ABSTRACT
Human endometrium regenerates on a cyclic basis from candidate stem/progenitors whose genetic programs are yet to be determined. A subpopulation of endometrial stromal cells, displaying key properties of mesenchymal stem cells (MSCs), has been characterized. The endometrial MSC (eMSC) is likely the precursor of the endometrial stromal fibroblast. The goal of this study was to determine the transcriptome and signaling pathways in the eMSC to understand its functional phenotype. Endometrial stromal cells from oocyte donors (n = 20) and patients undergoing benign gynecologic surgery (n = 7) were fluorescence-activated cell sorted into MCAM (CD146)+/PDGFRB+ (eMSC), MCAM (CD146)−/PDGFRB+ (fibroblast), and MCAM (CD146)+/PDGFRB− (endothelial) populations. The eMSC population contained clonogenic cells with a mesenchymal phenotype differentiating into adipocytes when cultured in adipogenic medium. Gene expression profiling using Affymetrix Human Gene 1.0 ST arrays revealed 762 and 1518 significantly differentially expressed genes in eMSCs vs. stromal fibroblasts and eMSCs vs. endothelial cells, respectively. By principal component and hierarchical clustering analyses, eMSCs clustered with fibroblasts and distinctly from endothelial cells. Endometrial MSCs expressed pericyte markers and were localized by immunofluorescence to the perivascular space of endometrial small vessels. Endometrial MSCs also expressed genes involved in angiogenesis/vasculogenesis, steroid hormone/hypoxia responses, inflammation, immunomodulation, cell communication, and proteolysis/inhibition, and exhibited increased Notch, TGFB, IGF, Hedgehog, and G-protein-coupled receptor signaling pathways, characteristic of adult tissue MSC self-renewal and multipotency. Overall, the data support the eMSC as a clonogenic, multipotent pericyte that displays pathways of self-renewal and lineage specification, the potential to respond to conditions during endometrial desquamation and regeneration, and a genetic program predictive of its differentiated lineage, the stromal fibroblast.
Keywords: cell fate, endometrium, immunomodulation, mesenchymal stem cells, microarray, pathways, pericytes, self-renewal
The multipotent perivascular endometrial mesenchymal stem cell expresses pathways involved in self-renewal and lineage commitment important for endometrial function and tissue regeneration.
INTRODUCTION
Human endometrium is a proliferative, angiogenic, and dynamically regenerated normal tissue that is the anatomic prerequisite for pregnancy [1]. At menses and postpartum the endometrium exhibits a proinflammatory environment, extensive extracellular matrix remodeling, vasospasm, hypoxia, cell death, and tissue desquamation, with wound repair mechanisms and cellular proliferation and differentiation participating in its regeneration without scarring [2–5]. Stem/progenitor cells (epithelial, mesenchymal, endothelial) likely contribute to the rapid endometrial cyclic growth and regeneration of 4–14 mm of mucosa per cycle during the more than 400 cycles in a woman's reproductive lifespan and endometrial regeneration/repair postpartum [6–8]. It is well known that bone marrow-derived mesenchymal stem cells (MSCs) in the circulation can home to sites of damaged tissues [9], and it has been postulated that bone marrow-derived MSCs participate in cyclic endometrial repair and regeneration and may be progenitors of stromal fibroblasts and perhaps epithelial cells in the endometrium [7]. The phenotype of the stem/progenitors, what regulates their potential homing to endometrium, and mechanisms underlying their roles in endometrial tissue regeneration remain unresolved and are the subject of much investigation [6, 8].
Stem cells in adult tissues are primarily quiescent, although they have enormous capacity to proliferate to achieve self-renewal (proliferation and maintenance of the undifferentiated state), and they also exhibit multipotency, differentiating along tissue-specific lineages and migrating within the tissue and replenishing their tissue constituents [10–12]. Mesenchymal stem cslls share these properties, are self-renewing, and differentiate in vitro and in vivo along chondrogenic osteogenic, adipogenic, myogenic, and tenogenic lineages [13–16]. In addition, they exhibit immunomodulatory properties [17–19]. Recently, a small population of clonogenic, self-renewing, multipotent MSCs with high proliferative potential has been identified in human endometrium [20–22]. The endometrial MSCs (eMSCs) coexpress melanoma cell adhesion molecule (MCAM; also known as cluster of differentiation 146 [CD146]) and beta-type platelet-derived growth factor receptor (PDGFRB) surface markers [6, 22, 23] and have a perivascular location [21, 22]. These eMSCs likely represent the reservoir of progenitors giving rise primarily to the stromal fibroblast lineage [23], which is also of mesenchymal origin. Recent studies using animal transplantation models are consistent with eMSC differentiation to the stromal fibroblast [24], supporting a common lineage of eMSCs and endometrial stromal fibroblasts.
Gene expression profiling has provided insights into molecular pathways involved in bone marrow and adipose MSC self-renewal and maintenance of multipotency [13, 15], and despite interest in the potential use of these MSCs in regenerative medicine [25], little is known about the genetic program of eMSCs. In this context, in the present study we prospectively isolated highly purified MCAM (CD146)+/PDGFRB+ eMSCs and report their clonogenic potential, multipotency, and perivascular location in adult human endometrium. Expression profiling results reveal that eMSCs have a phenotype consistent with self-renewal, multipotency, immunomodulation, and homing, and support a common lineage for eMSCs and endometrial stromal fibroblasts. Furthermore, as pericytes, the cells are well positioned to migrate into the stroma and differentiate and regenerate endometrial tissue when signaled by tissue breakdown processes, and their genetic program suggests they have the capacity to respond to tissue and matrix proteolysis, tissue hypoxia, inflammation, desquamation, and wound healing.
Understanding the biology of endometrial stem cell populations is important for defining normal and abnormal endometrial tissue regeneration and lineage cell commitment. Transmission of abnormalities across cell lineages may contribute to proliferative disorders, such as endometrial polyps, endometriosis, and endometrial hyperplasia/cancer; adenomyosis; and compromised interactions of lineage cells with the conceptus/placenta, contributing to infertility, implantation failure, or poor pregnancy outcomes. The genetic program of eMSCs described in the present study provides a fundamental understanding of pathways involved in self-renewal and multipotency, and candidate genes regulating the repair, regeneration, and function of human endometrium.
MATERIALS AND METHODS
Tissues and Cells
Endometrial tissues were obtained prospectively by biopsy using a Pipelle catheter (Cooper Surgical) from oocyte donors (n = 20) at the time of oocyte retrieval following gonadotropin stimulation and gonadotropin-releasing hormone agonist treatment, 36 h after administration of human chorionic gonadotropin (hCG). Thus, all of these samples were interval or early secretory endometrium by virtue of their timing to hCG. This phase determination was confirmed by histologic evaluation of the endometrial samples by two independent pathologists, using the criteria of Noyes et al. [26]. Endometrial tissues in the proliferative and mid secretory phases were also obtained from subjects who were undergoing benign gynecologic surgery (n = 7). These samples were used for additional quantitative RT-PCR, clonogenic, immunocytochemical, immunofluorescence, and cellular differentiation analyses. All subjects had regular menstrual cycles (25–35 days) and were documented as not being pregnant. Samples were obtained through the National Institutes of Health (NIH) University of California, San Francisco (UCSF) Human Endometrial Tissue and DNA Bank in accordance with the guidelines of the Declaration of Helsinki. Written, informed consent was obtained from all participants. The study was approved by the UCSF Committee on Human Research.
Fluorescence-Activated Cell Sorting Isolation of Endometrial Cell Populations
Endometrial tissue samples were dissociated with collagenase and hyaluronidase, and stromal and epithelial cell components were size fractionated by filtering [27]. The stromal cell fraction was washed with PBS and treated with DNAse (4 mg/ml). Contaminating erythrocytes were lysed by incubation for 5 min in 168 mM NH4Cl, 7 mM KHCO3, and 0.1 mM ethylene diamine tetraacetic acid (EDTA). Stromal cells were then collected by centrifugation and maintained at 4°C in Hypothermasol (BioLife Solutions) until they were processed for fluorescence-activated cell sorting (FACS) analysis. All cells were FACS analyzed within 24 h of tissue collection. The process initially used for FACS analysis was as previously described [20], involving removal of leukocytes and contaminating epithelial cells from the stromal cell fraction with magnetic-activated cell sorting magnetic bead-conjugated antibodies (Miltenyi Biotec Inc.) against protein tyrosine phosphatase receptor type C (PTPRC; also known as cluster of differentiation 45 [CD45]) [28] and epithelial cell adhesion molecule (EPCAM) [29], respectively, followed by FACS analysis of stromal cells using antibodies to MCAM (CD146) and PDGFRB. This procedure was successfully optimized, resulting in the four-antibody FACS process described below, which avoids nonspecific cell losses associated with the use of magnetic bead selection methods.
All of the samples analyzed by microarray in the current study were obtained using this new, optimized protocol as follows. All processing steps for FACS analysis were conducted at 4°C. A minimum of 106 cells per sample were incubated for 30 min in blocking buffer (PBS containing 40% human serum and 1% bovine serum albumin [BSA]), followed by incubation for 1 h in PBS containing 10% human serum and 1% BSA with the fluorochrome-conjugated antibodies (BD Biosciences) listed below. Antibodies were titrated and used at the indicated final dilutions: phycoerythrin (PE) anti-PDGFRB (1:5), fluorescein isothiocyanate anti-MCAM (CD146; 1:5), PE-Cy7 anti-CD45 (1:20), and allophycocyanin anti-EPCAM (1:20). Following incubation, cells were washed and resuspended in PBS containing 2% fetal bovine serum (FBS) and 1 mM EDTA at a concentration of 107 per milliliter and analyzed using 4′,6′-diamidino-2-phenylindole (DAPI; Sigma) as nuclear stain. Samples were analyzed on a FACS Aria II with FACS Diva software (BD Biosciences). Unstained, single-stain and fluorescence minus one controls were used to establish baselines and set compensation values. Trigger thresholding was done on forward scatter to exclude small debris. Instrument setup, laser delays, and sort calibration were done before each experiment using manufacturer bead controls (CST and Accudrop beads) to ensure optimal optical sensitivity and sort purity, and to minimize experimental variability.
RNA Isolation and Preparation for Microarray Analysis
Total RNA was isolated from sorted cell populations MCAM (CD146)+/PDGFRB+; MCAM (CD146)−/PDGFRB+; and MCAM (CD146)+/PDGFRB−, and purified using the Arcturus PicoPure RNA Isolation Kit (Applied Biosystems) following the manufacturer's protocol, including DNase treatment using the RNase-Free DNase Set (Qiagen). Isolated RNA was reverse transcribed/amplified into cDNA using the NuGEN FFPE (NuGen) kit, and sense-strand cDNA targets were created using the NuGEN WT-Ovation Exon Module, followed by fragmentation and labeling using NuGEN Encore Biotin Module for hybridization to Affymetrix Human Gene 1.0 ST arrays. The quality of the amplified cDNA and fragmented cDNA was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies), and individual samples meeting yields and quality standards were further processed and hybridized to Affymetrix Human Gene 1.0 ST arrays (Affymetrix), probing 19 492 genes. Arrays were scanned according to the protocol described in the WT Sense Target Labeling Assay Manual from Affymetrix (v4; FS450_0007).
Microarray Gene Expression Data Analysis
The intensity values of different probe sets (genes) in the GeneChip Operating Software (Affymetrix) were imported into GeneSpring GX 11.02 software (Agilent Technologies) and processed using the robust multiarray analysis algorithm for background adjustment, normalization, and log2 transformation of perfect match values [3, 30]. The data were normalized to the median of all samples. We performed two major comparisons: MCAM (CD146)+/PDGFRB+ (eMSC) vs. MCAM (CD146)−/PDGFRB+ (endometrial stromal fibroblasts), and MCAM (CD146)+/PDGFRB+ (eMSC) vs. MCAM (CD146)+/PDGFRB− (endometrial endothelial cells). The resulting gene lists generated include only genes with >2.0-fold change and P < 0.05 by two-way ANOVA with Benjamini-Hochberg multiple-testing correction for false discovery rate [31, 32]. Raw data files of the experiments have been uploaded to the National Center for Biotechnology Information Gene Expression Omnibus database under accession number GSE31152. Principal component analysis (PCA) and hierarchical clustering were performed as previously described [3, 30]. Briefly, the unbiased PCA algorithm in GeneSpring was applied to all samples, using all 19 492 genes on the Affymetrix Human Gene 1.0 ST array chip to identify similar expression patterns and underlying cluster structures. Hierarchical cluster analysis was conducted using only differentially expressed genes from all samples and among all experimental conditions. The smooth correlation distance measure algorithm (GeneSpring) was then used to identify samples with similar patterns of gene expression.
Biological Functions and Canonical Pathway Analyses
“Transcript cluster Id” and fold changes of up- and down-regulated genes in each pairwise comparison were imported into Ingenuity Pathway Analysis (IPA; Ingenuity Systems). Detailed pathway analysis was performed using the Core Analysis function on IPA to interpret data in the context of biological function, pathways, and networks. Biological functions are composed of molecular and cellular functions, and canonical pathways include signaling and metabolic pathways. Significance of the biological functions and the canonical pathways were tested by the Fisher exact test P value.
Quantitative RT-PCR
Ninety-two genes from the generated differential expression gene lists were chosen for validation based on their relevance to biological processes and steroid hormone response in endometrium [3, 30]. To this end, expression levels of these genes (and four internal controls) were analyzed on a total of 70 cDNA samples derived from FACS-isolated eMSC, endothelial cell, and stromal fibroblast populations, including 15 previously analyzed by microarray (technical validation) and 55 additional cell isolates (biological validation). Samples were analyzed by quantitative RT-PCR (Q-RT-PCR) using the Fluidigm 96.96 Dynamic Array Integrated Fluidic Circuits (IFC) and the Biomark System (www.fluidigm.com/biomark-system.html). Randomly assigned duplicate samples were placed in the remaining 26 array sample inlets.
Aliquots of cDNA were normalized to a concentration of 200 ng/μl (total RNA equivalent). Custom-designed forward (F) and reverse (R) primer pairs for the genes of interest were diluted to a working stock solution of 20 μM F+R for each gene. (Fluidigm; Supplemental Table S1; all Supplemental Data are available online at www.biolreprod.org). A pool of all 92 genes (F + R primer pairs) was generated by pooling 2 μl of each primer pair into a final volume of 200 μl, with a final primer concentration of 200 nM for each oligonucleotide in the primer pool. All cDNA samples were then preamplified by combining each sample with the primer pool and TaqMan Pre-Amp Mastermix (Applied Biosystems), in 5-μl reactions following the Fluidigm Specific Target Amplification (STA) protocol, enriching samples for loci of interest. Samples were subsequently treated with exonuclease 1 to remove any single-stranded material from the sample following the STA reaction. Finally, samples were diluted 1:5 in a Tris-EDTA dilution buffer (PN T0021; TekNova). A Fluidigm 96.96 Dynamic Array IFC was prepared according to the manufacturer's instructions. Quantitative PCR was performed using Evagreen binding dye (Biotium Inc., Hayward, CA), according to the protocol for the Biomark System. Reaction conditions were as follows: 50°C for 2 min and 95°C for 10 min, followed by 35 cycles of 95°C for 15 sec and 60°C for 60 sec. To assess amplicon integrity, melt curve analysis was performed following PCR amplification. Samples were heated at 1°C per second from 60°C to 95°C, with image capture every cycle. Data were processed by user detector threshold settings, allowing thresholds to be individually set for each gene, and linear baseline correction using Biomark Real-time PCR Analysis software (v.3.0.4). Melt curves were assessed using a melting temperature (Tm) threshold to allow filtration of Ct values with poor melt curve data.
The Q-RT-PCR data were analyzed using Microsoft Office Excel (2003). The comparative Ct method was used to obtain relative expression for each grouping comparison, where the amount of target normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and relative to a calculated calibrator is represented by 2–ΔΔCT. GAPDH has reproducibility and stable expression in endometrial tissue and was thus used as the reference gene in analysis.
Statistical Analysis of Correlations Between Microarray Data and Q-RT-PCR Data
The agreement between differential gene expressions derived by microarray and Q-RT-PCR was evaluated with two different nonparametric measures of association. Spearman rho measures the linear correlation between the ranks of the paired data. Kendall tau measures the probability of concordance between the pairs. For both of these association measures, a positive value indicates agreement between the microarray and Q-RT-PCR data. To assess the statistical significance of each association measure, a one-sided test with a null hypothesis of either negative or no association (i.e., less than or equal to zero) was performed. If the data offered sufficient evidence to reject this null hypothesis, then the positive association was statistically significant at the level given by the P value.
The base-2 logarithm of the Q-RT-PCR expression was calculated. The two measures of association were computed from these logarithmic Q-RT-PCR values, and the corresponding differential expression values were obtained by microarray analysis. The hypothesis test was applied to each association measure and a P value was derived. The results, obtained using R version 2.13.1 [33], were then compiled.
Clonogenic Potential of eMSCs
The clonogenicity of eMSCs was assessed by culturing FACS-isolated MCAM (CD146)+/PDGFRB+ cells from endometrial tissue samples (n = 3) seeded at clonal densities (2–18 cells per square centimeter) onto gelatin-coated culture dishes (Nunc) in Dulbecco modified Eagle medium (DMEM)/F12 (Life Technologies) or MCDB-105 (Sigma-Aldrich) media supplemented with 10% FBS (Gemini Bio-Products) and 5 μg/ml bovine insulin (Sigma-Aldrich). Colonies were monitored microscopically on a daily basis to ensure they were derived from single cells. The numbers of colonies were assessed after 25 days of culture, and the percent cloning efficiency (%CE) was calculated by the following formula: %CE = number of colonies > 50 cells/number of cells seeded × 100.
In Vitro Differentiation of eMSCs
FACS-isolated MCAM (CD146)+/PDGFB+ cells were cultured at clonal density as described above, and primary colonies were harvested and cells seeded into gelatin-coated 12-well plates at 2500 cells per well. To assess the differentiation potential of eMSCs into the adipogenic lineage, cells were then cultured in adipogenic differentiation medium (Millipore) following the manufacturer's instructions. Replicate control cultures were maintained in DMEM/F12 + 10% FBS. After 4 wk of culture in differentiation or control media, cells were fixed in 4% paraformaldehyde in PBS, rinsed in PBS, and stained with Oil Red O (Millipore), then rinsed in distilled water and the nuclei counterstained with hematoxylin. Stained cultures were examined by bright field microscopy to determine the presence of red-staining intracellular lipid deposits.
Immunocytochemistry
For immunostaining of vimentin, primary colonies derived from FACS-isolated MCAM (CD146)+/PDGFRB+ cells were harvested, and cells were cultured in Lab-Tek chamber slides (Nunc). All subsequent steps were carried out at room temperature, and all incubation steps were followed by three 5-min washes in PBS with 0.05% Tween 20. Subconfluent cultures were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) in PBS, and incubated in PBS containing 10% H2O2 and 0.1% saponin (Sigma) for 30 min, blocked with nonserum protein blocking solution (DAKO) for 30 min, and incubated with the primary antibody (mouse anti-vimentin clone V9; Sigma) at a 1:500 dilution in PBS containing 2% BSA for 1 h, followed by biotinylated goat anti-mouse immunoglobulin G (IgG; Jackson ImmunoResearch Laboratories) diluted in blocking solution at 1:1000 dilution for 1 h and horseradish peroxidase-conjugated streptavidin (Jackson ImmunoResearch Laboratories) at 1:500 in blocking solution for 30 min. Antibody binding was visualized using diaminobenzidine (Vector Laboratories) as chromogen and the reaction stopped by rinsing in water, and nuclei were counterstained with hematoxylin (Vector Laboratories). Slides were dehydrated through graded ethanol and histosol (National Diagnostics), mounted, and examined by bright field microscopy. For a negative control the primary antibody was substituted with nonimmune mouse IgG1 at the same concentration. Human endometrial stromal fibroblast cultures [30] were used as a positive control for vimentin immunoreactivity.
Immunofluorescence
Endometrial tissue sections (approximately 5-mm thickness, n = 3) from uteri obtained at hysterectomy were embedded in Tissue-Tek Optimal Cutting Temperature (OCT) compound (Miles Laboratories), snap frozen in liquid nitrogen, and stored at −80°C through the NIH UCSF Human Endometrial Tissue and DNA Bank. For immunofluorescence staining of frozen sections, primary antibodies included monoclonal mouse anti-human MCAM (CD146) IgG (ab24577; Abcam) and monoclonal rabbit anti-human PDGFRB (ab32570; Abcam). The secondary antibodies included goat anti-mouse IgG Alexa 488 (A11008; Invitrogen) and goat anti-rabbit IgG Alexa 594 (A11012; Invitrogen). Colocalization of antigens using double immunofluorescence was performed as previously described [34]. Briefly, frozen sections (∼8 μm) of human endometrium were cut with a cryostat and mounted on Superfrost/Plus microscope slides (Fisher Scientific). Sections were fixed in −20°C methanol for 10 min, permeabilized at room temperature with 0.3% Tween 20 in PBS (wash buffer), and blocked in antibody dilution buffer (two parts PBS, 1.0% BSA, and 0.3% Tween 20 [pH 8.0], and one part glycerol) containing 10% normal goat serum for 1 h at room temperature. Sections were then washed and incubated overnight at 4°C with 5 μg/ml initial primary antibody (anti-MCAM [CD146] IgG). Following three washes for 10 min each, sections were incubated with 8 μg/ml initial secondary antibody (anti-mouse IgG Alexa 488) for 4 h at room temperature, and washed six times for 10 min each. Sections were then incubated overnight at 4°C with 5 μg/ml second primary antibody (anti- PDGFRB IgG). Following six washes for 10 min each, sections were incubated with 8 μg/ml second secondary antibody (anti-rabbit IgG Alexa 594) for 4 h at 4°C, washed six times for 10 min each, and dipped in distilled-deionized H2O. Slides were overlaid with antifade mounting reagent containing DAPI (P36935; Invitrogen) and mounted with coverslips. Images were captured using a Leica microscope fitted with a high-resolution Leica digital camera with Leica software (Leica Microsystems Inc.).
RESULTS
Characterization of eMSCs
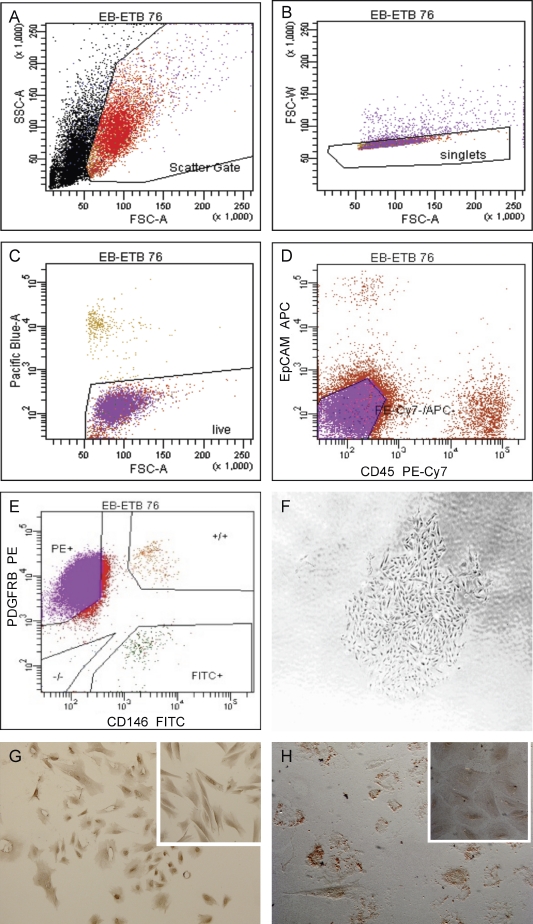
The stromal cell fraction obtained after enzymatic dissociation of endometrial tissue contained a mixture of cell populations that included CD45+ leukocytes and EPCAM+ epithelial cells. The FACS protocol for isolation of discrete endometrial stromal cell populations (Fig. 1, A–E) involved sorting single, viable, CD45−, EPCAM− cells into three populations: MCAM (CD146)+/PDGFRB+ (eMSCs); MCAM (CD146)−/PDGFRB+ (endometrial stromal fibroblasts); and MCAM (CD146)+/PDGFRB− (endometrial endothelial cells). The MCAM (CD146)−/PDGFRB+ (stromal fibroblast) cell population comprised the prevalent phenotype isolated from endometrial stromal cells, whereas the MCAM (CD146)+/PDGFRB+ (eMSC) and MCAM (CD146)+/PDGFRB− (endothelial) populations comprised a small portion of the stromal fraction (Fig. 1E and Table 1). The MCAM (CD146)+/PDGFRB+ cells comprised 0.04%–19.0% of the sorted stromal cells in hysterectomy samples and 0.8%–15.7% of endometrial biopsy samples. The MCAM (CD146)+/PDGFRB+ population isolated by FACS contained cells with clonogenic potential (Fig. 1F), with colony-forming efficiencies ranging from 0.2% to 4.0%, and had a mesenchymal phenotype, expressing vimentin (Fig. 1G). These cells differentiated under appropriate culture conditions into the adipogenic lineage (positive Oil red O revealing lipid inclusions; Fig. 1H), confirming the potential of the isolated endometrial cells to differentiate to nonendometrial mesenchymal cell lineages, characteristic of MSCs in general [16] and consistent with a previous report [21].
FIG. 1.
Isolation and characteristics of MCAM (CD146)+/PDGFRB+ cells from human endometrium. A–E) FACS analysis of endometrial stromal cells obtained by enzymatic dissociation of endometrial tissue labeled with DAPI and antibodies to CD45, EPCAM+, MCAM (CD146), and PDGFRB, showing single (B), viable (C), and CD45− and EPCAM− (D) cells sorted into three populations according to MCAM (CD146) and PDGFRB expression (E). F) Colony derived from FACS-sorted MCAM (CD146)+/PDGFRB+ stromal cells. G) Vimentin-positive cells derived from FACS-sorted MCAM (CD146)+/PDGFRB+ eMSC. Inset: vimentin-positive endometrial stromal fibroblasts (positive control). H) Oil red O-positive eMSC differentiated in vitro to adipogenic lineage. Inset: eMSC not cultured in adipogenic medium (negative control). Original magnifications ×40 (F) and ×100 (G, H, and insets in G and H).
TABLE 1.
FACS yields of eMSC, endometrial stromal fibroblast (eSF), and endothelial cell populations.
Transcriptome Analysis of eMSCs
PCA and hierarchical clustering analysis.
PCA distributes samples into three-dimensional space based on variance in gene expression; those with similar gene expression profiling cluster close together. Using all genes on the Affymetrix array and a completely unbiased approach, the sorted samples clustered into three major groups: MCAM (CD146)+/PDGFRB+ (eMSCs), MCAM (CD146)−/PDGFRB+ (stromal fibroblasts), and MCAM (CD146)+/PDGFRB− (endothelial cells), with the eMSCs clustering close to the stromal fibroblasts and separately from the endothelial cells (Fig. 2).
FIG. 2.
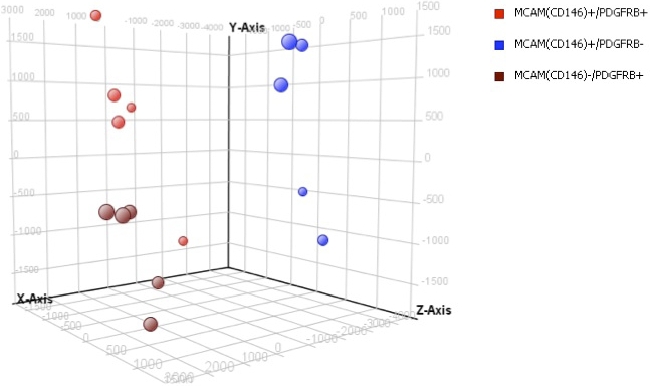
Clustering analysis of eMSCs. Principal component analysis clustering: eMSC (red), endometrial stromal fibroblast (brown), and endothelial cell (blue) populations.
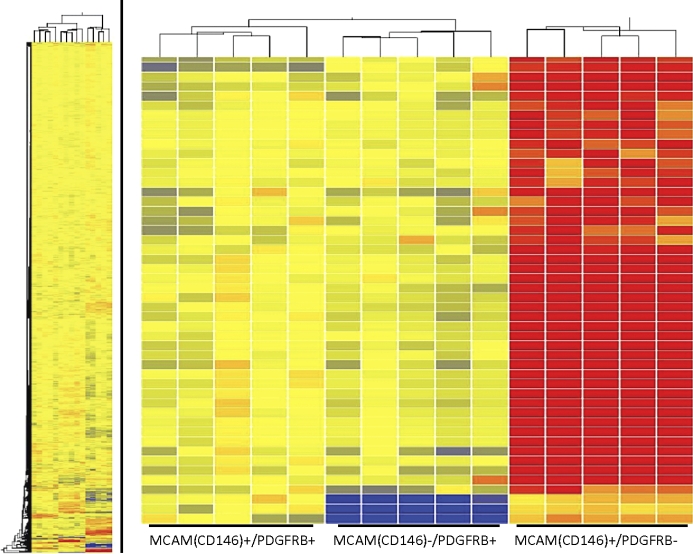
Unsupervised hierarchical clustering analyses based on the combined gene list derived from pairwise comparisons yielded a dendrogram of sample clustering and a heatmap of gene expression. The clustergram (Fig. 3) revealed two main branches: one branch consisting of only the endothelial cells and the other branch containing two subbranches: the eMSC and the stromal fibroblast populations. Collectively, these data demonstrate that samples cluster exclusively by cell type, with no crossover of another population in a given branch or subbranch, and that the eMSC and stromal fibroblast populations cluster closely and distinctly from the endothelial cell population.
FIG. 3.
Hierarchical clustering. Complete hierarchical clustering analysis between eMSC, endometrial stromal fibroblast, and endothelial cell populations (left). Magnified inset of most differentially expressed genes (right) taken from the complete clustering analysis.
Differentially expressed genes.
There were 762 significantly differentially expressed genes (378 up-regulated and 384 down-regulated) in eMSCs vs. endometrial stromal fibroblasts, using a 2-fold change threshold (Supplemental Table S2). There were 1518 significantly differentially expressed genes (883 up-regulated and 635 down-regulated) in eMSCs vs. endothelial cells, using a 2-fold change threshold (Supplemental Table S3). These data demonstrate that the eMSCs and endometrial stromal fibroblasts have more genes in common than do the eMSC and endometrial endothelial populations.
Differences in gene expression between the MCAM (CD146)+/PDGFRB+ (eMSC) and MCAM(CD146)−/PDGFRB+ (stromal fibroblast) populations.
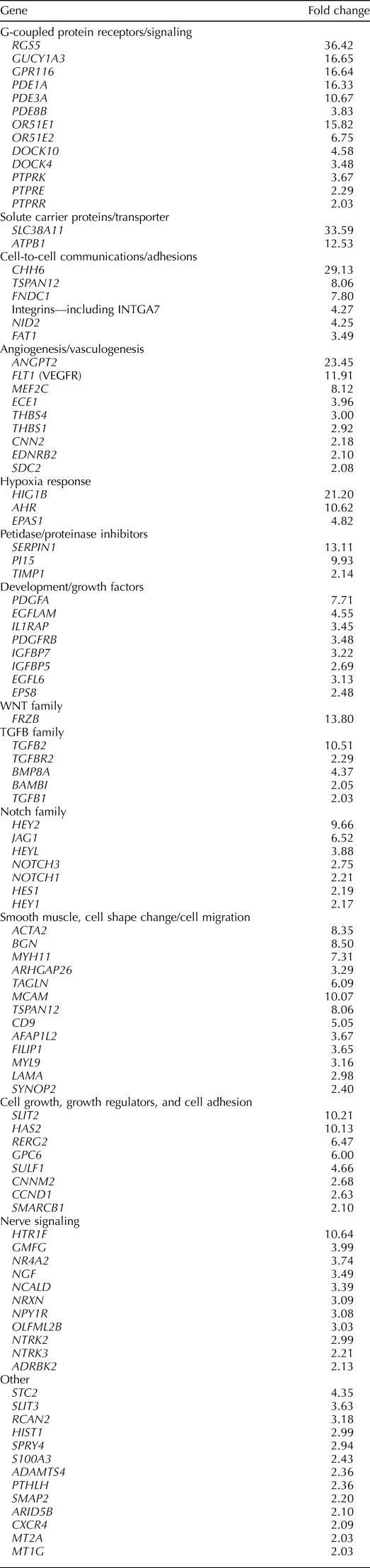
Genes that were significantly (P < 0.05) differentially expressed >2.0-fold in the MCAM (CD146)+/PDGFRB+ vs. MCAM (CD146)−/PDGFRB+ populations are shown in their entirety in Supplemental Table S2, and select genes/categories are shown in Tables 2 and 3.
TABLE 2.
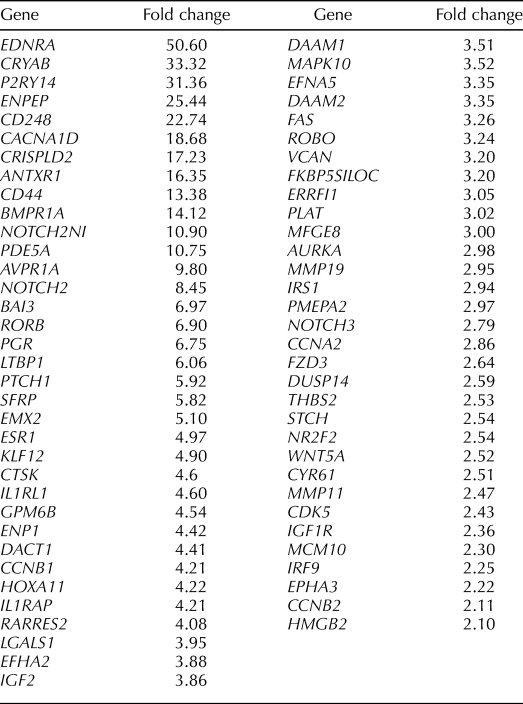
Select genes up-regulated in eMSC (MCAM (CD146)+/PDGFRB+) compared to endometrial stromal fibroblast populations (MCAM (CD146)−/PDGFRB+).
TABLE 3.
Select genes down-regulated in eMSC (MCAM (CD146)+/PDGFRB+) compared to endometrial stromal fibroblast populations (MCAM (CD146)−/PDGFRB+).
The most highly up-regulated gene in this comparison was regulator of G-protein signaling 5, RGS5, a pericyte marker that is coexpressed with PDGFRB and a master regulator of PDGFRB and G-protein-coupled receptor (GPCR)-mediated signaling pathways during fetal vascular development [35]. Other highly differentially expressed genes include cadherin 6 (CDH6, important in cell adhesion and epithelial integrity); other receptors and mediators in GPCR signaling (e.g., GPR116, olfactory receptor 51E1 and 51E2); phosphodiesterases; protein tyrosine kinase receptors; solute carrier proteins and transporters; genes involved in angiogenesis/vasculogenesis; cell-to-cell communication; and cell adhesion, growth, shape, and migration. Interestingly, the tumor suppressor gene ras-related and estrogen-regulated inhibitor 2 (RERG2) was up-regulated, as were genes involved in the response to hypoxia (hypoxia-inducible domain family 1B [HIGD1B], the aryl hydrocarbon receptor [AHR], and endothelial PAS domain protein [EPAS1]), and genes involved in proteinase/peptidase inhibition. Genes governing stem cell self-renewal and multipotency were up-regulated, including members of the WNT, transforming growth factor beta (TGFB), Notch, Hedgehog, insulinlike growth factor (IGF), and epidermal growth factor (EGF) families (Table 2 and Supplemental Table S2). As anticipated, MCAM (CD146) was up-regulated in the MCAM (CD146)+/PDGFRB+ eMSC compared with the MCAM (CD146)−/PDGFRB+ stromal fibroblast (Supplemental Table S2), providing strong validation of the robustness of the experimental protocol.
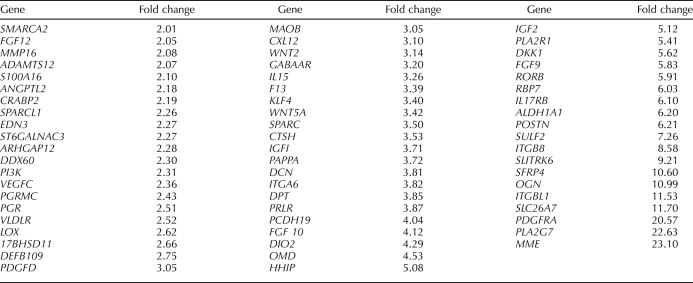
Genes significantly (P < 0.05) down-regulated ≥2.0-fold between the MCAM (CD146)+/PDGFRB+ and MCAM (CD146)−/PDGFRB+ populations (i.e., up-regulated in the MCAM (CD146)−/PDGFRB+ stromal fibroblast cells) included IGF1, IGF2, fibroblast growth factors (FGF9, FGF10, and FGF12), progesterone receptor (PGR), PGR membrane component 1 (PGRMC1), membrane metalloendopeptidase (MME), phospholipase A2 (PLA2G7) and its receptor (PLA2R1), secreted frizzled related protein 4 (SFRP4), dickkopf (DKK1), prolactin receptor (PRLR), pappalysin A (PAPPA), secreted protein acidic and rich in cysteine (SPARC) and SPARC-like 1 (SPARCL1), kruppellike factor 4 (KLF4), interleukin 15 (IL15), and tissue factor. Many of these genes are characteristic of human endometrial stromal fibroblasts in interval/early secretory endometrium [3, 36]. These data support the MCAM (CD146)−/PDGFRB+ population as the human endometrial stromal fibroblast (Table 3 and Supplemental Table S2).
Differences in gene expression between the MCAM (CD146)+/PDGFRB+ (eMSC) and the MCAM(CD146)+/PDGFRB− (endothelial) populations.
Genes that were significantly (P < 0.05) differentially expressed >2.0 -fold in the MCAM (CD146)+/PDGFRB+ vs. MCAM (CD146)+/PDGFRB− populations are shown in their entirety in Supplemental Table S3, and select genes are shown in Table 4.
TABLE 4.
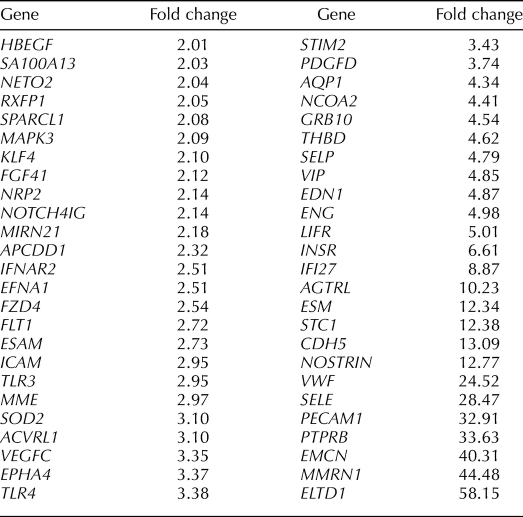
Select genes up-regulated in eMSC (MCAM (CD146)+/PDGFRB+) compared to endothelial cell populations (MCAM (CD146)+/PDGRFB−).
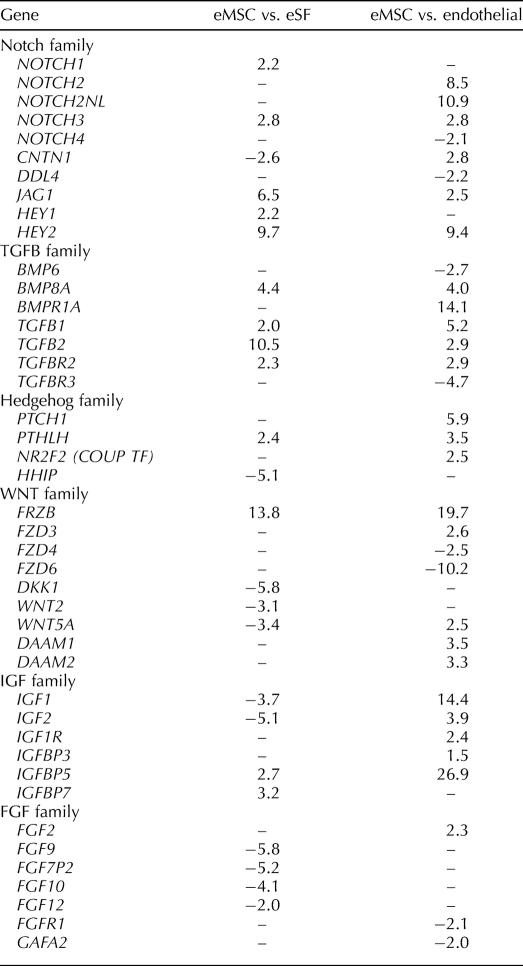
Among those most highly up-regulated genes are alpha smooth muscle actin (ACTA2), the endothelin receptor (EDNRA), protocadherin FAT tumor suppressor homolog 1 (FAT1; an adhesion molecule and/or signaling receptor important in cell communication), the arginine vasopressin receptor (AVPR1A), and ATP-binding cassette subfamily 3, member 9 (ABCC9; a member of a family of adult stem cell markers [37]), latent TGFB binding protein (LTBP1), Wnt family members and modulators of WNT (WNT5A, secreted frizzled-related protein 1 [SFRP1], disheveled associated activator of morphogenesis [DAAM1, DAAM2], frizzled homolog [FZD3]), insulinlike growth factor 1 (IGF1) and IGF binding protein 5 (IGFBP5), bone morphogenetic protein receptor 1A (BMPR1A), and ERBB receptor feedback inhibitor (ERRFI1) (Table 4 and Supplemental Table S3). HOXA11 and EMX2, an important regulator of HoxA10 signaling and uterine development [38], were among the up-regulated genes, as were ephrin A-5 (EFNA5) and its receptor (EPHA3), estrogen receptor alpha1 (ESR1); PGR; the cysteine-rich angiogenic inducer, CYR61; and a member of the high-mobility group protein family, HMGB2. Phosphodiesterases, members of the Notch family, and numerous GPCR genes were also up-regulated. Many of these genes up-regulated in eMSCs play important roles in stem cell self-renewal and also in differentiation of their lineage cell, the endometrial stromal fibroblast in response to progesterone (e.g., Notch, TGFB and BMP, IGF, Hedgehog, EGF, WNT family members, and HOXA10 signaling) (Table 6) [13, 15, 23, 39–42]. As anticipated, PDGFRB was up-regulated in the MCAM (CD146)+/PDGFRB+ eMSCs vs. the MCAM (CD146)+/PDGFRB− endothelial population (Supplemental Table S3).
TABLE 6.
Differentially expressed genes relevant to stem cell proliferation and differentiation.
Genes significantly (P < 0.05) down-regulated ≥2-fold between the MCAM (CD146)+/PDGFRB+ and MCAM (CD146)+/PDGFRB− populations (i.e., up-regulated in the endothelial population) were characteristic of endothelial cells, including genes associated with classical endothelial surface glycoproteins in other tissues [43–45], such as multimerin (MMRN1), endomucin (EMCN), selectin E (SELE), and vascular endothelial cadherin (CDH5), as well as other known endothelial-specific genes, including von Willebrand factor (VWF), platelet endothelial cell adhesion molecule (PECAM1; also known as cluster of differentiation 31 [CD31]), endothelial-specific molecule 1 (ESM1), endothelial cell-selective adhesion molecule (ESAM), nitric oxide synthase trafficker (NOSTRIN), and endothelin 1 (EDN1). Interestingly, micro-RNA 21 (MIR21) was up-regulated in endothelial cells vs. the eMSC population, whereas one of its targets, myocyte-specific enhancer factor 2C (MEF2C), was up-regulated in the eMSC compared with the stromal fibroblast and endothelial populations. (Table 5 and Supplemental Table S3).
TABLE 5.
Select genes down-regulated in eMSC (MCAM (CD146)+/PDGFRB+) compared to endothelial cell populations (MCAM (CD146)+/PDGFRB−).
The IPA summary is in Supplemental Tables S4 and S5, and full pathway analyses are in Supplemental Figure S1 and Supplemental Table S6. The major canonical pathways included hepatic fibrosis/hepatic stellate cell activation (including endothelin, PDE, cAMP, and GPCR signaling) [46]; cellular effects of sildenafil (phosphodiesterase mediated); sphingosine-1-phosphate signaling; axonal guidance signaling; endothelin-1 signaling; human embryonic stem cell pluripotency; Notch family, TGFB, VEGF, GPCR, and ephrin receptor signaling; calveolar-mediated endocytosis; chemokine (C-X-C motif) receptor 4 (CXCR4) signaling; cycle control of chromosomal replication; nuclear receptor subfamily 2 (LXR/RXR) activation; and others. The major biological functions (IPA summary of analysis in Supplemental Tables S4 and S5; full biological function analyses in Supplemental Fig. S2 and Supplemental Table S7) included: cancer; cell-to-cell signaling and interaction; cell cycle; cellular assembly and organization; cell morphology; DNA replication, recombination, and repair; genetic disorder; hepatic system disease; tissue development; developmental disorder, neurological disease; inflammatory response; cardiovascular system development and function; and organismal development. The scores and number of molecules involved are presented in Supplemental Table S7.
Validation Studies
Quantitative RT-PCR.
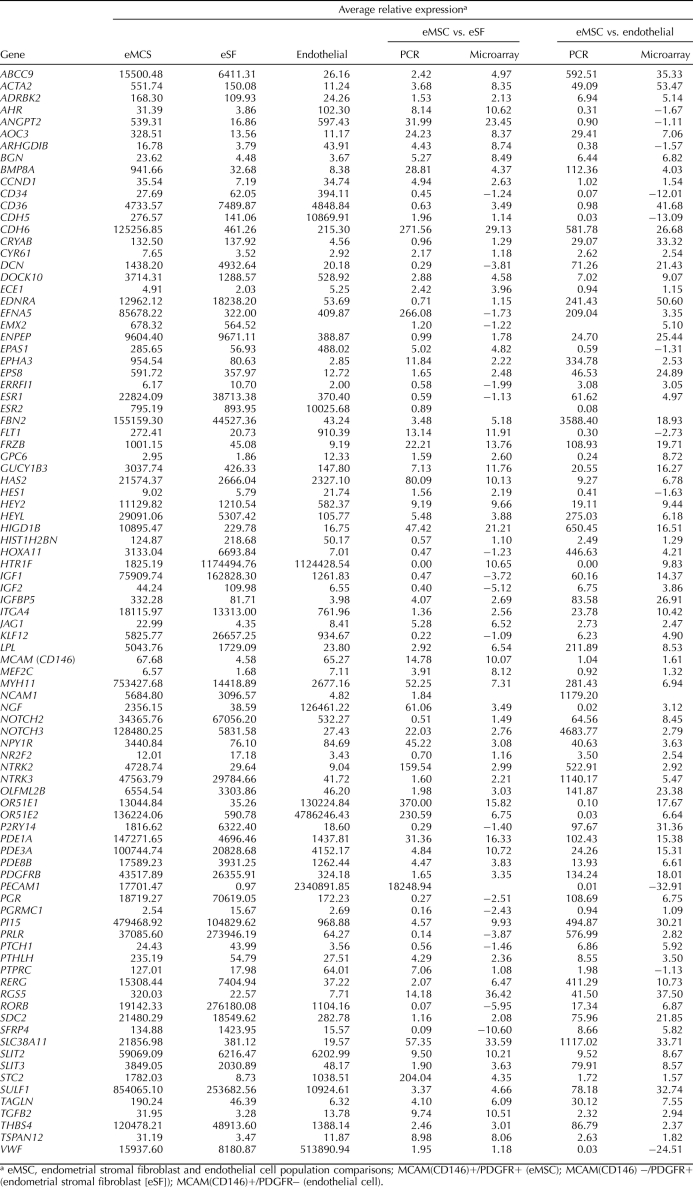
We used a microfluidic quantitative PCR approach that enabled simultaneous multigene analysis/validation in 92 cell isolates. The results for select gene expression in MCAM (CD146)+/PDGFRB+ cells and their relative expression compared with MCAM (CD146)−/PDGFRB+ and MCAM (CD146)+/PDGFRB− cells are shown in Table 7. Of note in the MCAM (CD146)+/PDGFRB+ eMSC is the high expression of members of pathways relevant to stem cell renewal and lineage specification, including: CCND1; BMP8A; NOTCH2; NOTCH3 and downstream effectors HES1 and HEY2; IGFs; WNT family members; Hedgehog mediator Jagged 1 (JAG1), receptor PTCH1, and downstream effector parathyroid hormonelike hormone (PTHLH); signaling pathways mediated by phosphodiesterases; GPCRs; retinoic acid receptor; neurotrophin receptor kinases; and other neuronally related receptors and signaling pathways. Also up-regulated in eMSCs were vascular factors (vascular adhesion protein, AOC3, angiopoietin 2 [ANGPT2], and EDNRA); members of the SLIT family important in (axonal) cell guidance; and genes important in cell-to-cell and cell-matrix communication (e.g., biglycan, CDH6, and thrombospondin 4). Interestingly, ESR1 was highly expressed in eMSCs and the stromal fibroblast population, whereas ESR2 was highly expressed in the endothelial population. PGR was highly expressed in both eMSCs and stromal fibroblasts but not endometrial endothelial cells.
TABLE 7.
Microfluidic Q-RT-PCR validation of select genes.
PDGFRB was down-regulated 18- and 134-fold in CD146+/PDGFRB− cells by microarray and Q-RT-PCR, respectively, and VWF, an exclusive marker of endothelial cells, was up-regulated 25- and 32-fold in CD146+/PDGFRB− cells by microarray and Q-RT-PCR, respectively (Table 7). Also, PECAM1 (CD31) was up-regulated 32.9-fold in the endothelial population by microarray analysis (Table 5) and was not appreciably expressed in either eMSCs or stromal fibroblasts isolated concomitantly by FACS. The Q-RT-PCR for PECAM1 (CD31) on all three cell populations confirmed that PECAM1 (CD31) is highly expressed in CD146+/PDGFRB− (endothelial cells) and not expressed in CD146−/PDGFRB+ (stromal fibroblasts) (Table 7). Together, these data provide objective and unequivocal evidence of the identity of the endothelial cell type.
We found very high and statistically significant positive association between differential gene expression data derived by microarray and Q-RT-PCR (Table 8). We performed technical validation, which included only samples analyzed by microarray, and biological validation, which included samples that had not undergone microarray analysis but were purified by identical standard operating procedures from additional participants' endometrial biopsy specimens. Technical validation for the differential expression of eMSCs vs. endometrial stromal fibroblasts produced a Spearman rho of 0.79 (P < 0.0001) and a Kendall tau of 0.64 (P < 0.0001) for the Q-RT-PCR/microarray association. The same validation for the differential expression of eMSCs vs. endothelial cell populations produced a Spearman rho of 0.61 (P < 0.0001) and a Kendall tau of 0.46 (P < 0.0001). Biological validation, using all samples, produced similar results (Table 8). For both validation methods, the data support a statistically significant positive association, as indicated by the reported P values. This substantiates the agreement between differential gene expression data derived by microarray and Q-RT-PCR.
TABLE 8.
Nonparametric association measures of agreement between microarray analysis and Q-RT-PCR for differential gene expression of eMSC vs. fibroblast and eMSC vs. endothelial and eMSC vs. endothelial.
Colocalization of MCAM (CD146) and PDGFRB in human endometrium.
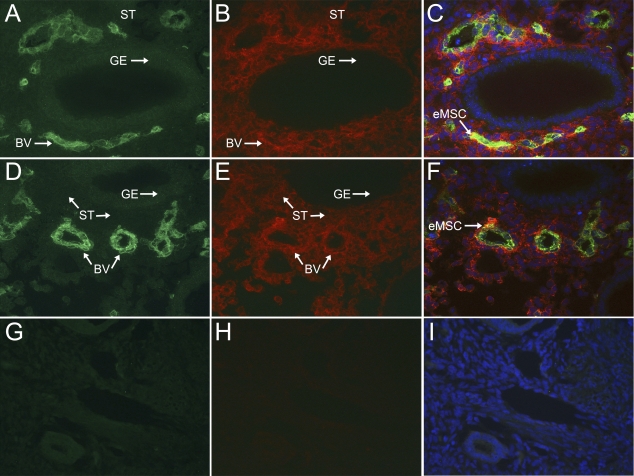
Analysis by FACS revealed a population of cells in the human endometrium that express MCAM (CD146) and PDGFRB (Fig. 4). Sections (5 mm) of full-thickness endometrium embedded in OCT compound were analyzed for the presence of MCAM (CD146)+/PDGFRB+ cells using indirect immunofluorescence. MCAM (CD146) localized to endothelial cells as well as perivascular cells in the endometrium (Fig. 4, A and D), whereas PDGRFB localized to endometrial stromal cells and cells in the perivascular regions of endometrium (Fig. 4, B and E). PDGFRB staining was not observed in endometrial glandular epithelium (Fig. 4, B and E). MCAM (CD146)+/PDGFRB+ cells were identified in the perivascular region around small blood vessels (Fig. 4, C and F). These results confirm the presence of MCAM (CD146)+/PDGFRB+ cells and their localization to the perivascular region of human endometrium, as previously reported by Schwab and Gargett [21], who used an indirect immunofluorescence method that used biotinylated IgG to amplify the MCAM (CD146) signal. We did not find differences in the distribution of MCAM (CD146)+/PDGFRB+ cells in the functionalis vs. the basalis regions of the endometrium (data not shown). The rarity of the MCAM (CD146)+/PDGFRB+ cells in the endometrium is characteristic of adult stem cells and consistent with the low yields of this population during their purification by FACS analysis (Table 1).
FIG. 4.
Colocalization of MCAM (CD146) and PDGFRB in human endometrium. Sections of full-thickness human endometrium were dual labeled for MCAM (CD146) and PDGFRB. MCAM (CD146) was localized to all endothelial cells and some perivascular cells in the basalis (A and D), whereas PDGFRB showed a wider distribution on endometrial stromal cells as well as some perivascular cells (B and E), but was absent in glandular epithelium (B and E). Colocalization (arrow) of MCAM (CD146) and PDGFRB was observed in a perivascular location (C and F). Nuclei in C, F, and I are stained with DAPI. Negative IgG controls for MCAM (CD146) (mouse; G) and PDGFRB (rabbit; H) and merged images (I) are shown. GE, glandular epithelium; BV, blood vessel; ST, stroma. Original magnification ×100.
DISCUSSION
The Functional Phenotype of eMSCs
Stem cells in adult tissues remain quiescent in a nonproliferative state until they are stimulated by signals such as tissue damage or tissue remodeling, resulting in their proliferation and maintenance of the undifferentiated state (self-renewal) and unipotent or multipotent lineage differentiation to cell type(s) characteristic of the adult tissue [10–12]. Mesenchymal stem cells share these properties and differentiate into cells of various lineages [6, 13–16, 25, 47]. Endometrial mesenchymal stem cells are also multipotent, as demonstrated by Schwab and Gargett [21], and confirmed in the present study.
Human endometrium regenerates on a cyclic basis from proposed progenitors of epithelial, fibroblast, and endothelial origins [6–8]. We found that eMSCs express markers of pericytes (e.g., RGS5, ACTA2, ANGPT2, IGFs, NGF and NGF high-affinity receptors, PDGF, PTHLH, TGFB, and syndecan) [48–51], and our immunofluorescence studies confirm the perivascular localization of these cells [21]. As pericytes, eMSCs in the vascular niche are well positioned in denuded vessels at menses or postpartum to respond to signals accompanying endometrial breakdown. We and others have shown that prior to and during menses, human endometrium displays genes and pathways reflecting a phenotype of extracellular matrix proteolysis, cell motility, hemostasis, vasoconstriction, inflammation, chemotaxis, and wound healing [3, 4, 52]. Indeed, eMSCs express genes involved in the responses to hypoxia, inflammation, proteolysis, and angiogenesis/vasculogenesis. Interestingly, MSC pericytes migrate in an in vitro culture model toward proteolytic digests of extracellular matrix, consistent with the important property of progenitors to migrate to the site of injured tissue for reconstruction [16]. Also, hypoxia enhances the proliferative capacity and the plasticity of human MSCs [53], which may be of relevance to endometrial repair and regeneration. It is not known whether the mechanical forces accompanying uterine contractions at menses or postpartum would influence eMSC behavior, as physical stress affects bone marrow-derived MSCs in bone repair [25].
The repair phase of endometrial regeneration is steroid hormone independent [54], and although we found that eMSCs express ESR1, ESR2, and PGR, it is unlikely that eMSCs respond to changing levels of circulating ovarian-derived steroid hormones throughout the cycle and still maintain their “stemness.” In support of this, we found in preliminary studies no differences in genes expressed by eMSCs from the endometrium of oocyte donors and eMSCs from a limited number of cycling women in the proliferative and mid secretory phases, suggesting that the eMSC transcriptome is not cycle dependent. Also, the high correlation coefficient in the biological validation of genes expressed in eMSCs isolated from oocyte donors and unstimulated participants in natural cycles (Supplemental Table S6) support this conclusion.
The location and activation of eMSCs resulting in regeneration of the endometrium cycle-to-cycle would necessitate self-renewal as well as migration into the remaining tissue (functionalis and basalis) and differentiation into lineage cells (stromal fibroblasts and perhaps other lineages) [6, 7]. Migration and differentiation of progenitors within tissues undergoing repair are governed largely by the physiologic conditions of the niche and involve molecular mechanisms, including chemoattractants, membrane receptors, paracrine factors, interactions with the extracellular matrix, and intercellular signaling molecules [55, 56]. A prototype of chemotactic factors and membrane receptors involved in the migration of MSCs in target tissues is CXC ligand 12 (CXCL12; also known as stromal derived factor 1 [SDF1]) and its receptor CXCR4, and involves matrix metalloproteinases (MMPs) [57]. It is of interest that we found that eMSCs have a high expression of CXCR4, whereas CXCL12 is highly up-regulated in the stromal fibroblast. Previous work from our group and others implicates extensive matrix degradation by MMPs in human endometrium under conditions of steroid hormone withdrawal [5, 58–60], consistent with proteolysis of the matrix for tissue desquamation. Further studies are needed to elucidate the functional roles of these molecules in eMSC migration into the endometrium during repair and regeneration.
The eMSCs express genes that may participate in subsequent differentiation, depending on the stimulus, and also have a cellular program that would endorse self-renewal (see below). In this regard, it is of interest that uncommitted adult stem cells have been proposed to maintain their multipotency by maintaining a reservoir of (suppressed) genes for diverse lineages prior to lineage differentiation [13, 61], and lineage-restricted stem cells express genes predictive of their differentiated lineage [62]. Genes expressed in eMSCs relevant to lineage cell differentiation (see below) would support this hypothesis.
Genes and Signaling Pathways, Self-Renewal, Multipotency, and Immunomodulation
Self-renewal and differentiation occur in the context of the niche in which the cells exist or to which they migrate [63]. Multiple gene families and signaling networks, interactions with neighboring cells and the extracellular matrix, and mechanical and chemical stress regulate these processes, which have been highly informed by transcriptome and pathway analyses [25]. In the present study, we pursued a new approach to isolate the eMSCs using classical markers of MSCs [16], and in particular eMSCs [21], and characterized the eMSC transcriptional program and biological and signaling pathways compared with simultaneously isolated, highly purified endometrial stromal fibroblasts and endothelial cells. This approach demonstrates expression in eMSCs of classical adult stem cell genes (e.g., the ATP-binding cassette family members [37]), and key gene families and pathways associated with both self-renewal and differentiation, including members of the Notch, TGFB, FGF, WNT, IGF, and Hedgehog families, and G-protein coupled receptor-mediated and cAMP-mediated signaling [10, 11, 13–16]. In addition, MSCs as a class are immunomodulatory/immunosuppressive [17–19], and our data are consistent with these properties.
Self-renewal.
The up-regulation in eMSCs of CCND1 and the IGF family, and cell division and chromosome replication as major networks, support the phenotype of eMSCs as capable of cell division as an adult stem cell. Furthermore, our findings of Notch, TGFB, and WNT signaling pathways in eMSCs are in striking agreement with genes and pathways involved in maintaining bone marrow-derived MSCs in the undifferentiated, proliferative state, when cultured in different media and analyzed on two different platforms [15], and also “stemness” genes and receptors after culture, differentiation down various lineages, and then dedifferentiation [13]. The latter include members of the Notch, TGFB, WNT, IGF, GPCR, and cAMP-mediated signaling pathways. These findings are remarkable in view of the fact that our analyses were on freshly isolated, uncultured eMSCs, compared with the cultured bone marrow-derived MSCs used in these other studies. In the present study, our gene expression analysis reflects a snapshot of eMSC gene expression and pathways and does not give information about changes occurring in eMSCs as cells are cultured, clonally expanded, passaged, and differentiated. It is known that characterization and functional performance of MSCs depend on tissue of origin, methods used to isolate them, and culturing conditions [25]. Interestingly, it was suggested by Song et al. [13] that under certain conditions, cultured bone marrow-derived MSCs dedifferentiate to a more stemlike population. This is consistent with our preliminary observations that cell populations isolated from different tissues (eMSCs and bone marrow-derived MSCs) retain their multipotency in culture and have similar basal gene expression patterns, whereas cultured lineage committed cells from one of these tissues (stromal fibroblasts) show by comparison a distinct set of differentially expressed genes (Irwin and Giudice, unpublished data).
With regard to the Notch and TGFB families, we found genes for Notch receptors, NOTCH1, NOTCH2, and NOTCH3, the NOTCH ligand, JAG1, and downstream effectors HEY1, HEY2, and HEYL to be up-regulated in eMSCs vs. endometrial stromal fibroblasts and/or endothelial cells, and TGFB family members (TGFB1, TGFB2, TGFBR2, BMPR1A, and BMP8A) also to be up-regulated in eMSCs vs. the other two cell types. There is a remarkable parallel between the Notch family expression in eMSCs and Notch-dependent gene expression in human endometrial stromal cells infected with a retroviral vector containing human JAG1 cDNA, reported by Mikhailik et al. [64]. Comparison of the corresponding gene lists demonstrated that 28 (67%) of the top 42 up-regulated JAG-target genes are identical. This strongly supports activation of the Notch system in the eMSC population and for renewal and is consistent with a common lineage between eMSCs and the stromal fibroblast.
WNT signaling plays a major role in differentiation of MSCs to chondrocytes and other lineages [14]. However, activation of the WNT signaling pathway is less certain in eMSCs in our study. Specifically, it is unclear whether WNT signaling is active or inhibited in eMSCs, because WNT ligands (WNT5A and WNT2) are down-regulated in eMSCs compared with the fibroblast population, but up-regulated compared with the endothelial population, and WNT inhibitors (DKK1, FRZB, FZD3, FZD4, and FZD6) are mixed in their regulation in the various cell types. Because there are 19 identified WNT ligands, 10 isoforms of the WNT coreceptor Frizzled, and at least four mechanisms for WNT signal transmission [47], further understanding of the role of the WNT family in eMSC biology awaits functional studies.
Differentiation and lineage specification.
Several reports on transcriptional signatures and pathway analyses have emerged in recent years with regard to MSCs, primarily those from bone marrow or adipose tissue [13, 15, 25], which have informed approaches to differentiate these progenitors down specific lineages for regenerative medicine [9]. These demonstrate that PDGF, TGFB, and FGF signaling are important for differentiation and growth of bone marrow-derived MSCs, and transcriptional profiling has identified markers and pathways important in MSC differentiation into adipogenic, chondrogenic, and osteogenic lineages [65]. In addition, TGFB and Notch signaling mediate bone marrow-derived MSC differentiation into smooth muscle cells [66], and Hedgehog and retinoic acid synergistically promote sensory fate specification from bone marrow-derived MSCs [67]. Whereas there are common pathways controlling differentiation and self-renewal, the molecular pathways and gene families found in the present study in eMSCs would suggest the potential for their participation in eMSC differentiation to its lineage cells. However, this is likely to be largely in the context of its niche and transcriptional program, and functional studies on various pathways hold great promise to dissect the importance of these on eMSC differentiation and regeneration of the endometrium.
Although the data obtained in the present study are consistent with the multipotency of eMSCs, we did not detect pluripotency markers (POU5F1 [OCT4], NANOG, SOX2) [10, 11] in this cell population, compared with their expression in controls (NIH-approved human embryonic stem cell line H7) using quantitative RT-PCR (data not shown) or in the gene lists (Supplemental Tables S2 and S3).
Immunomodulatory properties.
Mesenchymal stem cell immunomodulation and homing to sites of injury or ischemia have generated much interest in their clinical use as cell-based therapies in immunosuppressive disorders, and also in tissue repair and regeneration [17–19]. Targets of MSCs include members of the innate and adaptive immune systems, and through soluble factors secreted by MSCs and also cell-cell contact, MSCs induce an anti-inflammatory and tolerant immune phenotype [17, 18]. Mesenchymal stem cell-derived indoleamine 2,3 dioxygenase (IDO), prostaglandin E2 (PGE2), and TGFB1 suppress T-cell and natural killer (NK)-cell proliferation and activation, usually in response to interferon-gamma (IFNG) or interleukin 1 beta (IL1B), and ligation of Toll-like receptors (TLRs) on cultured MSCs induces IL6 secretion and activation of the nuclear factor kappa-light-chain-enhancer of activated B cells (NFKB) pathway involving JAG1 expression on MSCs and its cognate Notch receptor on T cells [17–19]. The transcriptome of eMSCs, including up-regulation of JAG1, TGFB1, CXCR4, IL1RAP, and IL1RL1 (Supplemental Tables S2 and S3) suggests an immunomodulatory potential for these cells. We did not observe up-regulation of IDO or prostaglandin synthases. Because these cells were isolated from tissue and directly analyzed and not cultured, it is possible that eMSCs exist in a quiescent state in endometrium with immunomodulatory functions induced by specific stimuli (e.g., influx of inflammatory cells during menstruation, hypoxia, tissue proteolysis) or the presence of a conceptus. The potential role of eMSCs as immunomodulators in early pregnancy (e.g., uterine NK cell function, cytotrophoblast function, immune tolerance) is largely unexplored and offers an innovative approach to understanding pregnancy establishment, maintenance, and outcome, and awaits functional studies. Furthermore, MSCs as a class display low expression of major histocompatibility complex (MHC) class I molecules (including HLAG) and lack MHC class II molecules, underscoring their low immunogenic potential [19]. Our data (Supplemental Tables S2 and S3) are consistent with these observations and underscore the potential use of these cells for tissue engineering applications [6].
The eMSC as a Precursor of the Endometrial Stromal Fibroblast
Evidence is mounting that the stromal fibroblast is a lineage cell of the eMSC. First, they are both of mesenchymal origin. Also, in the present study, we found that eMSCs cluster with the stromal fibroblast exclusive of the endothelial population in a completely unbiased approach (PCA), and also with unsupervised hierarchical clustering. Also, eMSCs and the stromal fibroblast have 50% less gene differences than do eMSCs and endothelial cells. A study from our group on differentiation of bone marrow-derived MSCs with 8-Br-cAMP revealed up-regulation of classical decidual markers and cell shape changes consistent with changes in stromal fibroblasts treated similarly [23]. Taken together, the data support a common lineage for the eMSC and the endometrial stromal fibroblast, a conclusion that is further supported by recent data demonstrating differentiation of eMSCs to endometrial stromal fibroblasts in an animal model [24].
Potential Communication with Other Cell Types and Regulation of Gene Expression
Because of its perivascular location, it is teleologically plausible that the eMSC would communicate with endothelial cells, vascular smooth muscle cells, and perhaps other cells. Interestingly, the eMSC population shows up-regulation of genes corresponding to receptors for ligands from endothelial cells. For example, eMSC express MEF2C, a target of MIR21 expressed by endothelial cells and an miRNA important in endometrial cell differentiation [68]. Compared with the fibroblast and endothelial populations, genes for ligands and receptors for neurons and axonal guidance are up-regulated in eMSCs, suggesting cross-talk between the pericytes and neuronal components. Importantly, eMSCs display up-regulation of numerous genes that govern cell shape, cell migration, cell adhesion, cell-to-cell communication, and shape-induced processes governing gene expression. Histone clusters, histone acetylases/deacetylases (HEY2 interacting with NCOR1, which recruits HDACs to DNA promoter regions), and high-mobility group protein genes suggest the potential for chromatin remodeling in this population, perhaps during differentiation.
Endothelial Cell Population
The transcriptome of the endothelial cell population has not been extensively characterized in human endometrium, and this gene list is valuable in defining the expression profile of this cell population compared with the other endometrial cellular constituents, at least in interval/early secretory phase of the cycle (when the parent endometrial tissue samples were obtained). These data also serve as a comparison of gene expression in this cell type in other phases of the menstrual cycle and disorders of the endometrium. The data can also give insight into the adhesive vs. nonadhesive functions of specific cell surface glycoproteins and cell-to-cell communication between endothelium, stromal fibroblasts, vascular smooth muscle, pericytes, and cells trafficking through the vasculature. Currently, our lab is pursuing further characterization of this cell type.
In summary, the data in the present study suggest a phenotype of the eMSC as an adult stem cell, a pericyte, with self-renewal, multipotency, and immunomodulatory potential that could be signaled by hypoxic, proteolytic, and inflammatory stimuli, with the ability to induce angiogenesis, communicate with other cells, and migrate and differentiate into lineage cells. These characteristics are important in endometrial menstrual shedding and in endometrial tissue regeneration, growth, and cellular differentiation, as well as endometrial cellular function. Elucidation of mechanisms involved in the self-renewal and differentiation of the eMSC to the stromal fibroblast (and perhaps other cell types) within the endometrium that involve the pathways and genes/gene families described in the present study and in the context of the endometrial niche, will be of great value in understanding normal and abnormal endometrial development in utero, and endometrial proliferation, differentiation, and regeneration in adult uterus, as well as endometrial cellular disorders implicated in infertility, poor pregnancy outcomes, and endometrial neoplasias.
Supplementary Material
Supplementary Material
ACKNOWLEDGMENT
The authors would like to acknowledge Tara Rambaldo from the UCSF Laboratory for Cell Analysis for her expertise and assistance in flow cytometric analysis and cell sorting, and Matthew Gormley for his helpful discussion and data analysis contributions.
Footnotes
Supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development/National Institutes of Health (NICHD/NIH) through cooperative agreement 1U54HD 055764-05 as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research (L.C.G.). Raw data files of the experiments have been uploaded to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database under accession number GSE31152.
REFERENCES
- Hess AP, Nayak NR, Giudice LC. Oviduct and endometrium: cyclic changes in primate oviduct and endometrium. In: Knobil E, Neill JD (eds.), The Physiology of Reproduction, 3rd ed. San Diego: Academic Press; 2005: 337 382 [Google Scholar]
- Okulicz WC. Cellular and molecular regulation of the primate endometrium: a perspective. Reprod Biol Endocrinol 2006; 4 (suppl 1): S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbi S, Hamilton AE, Vo KC, Tulac S, Overgaard MT, Dosiou C, Le Shay N, Nezhat CN, Kempson R, Lessey BA, Nayak NR, Giudice LC. Molecular phenotyping of human endometrium distinguishes menstrual cycle phases and underlying biological processes in normo-ovulatory women. Endocrinology 2006; 147 (3): 1097 1121 [DOI] [PubMed] [Google Scholar]
- Critchley HO, Kelly RW, Baird DT, Brenner RM. Regulation of human endometrial function: mechanisms relevant to uterine bleeding. Reprod Biol Endocrinol 2006; 4 (suppl 1): S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamonsen LA, Giudice LC. “The curse”: a 21st century perspective of models of its molecular basis. Endocrinology 2010; 151: 4092 4095 [DOI] [PubMed] [Google Scholar]
- Gargett CE, Masuda H. Adult stem cells in the endometrium. Mol Hum Reprod 2010; 16: 818 834 [DOI] [PubMed] [Google Scholar]
- Du H, Taylor HS. Contribution of bone marrow-derived stem cells to endometrium and endometriosis. Stem Cells 2007; 25: 2082 2086 [DOI] [PubMed] [Google Scholar]
- Alcaraz IC, Gil-Sanchis C, Perucho AM, Valles CS. Current understanding of endometrial stem cells. Expert Rev Obstet Gynecol 2009; 4: 273 282 [Google Scholar]
- Li L, Wang Z, Jiang J, Zhao M. Signaling pathways involved in migration of mesenchymal stem cells. Trends Biopharm Ind 2010; 6: 29 33 [Google Scholar]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science 2002; 298: 597 600 [DOI] [PubMed] [Google Scholar]
- Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science 2002; 298: 601 604 [DOI] [PubMed] [Google Scholar]
- Venezia TA, Merchant AA, Ramos CA, Whitehouse NL, Young AS, Shaw CA, Goodell MA. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS Biol 2004; 2: e301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Webb NE, Song Y, Tuan RS. Identification and functional analysis of candidate genes regulating mesenchymal stem cell self-renewal and multipotency. Stem Cells 2006; 24: 1707 1718 [DOI] [PubMed] [Google Scholar]
- Kolf CM, Cho E, Tuan RS. Mesenchymal stromal cells. Biology of adult mesenchymal stem cells: regulation of niche, self-renewal and differentiation. Arthritis Res Ther 2007; 9: 204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan NXW, Ng FSL, Tanavde V. Towards a serum-free medium: growth receptors and signaling pathways that regulate multipotency in human mesenchymal stem cells. : Proceedings of the World Congress on Engineering 2007, July 2–4, 2007, London, U.K. International Association of Engineers; 2007:1460–1465.
- Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, Park TS, Andriolo G, Sun B, Zheng B, Zhang L, Norotte C, Teng PN, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 2008; 3: 301 313 [DOI] [PubMed] [Google Scholar]
- Kode JA, Mukherjee S, Joglekar MV, Hardikar AA. Mesenchymal stem cells: immunobiology and role in immunomodulation and tissue regeneration. Cytotherapy 2009; 11: 377 391 [DOI] [PubMed] [Google Scholar]
- Yagi H, Soto-Gutierrez A, Parekkadan B, Kitagawa Y, Tompkins RG, Kobayashi N, Yarmush ML. Mesenchymal stem cells: mechanisms of immunomodulation and homing. Cell Transplant 2010; 19: 667 679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogduijn MJ, Popp F, Verbeek R, Masoodi M, Nicolaou A, Baan C, Dahlke MH. The immunomodulatory properties of mesenchymal stem cells and their use for immunotherapy. Int Immunopharmacol 2010; 10: 1496 1500 [DOI] [PubMed] [Google Scholar]
- Chan RW, Schwab KE, Gargett CE. Clonogenicity of human endometrial epithelial and stromal cells. Biol Reprod 2004; 70: 1738 1750 [DOI] [PubMed] [Google Scholar]
- Schwab KE, Gargett CE. Co-expression of two perivascular cell markers isolates mesenchymal stem-like cells from human endometrium. Hum Reprod 2007; 22: 2903 2911 [DOI] [PubMed] [Google Scholar]
- Gargett CE, Schwab KE, Zillwood RM, Nguyen HP, Wu D. Isolation and culture of epithelial progenitors and mesenchymal stem cells from human endometrium. Biol Reprod 2009; 80: 1136 1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghajanova L, Horcajadas JA, Esteban FJ, Giudice LC. The bone marrow-derived human mesenchymal stem cell: potential progenitor of the endometrial stromal fibroblast. Biol Reprod 2010; 82: 1076 1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao AP, Wang KH, Chang CC, Lee JN, Long CY, Chen HS, Tsai CF, Hsieh TH, Tsai EM. Comparative study of human eutopic and ectopic endometrial mesenchymal stem cells and the development of an in vivo endometriotic invasion model. Fertil Steril 2011; 95: 1308 1315 [DOI] [PubMed] [Google Scholar]
- Hwang NS, Zhang C, Hwang YS, Varghese S. Mesenchymal stem cell differentiation and roles in regenerative medicine. Wiley Interdiscip Rev Syst Biol Med 2009; 1: 97 106 [DOI] [PubMed] [Google Scholar]
- Noyes RW, Hertig AT, Rock J. Dating the endometrial biopsy. Am J Obstet Gynecol 1975; 122: 262 263 [DOI] [PubMed] [Google Scholar]
- Hess AP, Hamilton AE, Talbi S, Dosiou C, Nyegaard M, Nayak N, Genbecev-Krtolica O, Mavrogianis P, Ferrer K, Kruessel J, Fazleabas AT, Fisher SJ, et al. Decidual stromal cell response to paracrine signals from the trophoblast: amplification of immune and angiogenic modulators. Biol Reprod 2007; 76: 102 117 [DOI] [PubMed] [Google Scholar]
- Meye A, Bilkenroth U, Schmidt U, Fussel S, Robel K, Melchior AM, Blumke K, Pinkert D, Bartel F, Linne C, Taubert H, Wirth MP. Isolation and enrichment of urologic tumor cells in blood samples by a semi-automated CD45 depletion autoMACS protocol. Int J Oncol 2002; 21: 521 530 [PubMed] [Google Scholar]
- Trzpis M, McLaughlin PM, de Leij LM, Harmsen MC. Epithelial cell adhesion molecule: more than a carcinoma marker and adhesion molecule. Am J Pathol 2007; 171: 386 395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghajanova L, Horcajadas JA, Weeks JL, Esteban FJ, Nezhat CN, Conti M, Giudice LC. The protein kinase A pathway-regulated transcriptome of endometrial stromal fibroblasts reveals compromised differentiation and persistent proliferative potential in endometriosis. Endocrinology 2010; 151: 1341 1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc 1995; 57: 289 300 [Google Scholar]
- Reiner A, Yekutieli D, BenjaminI Y. Identifying differentially expresses genes using false discovery rate controlling procedures. Bioinformatics 2003; 19: 368 375 [DOI] [PubMed] [Google Scholar]
- R Development Core Team R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2008. World Wide Web (URL http://www.R-project.org). (September, 2010)
- Muniz JJ, Joyce MM, Taylor JD, II, Burghardt JR, Burghardt RC, Johnson GA. Glycosylation dependent cell adhesion molecule 1-like protein and L-selectin expression in sheep interplacentomal and placentomal endometrium. Reproduction 2006; 131: 751 761 [DOI] [PubMed] [Google Scholar]
- Cho H, Kozasa T, Bondjers C, Betsholtz C, Kehrl JH. Pericyte-specific expression of Rgs5: implications for PDGF and EDG receptor signaling during vascular maturation. FASEB J 2003; 17: 440 442 [DOI] [PubMed] [Google Scholar]
- Aghajanova L, Tatsumi K, Horcajadas JA, Zamah AM, Esteban FJ, Herndon CN, Conti M, Giudice LC. Unique transcriptome, pathways, and networks in the human endometrial fibroblast response to progesterone in endometriosis. Biol Reprod 2011; 84: 801 815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting KD. ABC transporters as phenotypic markers and functional regulators of stem cells. Stem Cells 2002; 20: 11 20 [DOI] [PubMed] [Google Scholar]
- Daftary GS, Taylor HS. Implantation in the human: the role of HOX genes. Semin Reprod Med 2000; 18: 311 320 [DOI] [PubMed] [Google Scholar]
- Takamoto N, Zhao B, Tsai SY, DeMayo FJ. Identification of Indian hedgehog as a progesterone-responsive gene in the murine uterus. Mol Endocrinol 2002; 16: 2338 2348 [DOI] [PubMed] [Google Scholar]
- Lee K, Jeong J, Kwak I, Yu CT, Lanske B, Soegiarto DW, Toftgard R, Tsai MJ, Tsai S, Lydon JP, DeMayo FJ. Indian hedgehog is a major mediator of progesterone signaling in the mouse uterus. Nat Genet 2006; 38: 1204 1209 [DOI] [PubMed] [Google Scholar]
- Simon L, Spiewak KA, Ekman GC, Kim J, Lydon JP, Bagchi MK, Bagchi IC, DeMayo FJ, Cooke PS. Stromal progesterone receptors mediate induction of Indian Hedgehog (IHH) in uterine epithelium and its downstream targets in uterine stroma. Endocrinology 2009; 150: 3871 3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daftary GS, Taylor HS. Pleiotropic effects of Hoxa10 on the functional development of peri-implantation endometrium. Mol Reprod Dev 2004; 67: 8 14 [DOI] [PubMed] [Google Scholar]
- Hayward CP, Cramer EM, Song Z, Zheng S, Fung R, Masse JM, Stead RH, Podor TJ. Studies of multimerin in human endothelial cells. Blood 1998; 91: 1304 1317 [PubMed] [Google Scholar]
- Huber P, Dalmon J, Engiles J, Breviario F, Gory S, Siracusa LD, Buchberg AM, Dejana E. Genomic structure and chromosomal mapping of the mouse VE-cadherin gene (Cdh5). Genomics 1996; 32: 21 28 [DOI] [PubMed] [Google Scholar]
- Strilic B, Kucera T, Eglinger J, Hughes MR, McNagny KM, Tsukita S, Dejana E, Ferrara N, Lammert E. The molecular basis of vascular lumen formation in the developing mouse aorta. Dev Cell 2009; 17 (4): 505 515 [DOI] [PubMed] [Google Scholar]
- Eng FJ, Friedman SL, Fibrogenesis I. New insights into hepatic stellate cell activation: the simple becomes complex. Am J Physiol Gastrointest Liver Physiol 2000; 279: G7 G11 [DOI] [PubMed] [Google Scholar]
- Gregory CA, Prockop DJ, Spees JL. Non-hematopoietic bone marrow stem cells: molecular control of expansion and differentiation. Exp Cell Res 2005; 306: 330 335 [DOI] [PubMed] [Google Scholar]
- Bondjers C, Kalen M, Hellstrom M, Scheidl SJ, Abramsson A, Renner O, Lindahl P, Cho H, Kehrl J, Betsholtz C. Transcription profiling of platelet-derived growth factor-B-deficient mouse embryos identifies RGS5 as a novel marker for pericytes and vascular smooth muscle cells. Am J Pathol 2003; 162: 721 729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M, Bergers G, Arnold B, Hammerling GJ, Ganss R. Regulator of G-protein signaling-5 induction in pericytes coincides with active vessel remodeling during neovascularization. Blood 2005; 105: 1094 1101 [DOI] [PubMed] [Google Scholar]
- Cai X, Lin Y, Friedrich CC, Neville C, Pomerantseva I, Sundback CA, Zhang Z, Vacanti JP, Hauschka PV, Grottkau BE. Bone marrow derived pluripotent cells are pericytes which contribute to vascularization. Stem Cell Rev 2009; 5: 437 445 [DOI] [PubMed] [Google Scholar]
- Sundberg C, Friman T, Hecht LE, Kuhl C, Solomon KR. Two different PDGF beta-receptor cohorts in human pericytes mediate distinct biological endpoints. Am J Pathol 2009; 175: 171 189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maybin JCH. Repair and regeneration of the human endometrium. Expert Rev Obstet Gynecol 2009; 4: 283 298 [Google Scholar]
- Grayson WL, Zhao F, Izadpanah R, Bunnell B, Ma T. Effects of hypoxia on human mesenchymal stem cell expansion and plasticity in 3D constructs. J Cell Physiol 2006; 207: 331 339 [DOI] [PubMed] [Google Scholar]
- Nayak NR, Brenner RM. Vascular proliferation and vascular endothelial growth factor expression in the Rhesus macaque endometrium. J Clin Endocrinol Metab 2002; 87: 1845 1855 [DOI] [PubMed] [Google Scholar]
- Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature 2008; 453: 314 321 [DOI] [PubMed] [Google Scholar]
- Stappenbeck TS, Miyoshi H. The role of stromal stem cells in tissue regeneration and wound repair. Science 2009; 324: 1666 1669 [DOI] [PubMed] [Google Scholar]
- Son BR, Marquez-Curtis LA, Kucia M, Wysoczynski M, Turner AR, Ratajczak J, Ratajczak MZ, Janowska-Wieczorek A. Migration of bone marrow and cord blood mesenchymal stem cells in vitro is regulated by stromal-derived factor-1-CXCR4 and hepatocyte growth factor-c-met axes and involves matrix metalloproteinases. Stem Cells 2006; 24: 1254 1264 [DOI] [PubMed] [Google Scholar]
- Rodgers WH, Matrisian LM, Giudice LC, Dsupin B, Cannon P, Svitek C, Gorstein F, Osteen KG. Patterns of matrix metalloproteinase expression in cycling endometrium imply differential functions and regulation by steroid hormones. J Clin Invest 1994; 94: 946 953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamonsen L. Matrix metalloproteinases and their tissue inhibitors in endocrinology. Trends Endocrinol Metab 1996; 7: 28 34 [DOI] [PubMed] [Google Scholar]
- Irwin JC, Kirk D, Gwatkin RB, Navre M, Cannon P, Giudice LC. Human endometrial matrix metalloproteinase-2, a putative menstrual proteinase. Hormonal regulation in cultured stromal cells and messenger RNA expression during the menstrual cycle. J Clin Invest 1996; 97: 438 447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury D, Reynolds K, Black IB. Adult bone marrow stromal stem cells express germline, ectodermal, endodermal, and mesodermal genes prior to neurogenesis. J Neurosci Res 2002; 69: 908 917 [DOI] [PubMed] [Google Scholar]
- Tanaka TS, Kunath T, Kimber WL, Jaradat SA, Stagg CA, Usuda M, Yokota T, Niwa H, Rossant J, Ko MS. Gene expression profiling of embryo-derived stem cells reveals candidate genes associated with pluripotency and lineage specificity. Genome Res 2002; 12: 1921 1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KA, Lemischka IR. Stem cells and their niches. Science 2006; 311: 1880 1885 [DOI] [PubMed] [Google Scholar]
- Mikhailik A, Mazella J, Liang S, Tseng L. Notch ligand-dependent gene expression in human endometrial stromal cells. Biochem Biophys Res Commun 2009; 388: 479 482 [DOI] [PubMed] [Google Scholar]
- Ng F, Boucher S, Koh S, Sastry KS, Chase L, Lakshmipathy U, Choong C, Yang Z, Vemuri MC, Rao MS, Tanavde V. PDGF. TGF-beta, and FGF signaling is important for differentiation and growth of mesenchymal stem cells (MSCs): transcriptional profiling can identify markers and signaling pathways important in differentiation of MSCs into adipogenic, chondrogenic, and osteogenic lineages. Blood 2008; 112: 295 307 [DOI] [PubMed] [Google Scholar]
- Kurpinski K, Lam H, Chu J, Wang A, Kim A, Tsay E, Agrawal S, Schaffer DV, Li S. Transforming growth factor-beta and notch signaling mediate stem cell differentiation into smooth muscle cells. Stem Cells 2010; 28: 734 742 [DOI] [PubMed] [Google Scholar]
- Kondo T, Johnson SA, Yoder MC, Romand R, Hashino E. Sonic hedgehog and retinoic acid synergistically promote sensory fate specification from bone marrow-derived pluripotent stem cells. Proc Natl Acad Sci U S A 2005; 102: 4789 4794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Luo X, Toloubeydokhti T, Chegini N. The expression profile of micro-RNA in endometrium and endometriosis and the influence of ovarian steroids on their expression. Mol Hum Reprod 2007; 13: 797 806 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.