Abstract
Objectives
Recent work shows promising associations between schizophrenia and polymorphisms in Neuregulin-1 (NRG1) and a large literature also finds strong familial relationships between schizophrenia and cognitive deficits. Given the role of NRG1 in glutamate regulation and glutamate’s effect on cognition, we hypothesized that cognitive deficits may be related to variation within NRG1, providing a possible mechanism to increase risk for schizophrenia.
Method
This study examined the associations between NRG1, cognition, and schizophrenia using a multigenerational multiplex family sample (total N = 419, 40 families), including 58 affected participants (schizophrenia or schizoaffective disorder-depressed type) and their 361 unaffected relatives. Participants were genotyped for 40 NRG1 single nucleotide polymorphisms (SNPs), chosen largely based on previous associations with schizophrenia. All participants completed structured diagnostic interviews and a computerized neurocognitive battery assessing eight cognitive domains. Variance component quantitative trait analyses tested for associations between individual NRG1 SNPs and cognitive performance in the total sample, a subsample of healthy participants with no DSM diagnosis, and using general intelligence as a covariate.
Results
Effect sizes (within-family beta coefficients) ranged from 0.08 to 0.73, and 61 of these associations were nominally significant (p≤.05), with 12 associations at p≤.01, although none achieved the modified Bonferroni significance threshold of p<.0003. Attention was the most frequently nominally associated domain and rs10503929, a non-synonymous SNP, was the most frequently nominally associated SNP.
Conclusions
Although not significant experiment-wise, these findings suggest that further study of the associations between variation in NRG1 and cognition may be productive.
Keywords: NRG1, attention, rs10503929
Introduction
Despite a great deal of research on schizophrenia, the details of its etiology remain largely unknown. Family, twin, and adoption studies have consistently suggested that genetic variation is the most important overall factor, with estimates of heritability in the range of 0.80–0.85 (Cardno and Gottesman, 2000, Sullivan et al., 2003). Yet, the genetic architecture of schizophrenia presents many complications, including a polygenic transmission (Gottesman and Shields, 1967), environmental effects (Pogue-Geile and Gottesman, 2006), and possible gene-environment and epistatic interactions. This complexity has slowed the elucidation of the molecular genetics of schizophrenia (Pogue-Geile and Yokley, 2010).
Convergent Validity
In order to identify true susceptibility genes and variants for schizophrenia rather than false positives, methods that examine the relationship between genes and important correlates of schizophrenia can be helpful. One strategy, convergent validity, may increase confidence in the small and inconsistent findings of previous molecular studies of schizophrenia and increase understanding of the effects of the candidate gene. Convergent validity utilizes a secondary measure to elaborate the putative relationship between two variables (Campbell and Fiske, 1959). For example, in order to evaluate convergent validity for a candidate gene that may be associated with schizophrenia, the gene variants would be examined for their association with another characteristic that is an important correlate of the diagnosis. Given an interest in genetic variants, this secondary phenotype should be a strong familial correlate of the diagnosis.
Familial correlates of diagnosis, often termed “endophenotypes”, may potentially aid susceptibility gene identification (Gottesman and Gould, 2003). The extent to which such phenotypes are more common among unaffected relatives of patients than the general population is presumably due to pleiotropic effects of genetic variation that affect both the phenotype of interest and schizophrenia, as well as the increased frequency of risk alleles among relatives of patients. These considerations strengthen the rationale for examining convergent validity among relatives of schizophrenia patients.
We utilized this strategy to assess the relationship between one of the strongest candidate genes in schizophrenia, neuregulin-1 (NRG1), and one of the strongest diagnostic and familial correlates of the disorder, cognitive deficits. Studies have consistently found that patients with schizophrenia have pervasive deficits in most areas of cognition, largely independent of symptom state, chronicity, and cognitive domain examined (Buchanan et al., 2005, Goldberg and Green, 2002, Heinrichs et al., 1997, Snitz et al., 2006). Unaffected relatives of patients also show significant cognitive impairments (Cannon et al., 1994, Snitz et al., 2006, Thompson et al., 2005). Similar to patient samples, these deficits are general across most tasks, although such hypotheses are difficult to evaluate. Together, these findings suggest that cognitive impairment is a strong familial correlate of schizophrenia, and as such, has the potential to be an excellent candidate for the convergent validity strategy.
Neuregulin-1 and Schizophrenia
Using a candidate gene strategy, only a few polymorphisms have consistently been associated with schizophrenia, including: NRG1, catechol-O-methyl transferase (COMT), dysbindin (DTNBP1), regulator of G-protein signaling 4 (RGS4), metabotropic glutamate receptor 3 (GRM3), disrupted-in-schizophrenia 1 (DISC1), and others (Harrison and Weinberger, 2005). Among these putative risk genes, NRG1 is supported by a positive linkage with a schizophrenia-related locus in the region and relatively consistent significant association findings (Harrison and Law, 2006), and is therefore the focus of the current study.
NRG1 was initially identified as a candidate gene for schizophrenia by a linkage study of Icelandic multiplex families and subsequent fine mapping of this locus (Stefansson et al., 2003). To date, nearly 50 replications have been conducted, with slightly more than half finding evidence of an association between schizophrenia and NRG1 (Allen et al., 2008). Overall, the estimates of relative risk (RR; risk of developing schizophrenia relative to having a particular genotype) lie between 1.0 and 2.2 for specific variants and haplotypes (Tosato et al., 2005). However, genome-wide association studies have not found significant genome-wide results for NRG1 (Purcell et al., 2009, Shi et al., 2009, Stefansson et al., 2009), and have instead pointed to genes of the major histocompatability complex (for a review, see Pogue-Geile and Yokley, 2010).
NRG1 is one of four members of the neuregulin family of genes, structurally-related glycoproteins that function as ligands for receptor tyrosine kinases of the ErbB family (Scolnick et al., 2006, Wolpowitz et al., 2000). NRG1 is a large gene, encompassing 1.3 million bases with at least 21 alternatively spliced exons (Steinthorsdottir et al., 2004). Approximately 0.3% of the gene codes for protein (Scolnick et al., 2006), and there are at least nine alternative promoters and 16 alternative splicing isoforms (Steinthorsdottir et al., 2004).
NRG1 is considered a pleiotropic growth factor with an integral role in the development, organization, and function of the central nervous system (CNS) (Li et al., 2006). At least twelve functions have been identified for this gene, including modulation of long-term potentiation (LTP) and regulation of N-methyl-D-aspartic acid receptor (NMDAR) (Harrison and Law, 2006). Because NRG1 is thought to modulate glutamate levels through its regulation of NMDAR, and because glutamate is thought to be important in multiple cognitive functions, a very common deficit in patients and their relatives, evaluating the relationship of NRG1 and cognition is important to providing a theoretical mechanism to explain NRG1’s potential role in schizophrenia.
Previous Studies of NRG1 and Cognition
Six previous studies assessing specific NRG1 variants and cognition in humans had mixed results. One NRG1 variant used in studies of cognition has been SNP8NRG221533 (renamed rs35753505). In healthy participants, rs35753505 had no association with working memory performance (Krug et al., 2008), but was associated with semantic verbal fluency (Kircher et al., 2009) and sustained attention (Stefanis et al., 2007). This SNP was also tested in patients with schizophrenia, finding significant effects on blood flow in several brain regions, but not task performance (Kircher et al., 2008).
A second NRG1 variant, SNP8NRG243177 (renamed rs6994992), was associated with premorbid IQ and fronto-temporal activation in patients (Hall et al., 2006), as well as verbal IQ and brain activation in verbal fluency tasks in participants at high-risk for developing the disorder (Hall et al., 2006), although non-significant findings for premorbid IQ in patients have also been reported (Crowley et al., 2008). A third study found that rs6994992 was moderately associated with spatial working memory in a general population sample (Stefanis et al., 2007). Microsatellite 433E1006 was modestly associated with sustained attention and verbal working memory in a study of Greek male military conscripts (Stefanis et al., 2007).
These six studies varied in their methodological strengths. Among patients, with the exception of one large study (Crowley et al., 2008) (N=738), all studies had sample sizes less than 40 (Hall et al., 2006, Kircher et al., 2008). Control samples were larger, including 2,000 (Stefanis et al., 2007), 733, and two of 429 participants. The only family design (Hall et al., 2006) used a small sample (N=79) of high-risk relatives. In addition, only one study used more than one genetic marker (i.e., five markers) and the studies assessed few cognitive domains (only one study assessed the maximum of three domains). None controlled for the role of intelligence in their analyses and only one study (Krug et al., 2008) controlled for non-schizophrenia psychopathology.
Aims and Rationale
To build upon the literature and improve upon past studies’ methodological constraints, the present study utilized a relatively large multigenerational, multiplex family sample with a multistage analytic technique, including multiple covariates, to assess associations between a large, literature-based NRG1 SNP set and multiple cognitive functions. To our knowledge, no previous study has used as comprehensive cognitive battery, as many SNPs, or as many relatives of schizophrenia patients as the current study.
For questions we address, multigenerational, multiplex family samples have several advantages over unrelated participants from the general population. To the extent that schizophrenia risk alleles are more common among relatives of patients than in the general population, studies of relatives increase power to detect effects due to the presumed increased frequency of risk alleles among patients’ relatives. Multiplex family samples composed of several schizophrenia patients per family are presumably even better. Family samples also allow genetic population stratification to be detected, and if present, allow association tests that are robust to such stratification. Studies of only schizophrenia patients may miss important genetic associations with cognition to the extent that other patient factors unrelated to NRG1 (e.g., symptom state, treatment history, drug abuse) affect performance and add nuisance variation.
The current study also examined associations between NRG1 variation and cognition with and without covarying for psychopathology and general intelligence. NRG1 may have important associations with cognition that are also shared with psychopathology and intelligence, thus it is important to measure these factors to better understand these relationships and to allow their effects on NRG1-cognition associations to be examined.
The specific questions that this study aimed to address were:
Is variation in NRG1 associated with cognitive performance in a combined group of patients and their relatives? Although generally a powerful design to test whether NRG1 variants cause variation in cognitive performance, findings here could also reflect a spurious effect if NRG1 is associated with schizophrenia and patients had cognitive deficits for reasons independent of NRG1 (e.g., medication side effects). In contrast, the presence of increased measurement error among patients due to symptoms or other factors could also serve to reduce true associations.
Is NRG1 variation associated with cognitive performance in a sub-sample of relatives with no psychiatric diagnoses? Although less statistically powerful than the analysis above due to reduced sample size (i.e., elimination of schizophrenia patients), associations here cannot be due to any spurious effects that schizophrenia or other psychopathology (e.g., substance abuse) might have on cognitive performance, thus providing a conservative assessment of the relationship between NRG1 variants and cognition in the context of multiplex schizophrenia families.
Is NRG1 variation associated with cognitive performance when controlling for general intelligence among relatives without diagnoses? Associations here suggest that the effect of NRG1 on cognition is not completely due to any effect NRG1 has on general intelligence. Although potentially important, joint NRG1 effects on both intelligence and cognition will be missed here.
Is NRG1 associated with schizophrenia? Although quite under-powered for this question, it is nonetheless useful to examine this in the same sample in which cognition is being investigated.
Methods
Participants - Pedigree Sample
Probands and their family members were recruited at two sites (University of Pittsburgh and University of Pennsylvania) through mental health and consumer organizations in Pennsylvania, New Jersey, Delaware, Ohio, West Virginia, Kentucky, Michigan, and Indiana (Gur et al., 2007). Probands were included if they had a diagnosis of schizophrenia, were of European-American origin, 18 years or older, competent to provide informed consent, and had one or more first degree relatives with a diagnosis of schizophrenia or schizoaffective disorder-depressed type and a large, multigenerational family with ten or more first and second degree relatives. Probands were excluded if they did not provide consent to contact their family members, their IQ was below 70, they were not proficient in English, or their diagnosis was complicated by substance use or medical conditions.
Relatives had to be 15 years or older and willing to provide signed consent (or assent with parental consent if younger than 18 years old). Exclusion criteria for this group included: IQ < 70; lack of English proficiency; or a neurological disorder that would interfere with the interpretation of cognitive measures. All relatives were of Euro-American origin, except one who reported mixed European and African-American parentage.
The sample for the present study consisted of a subset of participants from a larger study (Gur et al., 2007). Participants were chosen for the current sample if they had completed both the diagnostic and cognitive testing portions of the study, provided DNA, and were successfully genotyped for NRG1. This resulted in 419 pedigree members (23 index probands and 396 relatives) from 40 multiplex, multigenerational families. Family size within the current sample ranged from one to 38 members (mean members per family = 10.48), and the number of affected participants (with schizophrenia or schizoaffective disorder-depressed) per family ranged from zero to four (mean affected individuals per family = 1.45) and such individuals were frequently spread throughout the family pedigree structure. The current sample only includes individuals with complete data (including NRG1 genotypes), and thus despite coming from multigenerational, multiplex families, some of the individuals in the current sample had few relatives with complete data. Table 1 presents the family structure (with reference to the index, first identified proband), demographic, and clinical characteristics of the total sample successfully genotyped for NRG1. The last column of Table 1 provides the subsample of relatives who did not meet criteria for any diagnosis on the clinical measures and were analyzed as the “No Diagnosis” group (N=178, 40.5% male, mean age = 45.9 years, mean education = 13.5 years).
Table 1.
Demographic Information, pedigree relationships, and clinical composition
| N | Gender (% male) | Mean Age (SD) | Mean Education (SD) | Affected (%) | Spectrum (%) | Other Diagnosis (%) | No Diagnosis (%) | |
|---|---|---|---|---|---|---|---|---|
| Total Pedigree | 419 | 50.36% | 44.8 (16.6) | 13.2 (2.9) | 58 (13.8) | 36 (8.6) | 147 (35.1) | 178 (42.5) |
| Probands | 23 | 69.60% | 42.3 (10.0) | 12.5 (2.1) | 23 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Relatives | 396 | 49.20% | 44.9 (16.9) | 13.3 (2.9) | 35 (8.8) | 36 (9.1) | 147 (37.1) | 178 (44.9) |
| First-degree relatives | 86 | 47.70% | 47.7 (14.6) | 12.6 (3.0) | 28 (32.6) | 6 (6.9) | 25 (29.1) | 27 (31.4) |
| Second-degree relatives | 88 | 55.70% | 46.9 (20.9) | 12.9 (2.9) | 2 (2.3) | 12 (13.6) | 25 (28.4) | 49 (55.7) |
| Third-degree relatives | 97 | 49.50% | 44.7 (12.5) | 13.9 (2.9) | 2 (2.1) | 7 (7.2) | 40 (41.2) | 48 (49.5) |
| Extended relatives | 93 | 45.70% | 36.7 (16.6) | 13.4 (2.7) | 3 (3.2) | 7 (7.5) | 46 (49.5) | 37 (39.8) |
| Non-biological relatives | 32 | 43.80% | 55.9 (11.6) | 14.1 (2.9) | 0 (0.0) | 4 (12.5) | 11 (34.4) | 17 (53.1) |
Note: Family structure is with reference to the index (1st identified) proband in the family. Affected = schizophrenia or schizoaffective disorder-depressed; Spectrum = schizoaffective disorder-bipolar, bipolar disorder I or II, major depression with psychotic features, organic and non-organic psychoses, and cluster A personality disorder; Other Diagnosis = any other non-psychotic diagnosis, including substance disorders; No Diagnosis = no diagnosis as determined by DIGS assessment. Groups are mutually exclusive and hierarchical such that individuals with both affected (e.g., schizophrenia) and “other” (e.g., substance dependence) diagnoses were categorized in the affected group. Individuals with both spectrum and “other” diagnoses were categorized in the spectrum group.
Written informed consent for all participants was obtained in accordance with the Institutional Review Boards of the University of Pittsburgh, University of Pennsylvania, and Texas Biomedical Research Institute.
Clinical Assessment and Measures
Participants were assessed using the Diagnostic Interview for Genetic Studies, version 2.0 (DIGS) (Nurnberger et al., 1994) the Family Interview for Genetic Studies (FIGS) (Maxwell, 1992) and a review of medical records. Assessment was conducted by trained interviewers with established reliability under the supervision of investigators. Although interviewers were not blind to participant status (proband, relative, control), diagnoses were made using DSM-IV criteria by a consensus team of licensed psychiatrists and psychologists who were blind to participant identity. Diagnostic reliability across sites was demonstrated using videotaped interviews.
Neurocognitive Assessment
Computerized Neurocognitive Battery
Participants were administered a Computerized Neurocognitive Battery (CNB) previously used in both healthy and patient samples ( Gur et al., 2001a, Gur et al., 2001b, 2010). The battery took approximately 60 minutes to complete and was administered using desktop or laptop computers. The tests included training modules, were administered in a fixed order, and had automated scoring to ensure reliability of results. The battery assessed the following domains (Gur et al., 2007):
Abstraction and Mental Flexibility
The Penn Conditional Exclusion Test (Kurtz et al., 2004) is a measure of abstraction and concept formation. Subjects decide which of 4 objects does not belong with the other 3 and feedback is used to help the subject identify one of three sorting principles. The sorting principle shifts after its discovery is established.
Attention
The Penn Continuous Performance Test (Kurtz et al., 2001) uses a continuous performance test paradigm. The participant responds to a set of seven-segment displays presented 1/second whenever they form a digit (initial three minutes of the task) or letter (next three minutes).
Verbal Memory
The Penn Word Memory Test (Gur et al., 1993) presents 20 target words followed by an immediate recognition trial with targets intermixed with 20 distracter stimuli. Distracters were equated to targets for frequency, length, concreteness, and low imageability using Paivio’s norms. Delayed recognition is measured at 20 minutes.
Face Memory
The Penn Face Memory Test (Gur et al., 1993) presents 20 faces followed by an immediate recognition trial with targets intermixed with 20 distracters. Distracters were equated for age, gender, and ethnicity. Delayed recognition is measured at 20 minutes.
Spatial Memory
The Visual Object Learning Test (Glahn et al., 1997) presents 20 Euclidean shapes followed by an immediate recognition trial with targets intermixed with distracters. Delayed recognition is measured at 20 minutes.
Spatial Processing
Judgment of Line Orientation (Benton et al., 1975) includes stimuli of two lines at an angle. Participants indicate the corresponding lines on a simultaneously presented array.
Sensorimotor Dexterity
The task requires moving the mouse and clicking as quickly as possible on a green square that disappears after the click. The square gets increasingly small (Gur et al., 2001b).
Emotion Processing
Identification of facial affect was tested with two 40-item tasks. During the Penn Emotion Recognition Task, participants labeled faces as being happy, sad, angry, fearful, or neutral. During the second task, the Emotion Intensity Discrimination Test (Gur et al., 2006), each stimulus was comprised of two faces of the same individual showing the same emotion (happy or sad) with different intensities. The participant selects the more intense expression. Facial stimuli were balanced for gender, age, and ethnicity.
Domain Scoring
A control (N=199) sample with no Axis I disorders with psychotic features, recent psychotropic medication use, or first degree relatives with psychosis was used to standardize the cognitive data to enable comparisons between the multiplex sample and the general population’s cognitive function. Attempts were made to group match the control participants to the relatives based on age, sex, and education. Raw scores were converted to z-scores using the mean and standard deviation (SD) from the control group. For each of the eight domains, three performance indices were recorded: accuracy (number of correct responses), speed (median reaction time for correct responses), and efficiency (ratio of accuracy to the log of speed). Efficiency was analyzed in the current study because it is a single score that incorporates both accuracy and speed to provide an index of correct responses per unit of time that reflects general ideas of good performance (i.e., for a given level of accuracy, quicker responses are better and for a given level of speed, more accurate responses are better). In addition, focusing on the combined efficiency index also reduces the number of comparisons relative to analyzing both accuracy and speed separately. To be included in the current sample, all participants needed to have at least 14 of the 24 performance indices. The mean rate of missing data across domains in the current sample ranged from 2.1%–4.7% across the probands, relatives, and controls. The cognitive data for the current sample were checked for extreme outliers (≥6 standard deviations from the next most extreme score), which were Winsorized (i.e., the extreme outlier value was replaced with the next less extreme value) to reduce the effect the extreme value had on the analyses without eliminating useable data points.
Intelligence Estimate
All participants were administered the reading subtest of the Wide Range Achievement Test-III (WRAT) as an estimate of intelligence. This measure is commonly used to estimate crystallized verbal intelligence and is relatively robust to the effects of most psychiatric symptoms and brain injury. Raw scores were age-standardized based on published manual norms.
Genotyping
Forty-four NRG1 SNPs were initially genotyped in the current study. Of these, 19 SNPs had been reported to be associated with schizophrenia by at least one study, 17 were chosen based on proximity to haplotype blocks (e.g. HapICE, HapIRE, etc.) and associated microsatellites that had also been associated with schizophrenia, four were chosen from exonic and untranslated regions (UTR), and four were chosen as back-ups based on their high linkage disequilibrium (LD; r2≥0.80) with one or more previously associated SNP in the pool (rs10096573 and rs4298458: r2=0.99; rs13274954 and rs1081062: r2=0.95; rs13256173 and SNP8NRG444511/rs13268724: r2=1.0; rs13256173 and rs1354335: r2=0.98; rs2466044 and rs2466058: r2=0.98; rs2466044 and rs2466049: r2=0.99). These redundant SNPs (rs10096573, rs13274954, rs13256173, rs2466044) were used to provide internal validation of the genotyping in this study. Given that they provide very little unique information, they will not be considered further in these analyses. This SNP pool design was chosen to match the goal of testing convergent validity by using a literature-based approach to SNP selection.
SNPs were genotyped using the SNPlex Genotyping System (SNPlex, ABI Biosystems, Inc., Carlsbad, CA). Each DNA sample was assessed separately and participants needed to have more than 34 successful SNP genotypes and no Mendelian errors, as assessed by PedCheck (O’Connell and Weeks, 1998), to be included.
Three SNPs (rs1481747, rs776382, rs800501) were excluded for high failure rates (≥20%) and one SNP (SNP8NRG449280) was monomorphic in the sample and also excluded. After removing these SNPs and the redundant SNPs, the total SNP pool for analysis contained 36 SNPs, as shown in Table 2. Every SNP was in Hardy-Weinberg Equilibrium, except rs2919382 (p=0.0173), as calculated by the Sequential Oligogenic Linkage Analysis Routines (SOLAR) (Almasy and Blangero, 1998) program, with between 0% and 12.65% genotyping failure per SNP (mean failure = 1.09%). LD patterns, as calculated in SOLAR, were as expected, with very low LD (r2≤0.20) between most SNPs, with the exception of a few high LD SNPs that were built into the SNP pool for redundancy.
Table 2.
Line diagram of NRG1 with SNP Locations and Genotyping Information.
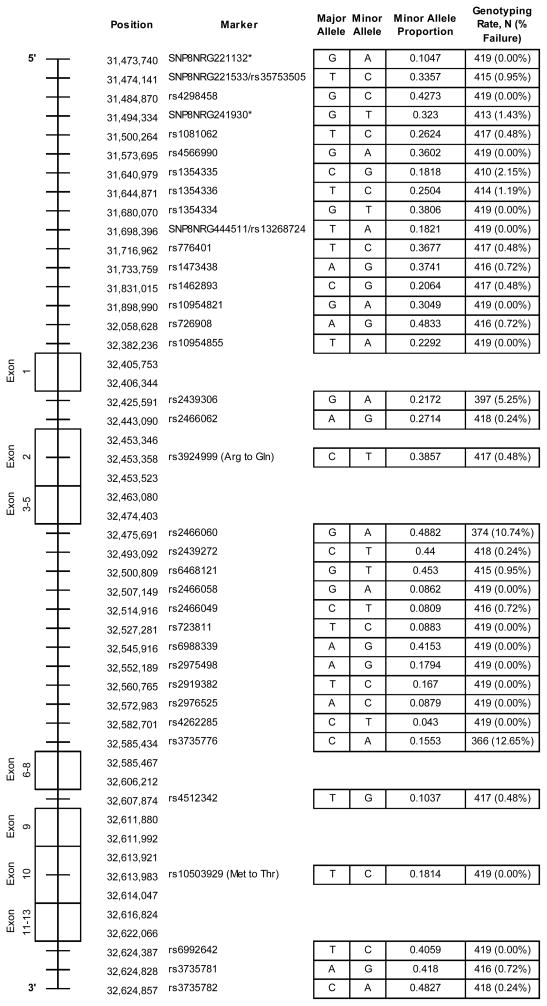
|
Note: Gene diagram not to scale. Exon locations based on Ensembl genome browser release 58 (May 2010) GRCh 37 (www.ensembl.org). SNP locations based on dbSNP build 131 GRCh37 (www.ncbi.nlm.nih.gov/projects/SNP/).
Position estimated as these deCODE SNPs do not have dbSNP ID.
Analyses
Genetic analyses were performed using the SOLAR program. SOLAR is a maximum likelihood variance component analytic program designed for multigenerational pedigrees of variable size and complexity (Almasy and Blangero, 1998). Polygenic models were estimated with ascertainment correction. Highly kurtotic variables were analyzed using a t-distribution. Potential covariates for all analyses included: age, sex, age2, age-by-sex, age2-by-sex, and handedness. Each covariate was screened separately and retained in the model if it was significant at p<.10. SNPs were tested for association with a trait (e.g., cognitive performance or affected status) by the quantitative trait linkage disequilibrium test (QTLD) (Havill et al., 2005), unless there was evidence of significant (p≤.05) stratification, in which case, the quantitative transmission disequilibrium test (QTDT) (Abecasis et al., 2000) was used. QTLD is a more powerful measure of association than QTDT but is less robust to population stratification, whereas QTDT is more conservative, but more appropriate when stratification is present. The QTDT reflects the correlation between SNP genotype (number of minor alleles 0,1,2) and cognitive domain within families and is thus robust to stratification effects, which occur between families. Stratification is detected in SOLAR when the between family association component (βbetween) is significantly different from the within family component (βwithin) (Fulker et al., 1999). Generally, the expected result of stratification is that βbetween> βwithin. The choice between QTDT and QTLD was based solely on the results of the stratification test and only one test was used for each SNP-domain combination.
Bonferroni correction to control experiment-wise alpha error is overly conservative when tests are performed on correlated variables as is the case here. In order to more accurately control experiment-wise alpha error, the pACT program (P-values Adjusted for Correlated Tests (Conneely and Boehnke, 2007)), which takes into account the intercorrelations among the cognitive domains and among the SNP set, was used. Based on the observed correlational structure among both the SNPs and cognitive domains, pACT estimated that the 288 (36 SNPs * 8 domains) correlated tests performed was equivalent to 160 independent tests. Thus, the significance level needed to yield an experiment-wise alpha error of .05 was .0003 (.05/160) (the corrected significance levels for the analyses in the no diagnosis sample and with intelligence as a covariate were also calculated to be p<.0003).
Results
Neurocognitive Measures
In previous analyses of this overall multiplex sample, affected participants performed significantly more poorly than controls on all eight CNB domains (Gur et al., 2007). In addition, except for spatial memory and verbal memory, all domains showed significant genetic correlations with affected status in the overall sample (Rg range: −0.27 to −0.56), indicating joint genetic effects on these traits and suggesting that the alleles that lower cognitive performance increase risk for schizophrenia (Hare et al., Under review). Each CNB domain has also shown significant heritability (h2 range: 0.21 to 0.72) (Hare et al., Under review). Analyses in the current sample replicated these findings (data not shown). The intercorrelations and a genome-wide linkage study with these domains have been published previously (Almasy et al., 2008, Hare et al., Under review).
Associations between NRG1 SNPs and CNB Domains
First, associations were estimated in the total pedigree sample (N=419), which included affected participants (N=58) and all other relatives (N=361). Affected members were included in this analysis to allow for cognitive effects on schizophrenia and thus to increase power. However, inclusion of affected individuals may also introduce spurious effects of schizophrenia on cognition and/or may increase environmental or error variance. To partial out any potential role of schizophrenia and psychopathology in the relationship between NRG1 and cognition, associations were also re-estimated in the sub-sample of healthy relatives who had no diagnosis on the DIGS (N=178). Next, to assess the associations between NRG1 SNPs and cognitive function independent of intelligence, WRAT score was entered as a covariate in analyses of the no diagnosis sub-group.
Associations in the Total Sample
As seen in Table 3, 21 nominally significant (p≤.05) associations between the efficiency measures of each CNB domain and NRG1 SNPs were detected in the total sample, which included both affected and unaffected participants. Every domain, except spatial memory, had at least one nominally significant association with a NRG1 SNP (range: 0–9 associations per domain; mean of 2.63). These associations encompassed 12 SNPs (range: 0–4 associations per SNP; mean of 0.58) and were spread across the gene, including UTR’s, introns, and exons. The domain of attention had the most nominally significant associations (9) and the largest number (3) of p≤.01 associations. However, none of these associations met the pACT adjusted significance level of p<.0003.
Table 3.
Significant associations (p-values, p<.05) and selected effect sizes by SNP, CNB domain, and analysis
| SNP | Analysis | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Abstraction & Mental Flexibility | Attention | Verbal Memory | Face Memory | Spatial Memory | Spatial Processing | Sensorimotor Dexterity | Emotion Processing | ||
| SNP8NRG221132 | Total | ||||||||
| NoDx | .015 | ||||||||
| NoDx & WRAT | .010 | ||||||||
|
| |||||||||
| SNP8NRG221533/rs35753505 | Total | ||||||||
| NoDx | .018 | ||||||||
| NoDx & WRAT | |||||||||
|
| |||||||||
| rs1081062° | Total | ||||||||
| NoDx | .022^ | .002^ | |||||||
| NoDx & WRAT | .002^ | ||||||||
|
| |||||||||
| rs1354335 | Total | .018^ | |||||||
| NoDx | |||||||||
| NoDx & WRAT | .041^ | ||||||||
|
| |||||||||
| rs1354336 | Total | .002^ | |||||||
| NoDx | .035^ | ||||||||
| NoDx & WRAT | |||||||||
|
| |||||||||
| SNP8NRG444511/rs13268724 | Total | .029^ | |||||||
| NoDx | |||||||||
| NoDx & WRAT | |||||||||
|
| |||||||||
| rs776401 | Total | .044^ | |||||||
| NoDx | |||||||||
| NoDx & WRAT | |||||||||
|
| |||||||||
| rs1473438 | Total | .032^ | |||||||
| NoDx | |||||||||
| NoDx & WRAT | |||||||||
|
| |||||||||
| rs726908 | Total | ||||||||
| NoDx | |||||||||
| NoDx & WRAT | .017 | ||||||||
|
| |||||||||
| rs10954855 | Total | .013 (+.31) | |||||||
| NoDx | .040 | .026 (+.26) | .015 | .002^ | |||||
| NoDx & WRAT | .027 (+.08) | .035 | |||||||
|
| |||||||||
| rs2439306 | Total | .041 | |||||||
| NoDx | |||||||||
| NoDx & WRAT | |||||||||
|
| |||||||||
| rs3924999 | Total | .031 (+.32) | .024 | ||||||
| NoDx | .030 (+.26) | ||||||||
| NoDx & WRAT | .014 (+.36) | ||||||||
|
| |||||||||
| rs2466060 | Total | .004 (+.42) | .026 | ||||||
| NoDx | .007 (+.51) | .037 | |||||||
| NoDx & WRAT | .005^ (+.73) | .024 | |||||||
|
| |||||||||
| rs2439272 | Total | .015 | .031 | .046 | .036 | ||||
| NoDx | .023 | ||||||||
| NoDx & WRAT | |||||||||
|
| |||||||||
| rs6468121 | Total | .049^ | .050 | ||||||
| NoDx | |||||||||
| NoDx & WRAT | |||||||||
|
| |||||||||
| rs2466058 | Total | ||||||||
| NoDx | .034 | ||||||||
| NoDx & WRAT | .021 | ||||||||
|
| |||||||||
| rs2466049 | Total | ||||||||
| NoDx | .035 | ||||||||
| NoDx & WRAT | .021 | ||||||||
|
| |||||||||
| rs2976525 | Total | ||||||||
| NoDx | .050 | .049 | |||||||
| NoDx & WRAT | |||||||||
|
| |||||||||
| rs4262285 | Total | ||||||||
| NoDx | |||||||||
| NoDx & WRAT | .047 | ||||||||
|
| |||||||||
| rs3735776 | Total | ||||||||
| NoDx | .035 | .021 | |||||||
| NoDx & WRAT | .004 | .015 | |||||||
|
| |||||||||
| rs4512342 | Total | ||||||||
| NoDx | |||||||||
| NoDx & WRAT | .024 | ||||||||
|
| |||||||||
| rs10503929 | Total | .032 (−.39) | .009^ | .029^ | .047 (−.18) | ||||
| NoDx | .035 (−.51) | .044 (−.32) | |||||||
| NoDx & WRAT | .006 (−.68) | .009^ | .020^ (−.36) | ||||||
|
| |||||||||
| rs3735781 | Total | ||||||||
| NoDx | |||||||||
| NoDx & WRAT | .047^ | ||||||||
Note: Only significant (p≤.05) associations are recorded. Effect sizes (within-family beta weight) for associations that were significant (p≤.05) for every analysis are provided in parentheses. A positive within-family beta weight value indicates that the minor allele confers a benefit to cognitive performance; a negative value indicates that the minor allele confers a detriment. Grey shading indicates associations that were significant (p≤.05) for every analysis.
Significant stratification and QTDT associations, all of which have greater within- than between-family beta weights.
The minor allele of rs1081062 was associated with reduced risk for affected status in the current study.
Controlling for Schizophrenia and Other Psychopathology
To partial out potential effects of schizophrenia or psychopathology in the association between NRG1 and cognitive function, associations were also estimated in the sub-sample of individuals who had no diagnosis on the DIGS. As seen in Table 3, 21 nominal associations were found in this sample. Every domain had at least one nominally significant association with a NRG1 SNP (range: 1–5 associations per domain; mean of 2.63). These associations encompassed 13 SNPs (range: 0–4 associations per SNP; mean of 0.58), and were spread across the gene. When compared to the nominal associations found in the total sample, seven associations remained, while 14 new associations were found in this diagnostically healthy sample and 14 previous associations were lost. Again however, no associations were significant when corrected for multiple testing by the pACT program.
Controlling for Intelligence
Associations were next re-estimated in the no diagnosis sub-sample using the WRAT score as a covariate. Nineteen nominally significant associations were found, as seen in Table 3. Every domain had at least one nominally significant association with a NRG1 SNP (range: 0–5 associations per domain; mean of 2.38), except emotional processing. These associations included 14 SNPs (range: 0–3 associations per SNP; mean of 0.53) and were spread across the gene. When compared to the associations found in the total sample, six associations remained nominally significant, while 13 new associations were found in this analysis and 15 previous associations from the total sample were lost. When compared to the associations in the no diagnosis sub-sample without intelligence as a covariate, 13 associations remained, while six new associations were found, and eight were lost. Overall, this suggests that variance in intelligence may account for some, but not all, of the relationship between NRG1 and cognitive functioning. No associations were significant however when corrected for multiple testing by the pACT program.
Association between NRG1 SNPs and Schizophrenia
Although under-powered due to the low number of schizophrenia patients, associations between NRG1 SNPs and affected status (i.e., diagnosis of schizophrenia or schizoaffective disorder-depressed) were also calculated to determine whether NRG1 was associated with schizophrenia in this sample. One association trended towards nominal significance: rs1081062 (p=0.0514; QTDT); however, it did not survive correction for multiple comparisons. The minor allele of rs1081062 was associated with decreased risk for schizophrenia.
Discussion
Summary of Findings and Previous Literature
A multi-step analysis strategy was utilized to understand better the associations between NRG1 and cognition in the context of schizophrenia, other psychopathology, and intelligence. At all three analytic steps, several variants in NRG1 were nominally associated with various cognitive domains, although none reached significance after correcting for multiple tests.
Given this context of many nominally significant but no experiment-wise significant associations, we will focus here on those associations that were nominally significant at all of the three analytic steps as potentially most promising for future study. Although statistically the most powerful test, associations in the total sample could be spurious due to secondary effects of schizophrenia or other psychopathology causing cognitive deficits, whereas analyses in the no-diagnosis sample eliminate this possibility and the analyses with intelligence as a covariate further narrow the field to those SNPs whose associations with cognition are not completely due to effects on general intelligence. Although some promising associations might not meet these criteria, those that do seem most likely to be of interest.
Associations that were nominally significant at all steps include four cognitive domains (abstraction and mental flexibility; attention; verbal memory; and sensorimotor dexterity) and four SNPs (rs10954855, rs3924999, rs2466060, and rs10503929). The LD within this group of SNPs was less than or equal to an r2 of 0.2, except for the following: rs3924999 and rs10954855 (r2=0.3), and rs3924999 and rs2466060 (r2=0.5). These associations are discussed below in order of SNP location within NRG1 (5′ to 3′). Effect sizes are shown in Table 3. In addition, previous findings from the literature for each of these SNPs are summarized.
Marker rs10954855 was nominally associated with verbal memory performance in each of the analyses, with the minor allele (A) always conferring a benefit. This SNP lies less than 1000bp from microsatellite 317J8-2123, which was associated with schizophrenia in a Chinese sample (Li et al., 2004). To our knowledge, no studies of rs10954855 and cognition exist, although one previous study (Duan et al., 2005) failed to find any significant association with schizophrenia.
SNP rs3924999 was nominally associated with attention in all analyses with the minor allele (T) improving performance. This marker is a non-synonymous exonic SNP that results in a change from arginine (R) to glutamine (Q) if a C to T (minor) allele substitution occurs. This SNP’s minor allele has been associated with schizophrenia (Yang et al., 2003), as well as perceptual aberrations in adolescents (Lin et al., 2005), although, another study failed to find association with schizophrenia (Hong et al., 2004). To our knowledge, no studies of this SNP and cognition have been performed. In contrast to these previous studies, the minor allele of rs3924999 was associated with an improvement in cognitive performance in the current study.
SNP rs2466060 was also nominally associated with attention in all analyses with the minor allele (A) improving performance. Although not significant after correction for multiple testing, rs2466060 and attention produced the strongest consistent associations across analyses (uncorrected ps <.007) in this study. This marker is located near microsatellite 317J8-4858, which was associated with schizophrenia in a Chinese sample (Li et al., 2004). To our knowledge, no studies of rs2466060 and cognition exist, although one previous study (Petryshen et al., 2005) failed to find any significant association with schizophrenia.
Finally, rs10503929 was nominally associated with both abstraction and mental flexibility performance and sensorimotor dexterity in all analyses, where the minor allele (C) always reduced performance. This marker is a missense non-synonymous SNP that leads to the substitution from a methionine (M) to threonine (T) amino acid if a T to C (minor) allele change occurs. A recent meta-analysis (Allen et al., 2008) of case-control studies found that rs10503929 was the only NRG1 SNP of 13 assessed that was significantly related to schizophrenia (odds ratio: 0.88), suggesting a protective effect of the minor allele. In contrast, the minor allele of rs10503929 was associated with a decrease in cognitive performance in the current study.
Other NRG1 Markers Associated with Cognition in Previous Literature
Despite some significant associations between rs6994992 and cognition in previous studies, this marker consistently failed the SNPlex Assay Design algorithm that was used to create the primer pool and could not be analyzed in the current study. Marker rs35753505 had been associated with sustained attention in a general population sample (Stefanis et al., 2007), but in the current study it was associated with abstraction and mental flexibility in the “no diagnosis” sample (p=0.0178) but not with attention in any sample.
Association between NRG1 SNPs and Schizophrenia
There was one nominally significant association between NRG1 SNPs and the diagnosis of schizophrenia or schizoaffective disorder-depressed in the current study, although it was non-significant after correction. This relative lack of findings is likely due to the small sample of affected participants leading to low power to detect associations between the disorder and individual SNPs.
Strengths & Limitations
Relative to other studies on this topic, this study utilized a large, multiplex, multigenerational family sample of schizophrenia to assess the association between the largest number of NRG1 SNPs and most cognitive domains to date. In addition, a strategy of sequential analyses was employed in order to control for factors such as schizophrenia, general psychopathology, and intelligence that can significantly affect analyses.
In addition to these strengths, it also has some limitations. First, the number of participants with schizophrenia or schizoaffective disorder-depressed in the sample was relatively small, which prevented us from testing for associations with NRG1 and cognition in the patient-only sample. The general lack of associations between NRG1 and affected status also likely reflects this low power. Furthermore, although the largest such study to date, the number of relatives was not as large as would be ideal. At the nominal significance level of .05, statistical power was good in the total sample, being able to detect SNPs accounting for possibly only 1% of trait variance (N=419, power = .80, assuming complete LD between SNP and causal variant), and in the No Diagnosis sample, being able to detect SNP effects accounting for 4.3% of trait variance (N=178, power = .80). As expected, power was reduced when correcting for multiple comparisons (p<.0003), being able to detect effects accounting for 3.4% of trait variance in the total sample but only 7.7% in the No Diagnosis sample. Finally, although a literature based SNP set was utilized in this study, a more comprehensive tag SNP (tSNP) design may further elucidate the individual SNPs that are important in cognitive performance (estimated 348 tag SNPs needed due to the size of NRG1).
Many of the SNPs with significant associations with cognition exhibited significant stratification. Although stratification is generally expected to lead to greater associations between- than within-families, in the current study, the between-family component was typically less than the within-family. Such results may indicate that some between-family factor, such as socioeconomic status, is associated, perhaps due to the recruitment process, with the allele, leading to significant stratification in which the direction of the association is different between compared to within families. Regardless of the nature of such effects, the QTDT analyses and within-family beta weights reported here are robust to any stratification effects (Abecasis et al., 2000).
It is also possible that these nominal associations with cognition are irrelevant to schizophrenia, but the literature on NRG1 and schizophrenia suggests otherwise. In addition, although several of the alleles that were previously associated with schizophrenia in the literature were associated with reduced cognitive function in the current study, some alleles previously associated with affected status were associated with improved cognition here. The causes of such inconsistencies or “allele-flipping” are frequently unknown, but recent studies (Clarke and Cardon, 2010, Goldberg and Green, 2002) suggest that these inconsistencies are not always merely alpha error but may be due to allelic and locus heterogeneity, population-related and sample variation-related differences in LD structure between markers, and environmental exposures.
Conclusions
None of the associations met the criteria for significance after correcting for multiple testing, although each sample had between 19–21 nominally significant (p≤.05) associations, encompassing multiple SNPs and multiple cognitive domains. The large effect sizes of these associations with cognition (absolute value βwithin across all analyses: 0.08–0.73) are noteworthy given the usually reported modest size of the association between NRG1 and schizophrenia (odds ratio: 1–2.2). The numerous nominally significant findings in progressively healthier sub-samples also may suggest that any possible NRG1 effects are not solely secondary to an effect of schizophrenia or psychopathology. If replicated and found significant experiment-wise, these associations may suggest a role for NRG1 in cognition that is a mediating risk factor for schizophrenia or psychopathology more generally, although it is possible that NRG1 could affect schizophrenia and cognitive function independently. Although the current lack of significant experiment-wise results does not provide evidence for convergent validity for NRG1 as a risk gene for schizophrenia, the consistent “nominally significant” findings may suggest hypotheses for future studies that focus on a few specific promising SNPs and cognitive functions and thus are more statistically powerful given a reduced penalty for multiple tests.
In particular, when considering the cognitive domains separately from individual SNPs, attention consistently had the most nominally significant associations over all of the samples assessed, with abstraction and mental flexibility, verbal memory, and sensorimotor dexterity also having interesting results, suggesting that future studies could focus on these cognitive processes specifically. Similarly, SNPs rs10954855, rs3924999, rs2466060, and rs10503929 appear to be the most promising for future study. Overall, although not significant experiment-wise, these findings may suggest more specific hypotheses that warrant future study concerning the relationships among NRG1, schizophrenia and cognition. In addition, studies of the NRG1-ErbB signaling pathway, with particular attention paid to ErbB4, may further elucidate the pathophysiology of schizophrenia-related cognitive dysfunction.
Acknowledgments
The authors wish to thank Lora McClain and Jan Richards for their help in laboratory methods and data management.
Funding: This work was supported by the National Institute of Mental Health (MH42191 to R.E.G.; MH063480 to V.L.N and M.F.P-G.; MH061622 to L.A.; MH084856 to R.C.G; and MH72995 to K.M.P.). Development of SOLAR is supported by the National Institute of Mental Health (MH59490).
References
- ABECASIS GR, CARDON LR, COOKSON WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–92. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALLEN N, BAGADE S, MCQUEEN M, IOANNIDIS J, KAVVOURA F, KHOURY M, et al. Systematic Meta-Analyses and Field Synopsis of Genetic Association Studies in Schizophrenia: The SzGene Database. Nat Genet. 2008;40:827–34. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- ALMASY L, BLANGERO J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALMASY L, GUR RC, HAACK K, COLE SA, CALKINS ME, PERALTA JM, et al. A genome screen for quantitative trait loci influencing schizophrenia and neurocognitive phenotypes. Am J Psychiatry. 2008;165:1185–92. doi: 10.1176/appi.ajp.2008.07121869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BENTON A, VARNEY N, HAMSHER K. Judgment of Line Orientation, Form V. Iowa City, IA: University of Iowa Hospitals; 1975. [Google Scholar]
- BUCHANAN RW, DAVIS M, GOFF D, GREEN MF, KEEFE RS, LEON AC, et al. A summary of the FDA-NIMH-MATRICS workshop on clinical trial design for neurocognitive drugs for schizophrenia. Schizophr Bull. 2005;31:5–19. doi: 10.1093/schbul/sbi020. [DOI] [PubMed] [Google Scholar]
- CAMPBELL DT, FISKE DW. Convergent and discriminant validation by the multitrait-multimethod matrix. Psychol Bull. 1959;56:81–105. [PubMed] [Google Scholar]
- CANNON TD, ZORRILLA LE, SHTASEL D, GUR RE, GUR RC, MARCO EJ, et al. Neuropsychological functioning in siblings discordant for schizophrenia and healthy volunteers. Arch Gen Psychiatry. 1994;51:651–61. doi: 10.1001/archpsyc.1994.03950080063009. [DOI] [PubMed] [Google Scholar]
- CARDNO AG, GOTTESMAN II. Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am J Med Genet. 2000;97:12–7. [PubMed] [Google Scholar]
- CLARKE GM, CARDON LR. Aspects of observing and claiming allele flips in association studies. Genet Epidemiol. 2010;34:266–74. doi: 10.1002/gepi.20458. [DOI] [PubMed] [Google Scholar]
- CONNEELY KN, BOEHNKE M. So Many Correlated Tests, So Little Time! Rapid Adjustment of P Values for Multiple Correlated Tests. Am J Hum Genet. 2007:81. doi: 10.1086/522036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CROWLEY JJ, KEEFE RS, PERKINS DO, STROUP TS, LIEBERMAN JA, SULLIVAN PF. The neuregulin 1 promoter polymorphism rs6994992 is not associated with chronic schizophrenia or neurocognition. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1298–300. doi: 10.1002/ajmg.b.30727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DUAN J, MARTINEZ M, SANDERS AR, HOU C, KRASNER AJ, SCHWARTZ DB, et al. Neuregulin 1 (NRG1 ) and schizophrenia: analysis of a US family sample and the evidence in the balance. Psychol Med. 2005;35:1599–610. doi: 10.1017/S0033291705005428. [DOI] [PubMed] [Google Scholar]
- FULKER DW, CHERNY SS, SHAM PC, HEWITT JK. Combined linkage and association sib-pair analysis for quantitative traits. Am J Hum Genet. 1999;64:259–67. doi: 10.1086/302193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GLAHN DC, GUR RC, RAGLAND JD, CENSITS DM, GUR RE. Reliability, performance characteristics, construct validity, and an initial clinical application of a visual object learning test (VOLT) Neuropsychology. 1997;11:602–12. doi: 10.1037//0894-4105.11.4.602. [DOI] [PubMed] [Google Scholar]
- GOLDBERG TE, GREEN MF. Neurocognitive Functioning in Patients with Schizophrenia: An Overview. In: DAVIS KL, CHARNEY D, COYLE JT, NEMEROFF C, editors. Neuropsychopharmacology: The Fifth Generation of Progress. Lippincott Williams & Wilkins; 2002. [Google Scholar]
- GOTTESMAN II, GOULD TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–45. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- GOTTESMAN II, SHIELDS J. A polygenic theory of schizophrenia. Proc Natl Acad Sci U S A. 1967;58:199–205. doi: 10.1073/pnas.58.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUR RC, JAGGI JL, RAGLAND JD, RESNICK SM, SHTASEL D, MUENZ L, et al. Effects of memory processing on regional brain activation: cerebral blood flow in normal subjects. Int J Neurosci. 1993;72:31–44. doi: 10.3109/00207459308991621. [DOI] [PubMed] [Google Scholar]
- GUR RC, RAGLAND JD, MOBERG PJ, BILKER WB, KOHLER C, SIEGEL SJ, et al. Computerized neurocognitive scanning: II. The profile of schizophrenia. Neuropsychopharmacology. 2001a;25:777–88. doi: 10.1016/S0893-133X(01)00279-2. [DOI] [PubMed] [Google Scholar]
- GUR RC, RAGLAND JD, MOBERG PJ, TURNER TH, BILKER WB, KOHLER C, et al. Computerized neurocognitive scanning: I. Methodology and validation in healthy people. Neuropsychopharmacology. 2001b;25:766–76. doi: 10.1016/S0893-133X(01)00278-0. [DOI] [PubMed] [Google Scholar]
- GUR RC, RICHARD J, HUGHETT P, CALKINS M, MACY L, BILKER W, et al. A cognitive neuroscience based computerized battery for efficient measurement of individual differences: standardization and initial construct validation. Journal of Neuroscience Methods. 2010;187:254–62. doi: 10.1016/j.jneumeth.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUR RE, KOHLER CG, RAGLAND JD, SIEGEL SJ, LESKO K, BILKER WB, et al. Flat affect in schizophrenia: relation to emotion processing and neurocognitive measures. Schizophr Bull. 2006;32:279–87. doi: 10.1093/schbul/sbj041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUR RE, NIMGAONKAR VL, ALMASY L, CALKINS ME, RAGLAND JD, POGUE-GEILE MF, et al. Neurocognitive endophenotypes in a multiplex multigenerational family study of schizophrenia. Am J Psychiatry. 2007;164:813–9. doi: 10.1176/ajp.2007.164.5.813. [DOI] [PubMed] [Google Scholar]
- HALL J, WHALLEY HC, JOB DE, BAIG BJ, MCINTOSH AM, EVANS KL, et al. A neuregulin 1 variant associated with abnormal cortical function and psychotic symptoms. Nat Neurosci. 2006;9:1477–8. doi: 10.1038/nn1795. [DOI] [PubMed] [Google Scholar]
- HARE E, GUR RC, POGUE-GEILE MF, CALKINS ME, PERALTA JM, PRASAD K, et al. Genetic correlations between neurocognitive measures in a multiplex, multigenerational study of schizophrenia. Under review. [Google Scholar]
- HARRISON PJ, LAW AJ. Neuregulin 1 and schizophrenia: genetics, gene expression, and neurobiology. Biol Psychiatry. 2006;60:132–40. doi: 10.1016/j.biopsych.2005.11.002. [DOI] [PubMed] [Google Scholar]
- HARRISON PJ, WEINBERGER DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- HAVILL LM, DYER TD, RICHARDSON DK, MAHANEY MC, BLANGERO J. The quantitative trait linkage disequilibrium test: a more powerful alternative to the quantitative transmission disequilibrium test for use in the absence of population stratification. BMC Genet. 2005;6(Suppl 1):S91. doi: 10.1186/1471-2156-6-S1-S91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEINRICHS RW, RUTTAN L, ZAKZANIS KK, CASE D. Parsing schizophrenia with neurocognitive tests: evidence of stability and validity. Brain Cogn. 1997;35:207–24. doi: 10.1006/brcg.1997.0938. [DOI] [PubMed] [Google Scholar]
- HONG CJ, HUO SJ, LIAO DL, LEE K, WU JY, TSAI SJ. Case-control and family-based association studies between the neuregulin 1 (Arg38Gln) polymorphism and schizophrenia. Neurosci Lett. 2004;366:158–61. doi: 10.1016/j.neulet.2004.05.027. [DOI] [PubMed] [Google Scholar]
- KIRCHER T, KRUG A, MARKOV V, WHITNEY C, KRACH S, ZERRES K, et al. Genetic variation in the schizophrenia-risk gene neuregulin 1 correlates with brain activation and impaired speech production in a verbal fluency task in healthy individuals. Hum Brain Mapp. 2009;30:3406–16. doi: 10.1002/hbm.20761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIRCHER T, THIENEL R, WAGNER M, RESKE M, HABEL U, KELLERMANN T, et al. Neuregulin 1 ICE-single nucleotide polymorphism in first episode schizophrenia correlates with cerebral activation in fronto-temporal areas. Eur Arch Psychiatry Clin Neurosci. 2008 doi: 10.1007/s00406-008-0837-4. [DOI] [PubMed] [Google Scholar]
- KRUG A, MARKOV V, EGGERMANN T, KRACH S, ZERRES K, STOCKER T, et al. Genetic variation in the schizophrenia-risk gene neuregulin1 correlates with differences in frontal brain activation in a working memory task in healthy individuals. Neuroimage. 2008;42:1569–76. doi: 10.1016/j.neuroimage.2008.05.058. [DOI] [PubMed] [Google Scholar]
- KURTZ MM, RAGLAND JD, BILKER W, GUR RC, GUR RE. Comparison of the continuous performance test with and without working memory demands in healthy controls and patients with schizophrenia. Schizophr Res. 2001;48:307–16. doi: 10.1016/s0920-9964(00)00060-8. [DOI] [PubMed] [Google Scholar]
- KURTZ MM, RAGLAND JD, MOBERG PJ, GUR RC. The Penn Conditional Exclusion Test: a new measure of executive-function with alternate forms of repeat administration. Arch Clin Neuropsychol. 2004;19:191–201. doi: 10.1016/S0887-6177(03)00003-9. [DOI] [PubMed] [Google Scholar]
- LI D, COLLIER DA, HE L. Meta-analysis shows strong positive association of the neuregulin 1 (NRG1) gene with schizophrenia. Hum Mol Genet. 2006;15:1995–2002. doi: 10.1093/hmg/ddl122. [DOI] [PubMed] [Google Scholar]
- LI T, STEFANSSON H, GUDFINNSSON E, CAI G, LIU X, MURRAY RM, et al. Identification of a novel neuregulin 1 at-risk haplotype in Han schizophrenia Chinese patients, but no association with the Icelandic/Scottish risk haplotype. Mol Psychiatry. 2004;9:698–704. doi: 10.1038/sj.mp.4001485. [DOI] [PubMed] [Google Scholar]
- LIN HF, LIU YL, LIU CM, HUNG SI, HWU HG, CHEN WJ. Neuregulin 1 gene and variations in perceptual aberration of schizotypal personality in adolescents. Psychol Med. 2005;35:1589–98. doi: 10.1017/S0033291705005957. [DOI] [PubMed] [Google Scholar]
- MAXWELL M. Manual for the family interview for genetic studies (FIGS) Bethesda, MD: National Institute of Mental Health; 1992. [Google Scholar]
- NURNBERGER JI, JR, BLEHAR MC, KAUFMANN CA, YORK-COOLER C, SIMPSON SG, HARKAVY-FRIEDMAN J, et al. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry. 1994;51:849–59. doi: 10.1001/archpsyc.1994.03950110009002. discussion 863-4. [DOI] [PubMed] [Google Scholar]
- O’CONNELL JR, WEEKS DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–66. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PETRYSHEN TL, MIDDLETON FA, KIRBY A, ALDINGER KA, PURCELL S, TAHL AR, et al. Support for involvement of neuregulin 1 in schizophrenia pathophysiology. Mol Psychiatry. 2005;10:366–74. 328. doi: 10.1038/sj.mp.4001608. [DOI] [PubMed] [Google Scholar]
- POGUE-GEILE MF, GOTTESMAN II. Schizophrenia: Study of a genetically complex phenotype. In: JONES BC, MOREMEDE P, editors. Neurobehavioral Genetics: Methods and Applications. 2. Boca Raton, FL: CRC Press; 2006. [Google Scholar]
- POGUE-GEILE MF, YOKLEY JL. Current Research on the Genetic Causes of Schizophrenia. Current Directions in Psychological Science 2010 [Google Scholar]
- PURCELL SM, WRAY NR, STONE JL, VISSCHER PM, O’DONOVAN MC, SULLIVAN PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCOLNICK EM, PETRYSHEN T, SKLAR P. Schizophrenia: do the genetics and neurobiology of neuregulin provide a pathogenesis model? Harv Rev Psychiatry. 2006;14:64–77. doi: 10.1080/10673220600642960. [DOI] [PubMed] [Google Scholar]
- SHI J, LEVINSON DF, DUAN J, SANDERS AR, ZHENG Y, PE’ER I, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–7. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SNITZ BE, MACDONALD AW, 3RD, CARTER CS. Cognitive deficits in unaffected first-degree relatives of schizophrenia patients: a meta-analytic review of putative endophenotypes. Schizophr Bull. 2006;32:179–94. doi: 10.1093/schbul/sbi048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEFANIS NC, TRIKALINOS TA, AVRAMOPOULOS D, SMYRNIS N, EVDOKIMIDIS I, NTZANI EE, et al. Impact of schizophrenia candidate genes on schizotypy and cognitive endophenotypes at the population level. Biol Psychiatry. 2007;62:784–92. doi: 10.1016/j.biopsych.2006.11.015. [DOI] [PubMed] [Google Scholar]
- STEFANSSON H, OPHOFF RA, STEINBERG S, ANDREASSEN OA, CICHON S, RUJESCU D, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–7. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEFANSSON H, SARGINSON J, KONG A, YATES P, STEINTHORSDOTTIR V, GUDFINNSSON E, et al. Association of neuregulin 1 with schizophrenia confirmed in a Scottish population. Am J Hum Genet. 2003;72:83–7. doi: 10.1086/345442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEINTHORSDOTTIR V, STEFANSSON H, GHOSH S, BIRGISDOTTIR B, BJORNSDOTTIR S, FASQUEL AC, et al. Multiple novel transcription initiation sites for NRG1. Gene. 2004;342:97–105. doi: 10.1016/j.gene.2004.07.029. [DOI] [PubMed] [Google Scholar]
- SULLIVAN PF, KENDLER KS, NEALE MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–92. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- THOMPSON JL, WATSON JR, STEINHAUER SR, GOLDSTEIN G, POGUE-GEILE MF. Indicators of genetic liability to schizophrenia: a sibling study of neuropsychological performance. Schizophr Bull. 2005;31:85–96. doi: 10.1093/schbul/sbi009. [DOI] [PubMed] [Google Scholar]
- TOSATO S, DAZZAN P, COLLIER D. Association between the neuregulin 1 gene and schizophrenia: a systematic review. Schizophr Bull. 2005;31:613–7. doi: 10.1093/schbul/sbi043. [DOI] [PubMed] [Google Scholar]
- WOLPOWITZ D, MASON TB, DIETRICH P, MENDELSOHN M, TALMAGE DA, ROLE LW. Cysteine-rich domain isoforms of the neuregulin-1 gene are required for maintenance of peripheral synapses. Neuron. 2000;25:79–91. doi: 10.1016/s0896-6273(00)80873-9. [DOI] [PubMed] [Google Scholar]
- YANG JZ, SI TM, RUAN Y, LING YS, HAN YH, WANG XL, et al. Association study of neuregulin 1 gene with schizophrenia. Mol Psychiatry. 2003;8:706–9. doi: 10.1038/sj.mp.4001377. [DOI] [PubMed] [Google Scholar]


