progression.
Keywords: meiosis, mutagenesis, oogenesis, Spata22, spermatogenesis
An unbiased mutagenesis strategy identified a novel gene, Spata22, that is required for mammalian meiotic progression.
INTRODUCTION
Meiosis is a defining event of gametogenesis, leading to the production of haploid gametes capable of supporting embryonic development. Failure to execute meiosis correctly frequently causes infertility and/or aneuploidy. Approximately 15% of human couples worldwide suffer from infertility, and much of this involves meiotic failure [1]. Thus, greater knowledge of genes responsible for successful meiotic progression would significantly increase our understanding of male- and female-factor infertility.
Meiosis involves pairing of homologous chromosomes, recombination to ensure links (chiasmata) between homologs, and two division phases: one reductional to separate homologs and the other equational to separate sister chromatids. Assembly of the synaptonemal complex (SC), the proteinaceous scaffold postulated to facilitate synapsis and homologous recombination, is a hallmark of meiosis [2, 3]. The SC is a tripartite structure consisting of two lateral elements (LEs) and a central element (CE) [2, 4]. Proteins unique to the SC have been defined in a variety of organisms and are required for fertility. Events taking place during the lengthy first meiotic prophase ensure that homologous chromosomes pair, synapse, and undergo reciprocal recombination. In fungi, mammals, and plants, these events are intrinsically linked, and sterility is commonly observed in mouse models harboring mutations disrupting any of these processes [5, 6].
Although the basic features of homologous chromosome pairing, meiotic recombination, and reductional division occur in both males and females, in many species, and notably in mammals, there is sexual dimorphism of timing and events of meiosis [7, 8]. In males, creation of spermatozoa through the process of spermatogenesis requires the coordinated proliferation of spermatogonia, meiotic division of spermatocytes, and postmeiotic maturation of spermatids continuously throughout the lifespan of the individual. Meiosis is an entirely postnatal process, first beginning around Day 10 after birth in male mice. In mammalian females, both oogonial proliferation and initiation of meiosis occur during fetal development [9]. Shortly after birth, oocytes enter meiotic arrest prior to completion of meiosis I and remain arrested until ovulation. In humans, differences in the rate of aneuploidy are also noted between male and female germ cells. Oocytes are far more susceptible to chromosomal abnormalities, suggesting differences in the stringency of mechanisms monitoring the quality of events during meiosis [10].
Although meiosis is a remarkably conserved process, many meiotic genes are not evolutionarily conserved [6]. Studies of model organisms such as yeast have largely contributed to our understanding of the genetic control of meiosis in mammals. Mice carrying null mutations in orthologs of meiosis genes have identified conserved pathways from yeast to mammals, as meiotic phenotypes very similar to those observed in yeast are often uncovered in mice. However, it has become clear that certain critical factors in yeast are dispensable in mice, other genes required in yeast are not conserved in mammals, and many genes required for progression of meiosis in mammals do not have orthologs in yeast. Additionally, model organisms such as yeast provide little to no insight into the basis of sex-specific phenotypes of mutations in meiotic genes. These issues prompted us to use an unbiased forward genetics strategy to identify novel genes involved in meiotic progression in mice [11–13]. Here we report on the characterization of a mutation, repro42, which causes infertility in both males and females due to meiotic arrest during prophase I, thereby identifying a common, not sexually dimorphic, process. Positional cloning of repro42 led to the discovery of a nonsense mutation in spermatogenesis associated 22 (Spata22), a previously uncharacterized vertebrate-specific gene of unknown function. Synapsis and DNA double-strand break (DSB) repair are both severely impaired in mutant oocytes and spermatocytes, suggesting that SPATA22 is a novel germ cell-specific factor required for progression of meiotic prophase in mice.
MATERIALS AND METHODS
Mice
The repro42 mutation was induced in a C57BL6/J (B6) background and carriers were outcrossed to C3HeB/FeJ (C3H) or CAST/EiJ (CAST) to define the critical interval containing the mutation. Once the interval was identified, the mutation was maintained on a mixed B6/C3H background by mating heterozygous animals. Some experiments were conducted using B6SJL F1 mice as wild-type controls. Day of birth was designated as day postpartum (dpp) 0, and the day a vaginal plug was found was designated as day postcoitum (dpc) 0.5. Mice were maintained under standard conditions by the investigators at The Jackson Laboratory (JAX) (Bar Harbor, ME) in accordance with the National Institutes of Health and U.S. Department of Agriculture standards; all procedures conducted were approved by the JAX Animal Care and Use Committee.
Fine Mapping and Sequencing
Adult B6 males were mutagenized with N-ethyl-N-nitrosourea (ENU) and the repro42 infertility phenotype was identified in a standard three-generation breeding scheme as previously described [11]. To establish chromosomal linkage of the repro42 gene mutation, genome scans using two to three polymorphic satellite markers per autosomal chromosome were performed on DNA obtained from affected (infertile) and unaffected (fertile) G3 mice. A region of 18.36 Mb on Chromosome (Chr) 11, located between D11Mit208 and D11Mit245, was identified as the candidate region for the repro42 mutation. For fine mapping, heterozygous B6/C3H progeny carrying the mutation were intercrossed for several generations, and recombinant individuals were analyzed for fertility and typed for additional polymorphic markers within the region. Heterozygous repro42 carriers were also crossed with CAST males and females, and F1 offspring intercrossed to produce the F2 recombinants used to narrow the crucial interval. For genetic fine mapping, repro42/+ mice were also crossed to CAST males and females, and F2 individuals were tested for phenotype and genotyped with additional polymorphic markers. Mice used for experiments were genotyped by PCR amplification of tail DNA with the polymorphic markers D11Jmp3 and D11Nds1.
For sequencing of candidate genes, genomic DNA was isolated from tail tips of repro42/+ and repro42/repro42, as well as the founder strain B6. Primer pairs were designed to flank exons and surrounding intronic sequences of each of the candidate genes (sequences are available upon request). Following PCR amplification, products were electrophoresed on 2% SeaKem agarose gels (Lonza, Rockland, ME); suitable bands were cut and DNA was extracted using the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA). DNA sequencing of exons and exon-intron boundaries was performed by the JAX Genome Sciences Service using standard methods.
Bioinformatic Analysis
Information related to the genomic structure, mRNA transcripts, and protein products of Spata22 was obtained through the Ensembl Genome Browser, Release 47 (http://uswest.ensembl.org/index.html), the University of California Santa Cruz Genome Browser (http://genome.ucsc.edu/), the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/), and Mouse Genome Informatics (MGI) resources (http://www.informatics.jax.org/), using the Build 37 mouse genome assembly by NCBI (NCBI37/mm9). Orthology (gene conservation through speciation) and paralogy (homology associated with gene duplication) were also assessed using these platforms. Multiple sequence alignment of identified orthologs (proven or predicted) was conducted using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) to uncover conserved regions and establish the phylogenic relationship. Predictive tools from ExPASy Proteomics Server (http://au.expasy.org/sprot/), Eukaryotic Linear Motif (http://elm.eu.org/), and the graphical tool GlobPlot (http://globplot.embl.de/) were used to analyze the SPATA22 protein sequence and determine putative posttranslational modifications.
Histology and Immunohistochemistry
Testes and ovaries were fixed by immersion in Bouin fixative (Rowley Biochemical Institute, Danvers, MA) (2–5 h for prepubertal testes and ovaries; overnight for adult testes), dehydrated, and paraffin embedded. Sections (5 μm thick) were cut, mounted on glass slides, deparaffinized with xylene, and stained with periodic acid-Schiff following standard procedures. A Leica Leitz DMRD upright microscope was used to visualize the slides, and images were acquired using a DCF 300FXRI camera and Leica FireCam software (Leica Microsystems, Bannockburn, IL).
For quantification of germ cell numbers, the paraffin-embedded testes used for histological analysis were cut into serial sections and every fifth section was analyzed. The monoclonal germ cell nuclear antigen 1 (GCNA1) antibody was used to identify germ cells [14]. Following deparaffinization and rehydration of sections, antigen retrieval was performed using sodium citrate buffer and heating in a microwave for 10 min (until boiling). Following quenching of endogenous peroxidase activity with 3% hydrogen peroxide (Sigma Aldrich, St. Louis, MO), sections were blocked for 1 h in antibody dilution buffer (ADB) (3% bovine serum albumin [BSA], 10% goat serum, and 0.05% Triton X-100; Sigma) and then incubated with the anti-GCNA1 antibody (1:2 dilution in ADB; a gift from G. Enders) at 37°C for 1 h in a humidified chamber. Antigens were localized using SuperPicTure Polymer Detection Kit according to the manufacturer's instructions (Invitrogen, Camarillo, CA). Sections were counterstained with Mayer hematoxylin (Sigma) and mounted in ClearMount (Invitrogen). Slides were viewed and images captured as described above. Alternatively, some slides were scanned with the NanoZoomer Digital Pathology system (Aperio Technologies, Vista, CA). The number of germ cells in 200 tubules was assessed for a minimum of two animals per genotype at each time point (except for 8 dpp) by an individual blinded to sample genotypes. Results are reported as mean number of germ cells per 200 tubules.
For localization of SPATA22, testes fixed by immersion in 4% paraformaldehyde were sectioned and processed as described above, except for incubation with the primary antibody, where sections were incubated with a commercial rabbit polyclonal antibody raised against full-length human recombinant SPATA22 (Proteintech, Chicago, IL) in 10% ADB (1:10 dilution; see Table 1).
TABLE 1.
Primary antibodies used in this study.
Preparation of Enriched Male Germ Cell Populations
Mixed germ cell preparations were obtained from juvenile mice as previously described using mice of different ages [15]. Detunicated testes were digested in 0.5 mg/ml collagenase (Sigma) and then 0.5 mg/ml trypsin (Sigma) supplemented with DNaseI (USB, Cleveland, OH) in Krebs-Ringer bicarbonate (KRB) media; germ cells were subsequently released by pipetting. The single-cell suspensions were filtered through 80-μm Nitex mesh (Sefar America, Depew, NY) and washed three times in 0.5% BSA (Sigma) in KRB. After the final wash, cells were counted and resuspended in KRB media prior to being processed in subsequent applications.
Enriched populations of germ cells were obtained from the testes of 17- and 70-dpp B6SJL F1 male mice according to the sedimentation velocity cell separation method [16, 17]. Mixed germ cells suspensions were prepared as described above from males of each age group, but after the three 0.5% BSA/KRB washes, cells were allowed to separate by cellular sedimentation at unit gravity in a 2%–4% BSA gradient generated in a STA-PUT apparatus (ProScience Inc., GlassShop, Toronto, ON, Canada). Following sedimentation, fractions were collected and examined using light microscopy Normarski optics. Cells were identified on the basis of morphological criteria and size as described previously [16]. Populations of leptotene/zygotene spermatocytes (average purity = 81%) and prepubertal pachytene spermatocytes (average purity = 85%) were obtained from the testes of 17-dpp mice; adult pachytene spermatocytes (average purity = 84%) and round spermatids (average purity = 90%) were obtained from 70-dpp mice. For every cell separation, and for each population of germ cells collected, an aliquot of cells was snap frozen for subsequent RNA extraction, and the rest of the cells were processed immediately for protein extraction as described below.
Surface-Spread Chromatin and Indirect Immunofluorescence Microscopy
Enriched germ cells from 15- and 18-dpp wild-type and repro42 mutant testes were used in the preparation of spread chromatin. Surface-spread chromatin was prepared as previously described [15]. Slides were allowed to completely air dry at room temperature and were stored at −20°C before being processed for immunofluorescence labeling.
For the preparation of female spread chromatin, timed pregnancies were established after mating repro42/+ heterozygous females and males. Ovaries were collected from female fetuses at either 15.5 or 16.5 dpc and rinsed in sterile PBS. They were digested in hypotonic extraction buffer pH 8.2 (15 mM Tris, 50 mM sucrose, 20 mM trisodium citrate, 5 mM EDTA, and a protease inhibitor cocktail tablet [Roche, Indianapolis, IN]) supplemented with 0.5 mg/ml collagenase (Sigma) for 30 min. Digested ovaries were disrupted by pipetting in 100 mM sucrose, pH 8.2, to form a single-cell suspension that was subsequently dispensed onto glass slides covered with 1% paraformaldehyde containing 0.1% Triton X-100, pH 9.2. The slides were placed in a humidified chamber for 3–5 h until the PFA solution was almost completely evaporated. Slides were washed in 0.4% Photo-Flo (Electron Microscopy Sciences, Hatfield, PA), allowed to air dry at room temperature, and stored at −20°C prior to being processed for indirect immunofluorescence labeling.
Solutions used for immunolabeling of surface-spread chromatin were prepared in PBS, pH 7.4, and procedures were carried out at room temperature as previously described [15] with appropriate primary antibodies (listed in Table 1) diluted in ADB. At least one well per slide received only ADB to serve as a negative control for primary antibody specificity. Following primary antibody incubation, slides were incubated with a 1:1000 dilution of the appropriate secondary antibodies tagged with Alexa Fluor 488 or 594 (Molecular Probes/Invitrogen, Eugene, OR) for 1–2 h in a humidified dark chamber. Slides were next passed through a series of washes, one of which was supplemented with 4′,6-diamidino-2-phenylindole (Sigma), and were mounted with Prolong Antifade (Molecular Probes/Invitrogen). Slides were analyzed using a Zeiss AxioObserver.Z1 inverted microscope and pictures were taken using a Zeiss AxioCam MRm camera with the AxioVision 4 software (Carl Zeiss Microimaging, Thornwood, NY). All experiments were repeated at least three times using different sets of samples.
RNA Extraction and RT-PCR
Total RNA was extracted from snap-frozen pellets of enriched populations of male germ cells or from whole testes initially submerged in RNAlater using the RNeasy extraction kit with DNaseI treatment as described by the manufacturer (Qiagen). Samples were diluted to a concentration of 5 ng/μl, dispensed into single-use aliquots, and stored at −80°C. Transcript-specific primers were designed to span introns and the repro42 mutation site and used in one-step RT-PCR reactions (OneStep RT-PCR kit, Qiagen). Primers for Spata22-001 were as follows: (5′–3′): F = ATGAAGTATTGGCGAGCACA, R = TGTCTGGGTCGTAGTGTCCA, giving an amplicon size of 220 bp and requiring an annealing temperature of 59°C. For amplification of Spata22-002, the same forward primer as for Spata22-001 was used in combination with the following reverse primer: R = ACAGTTTGGTTCCCTTCTGC; we were unable to amplify the Spata22-002 product under any experimental conditions used. 18S primer sequences are as described previously [18]. Reactions were performed using 10 ng of RNA for Spata22-001 and Spata22-002 or 100 pg of RNA for 18S according to the manufacturer's instruction with the following modifications: denaturation for 15 sec, annealing for 30 sec, and extension for 30 sec, with no final extension step, for a total of 30 cycles. Products were separated by electrophoresis following standard procedures.
Protein Extraction and Immunoblotting
Protein lysates were prepared from freshly isolated male germ cells or whole testes using RIPA buffer (Santa Cruz Biotechnology Inc., Santa Cruz, CA) supplemented with a protease inhibitor cocktail (Roche) according to the manufacturer's recommendations. Protein concentration was determined using the BCA protein assay following the manufacturer's instructions (Pierce/Thermo Scientific, Rockford, IL). Total protein in reducing sample buffer (10–15 μg per lane) was fractionated by SDS-PAGE and transferred to Immobilon-P polyvinylidene fluoride membranes (Millipore, Billerica, MA). Membranes were blocked overnight at 4°C in 5% nonfat dried milk diluted in PBS with 0.1% Tween 20 (PBST), and incubated with the anti-SPATA22 antibody (1:2500) diluted in blocking buffer for 1 h at room temperature. Membranes were washed with PBST according to the instructions provided with the ECL Plus Western Blotting Detection Reagent kit (Amersham, GE Healthcare, Piscataway, NJ), followed by incubation with a goat anti-rabbit IgG horseradish peroxidase-conjugated antibody (1:50 000; Molecular Probes/Invitrogen). Membranes were washed with PBST and exposed to the ECL Plus Western Blotting detection solution according to the manufacturer's instructions (Amersham). Chemiluminescence was revealed on Kodak X-Omatic film (Kodak, Rochester, NY). Following immunoblotting, membranes were stained with GelCode Blue Stain (Pierce) according to the manufacturer's instructions to confirm equal loading of proteins and consistency of transfer.
RESULTS
The repro42 Phenotype: Male and Female Infertility
The autosomal recessive repro42 phenotype was identified by the Reproductive Genomics Program at JAX in a screen following ENU induction of germline mutations in B6 male mice. Intercrosses of heterozygous repro42/+ animals generated infertile homozygous offspring at a frequency of 22.8% (188/826), slightly but not significantly lower than 25%, as expected for an autosomal Mendelian trait.
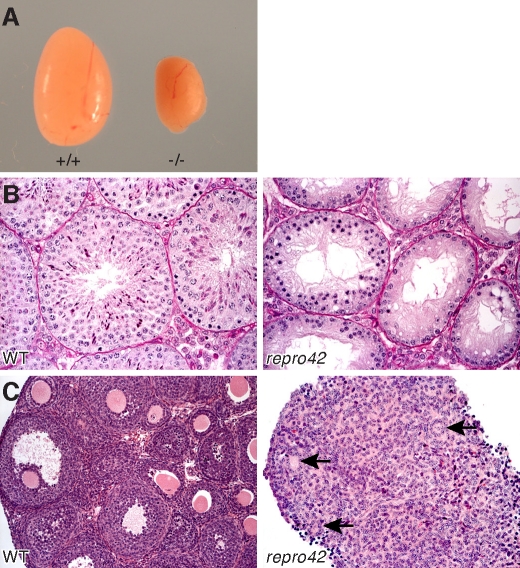
Initial phenotype analysis revealed that homozygous repro42/repro42 males and females (herein referred to as repro42 mutants) were overtly normal and presented no apparent gross phenotypic differences other than infertility. Epididymal sperm were absent in adult repro42 mutant males and there were no ovulated oocytes produced by hormonal stimulation of mutant females. However, plugs were observed in wild-type females mated to repro42 mutant males, and plugs were observed in repro42 mutant females mated with wild-type males, suggesting normal mating behavior. The average paired testis weight of mutant adults was approximately 20% of wild-type littermate controls (Fig. 1A and Table 2). Histological examination of adult repro42 mutant testes revealed complete lack of spermatids and spermatozoa as well as some stages of spermatocytes, whereas all stages of spermatogenesis were observed in seminiferous tubules of wild-type littermate controls (Fig. 1B). Adult repro42 mutant ovaries were smaller, were devoid of oocytes, and contained degenerated follicles and hemorrhagic cysts; in contrast, all stages of follicular development were observed in wild-type littermate control ovaries (Fig. 1C). This phenotype of germ cell depletion in repro42 mutant males and females is typical of meiotic arrest mutations [6, 19].
FIG. 1.
Absence of mature germ cells in adult repro42 mutant mice. A) Homozygous repro42 testes (right) are significantly smaller than those of wild-type littermate controls (left). B) Spermatogonia, spermatocytes, and spermatids are observed in testis sections from adult wild-type males (left), whereas only spermatogonia and spermatocytes up to the zygotene stage are visible in repro42 mutant testis sections (right). C) In adult wild-type females, all stages of follicular development are observed (left), whereas only degenerated follicles (black arrows) are present in repro42 mutant ovaries (right). Original magnification ×400.
TABLE 2.
Testis and body weights of adult wild-type and repro42/repro42 males.
Mapping of the repro42 Mutation to Spata22, a Novel Gene of Unknown Function
The repro42 mutation was mapped to 18.36 Mb on Chr 11, located between D11Mit208 and D11Mit245. Analysis of 3860 meioses identified 50 informative recombinant males and females, narrowing the repro42 critical region to 1.74 Mb between D11Jmp11 and D11Nds1, comprising 76 genes. The proximal boundary of the interval was defined by five individual recombination events (three from females, two from males) and the distal boundary was defined by four individual events (one from a female, three from males).
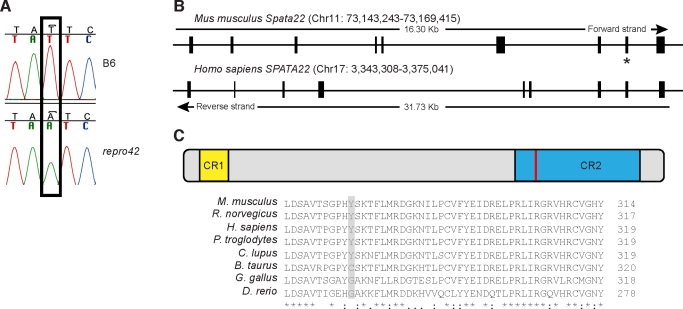
Of the 76 genes present in the interval, more than half (44/76) were members of the olfactory receptor gene family. Searches of the literature and public gene expression databases were conducted to determine likely candidates among the other 32 genes. Genes not expressed in the gonads were assigned low priority, leading to five remaining candidate genes: Gsg2 (germ cell-specific gene 2; MGI id: 1194498), Spata22 (spermatogenesis associated 22; MGI id: 2685728), n-R5s71 (nuclear encoded rRNA 5S 71; MGI id: 4421916), Mettl16 (methyltransferase like 16; MGI id: 1914743), and Mnt (max binding protein; MGI id: 109150). Exons and intron-exon junctions of these genes were sequenced using genomic DNA isolated from tail tissue of repro42 heterozygous and mutant mice, as well as from the B6 parental strain. Only one mutation was identified: a T-to-A transversion in exon 8 of Spata22, resulting in a nonsense mutation from a tyrosine to a premature stop codon at amino acid position 275 (Y275X) (Fig. 2A). Examination of available single-nucleotide polymorphism (SNP) information within 2 kb of the gene (dbSNP Build 128) confirmed that there were no known SNPs in the C57BL/6J strain corresponding to the identified mutation site.
FIG. 2.
A mutation in the Spata22 gene causes the repro42 phenotype. A) Sequencing of homozygous repro42 mutant DNA identified a T-to-A transversion in exon 8 of Spata22. The mutation, marked by an open box, is not present in control B6 DNA. B) Genomic structure of the mouse Spata22 and human SPATA22 genes. The exon containing the mutation, which results in a premature stop codon, is indicated by a star. C) A multiple sequence alignment performed with ClustalW2 identified two conserved regions (CR) in SPATA22. CR1 is located in the proximal end, whereas CR2 spans ∼100 amino acids and is closer to the C-termini. The identified mutation in mouse, situated at amino acid position 275, is located in CR2. A portion of the amino acid sequence of SPATA22 CR2 in eight different species is shown at the bottom. Fully conserved amino acids are marked by asterisks (*), colons (:) indicate amino acids with strongly similar physiochemical properties, and periods (.) indicate amino acids with weakly similar physiochemical properties. The position of the mutated tyrosine is marked by a red line (top) or by a pale gray box (bottom). The name of the species and the corresponding position of the last amino acid illustrated are indicated on the left and right, respectively.
Spata22 (also known as LOC380709 and as NYD-SP20 in humans) is an uncharacterized gene first identified in human testis as part of a large screen for developmentally regulated testis-specific alternatively spliced transcripts [20, 21]. The mouse and human genes share similar structure, both being comprised of nine exons encoding for proteins of 358 and 363 amino acids, respectively (Fig. 2B). No paralogs have been identified in either the mouse or human genomes. Presence of orthologous genes has been confirmed or predicted in a number of species (NCBI and Ensembl Genome Browsers), including Pan troglodytes (chimpanzee), Sus scrofa (pig), Gallus gallus (chicken), Thamnophis elegans (garter snake; [22]), and Danio rerio (zebrafish). Interestingly, evolutionary conservation does not appear to extend beyond Euteleostomes (bony vertebrates), which include mammals, birds, reptiles, amphibians, and bony fish. A phylogenic analysis was conducted with 36 confirmed or predicted SPATA22 protein sequences to establish evolutionary relationship among sequences (data not shown). This analysis showed that very little amino acid substitution has occurred in closely related species such as members of the Primates order, but divergence in composition is observed for distantly related species. Homo sapiens SPATA22 has 97% amino acid identity with P. troglodytes SPATA22, but shares only 72% and 31% identity with its Mus musculus and D. rerio counterparts, respectively. This multiple sequence alignment also identified two highly conserved regions in SPATA22: one smaller region in the N-termini (CR1) corresponding to amino acids 11–36 in the mouse sequence, and a second larger conserved area (CR2) positioned between amino acids 238 and 332 (Fig. 2C). The repro42 mutation is situated in this second highly conserved area.
To infer function of SPATA22 protein (UniProtKD id: Q5SV06), a search for functional domains was conducted with a number of predictive tools, but no significant matches were detected. An analysis of protein structure was conducted to determine the placement of regions of order and disorder, as these are often correlated with placement of functional sites [23]. Seven short segments between amino acids 1 and 190 and one short segment between amino acids 351 and 355 were identified as disordered regions, whereas a large region from amino acids 191–350 was recognized as a globular domain (Supplemental Table S1, available online at www.biolreprod.org). Sequence analysis revealed numerous functional sites across the SPATA22 protein sequence, including nuclear localization signals, docking motifs, and phosphorylation sites, as well as potential protein-protein interaction motifs (Supplemental Table S1). These motifs included two phosphopeptide motifs that interact with the BRCT domain of BRCA1 (breast cancer 1, Brca1), four Polo-like kinase phosphorylation sites, as well as two phosphoinositide-3-OH-kinase related kinases (PIKK) phosphorylation motifs. The latter are substrates for the PIKK family members ATM (ataxia telangiectasia mutated, Atm) and ATR (ATM and Rad3 related, Atr), proteins known to play important roles in DNA DSB repair or progression of meiosis.
Expression of Spata22 Transcript and Protein
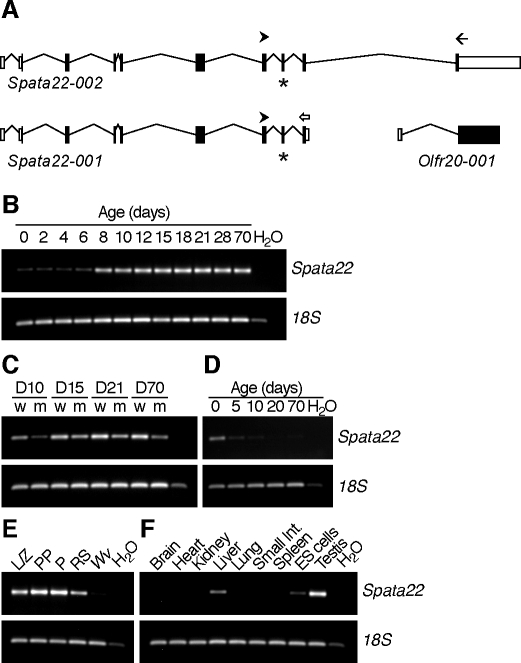
A previous study [21] identified four alternatively spliced transcripts of the SPATA22 gene in adult human testis, and the Ensembl Genome Browser predicted two transcripts for the mouse Spata22 gene: Spata22-001 (ENSMUST00000092926) and Spata22-002 (ENSMUST00000117445). Spata22-001 is encoded by exons 1–9, whereas Spata22-002 is predicted to contain an additional 10th exon shared with olfactory receptor 20 gene (Olfr20) (Fig. 3A). To determine which transcripts are present in mouse testis, an expression analysis using total testis RNA extracts from a developmental series was performed with primers designed to detect each of the two transcripts. The developmental time points chosen represent progression through the first wave of spermatogenesis, including appearance of spermatogonia, spermatocytes, and spermatids [24]. Spata22-001 was detected at all time points evaluated, from day of birth onward, but there was a marked increase in expression between 6 and 8 dpp (Fig. 3B). The Spata22-002 transcript could not be amplified under any of the experimental conditions tested, or in any of the tissues sampled (data not shown). These results suggested that Spata22-001 is the primary transcript in the testis, and it will be referred to as Spata22 henceforward. The T-to-A transversion identified in exon 8 translates as a premature stop codon, raising the possibility that Spata22 transcripts might not be detectable in mutant testis extracts. Primers designed to span the mutation site, thus detecting both wild-type and mutant mRNAs, were used to evaluate Spata22 expression in a developmental series of wild-type and repro42 mutant testes. Although a slight decrease in mRNA was apparent, Spata22 transcripts could be amplified in repro42 mutant testis extracts (Fig. 3C). This apparent decrease in mRNA could be caused by reduced transcription of Spata22 or by nonsense-mediated decay.
FIG. 3.
Spata22 transcripts are primarily expressed in gonads in a germ cell-specific manner. A) Diagram of postulated Spata22 transcripts. The Spata22 gene is predicted to encode two transcripts: Spata22-001, which contains nine exons, and Spata22-002, which contains an additional 10th exon shared with Olfr20. The position of the primers used in the RT-PCR analysis is indicated by arrowheads for the common forward primer, an open arrow for the reverse primer for Spata22-001, and a black arrow for the reverse primer for Spata22-002. The exon containing the mutation is indicated by a star. Adapted from the Spata22 gene summary of Ensembl Genome Browser (ENSMUSG00000069825). B) Expression of Spata22 in RNA extracted from developmental series of testes collected between Postnatal Day 0 to adulthood (70 dpp). Spata22-001 was detected at all time points tested and appeared to be the sole transcript in the testis. C) Spata22 transcripts were amplified by RT-PCR in wild-type (w) and repro42 mutant (m) testes at different developmental stages. D, postnatal day. D) RT-PCR amplified Spata22 transcripts primarily in newborn (0) and 5-dpp ovaries. E) Spata22 expression was detected in meiotic and postmeiotic germ cells, but not in an extract of RNA prepared from KitW/KitW-v testes, which are devoid of differentiating germ cells. L/Z, leptotene/zygotene spermatocytes; PP, prepubertal pachytene spermatocytes; P, adult pachytene spermatocytes; RS, round spermatids; Wv, KitW/KitW-v. F) Spata22 expression in adult mice is predominantly in the testis, but also detected in liver and ES cells. Int., intestine; H20, water control.
Spata22 transcripts were detected in ovaries from newborn mice and to a lower extent in 5-dpp ovaries, but could barely be amplified in 10-dpp ovaries and were not detectable thereafter (Fig. 3D). These results are consistent with a recently published microarray study confirming Spata22 gene expression during prophase I in fetal female germ cells [25].
Expression of Spata22 mRNA was assessed in highly enriched preparations of meiotic and postmeiotic male germ cells, as well as in RNA extracted from testes of KitW/KitW-v mice, which are deficient in differentiating germ cells. Spata22 transcripts were detected in leptotene/zygotene spermatocytes, in prepubertal and adult pachytene spermatocytes, and to a lower extent in round spermatids, but were not detected in KitW/KitW-v testis RNA (Fig. 3E). To further verify gonad specificity, expression was determined in a variety of adult somatic tissues as well as in mouse embryonic stem (ES) cells. Spata22 transcript was not found in most tissues, except for liver and ES cells, but expression appeared lower than in the testis (Fig. 3F). Overall, the data presented here indicate that Spata22 mRNA is expressed primarily in gonadal tissues in a germ cell-specific manner, at times when male and female germ cells progress through prophase of meiosis I.
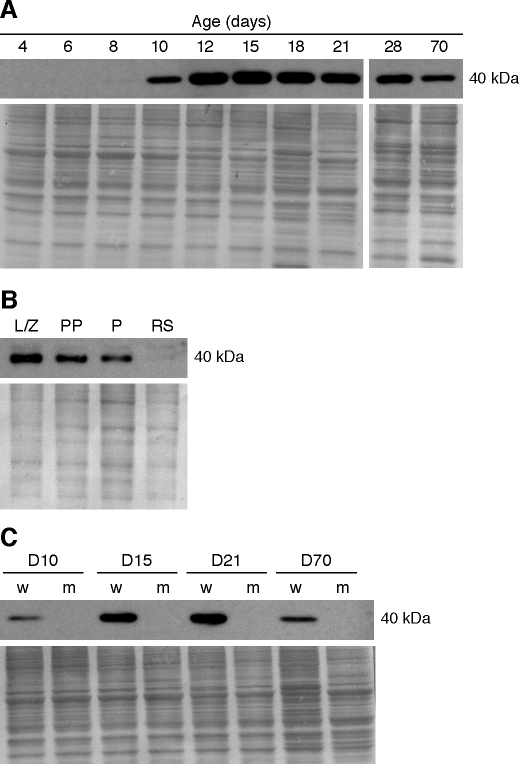
Proteins extracted from testes at various developmental ages were analyzed by immunoblotting. A single band of approximately 40 kDa, consistent with the predicted molecular weight of SPATA22 encoded by transcript Spata22-001, was detected in testis extracts (Fig. 4A). No band corresponding to the predicted molecular weight of the postulated protein encoded by Spata22-002 was detected, further suggesting that Spata22-001 is the sole transcript variant and protein isoform in the testis. SPATA22 protein expression was not detectable in testis extracts at 4 and 6 dpp, was only faintly visible at 8 dpp, and by 10 dpp became readily detectable, with expression sustained through adulthood. Lysates of purified germ cells were examined to determine if SPATA22 is present in germ cells. SPATA22 was found in all populations of spermatocytes tested, consistent with the RT-PCR analysis, but not in round spermatid extract (Fig. 4B). Because the mutation causes a premature stop codon at position 275, which could lead to either complete absence of SPATA22 or production of a truncated protein, an immunoblot analysis was conducted on wild-type and repro42 mutant testis extracts (Fig. 4C). The commercial antibody detected no SPATA22 in all mutant extracts tested, and likewise, there was no evidence of a truncated product.
FIG. 4.
SPATA22 protein is predominantly expressed in spermatocytes, but is absent in repro42 mutant testes. In each panel, the immunoblot results are presented at the top, and below is the corresponding stained membrane that confirms consistent protein loading and transfer. A) Immunoblotting of SPATA22 in protein extracts prepared from a developmental series of postnatal testes. SPATA22 is undetectable between 4 and 6 dpp and is faintly visible at 8 dpp, but can be readily detected from 10 dpp onwards. B) Protein extracts prepared from enriched populations of male germ cells were analyzed by Western blot. SPATA22 is present in spermatocytes but not in round spermatids. L/Z, leptotene/zygotene spermatocytes; PP, prepubertal pachytene spermatocytes; P, adult pachytene spermatocytes; RS, round spermatids. C) Analysis of protein extracts prepared from wild-type (w) and repro42 mutant (m) testes. The mutant testes lack SPATA22 detected by this antibody. D, postnatal day. The molecular weight (kDa) of SPATA22 is indicated on the right of each immunoblot.
The expression analyses provided evidence supporting SPATA22 as a germ cell-specific protein (Figs. 3 and 4), which was confirmed by immunohistochemistry. In wild-type adult testis cross sections, SPATA22 was consistently observed in a single layer of cells consistent with leptotene-, zygotene-, and early pachytene-stage spermatocytes, but was not detected in repro42 mutant testis sections (Fig. 5). Taken together, these data establish SPATA22 as a meiotic cell-specific protein and link the repro42 mutation to absence of SPATA22 protein production.
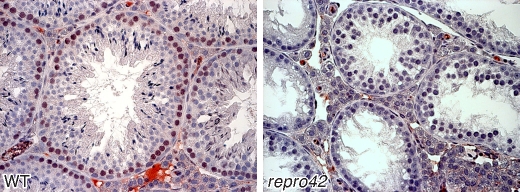
FIG. 5.
SPATA22 protein is detected in spermatocytes and is absent in repro42 mutant testes. The brown stain in the left panel reveals expression of SPATA22 in peripheral germ cells of seminiferous tubule cross sections from wild-type (WT) adult testes, and the right panel shows absence of expression in germ cells of repro42 mutant testes. Original magnification ×400.
Germ Cell Loss During Prophase I of Meiosis in repro42 Mutants
To determine stage of arrest in repro42 mutant males, a histological examination of repro42 testes during the first wave of spermatogenesis was conducted. At 8 and 10 dpp, there were no obvious differences in morphology, cell composition, or cellular relationships between mutant and wild-type littermate control testes (Fig. 6A and data not shown); spermatogonia and spermatocytes up to the zygotene stage were present in seminiferous tubules of animals of both genotypes. However, differences in cellular composition were observed by 12 dpp (Fig. 6B). Spermatocyte nuclei with highly condensed chromatin (heteropycnotic nuclei) were observed in mutant animals but not in wild-type littermate controls. Although pachytene spermatocytes were clearly identifiable in seminiferous tubules of wild-type littermate controls, none were detected in seminiferous tubules of repro42 mutant testes, suggesting that developmental arrest occurs before the pachytene stage. Tubules showing cellular depletion typical of stage IV arrest were present in mutants, but not in wild-type littermates.
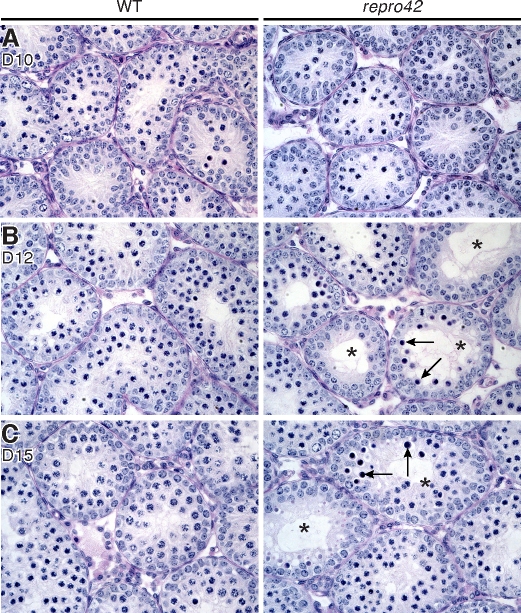
FIG. 6.
Meiotic arrest and loss of spermatocytes by 12 dpp in repro42 mutant testes. A) At 10 dpp, histological differences are not detectable in cross sections of repro42 mutant testes (right) when compared to cross sections of wild-type (WT) testes (left). B) At 12 dpp, spermatogonia and spermatocytes up to the pachytene stage are visible in wild-type testes (left), but differences in germ cell content become visible in mutant testes (right), where spermatocytes with condensed nuclei (arrows) and tubules with stage IV arrest (stars) are observed. C) By 15 dpp, repro42 mutant testes exhibit more spermatocytes with condensed nuclei and an increased number of tubules with stage IV arrest (right) compared to wild-type littermate controls (left). D, postnatal day. Original magnification ×400.
Germ cells numbers were quantified in control and repro42 mutant testes using the germ cell-specific marker GCNA1. GCNA1 can be detected in fetal gonocytes, in spermatogonia, and in spermatocytes until the zygotene stage, after which point expression becomes restricted to specific chromatin domains in pachytene spermatocytes and round spermatids [14]. There were no significant differences in germ cell numbers between genotypes at 8 and 10 dpp, but by 12 dpp, germ cell numbers were one-third lower in repro42 mutant seminiferous tubules than in wild-type littermate controls (Table 3). At 15 and 18 dpp, pachytene spermatocytes were clearly identifiable in wild-type littermate controls, but were not observed in mutant testes, and heteropycnotic spermatocyte nuclei and stage IV arrest were widespread in mutant seminiferous tubules (Fig. 6C and data not shown). Analysis of 21- and 28-dpp repro42 mutant testes confirmed the absence of spermatocytes beyond the zygotene stage, as well as the complete absence of spermatids and spermatozoa (data not shown). Taken together, this histological assessment pinpointed the 10–12-dpp time period for the onset of the defects observed in repro42 mutant males, implying meiotic prophase I arrest.
TABLE 3.
Numbers of germ cells in wild-type and repro42 mutant juvenile males.
Histological assessment of fetal ovaries at 17.5 dpc revealed germ cells in various stages of meiotic prophase (Fig. 7A). However, newborn and 10-dpp mutant ovaries showed fewer oocytes than wild-type ovaries, even as early as 0 dpp (Fig. 7B), and an almost complete absence of oocytes by 10 dpp (Fig. 7C). Consistent with germ cell demise in excess of that in wild-type ovaries, the number of degenerating or degenerated oocyte-containing follicles appeared to increase between 0 and 10 dpp. These observations are consistent with dramatic oocyte depletion shortly before or shortly after birth in repro42 mutant ovaries, a typical meiotic arrest phenotype [8, 19].
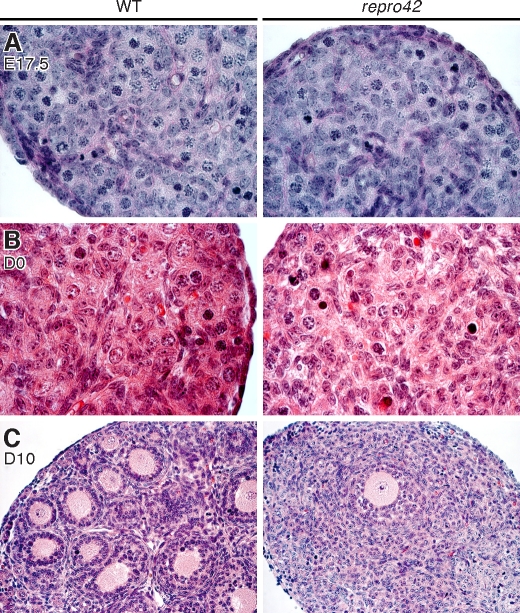
FIG. 7.
Meiotic arrest and loss of female germ cells in repro42 mutant ovaries after birth. A) Histology of wild-type (WT) (left) and repro42 mutant (right) ovaries at 17.5 dpc reveals the presence of numerous meiotic germ cells in mutant ovaries. Original magnification ×1000. B) Histological analysis of wild-type (left) and repro42 mutant (right) newborn (D0) ovaries reveals loss of germ cells in mutant ovaries. Original magnification ×1000. C) In 10-dpp ovaries, multiple primary and secondary follicles are visible in cross sections from wild-type mice (left), but only a few follicles remain in cross sections of repro42 mutant mice. Original magnification ×400.
Impaired Synapsis, DNA Repair, and Meiotic Arrest in repro42 Germ Cells
To confirm meiotic substage of arrest, spread chromatin from mutant oocytes and spermatocytes was examined with respect to prophase substages (leptonema, zygonema, pachynema, diplonema, and diakinesis) characterized by the expression and localization of specific markers [8]. To confirm spermatogenic arrest prior to the midpachytene stage (Fig. 6), we assessed the presence of histone HIST1H1T (more commonly known as H1t), a midpachynema marker, and phosphorylated histone H3, a marker for meiotic division [26–28]. Neither histone H1t nor phosphorylated histone H3 were detected in repro42 mutant spermatocytes (data not shown), confirming arrest before the midpachytene stage of meiotic prophase, also supported by failure to detect two markers of the XY body, SUMO1 and SUMO2/3 [15] (data not shown).
Assembly of the SC and the extent of synapsis were assessed by determining the labeling pattern of SC LE and CE proteins. Localization of SYCP2 and SYCP3 established that fully formed LEs were present in repro42 mutant germ cells (Fig. 8, A and B, and data not shown), indicating that repro42 mutant germ cells progressed to the zygotene stage. However, SYCP1 immunolabeling revealed that formation of the SC and synapsis were severely compromised in mutant germ cells of both sexes (Fig. 8, A and B). Very limited profiles of CE-associated SYCP1 were observed in repro42 mutant spermatocytes, detectable only in short discontinuous stretches. Unlike wild-type spermatocytes, fully synapsed homolog pairs indicative of pachynema were never identified in mutant spermatocytes (Fig. 8A). Interestingly, some repro42 mutant oocytes exhibited long continuous segments of SYCP1 (Fig. 8B), suggesting greater synapsis than in spermatocytes. However, although some pairs of chromosomes appeared to be almost fully synapsed, full synapsis of all homolog pairs was never observed in mutant oocytes. To confirm stage of arrest, the number of oocytes at each substage of prophase I was evaluated based on SYCP3 immunolabeling using criteria previously described [5, 8, 28]. Zygotene oocytes were further subclassified according to SYCP1 immunolabeling pattern: SYCP1 was not visible in early zygotene oocytes; SYCP1 had a punctate appearance in early- to midzygotene oocytes; at midzygonema, SYCP1 accumulated along short stretches of synapsis; and finally, by late zygonema, long regions of synapsis were marked by SYCP1. At 15.5 dpc, there were no obvious differences between mutant and repro42/+ (control) ovaries in the numbers of cells up to the midzygotene stage, but more control oocytes reached late zygonema (Table 4). By 16.5 dpc, a considerable number of oocytes had reached the pachytene stage in the repro42/+ control sample, whereas there were no pachytene oocytes visible in repro42 mutant ovaries (Table 4). Interestingly, in the populations of oocytes analyzed at both time points, a small proportion of midzygotene repro42 mutant oocytes (ranging from 2% to 6%) were completely devoid of any SYCP1 even though they presented fully formed LEs (data not shown). Together these observations reveal that the repro42 mutation prevents progression to the pachytene stage in both male and female germ cells, with arrest at a late zygotene-like stage.
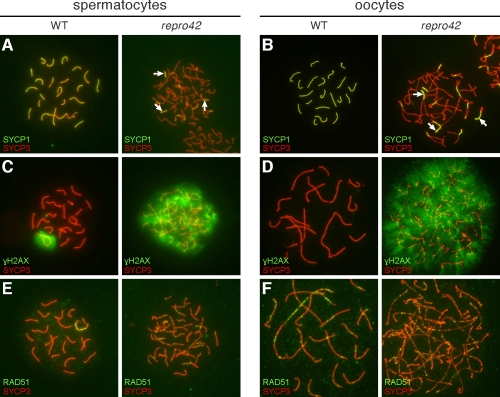
FIG. 8.
Synapsis and DNA repair are impaired in repro42 mutant oocytes and spermatocytes. A, B) Colocalization of SYCP1 and SYCP3 in wild-type (WT) (left) and repro42 mutant (right) spermatocytes (A) and oocytes (B). The extent of synapsis is reflected by the pattern of yellow color representing colabeling with anti-SYCP1 (green) and anti-SYCP3 (red), detecting proteins of the CE and LEs of the SC, respectively. C, D) Labeling of γH2AX and SYCP3 in wild-type (left) and repro42 mutant (right) spermatocytes (C) and oocytes (D). The pattern of γH2AX (green) reflects DSBs and repair; the pattern of SYCP3 (red) reveals the pachytene stage of the wild-type germ cells and prepachytene of the repro42 mutant germ cells. Persistence of γH2AX suggests DSBs are induced but not repaired in mutant germ cells. E, F) Colocalization of RAD51 and SYCP3 in wild-type (left) and repro42 mutant (right) spermatocytes (E) and oocytes (F). RAD51 (green) accumulates at sites of DSBs, frequently colocalizing with SYCP3 (red), and is an early marker of meiotic recombination. In wild-type spermatocytes it is restricted to the sex chromosomes in the pachytene stage (left panel), but is found associated with autosomal chromosomes in the mutant spermatocytes. Original magnification ×630.
TABLE 4.
Number of oocytes at various meiotic prophase substages in control and mutant repro42 fetal ovaries.
DNA DSBs associated with meiotic recombination were assessed in repro42 mutant germ cells using two markers: the phosphorylated form of histone H2AFX (commonly known as γH2AX), which accumulates at sites of DSBs, and the early recombination repair protein RAD51. In wild-type pachytene spermatocytes, γH2AX labeling became restricted to the XY body (Fig. 8C). In contrast, γH2AX labeling persisted across the chromatin of repro42 zygotene-like spermatocytes, confirming that DSBs were induced at the onset of meiosis, but were not repaired before meiotic arrest. Similar observations were made of mutant oocytes, where γH2AX labeling persisted across the nucleoplasm (Fig. 8D). Accumulation of RAD51 at break sites was visible in both wild-type and mutant spermatocytes and oocytes (Fig. 8, E and F). However, the number of RAD51 foci did not decline prior to arrest in mutant spermatocytes and oocytes, contrary to the control cells that progressed to pachynema and proceeded with break repair, which results in gradual removal of RAD51 from break sites. These observations are consistent with the repro42 mutation causing meiotic arrest at a late zygotene-like stage in both the male and female germ lines.
DISCUSSION
An unbiased mutagenesis and fertility screen identified the mouse mutation repro42, thereby uncovering a novel germ cell-specific factor essential to both male and female gametogenesis. Sequencing of candidate genes identified a nonsense mutation in the Spata22 gene, and no SPATA22 protein was detected in repro42 mice. Analysis of the repro42 phenotype revealed that Spata22 plays a key role during meiosis: mutant mice are infertile because of meiotic prophase arrest. Germ cell loss occurs as early as 12 dpp in repro42 mutant males, and histological changes are clearly visible in newborn ovaries of mutant females. Although initiation of meiotic recombination is not obviously impaired in repro42 mutant oocytes and spermatocytes, downstream key events such as processing of DNA DSBs and synapsis are compromised, and mutant meiocytes do not reach pachynema. A role for SPATA22 at an early stage of prophase I is further corroborated by the restricted expression and localization of Spata22 transcript in male germ cells.
Expression analysis of Spata22 strongly suggests a role in meiotic progression in both male and female germ cells. In the male germ line, Spata22 transcripts are present throughout testis development, from basal levels to more pronounced levels between 6 and 8 dpp. Subsequently, expression of Spata22 appears relatively constant. We took advantage of published microarray data deposited at NCBI in the Gene Expression Omnibus (accession number GSE23322) to establish expression of Spata22 in pure populations of oocytes isolated daily between 12.5 and 18.5 dpc [25]. Analysis of these data showed that Spata22 transcript levels increase sharply between 12.5 and 13.5 dpc, peak by 15.5 dpc, and remain elevated until 18.5 dpc. Together, these male and female germ line Spata22 expression profiles are similar to those of genes with known meiotic roles, such as Spo11, Dmc1, and Sycp3 (see below). Spata22 transcripts are predominantly expressed in germ cells, and antibody localization of SPATA22 confirmed its presence in leptotene and zygotene to early pachytene spermatocytes in testis sections. Interestingly, Spata22 transcripts were detected in wild-type round spermatids, albeit at lower levels, but SPATA22 protein was undetectable in these cells. Although the mechanisms governing Spata22 meiotic expression have yet to be determined, examination of published regulatory features has revealed two transcription factor binding sites—a ZFX site (zinc finger protein X-linked, Zfx) and a TCFCP2L1 site (transcription factor CP2-like 1, Tcfcp2l1), as well as a DNaseI hypersensitivity site in the promoter area of the Spata22 gene. These data emerged from mapping transcription factor binding sites in mouse ES cells using chromatin immunoprecipitation and high-throughput DNA sequencing [29]. ZFX is highly expressed in both the testis and the ovary, and thus could be involved in controlling meiotic Spata22 expression.
Because the mutant phenotypes and gene expression profiles are similar in male and female gonads, SPATA22 appears to belong to a group of proteins contributing to sexually conserved features of meiosis. Although induction of DNA DSBs does not appear to be impaired, both synapsis and progress of recombination are affected in mutant mice. Our data indicate SPATA22 is required for both spermatocytes and oocytes to progress past late zygonema of prophase I. The repro42 mutant germ cells could be eliminated because of a DNA damage-dependent response, which results in elimination of spermatocytes at stage IV and of oocytes shortly after birth prior to the formation of primordial follicles, as observed in repro42 mutant ovaries [19, 30]. In mammals, effects on progress of meiosis are observed for mutations in genes encoding proteins involved in initiation and progression through the early steps of meiotic recombination in both germ lines. Mice deficient in SPO11, the enzyme responsible for inducing genetically programmed DSBs at the onset of meiosis, are infertile and present severe chromosome synapsis defects [31–33]. However, although there is a reduction in oocyte numbers, it is not a severe depletion [19]. A nearly identical phenotype is observed in germ cells of mutants interrupted in the Mei1 (meiosis defective 1) gene, encoding a protein postulated to be involved in induction of DSBs [34, 35].
Proteins involved in early DSB processing and repair are also required for synapsis and meiotic progress. Two RecA homologs, RAD51 and its meiosis-specific counterpart DMC1, mediate homologous chromatid invasion following 5′ to 3′ resection of DSBs and promote homologous chromosome pairing and synapsis in both oocytes and spermatocytes [36, 37]. Interestingly, the depletion of oocytes in Dmc1 mutants is more severe than in Spo11 or Mei1 mutants [19]; in this respect the oocyte depletion phenotype of repro42 mutants is more similar to that of Dmc1 mutants, suggesting that Spata22 function is required for meiotic DNA DSB repair. Additionally, there is sexual dimorphism in mutant phenotypes of genes encoding proteins of the SC. Mutations in genes encoding CE proteins exhibit sexually similar phenotypes, e.g., knockouts of Sycp1 [38], Syce1 [39], Syce2 [40], Syce3 [41], and Tex12 [42] all produce male and female infertility with an arrest during prophase exhibiting similarities to the repro42 phenotype. Conversely, ablation of genes encoding LE proteins, e.g., Sycp2 [43] and Sycp3 [44], produce sexually dimorphic phenotypes with male-only infertility, in marked contrast to the repro42 phenotype. Thus, by comparison to these meiotic mutant phenotypes, it could be postulated that SPATA22 plays a role in early events of meiotic prophase, perhaps in assembly and/or function of the SC. However, a nonmeiotic role of SPATA22 is not excluded by the available data. It could be involved in a non-sexually dimorphic aspect of germ cell survival and function required for meiotic progress.
Available information about other Spata genes does not shed much light on possible function of the Spata22 gene. A number of novel postulated testis development and spermatogenesis-related genes were identified using microarray screens [20, 21], including Spata22 (originally known as NYD-SP20). Twenty-four genes containing the Spata acronym in their symbol are currently listed in MGI, but these are widely distributed across the genome and do not appear to present any conserved sequence features. Functional information exists for very few of the Spata genes and so far, no mechanistic link has been uncovered among them. For example, Spatc1 (SPATA15 in humans) encodes for a centrosomal protein detected in the connecting piece of both human and mouse spermatozoa and is a germ cell-specific binding partner of CDC20 [45, 46]. Rnf17 (SPATA23 in humans) encodes for a protein containing a RING finger motif and tudor domains that assembles into RNF17 granules, a novel mouse nuage distinct from the chromatoid body [47]. Male mice deficient in Rnf17 exhibit complete spermatogenesis arrest at the round spermatid stage. Additionally, a homozygous mutation in SPATA16 is associated with globozoospermia in some infertile male patients [48]. In spite of this information, the role of Spata genes is not limited to testis development or spermatogenesis. In the mouse, Spata7 transcripts can be detected in multiple layers of the adult mouse retina in addition to spermatocytes [49], and human SPATA genes are implicated in multiple somatic roles. Thus Spata is a misleading symbol, not reflective of tissue or function, and it cannot be assumed that every Spata gene has a role in testis development and spermatogenesis. Characterization of repro42 mutant mice did not reveal the presence of defects in somatic tissues, suggesting the role of Spata22 is restricted to the germ line. However, because increased incidence of cancer is often observed in mice deficient for proteins involved in meiotic prophase progression, we analyzed males and females of all genotypes over a period of 18 mo. We did not observe any significant differences among the animals in either phenotype or tumor incidence (data not shown), further suggesting the role of SPATA22 is restricted to the germ line.
Until now, very little has been reported on the Spata22 gene, its regulation, or its postulated role. The relationship between alternative splice variants of human SPATA22 and developmental stage examined by cDNA microarray showed a dynamic expression profile of all four variants in fetal and adult testis extracts [21]. Our results showed only one Spata22 transcript is expressed in mouse testes, suggesting species-specific differences in regulation or mode of action. Human SPATA22 transcripts were detected in pediatric primary central nervous system germ cell tumors (GCTs) classified as germinomas but not in nongerminomatous GCTs [50]. Of more relevance, human SPATA22 expression was induced by cisplatin treatment of testicular GCT cell lines [51]. Recently Spata22 was identified as a candidate meiosis-specific gene in an expression study monitoring effects of exposure to bisphenol A (BPA) on the developing fetal ovary [52]. In this study, Spata22 ranked among the genes most affected by low-dose BPA exposure, similar to other known meiotic factors, e.g., Sycp2, Tex11, Syce1, and Dazl.
Although lack of recognizable domains in the SPATA22 protein sequence precludes prediction of function, identification of predicted linear motifs provides some insight into potential interacting partners. The presence of two phosphopeptide motifs known to interact with the BRCT domain of BRCA1 suggests SPATA22 could be involved in a signaling cascade leading to DSB repair in oocytes and spermatocytes. In spermatocytes, BRCA1 directs ATR to the sex chromosomes for phosphorylation of H2AFX and initiation of meiotic sex chromosome inactivation [53]. BRCA1 is also involved in the more general response to unsynapsed autosomal chromosome axes, meiotic silencing of unsynapsed chromatin (MSUC), in both oocytes and spermatocytes [54–56]. There are also two putative ATR and ATM phosphorylation sites along the sequence of SPATA22. ATR is required for XY body formation and MSUC, and ATM is involved in signaling DSB repair following initiation of meiotic recombination [53, 56–58]. However, whether SPATA22 is a bona fide target of ATM signaling, such as, for example, BTBD12 [59], is difficult to unravel because of diverse fertility phenotypes of these proteins.
Despite conservation of the most salient characteristics of meiosis between yeast and vertebrates, functional differences are noted for individual protein players. Additionally, the generation of distinct sexes in higher eukaryotes is linked to the elaboration of complex regulatory systems to accommodate physiological and endocrine constraints, and thus meiosis is not an autonomous process. In mammals, the complexity of gametogenesis imposes both temporal and spatial constraints on the progression of meiosis, requiring regulatory mechanisms to accommodate gametogenic differentiation unfolding in parallel with meiosis. The most novel aspect of the repro42 mutation is that it identifies a vertebrate-specific gene of unknown function, Spata22, that is required early in meiotic prophase in both male and female germ cells. Absence of homologs in organisms such as Saccharomyces cerevisiae, Drosophila melanogaster, and Caenorhabditis elegans suggests that the role of Spata22 is related to molecular interactions that are not present in these model organisms. The Spata22 gene would likely have remained uncharacterized had it not been for the unbiased approach taken here. Future studies should define the interacting partners of SPATA22 as well as determine the precise spatiotemporal requirement for this novel protein in the hierarchy of meiotic events.
Supplementary Material
Linear motifs predicted by GlobPlot and ELM
ACKNOWLEDGMENT
We appreciate the assistance of Heather Lothrop in maintaining mice, and thank Sheila Bornstein and Dr. Janice Pendola for identification of the repro42 line and Lucy Rowe and Mary Barter for their expert assistance and advice in fine mapping. We thank Dr. Dirk de Rooij for valuable consultation in determining stage of arrest in mutant testes, and are grateful to Drs. George Enders (Kansas University Medical Center) and Jeremy Wang (University of Pennsylvania College of Veterinary Medicine) for the gifts of GCNA1 and SYCP2 antibodies, respectively. We acknowledge the Scientific Services of the Jackson Laboratory for outstanding support. We thank Drs. Greg Cox and Laura Reinholdt for critical comments on the manuscript, and the Handel laboratory for helpful discussion throughout this study.
Footnotes
Supported by NIH grant HD42137 to J.J.E., M.A.H., and J.C.S.; a Cancer Center Core Grant CA34196 to the Jackson Laboratory; and a fellowship from the Canadian Institutes of Health Research (CIHR) to S.L.
REFERENCES
- Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med 2008; 14: 1197 1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Y, Cooke HJ. Dissecting the mammalian synaptonemal complex using targeted mutations. Chromosome Res 2007; 15: 579 589 [DOI] [PubMed] [Google Scholar]
- de Boer E, Heyting C. The diverse roles of transverse filaments of synaptonemal complexes in meiosis. Chromosoma 2006; 115: 220 234 [DOI] [PubMed] [Google Scholar]
- Page SL, Hawley RS. The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol 2004; 20: 525 558 [DOI] [PubMed] [Google Scholar]
- Cohen PE, Pollack SE, Pollard JW. Genetic analysis of chromosome pairing, recombination, and cell cycle control during first meiotic prophase in mammals. Endocr Rev 2006; 27: 398 426 [DOI] [PubMed] [Google Scholar]
- Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet 2010; 11: 124 136 [DOI] [PubMed] [Google Scholar]
- Hunt PA, Hassold TJ. Sex matters in meiosis. Science 2002; 296: 2181 2183 [DOI] [PubMed] [Google Scholar]
- Morelli MA, Cohen PE. Not all germ cells are created equal: aspects of sexual dimorphism in mammalian meiosis. Reproduction 2005; 130: 761 781 [DOI] [PubMed] [Google Scholar]
- Pepling ME. From primordial germ cell to primordial follicle: mammalian female germ cell development. Genesis 2006; 44: 622 632 [DOI] [PubMed] [Google Scholar]
- Hunt PA, Hassold TJ. Human female meiosis: what makes a good egg go bad? Trends Genet 2008; 24: 86 93 [DOI] [PubMed] [Google Scholar]
- Handel MA, Lessard C, Reinholdt L, Schimenti JC, Eppig JJ. Mutagenesis as an unbiased approach to identify novel contraceptive targets. Mol Cell Endocrinol 2006; 250: 201 205 [DOI] [PubMed] [Google Scholar]
- Schimenti J, Bucan M. Functional genomics in the mouse: phenotype-based mutagenesis screens. Genome Res 1998; 8: 698 710 [DOI] [PubMed] [Google Scholar]
- Ward JO, Reinholdt LG, Hartford SA, Wilson LA, Munroe RJ, Schimenti KJ, Libby BJ, O'Brien M, Pendola JK, Eppig J, Schimenti JC. Toward the genetics of mammalian reproduction: induction and mapping of gametogenesis mutants in mice. Biol Reprod 2003; 69: 1615 1625 [DOI] [PubMed] [Google Scholar]
- Enders GC, May JJ., II Developmentally regulated expression of a mouse germ cell nuclear antigen examined from embryonic day 11 to adult in male and female mice. Dev Biol 1994; 163: 331 340 [DOI] [PubMed] [Google Scholar]
- La Salle S, Sun F, Zhang XD, Matunis MJ, Handel MA. Developmental control of sumoylation pathway proteins in mouse male germ cells. Dev Biol 2008; 321: 227 237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellvé AR. Purification, culture and fractionation of spermatogenic cells. Methods Enzymol 1993; 225: 84 113 [DOI] [PubMed] [Google Scholar]
- La Salle S, Sun F, Handel MA. Isolation and short-term culture of mouse spermatocytes for analysis of meiosis : Keeney S. (ed.), Methods in Molecular Biology, Molecular Medicine and Biotechnology: Meiosis Protocols, vol. 558. New York: Humana Press; 2009: 279 297 [DOI] [PubMed] [Google Scholar]
- La Salle S, Mertineit C, Taketo T, Moens PB, Bestor TH, Trasler JM. Windows for sex-specific methylation marked by DNA methyltransferase expression profiles in mouse germ cells. Dev Biol 2004; 268: 403 415 [DOI] [PubMed] [Google Scholar]
- Di Giacomo M, Barchi M, Baudat F, Edelmann W, Keeney S, Jasin M. Distinct DNA-damage-dependent and -independent responses drive the loss of oocytes in recombination-defective mouse mutants. Proc Nat Acad Sci U S A 2005; 102: 737 742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha J, Zhou Z, Li J, Yin L, Yang H, Hu G, Luo M, Chan HC, Zhou K. Identification of testis development and spermatogenesis-related genes in human and mouse testes using cDNA arrays. Mol Hum Reprod 2002; 8: 511 517 [DOI] [PubMed] [Google Scholar]
- Huang X, Li J, Lu L, Xu M, Xiao J, Yin L, Zhu H, Zhou Z, Sha J. Novel development-related alternative splices in human testis identified by cDNA microarrays. J Androl 2005; 26: 189 196 [DOI] [PubMed] [Google Scholar]
- Schwartz TS, Tae H, Yang Y, Mockaitis K, Van Hemert JL, Proulx SR, Choi JH, Bronikowski AM. A garter snake transcriptome: pyrosequencing, de novo assembly, and sex-specific differences. BMC Genomics 2010; 11: 694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res 2003; 31: 3701 3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellvé AR, Cavicchia JC, Millette CF, O'Brien DA, Bhatnagar YM, Dym M. Spermatogenic cells of the prepuberal mouse. J Cell Biol 1977; 74: 68 85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabour D, Arauzo-Bravo MJ, Hubner K, Ko K, Greber B, Gentile L, Stehling M, Scholer HR. Identification of genes specific to mouse primordial germ cells through dynamic global gene expression. Hum Mol Genet 2011; 20: 115 125 [DOI] [PubMed] [Google Scholar]
- Cobb J, Cargile B, Handel MA. Acquisition of competence to condense metaphase I chromosomes during spermatogenesis. Dev Biol 1999; 205: 49 64 [DOI] [PubMed] [Google Scholar]
- Drabent B, Bode C, Doenecke D. Structure and expression of the mouse testicular H1 histone gene (H1t). Biochim Biophys Acta 1993; 1216: 311 313 [DOI] [PubMed] [Google Scholar]
- Inselman A, Eaker S, Handel MA. Temporal expression of cell cycle-related proteins during spermatogenesis: establishing a timeline for the onset of the meiotic divisions. Cytogenet Genome Res 2003; 103: 277 284 [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008; 133: 1106 1117 [DOI] [PubMed] [Google Scholar]
- Barchi M, Mahadevaiah S, Di Giacomo M, Baudat F, de Rooij DG, Burgoyne PS, Jasin M, Keeney S. Surveillance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Mol Cell Biol 2005; 25: 7203 7215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell 2000; 6: 989 998 [DOI] [PubMed] [Google Scholar]
- Mahadevaiah SK, Turner JMA, Baudat F, Rogakou EP, de Boer P, Blanco-Rodriguez J, Jasin M, Keeney S, Bonner WM, Burgoyne PS. Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet 2001; 27: 271 276 [DOI] [PubMed] [Google Scholar]
- Romanienko PJ, Camerini-Otero RD. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 2000; 6: 975 987 [DOI] [PubMed] [Google Scholar]
- Libby BJ, De La Fuente R, O'Brien MJ, Wigglesworth K, Cobb J, Inselman A, Eaker S, Handel MA, Eppig JJ, Schimenti JC. The mouse meiotic mutation mei1 disrupts chromosome synapsis with sexually dimorphic consequences for meiotic progression. Dev Biol 2002; 242: 174 187 [DOI] [PubMed] [Google Scholar]
- Libby BJ, Reinholdt LG, Schimenti JC. Positional cloning and characterization of Mei1, a vertebrate-specific gene required for normal meiotic chromosome synapsis in mice. Proc Natl Acad Sci U S A 2003; 100: 15706 15711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman DL, Cobb J, Schimenti KJ, Wilson LA, Cooper DM, Brignull E, Handel MA, Schimenti JC. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell 1998; 1: 697 705 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Kondoh G, Matsuda Y, Habu T, Nishimune Y, Morita T. The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol Cell 1998; 1: 707 718 [DOI] [PubMed] [Google Scholar]
- de Vries FA, de Boer E, van den Bosch M, Baarends WM, Ooms M, Yuan L, Liu JG, van Zeeland AA, Heyting C, Pastink A. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev 2005; 19: 1376 1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcun-Filas E, Speed R, Taggart M, Grey C, de Massy B, Benavente R, Cooke HJ. Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLoS Genet 2009; 5: e1000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcun-Filas E, Costa Y, Speed R, Taggart M, Benavente R, De Rooij DG, Cooke HJ. SYCE2 is required for synaptonemal complex assembly, double strand break repair, and homologous recombination. J Cell Biol 2007; 176: 741 747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm S, Fraune J, Naumann R, Hernandez-Hernandez A, Hoog C, Cooke HJ, Alsheimer M, Benavente R. A novel mouse synaptonemal complex protein is essential for loading of central element proteins, recombination, and fertility. PLoS Genet 2011; 7: e1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer G, Wang H, Bolcun-Filas E, Cooke HJ, Benavente R, Hoog C. Progression of meiotic recombination requires structural maturation of the central element of the synaptonemal complex. J Cell Sci 2008; 121: 2445 2451 [DOI] [PubMed] [Google Scholar]
- Yang F, De La Fuente R, Leu NA, Baumann C, McLaughlin KJ, Wang PJ. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J Cell Biol 2006; 173: 497 507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Liu JG, Zhao J, Brundell E, Daneholt B, Hoog C. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol Cell 2000; 5: 73 83 [DOI] [PubMed] [Google Scholar]
- Goto M, Eddy EM. Speriolin is a novel spermatogenic cell-specific centrosomal protein associated with the seventh WD motif of Cdc20. J Biol Chem 2004; 279: 42128 42138 [DOI] [PubMed] [Google Scholar]
- Goto M, O'Brien DA, Eddy EM. Speriolin is a novel human and mouse sperm centrosome protein. Hum Reprod 2010; 25: 1884 1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Goodheart M, Chuma S, Nakatsuji N, Page DC, Wang PJ. RNF17, a component of the mammalian germ cell nuage, is essential for spermiogenesis. Development 2005; 132: 4029 4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dam AH, Koscinski I, Kremer JA, Moutou C, Jaeger AS, Oudakker AR, Tournaye H, Charlet N, Lagier-Tourenne C, van Bokhoven H, Viville S. Homozygous mutation in SPATA16 is associated with male infertility in human globozoospermia. Am J Hum Genet 2007; 81: 813 820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, den Hollander AI, Moayedi Y, Abulimiti A, Li Y, Collin RW, Hoyng CB, Lopez I, Abboud EB, Al-Rajhi AA, Bray M, Lewis RA, et al. Mutations in SPATA7 cause Leber congenital amaurosis and juvenile retinitis pigmentosa. Am J Hum Genet 2009; 84: 380 387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HW, Wu YH, Hsieh JY, Liang ML, Chao ME, Liu DJ, Hsu MT, Wong TT. Pediatric primary central nervous system germ cell tumors of different prognosis groups show characteristic miRNome traits and chromosome copy number variations. BMC Genomics 2010; 11: 132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duale N, Lindeman B, Komada M, Olsen AK, Andreassen A, Soderlund EJ, Brunborg G. Molecular portrait of cisplatin induced response in human testis cancer cell lines based on gene expression profiles. Mol Cancer 2007; 6: 53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson C, Gieske M, Murdoch B, Ye P, Li Y, Hassold T, Hunt PA. Gene expression in the fetal mouse ovary is altered by exposure to low doses of bisphenol a. Biol Reprod 2011; 84: 79 86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JMA, Aprilikova O, Xu X, Wang RS, Kim SJ, Chandramouli GVR, Barrett JC, Burgoyne PS, Deng C-X. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr Biol 2004; 14: 2135 2142 [DOI] [PubMed] [Google Scholar]
- Kouznetsova A, Wang H, Bellani M, Camerini-Otero RD, Jessberger R, Hoog C. BRCA1-mediated chromatin silencing is limited to oocytes with a small number of asynapsed chromosomes. J Cell Sci 2009; 122: 2446 2452 [DOI] [PubMed] [Google Scholar]
- Mahadevaiah SK, Bourc'his D, de Rooij DG, Bestor TH, Turner JM, Burgoyne PS. Extensive meiotic asynapsis in mice antagonises meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation. J Cell Biol 2008; 182: 263 276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JMA, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, Deng C-X, Burgoyne PS. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet 2005; 37: 41 47 [DOI] [PubMed] [Google Scholar]
- Barchi M, Roig I, Di Giacomo M, de Rooij DG, Keeney S, Jasin M. ATM promotes the obligate XY crossover and both crossover control and chromosome axis integrity on autosomes. PLoS Genet 2008; 4: e1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow C, Liyanage M, Moens PB, Tarsounas M, Nagashima K, Brown K, Rottinghaus S, Jackson SP, Tagle D, Ried T, Wynshaw-Boris A. Atm deficiency results in severe meiotic disruption as early as leptonema of prophase I. Development 1998; 125: 4007 4017 [DOI] [PubMed] [Google Scholar]
- Holloway JK, Mohan S, Balmus G, Sun X, Modzelewski A, Borst PL, Freire R, Weiss RS, Cohen PE. Mammalian BTBD12 (SLX4) protects against genomic instability during mammalian spermatogenesis. PLoS Genetics 2011; 7: e1002094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaker S, Pyle A, Cobb J, Handel MA. Evidence for meiotic spindle checkpoint from analysis of spermatocytes from Robertsonian-chromosome heterozygous mice. J Cell Sci 2001; 114: 2953 2965 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Linear motifs predicted by GlobPlot and ELM