Abstract
The progesterone receptor (PR) interacts with chromatin in a highly dynamic manner that requires ongoing chromatin remodeling, interaction with chaparones and activity of the proteasome. Here we discuss dynamic interaction of steroid receptor with chromatin, with special attention not only to PR but also to the glucocorticoid receptor (GR), as these receptors share many similarities regarding interaction with, and remodeling of, chromatin. Both receptors can bind nucleosomal DNA and have accordingly been described as pioneering factors. However recent genomic approaches (ChIP-seq and DHS-seq) show that a large fraction of receptor binding events occur at pre-accessible chromatin. Thus factors which generate and maintain accessible chromatin during development, and in fully differentiated tissue, contribute a major fraction of receptor tissue specificity. In addition, chromosome conformation capture techniques suggest that steroid receptors preferentially sequester within distinct nuclear hubs. We will integrate dynamic studies from single cells and genomic studies from cell populations, and discuss how genomic approaches have reshaped our current understanding of mechanisms that control steroid receptor interaction with chromatin.
Introduction
The progesterone receptor (PR) belongs to the steroid receptor family of transcription factors. In the absence of ligand it is restricted from chromatin interaction through interaction with specific chaperone proteins (Botos et al., 2004). Binding of ligand results in phosphorylation of the receptor and strong nuclear localization, where the receptor engages the chromatin embedded genome to bind specific DNA response elements. DNA bound receptors recruit transcriptional co-regulator complexes though direct protein-protein interaction. These complexes in turn modify the chromatin status and facilitate auxiliary transcription factor and co-regulator recruitment, thus promoting either activation or repression of target genes.
PR manifests a high sequence homology with other steroid receptors, mineralocorticoid receptor (MR), glucocorticoid receptor (GR) and androgen receptor (AR). The DNA binding domain in particular has very high sequence conservation, and these receptors bind to very similar DNA elements, with the general consensus ACANNNTGT (Bain et al., 2007). Although the genome harbors hundreds of thousands of DNA sequences that resemble this consensus sequence, only a few thousands of these sites are occupied by receptor in a given cell type at a given time (John et al., 2011). The structure of chromatin presents a barrier for transcription factor access to DNA. Thus, in addition to the recognition of DNA sequences, the organization of chromatin at specific DNA elements in the genome is believed to be a major contributor to specific transcription factor accessibility of the genome (Jiang and Pugh 2009).
The structural core of chromatin consists of a nucleosome with ~146 bp of DNA wrapped around a histone octamer containing two each of H2A H2B H3 and H4 (Luger et al., 1997). H1 occupies the linker DNA between nucleosomes and further condenses chromatin (Bustin et al., 2005). Histones are heavily modified by acetylation methylation ubiquitination sumolation phosphorylation and glycosylate, and the pattern of modification correlates with the accessibility of chromatin and the transcriptional potential of nearby genes(Campos and Reinberg, 2008; Li et al., 2007). Histone modifications are catalyzed by a large number of protein complexes, which are recruited to chromatin by direct protein-protein interactions with transcription factors, and through high affinities for specific histone modifications (Campos and Reinberg, 2008; Bartke et al., 2010). Moreover nucleosomes can be modified by incorporation of histone variants (Talbert and Henikoff, 2010) and reorganized by chromatin remodeling enzymes (Wu et al., 2009). Finally DNA modification by methylation has profound effects of the chromatin structure (Chodavarapu et al., 2010; Schubeler, 2009) and affects transcription factor binding to DNA in a tissue specific manner (Rishi et al., 2010). Collectively, modifications of the nucleosome and the DNA fiber contribute to local chromatin accessibility.
When bound to agonist, steroid receptors such as PR, GR, and ER are able to bind nucleosomal DNA (Sun et al., 1983) and cause local chromatin remodeling. GR and PR in particular have been characterized as pioneering factors promoting recruitment of other transcription factors to specific sites of the genome. When bound to DNA the steroid receptor-DNA complexes are classically viewed as stable complexes with residence times of several minutes, creating a persistent signal for gene transcription in the presence of agonist. This model is typically based on chromatin immunoprecipitation (ChIP) experiments with poor temporal resolution, averaging signals over large populations of cells. In contrast, experimental systems using FRAP analysis on single cells, together with in vitro laser assisted crosslinking, have shown that steroid receptor interactions with DNA are considerably more dynamic. Receptors typically manifest residence times on DNA on a time scale of seconds. Several molecular processes are thought to contribute to this rapid mobility, including continuous remodeling of chromatin (Nagaich et al., 2004; McNally et al., 2000; Rayasam et al., 2005). Here we discuss the dynamic interaction of steroid receptors with chromatin, with special attention not only to PR but also to GR, as these receptors share many similarities regarding interaction with, and remodeling of, chromatin (Rayasam et al., 2005; McNally et al., 2000).
In recent years methods for genome wide identification of chromatin accessibility and higher order chromatin structure have been developed. We also briefly introduce these techniques, and discuss how studies using these methodologies have reshaped our understanding of the mechanisms involved in steroid receptor access to chromatin.
The MMTV promoter as a model system
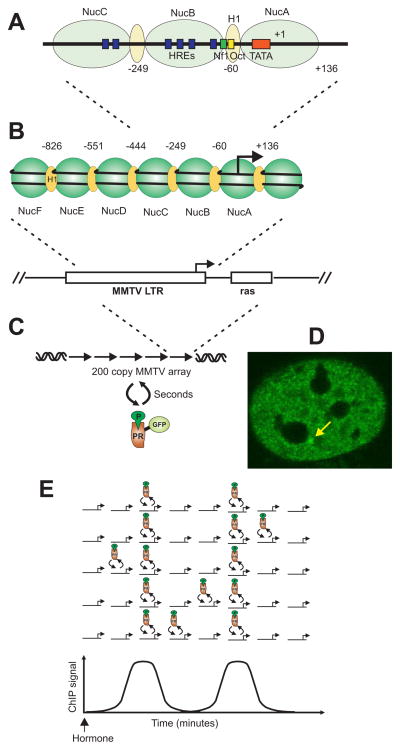
The mouse mammary tumor virus (MMTV) LTR has been widely used a model system for steroid receptor mediated chromatin remodeling. When the LTR is reconstituted with nucleosomes in vitro (Perlmann and Wrange, 1988; Pina et al., 1990; Venditti et al., 1998), or stably introduced into the genome of replicating cells, it assembles in an ordered nucleosomal structure containing six nonrandomly positioned nucleosome families designated A–F (Richard-Foy and Hager, 1987), interspersed by H1 occupancy of the linker regions (Bresnick et al., 1992) (figure 1B). Each of these nucleosome positions is occupied by a family of closely spaced octamers (Fragoso et al., 1998), referred to as a “frequency-biased nucleosome distribution.” The LTR contains six hormone response elements (HRE) located on nucleosome families B and C (Fletcher et al., 2002)(figure 1A). These elements bind GR, PR, MR and AR, and the promoter is strongly activated by each of these receptors. Moreover functional binding sites for Oct1 and NF1 have been reported, and binding of these factors is essential for full MMTV activation by steroid receptors (figure 1A). In the absence of steroid receptor agonists, MMTV chromatin structure is resistant to cleavage by DNAsel and this closed chromatin conformation prevents binding of NF1, Oct1 and the basal transcription machinery (Archer et al., 1992).
Figure 1. PR rapidly and dynamically interacts with the MMTV LTR.
(A) Relative position of regulatory elements in the MMTV LTR. DNA elements reported to bind PR Nfl and Oct are marked by blue, green and red respectively. (B) Relative position of nucelosmes at reconstituted MMTV LTR. (C and D) Array of two hundred copies head to tail MMTV LTR incorporated into chromosome four. Activated GFP tagged PR binds the MMTV array and give rise to an intense fluorescence signal (marked by yellow arrow) that can be detected in the nucleus above background. FRAP studies of GFP-PR bound MMTV array show that PR dynamically interacts with the MMTV array with on/off rates in a matter of seconds. (E) Integrative analysis of receptor occupancy of MMTV from single cells correlates with ChIP analysis on a population of cells. Model shows alleles with a single PR binding site from different cells in a cell population. After activation with hormone, interaction frequency of receptor and binding site increases in the majority of cells in the population. When a population of cells are analyzed by ChIP this dynamic interactions give rise to a signal above background. When a population of cells are synchronized waves spaced by minutes of receptor interaction with DNA can be detected. However at any time when focusing on single cells the receptor rapidly interacts (seconds) with chromatin.
The nucleosome organization on DNA forms a barrier for transcription factor access by two central mechanisms; the translational positioning of nucleosomes determines the DNA compartment occupied by nucleosomes, whereas rotational positioning dictates sequences accessible on the surface of the nucleosome. Steroid receptors, including PR and GR, are able to occupy nucleosomal DNA (Perlmann and Wrange, 1988; Sun et al., 1983). Binding is regulated by rotational positioning, wherein response elements positioned at the dyad and 180 degrees from the dyad are favorable (Li and Wrange, 1995). It is likely that PR initially engages an accessible HRE (figure 1B), promoting initial remodeling that allows occupancy of the other HREs (Pina et al., 1990). In contrast, NF1 binding is regulated by translational positioning where binding sequences positioned in the nucleosome blocks NF1 occupancy and DNA positioned in nucleosome free regions favor NF1 binding (Cordingley et al., 1987; Archer et al., 1992) (Blomquist et al., 1996). This intrinsic difference between steroid receptors and NF1 can be explained by the different DNA binding interface of the proteins. Steroid receptors such as PR and GR bind a few bases at the same surface of the DNA helix whereas NF1 engulfs both sides of the helix and some DNA recognition sequences is evidently facing the nucleosomal core and restricted from NF1 interaction.
The molecular events followed by PR engagement with chromatin have been studied extensively using the MMTV promoter (see Beato et al., this issue). In the absence of hormone the MMTV promoter is retained in an inaccessible conformation through occupancy of linker H1 and repressive complexes containing HP1 mediated by histone modifications such as H3K9Me3 (Vicent et al., 2004). Shortly after hormone addition PR interacts with activated Erk and Msk1, which phosphorylates PR. Ligand bound PR-P, Erk and Msk1 subsequently interacts with response elements occupied by nucleosome B (Vicent et al., 2006). This leads to a series of events where H3S10 is phosphorylated by Msk1, HP1 is displaced, H1 is evicted and H3K14 and H4K8 are acetylated (Koop et al., 2003; Vicent et al., 2006). This facilitates SWI/SNF recruitment and subsequently chromatin remodeling (Vicent et al., 2009), which produces a chromatin structure prone to DNAse I cleavage. The structural transitions responsible for this increase susceptibility are likely to be quite complex (Fragoso et al., 1995), but include remodeling nucleosome B and C (Fletcher et al., 2000; Fragoso et al., 1998) and eviction of H1 from the chromatin template (Bresnick et al., 1992). This collectively increases accessibility of auxiliary transcription factors such as NF1 and Oct1 (Lee and Archer, 1994). Once chromatin has been remodeled these factors act synergistically with PR to maintain an activated MMTV (Vicent et al., 2010). These accumulated events, and probably many more to be identified, attract the basal transcription machinery. Disruption of remodeling enzymes in the SWI/SNF complex collapses many of these events (Vicent et al., 2009) emphasizing the importance of chromatin remodeling during PR mediated gene activation.
Dynamic interaction with chromatinized MMTV
Transcription factors diffuse rapidly through the nucleus with frequent non specific interaction with chromatin with retention times in the order of a few hundred ms. In this scanning mode, a transcription factor will occasionally interact with specific accessible binding sites and residence time is somewhat increased. By this so called genome scanning mechanism activated transcription factors are able find and interact with specific target sites within seconds after activation (Hager et al., 2009).
Accordingly, unliganded steroid receptors have been found to diffuse rapidly in the nucleus. Upon ligand binding the receptors localize in subnuclear compartments (Htun et al., 1999; Baumann et al., 2001; Prufer et al., 2000; Tomura et al., 2001). The widely employed techniques of ChIP and chromatin accessibility have led to a model of steroid receptor interaction with chromatin that visualizes relative stable complexes, with residence times of minutes to hours, which in turn stimulate the assembly of a productive transcription complex. A markedly different view of receptor action emerged with the ability to visualize receptor action in living cells [the “green revolution” (Stearns, 1995)]. These techniques now permit the characterization of receptor-chromatin interaction with very high temporal resolution on a single cell level. The first identification of highly dynamic interaction of a transcription factor with chromatin came from studies using a genome integrated array of 200 copies of the MMTV promoter (figure 1C).
This permitted for the first time real time kinetic analysis of factor residency times in living cells, using photobleaching methods (fluorescence recovery after photobleaching, FRAP) coupled with visualization of receptor binding to authentic response elements. It was shown that GR (McNally et al., 2000) and PR (Rayasam et al., 2005) undergoes rapid exchange (few seconds) between the nucleoplasm and the chromatin embedded MMTV promoter array (figure 1C and D). Using similar strategies, other classes of transcription factors have also been shown to interact with chromatin in a highly dynamic manner (Bosisio et al., 2006; Sharp et al., 2006). Thus within the subnuclear compartment, steroid receptors undergo a repeated and rapid association and disassociation from chromatin. The standard technique of ChIP analysis, wherein a population of cells are crosslinked and analyzed, is insensitive to this dynamic association with chromatin (Voss et al., 2006; Voss et al., 2009). The ChIP signals obtained with this methodology represent population averages of the highly transient binding events observed in living cells (figure 1E). Further time complexity in receptor action occurs during the activation phase. Levels of promoter activity can fluctuate on time scales of minutes to hours, a phenomenon designated “promoter progression” (Voss et al., 2009; Hager et al., 2006). In a synchronized population of cells, estrogen receptor interaction with chromatin has been shown to occur in waves separated by a few minutes, which leads to cycles of co-regulator recruitment and chromatin remodeling (Metivier et al., 2003)(figure 1E).
Dynamic interaction of receptor with chromatin requires energy (Elbi et al., 2004; Stavreva et al., 2004), emphasizing that the dynamic processes of steroid receptor interaction with chromatin are not explained by simple diffusion. During the process of chromatin remodeling, PR is transiently retarded on the MMTV array (Rayasam et al., 2005), suggesting that nucleosome remodeling is important for the dynamic interaction of steroid receptor with chromatin in vivo. This is supported by studies of the MMTV promoter reconstituted in vitro, where laser UV assisted crosslinking shows that GR periodically interacts with the MMTV promoter (Nagaich et al., 2004). The binding event of GR is less than one minute and coincides with an initial increased BRG1 recruitment, followed by remodeling of nucleosome B and C, eviction of BRG1 and then loss of GR binding to the chromatin template. Importantly, the periodically binding event of GR and BRG1 is dependent on BRG1 remodeling activity, highlighting a mechanistic link between active chromatin remodeling and dynamic interaction between GR and chromatin. Rayasam et al (Rayasam et al., 2005) also reported ligand specific effects on progesterone receptor recruitment of remodeling activity in vitro that correlated with mobility observed in living cells, further emphasizing the link between remodeling and receptor exchange. Finally, binding of receptors such as GR to some endogenous promoters requires active BRG1 (Fryer and Archer, 1998; John et al., 2008) and genome wide binding to chromatin occurs predominantly at DNAsel hypersensitive sites (DHS), a hallmark for disordered chromatin structure.
In addition to active chromatin remodeling, chaparones and the proteasome also play an important role in dynamic interaction of steroid receptors with chromatin. Chaparones such as p23 and Hsp90 are recruited to steroid receptor responsive promoters and facilitate disassembly of the receptor from chromatin (Elbi et al., 2004; Freeman and Yamamoto 2002). In agreement, inhibition of chaparones leads to stabilization of GR at MMTV array, whereas addition of chaparones increases GR mobility in the nucleus. Likewise inhibition of the proteasome leads to retention of GR at MMTV array, suggesting that chaparones and the proteasome together with chromatin remodeling contributes to the highly dynamic assembly and disassembly of steroid receptors on chromatin.
Steroid receptors preferably engage accessible chromatin
With the development of second generation sequence technology, techniques traditionally used to characterize local chromatin structures have been optimized to precisely map genome wide chromatin organization. Each of these techniques provides information on the chromatin structure; however, due to technical differences they give rise to very different interpretations of chromatin accessibility. Deep sequencing of MNase digested native or crosslinked chromatin maps positions of nucleosomes. The DNAsel chromatin accessibility, assay combined with massive parallel sequencing (termed DHS-seq or DNAse-seq), identifies all accessible sites within the genome irrespective of nucleosome positioning. Thus either nucleosome free or nucleosome “reorganized” regions (i.e PR binding sites in MMTV promoter;) figure 2A), can be characterized on the global scale. DHS-seq typically identifies ~100,000 accessible sites within the genome of mammalian cell lines and tissue representing about ~2% of the genome, with most of the sites distributed within promoters and intergenic regions (John et al., 2011; Bernstein et al., 2010). Importantly extensive sequencing of the DHS’s to great depth can provide information of footprints of transcription factors that occupy accessible sites and thus offer an unbiased approach to identify regulatory proteins that maintains chromatin accessibility (Hager, 2010; Boyle et al., 2010; Hesselberth et al., 2009).
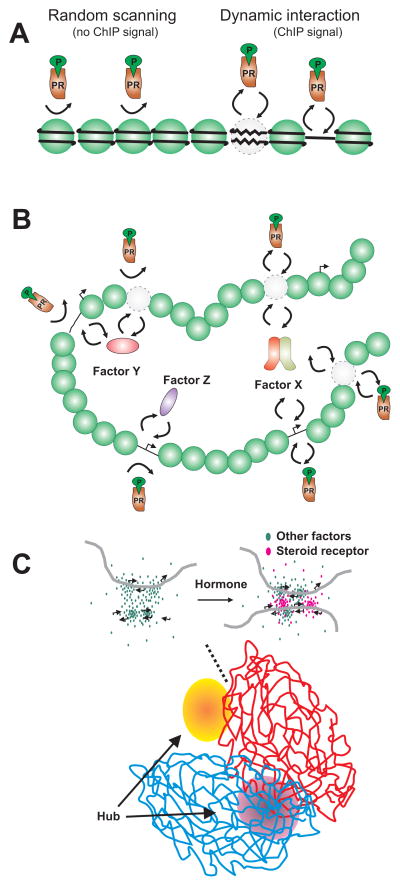
Figure 2. Model illustrating mechanisms that contribute to specific PR interaction with chromatin.
(A) Dynamic receptor interaction with chromatin occurs almost exclusively at an accessible chromatin conformation either at sites where nucleosomes are remodeled or at nuclesome free regions. At regions of inaccessible chromatin the receptor scans for accessible sites but binding is not observed. (B) Specific transcription factors determine receptor interaction with accessible chromatin. Interaction with specific transcription factors (e.g. factor X) at accessible chromatin regions facilitates receptor interaction. In contrast, at accessible sites where other factors interact (e.g. factor Y and Z), receptor interaction with chromatin is not necessarily favored. (C) Chromatin regions harboring steroid receptor specific binding sites are clustered in sub-nuclear regions designated hubs. These hubs are enriched for accessible chromatin and factors that maintain accessibility. Within these hubs, activation of receptor by hormone leads to increased frequency of dynamic interaction with chromatin.
Based upon studies of the MMTV promoter, PR and GR have traditionally been characterized as pioneering factors that bind to chromatin embedded DNA, recruit remodeling proteins, and facilitate binding of additional transcription factors. This model was initially challenged by the observation that GR not only binds to inaccessible chromatin but also to preexisting remodeled chromatin (John et al., 2008). This concept has been further explored in genome wide studies combining DHS-seq with GR ChlP-seq (John et al., 2011). Here it was shown that after ligand activation GR is bound primarily in the preexisting accessible chromatin landscape. Only a fraction (10–15%) of GR binding takes place within preexisting inaccessible chromatin. Binding of GR to the inaccessible chromatin compartment leads to subsequent chromatin remodeling and increased accessibility, leaving only a tiny fraction of the total GR binding sites inaccessible. Thus occupancy of chromatin by GR is almost exclusively associated with chromatin remodeling events. Importantly, DHS-seq revealed that the accessible genome is highly cell type specific and accordingly the genome wide GR binding profile between cell types is cell selective. This important finding emphasizes that the accessible chromatin landscape is, along with specific DNA sequences, a major determinant for cell type specific GR signaling. Future studies using DHS-seq combined with ChlP-seq for other steroid receptors such as PR in different cell types and tissue will reveal if this is a general phenomenon among the nuclear receptor family.
Given that steroid receptors to a large extend occupy preexisting accessible sites in the genome, it will be important to identify regulatory factors that maintain chromatin accessibility. These preexisting factors are likely to contribute to specific binding of a steroid receptor with similar sequence preference (i.e PR, GR, MR and AR) to sites in the same cell type, and to determine cell specific binding profiles of a specific transcription factor (figure 2B). Indeed de novo motif analysis of DNA sequences surrounding GR binding sites identified by ChlP-seq from two different cell lines have indicated that API is important for GR binding in one cell type and FOXA1 in another. Genome wide profiling of GR in differentiating adipocytes have suggested that C/EBPbeta and STAT5 controls GR binding specificity in adipocytes (Siersbaek et al., 2011; Steger et al., 2010). In agreement with this concept, PR dependent activation of target genes in breast cancer cells has been reported to be dependent on STAT5 (Subtil-Rodriguez et al., 2008) and NF1 (Vicent et al., 2010), and FOXA1 has been shown to increase accessibility of the MMTV promoter without the presence of steroid hormone, facilitating steroid receptor binding and hormone dependent transcription (Belikov et al., 2009). A series of studies have described FOXA1 as a pioneering factor for ER and AR binding to chromatin (Lupien et al., 2008; Wang et al., 2007). The FOXA1 binding motif is enriched at ER and AR binding sites and FOXA1 genome wide occupancy highly correlates with ER and AR binding. Depletion of FOXA1 dramatically reduces ER occupancy and FOXA1 over expression can reprogram ER binding (Hurtado et al., 2010). Moreover FOXA1 binding is different between cell types and highly correlates with cell type specific binding of ER (Eeckhoute et al., 2006; Hurtado et al., 2010). Concurrently during development, FOXA1 has been suggested to bind methylated DNA, promote DNA demethylation and increase H3K4 methylation illustrating that FOXA1 is able to reprogram the chromatin status (Serandour et al., 2011).
A recent comprehensive study has addressed transcription factor cooperation during drosophila development and found a staggering number of potential transcription factor interactions at accessible sites of the genome (Negre et al., 2011), illustrating the biological significance of multiple interaction between transcription factors at specific accessible sites of the genome. Thus the accessible genome to which steroid receptors bind is established during development and maintained by a variety of cell type specific transcription factors. Genome wide interaction with specific transcription factors determine where in the genome steroid receptors such as PR potentially will bind (figure 2B). Identification of these factors will be crucial in order to fully understand tissue specific transcriptional regulation by PR.
Impact of chromatin 3D structure
Imaging of chromosome structures (Cremer and Cremer, 2001) and chromatin conformation capture studies (Lieberman-Aiden et al., 2009) have shown that chromosome territories are spatially organized in the nucleus, allowing long range (hundreds of kilobases) intrachromosomal as well as interchromosomal interactions (Hakim et al., 2010). Gene rich regions tend to be more internally localized compared to gene poor regions (Hepperger et al., 2008) and transcriptionally active chromatin has been suggested to be localized in RNA polymerase “factories” (Osborne et al., 2004). Several components contribute to these subnuclear states, including structural proteins such as insulator binding factors (Splinter et al., 2006; Lanzuolo et al., 2007), cohesin (Hadjur et al., 2009) and lamin (Guelen et al., 2008). Transcriptional components such as transcription factors [including steroid receptors (Fullwood et al., 2009)], the mediator complex (Kagey et al., 2010) and polycomb (Lanzuolo et al., 2007) have also been implicated in the regulation of chromatin spatial organization, emphasizing that the transcription process is central for maintenance of chromatin conformational structures.
Long range chromatin interactions can be identified by the chromatin conformation capture (3C) assay which allows detection of interaction frequency between regions of interest (Simonis et al., 2007). Combined with micro arrays or second generation sequencing, all regions in the genome interacting with a specific region of interest (bait) can mapped (4C) or (5C) at relative high resolution (Simonis et al., 2007). Another approach termed ChlA-PET combines the conformation assay with ChIP to identify interaction frequency between regions that are bound by a specific transcription factor of interest (Fullwood et al., 2009).
To date, a limited number of studies have analyzed the impact of steroid receptor activation on chromosome conformation. One study, using ChlA-PET with ER found that ER bound chromatin is organized in hubs around promoters of estrogen responsive genes (Fullwood et al., 2009) through long range looping. Using the 3C method this study and others also suggested that some of these long range intra-chromosomal interactions are dependent on ER activation(Fullwood et al., 2009). Another study suggested that long distance interchromosomal interactions are actively created after ER stimulation (Hu et al., 2008); however these studies failed to be reproduced by others (Kocanova et al., 2010). The near term resolution of this controversy is important to the field (Belmont, 2010).
In agreement with the Kocanova et al. observations, GR activation does not lead to increased interaction frequency between long distance genomic regions (Hakim et al., 2009). Interestingly, long range interaction regions are highly correlated with chromatin accessible regions identified by DHS-seq. Studies using GR binding regions as bait (4C) suggest that accessible GR binding regions are clustered in chromatin territories facilitating GR recruitment (Hakim et al., 2011). Thus clustered accessible regions of chromatin provide a sub environment that attracts activated transcription factors (figure 2C). Binding within these regions increases recruitment of transcriptional co-regulators which change transcription of nearby genes.
Conclusions and perspectives
Decades of studies using model systems such as the MMTV promoter have provided profound knowledge on the mechanisms that control PR interaction with chromatin. Importantly PR interacts with chromatin in a highly dynamic manner that is dependent on chromatin remodeling, chaparones, the proteasome and occupancy of other transcription factors such as NF1. In its role as an inducer of chromatin remodeling, it was originally proposed that PR primarily functions as pioneering factor that facilitates the generation of accessible chromatin for auxiliary transcription factors to bind. However, cutting edge genomic based chromatin accessibility studies with the closely related steroid receptors GR and ER have reshaped our understanding of chromatin binding. It is now clear that the ability of steroid receptors to engage chromatin is determined to a great extent by preset accessible chromatin states. These states are highly cell specific, and determine to a great extent the tissue selectivity of receptor function. Future genomic studies of PR based model systems will be essential to explore the role of these mechanisms in PR function.
Finally, studies on the subnuclear architecture of chromosome elements in mammalian cells are rapidly moving to center stage. There are early indications that these organizational patterns contribute to the genomic action of steroid receptors, and it is likely that these mechanisms will be important for PR action as well.
Acknowledgments
L.G was supported by a research grant from the Lundbeck Foundation. This work was supported by the intramural Research Program of the NIH National Cancer Institute, Center for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Archer TK, Lefebvre P, Wolford RG, Hager GL. Transcription factor loading on the MMTV promoter: A bimodal mechanism for promoter activation. Science. 1992;255:1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- Bain DL, Heneghan AF, Connaghan-Jones KD, Miura MT. Nuclear receptor structure: implications for function. Annu Rev Physiol. 2007;69:201–220. doi: 10.1146/annurev.physiol.69.031905.160308. [DOI] [PubMed] [Google Scholar]
- Bartke J, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann CT, Maruvada P, Hager GL, Yen PM. Nuclear-cytoplasmic shuttling by thyroid hormone receptors: Multiple protein interactions are required for nuclear retention. JBiol Chem. 2001;276:11237–11245. doi: 10.1074/jbc.M011112200. [DOI] [PubMed] [Google Scholar]
- Belikov S, Astrand C, Wrange O. FoxA1 binding directs chromatin structure and the functional response of a glucocorticoid receptor-regulated promoter. Mol Cell Biol. 2009;29:5413–5425. doi: 10.1128/MCB.00368-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont AS. Estrogen fueled, nuclear kiss: Did it move for you? Nucleus. 2010;1:440–443. doi: 10.4161/nucl.1.5.13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, Farnham PJ, Hirst M, Lander ES, Mikkelsen TS, Thomson JA. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomquist P, Li Q, Wrange O. The affinity of nuclear factor 1 for its DNA site is drastically reduced by nucleosome organization irrespective of its rotational or translational position. J Biol Chem. 1996;271:153–159. doi: 10.1074/jbc.271.1.153. [DOI] [PubMed] [Google Scholar]
- Bosisio D, Marazzi I, Agresti A, Shimizu N, Bianchi ME, Natoli G. A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-kB-dependent gene activity. EMBO J. 2006;25:798–810. doi: 10.1038/sj.emboj.7600977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botos J, Xian W, Smith DF, Smith CL. Progesterone receptor deficient in chromatin binding has an altered cellular state. J Biol Chem. 2004;279:15231–15239. doi: 10.1074/jbc.M309718200. [DOI] [PubMed] [Google Scholar]
- Boyle AP, Song L, Lee BK, London D, Keefe D, Birney E, Iyer VR, Crawford GE, Furey TS. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 2010;21:456–464. doi: 10.1101/gr.112656.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnick EH, Bustin M, Marsaud V, Richard-Foy H, Hager GL. The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res. 1992;20:273–278. doi: 10.1093/nar/20.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M, Catez F, Lim JH. The dynamics of histone H1 function in chromatin. Mol Cell. 2005;17:617–620. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Campos E, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2008;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, Casero D, Bernal M, Huijser P, Clark AT, Kramer U, Merchant SS, Zhang X, Jacobsen SE, Pellegrini M. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordingley MG, Riegel AT, Hager GL. Steroid-dependent interaction of transcription factors with the inducible promoter of mouse mammary tumor virus in vivo. Cell. 1987;48:261–270. doi: 10.1016/0092-8674(87)90429-6. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin Dl and cell cycle progression in breast cancer. Genes Dev. 2006;20:2513–2526. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbi C, Walker DA, Romero G, Sullivan WP, Toft DO, Hager GL, DeFranco DB. Molecular chaperones function as steroid receptor nuclear mobility factors. Proc Natl Acad Sci USA. 2004;101:2876–2881. doi: 10.1073/pnas.0400116101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher TM, Ryu BW, Baumann CT, Warren BS, Fragoso G, John S, Hager GL. Structure and dynamic properties of the glucocorticoid receptor-induced chromatin transition at the MMTV promoter. Mol Cell Biol. 2000;20:6466–6475. doi: 10.1128/mcb.20.17.6466-6475.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher TM, Xiao N, Mautino G, Baumann CT, Wolford RG, Warren BS, Hager GL. ATP-dependent mobilization of the glucocorticoid receptor during chromatin remodeling. Mol Cell Biol. 2002;22:3255–3263. doi: 10.1128/MCB.22.10.3255-3263.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso G, John S, Roberts MS, Hager GL. Nucleosome positioning on the MMTV LTR results from the frequency-biased occupancy of multiple frames. Genes Dev. 1995;9:1933–1947. doi: 10.1101/gad.9.15.1933. [DOI] [PubMed] [Google Scholar]
- Fragoso G, Pennie WD, John S, Hager GL. The position and length of the steroid-dependent hypersensitive region in the mouse mammary tumor virus long terminal repeat are invariant despite multiple nucleosome B frames. Mol Cell Biol. 1998;18:3633–3644. doi: 10.1128/mcb.18.6.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman BC, Yamamoto KR. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science. 2002;296:2232–2235. doi: 10.1126/science.1073051. [DOI] [PubMed] [Google Scholar]
- Fryer CJ, Archer TK. Chromatin remodeling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EG, Huang PY, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KD, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, Karuturi RK, Herve T, Bourque G, Stunnenberg HG, Ruan X, Cacheux-Rataboul V, Sung WK, Liu ET, Wei CL, Cheung E, Ruan Y. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, van Steensel B. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager GL. Footprints by deep sequencing. Nat Methods. 2010;6:254–255. doi: 10.1038/nmeth0409-254. [DOI] [PubMed] [Google Scholar]
- Hager GL, Elbi C, Johnson JA, Voss JC, Nagaich AK, Schiltz RL, Qiu Y, John S. Chromatin dynamics and the evolution of alternate promoter states. Chromosome Res. 2006;14:107–116. doi: 10.1007/s10577-006-1030-0. [DOI] [PubMed] [Google Scholar]
- Hager GL, McNally JG, Misteli J. Transcription dynamics. Mol Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim O, John S, Ling JA, Biddie SC, Hoffman AR, Hager GL. Glucocorticoid receptor activation of the Cizl-Lcn2 locus by long range interactions. J Biol Chem. 2009;284:6048–6052. doi: 10.1074/jbc.C800212200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim O, Sung MH, Hager GL. 3D Shortcuts to Gene Regulation. Curr Opin Cell Biol. 2010;22:305–313. doi: 10.1016/j.ceb.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim O, Sung MH, Voss TC, John S, Splinter E, Sabo PJ, Thurman RE, Stamatoyannopoulos JA, de Laat W, Hager GL. Diverse gene reprogramming events occur in the same spatial clusters of distal regulatory elements. Genome Res. 2011;21:697–706. doi: 10.1101/gr.111153.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepperger C, Mannes A, Merz J, Peters J, Dietzel S. Three-dimensional positioning of genes in mouse cell nuclei. Chromosoma. 2008;117:535–551. doi: 10.1007/s00412-008-0168-2. [DOI] [PubMed] [Google Scholar]
- Hesselberth JR, Zhang Z, Sabo PJ, Chen X, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, Fields S, Stamatoyannopoulos JA. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htun H, Holth LT, Walker D, Davie JR, Hager GL. Direct visualization of ligand occupied and unoccupied human estrogen receptor alpha reveals a role for ligand in the nuclear distribution of the receptor. Mol Biol Cell. 1999;10:471–486. doi: 10.1091/mbc.10.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, Fu XD. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSDl-dependent gene networking in interchromatin granules. Proc Natl Acad Sci USA. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-lnnes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2010;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nature Reviews Genetics. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, Voss JC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, Hager GL. Interaction of the glucocorticoid receptor with the global chromatin landscape. Mol Cell. 2008;29:611–624. doi: 10.1016/j.molcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson JA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocanova S, Kerr EA, Rafique S, Boyle S, Katz E, Caze-Subra S, Bickmore WA, Bystricky K. Activation of estrogen-responsive genes does not require their nuclear co-localization. PLoS Genet. 2010;6:e1000922. doi: 10.1371/journal.pgen.1000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koop R, Di CL, Beato M. Histone H1 enhances synergistic activation of the MMTV promoter in chromatin. EMBO J. 2003;22:588–599. doi: 10.1093/emboj/cdg052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol. 2007;9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- Lee H-L, Archer JK. Nucleosome mediated disruption of transcription factor:chromatin initiation complexes at the mouse mammary tumour virus long terminal repeat in vivo. Mol Cell Biol. 1994;14:32–41. doi: 10.1128/mcb.14.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li Q, Wrange O. Accessibility of a glucocorticoid response element in a nucleosome depends on its rotational positioning. Mol Cell Biol. 1995;15:4375–4384. doi: 10.1128/mcb.15.8.4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy J, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond JJ. Crystal structure of the nucleosome core particle at 2.8 A resolution [see comments] Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally JG, Mueller WG, Walker D, Wolford RG, Hager GL. The glucocorticoid receptor: Rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Nagaich AK, Walker DA, Wolford RG, Hager GL. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol Cell. 2004;14:163–174. doi: 10.1016/s1097-2765(04)00178-9. [DOI] [PubMed] [Google Scholar]
- Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, Li Z, Ishii H, Spokony RF, Chen J, Hwang L, Cheng C, Auburn RP, Davis MB, Domanus M, Shah PK, Morrison CA, Zieba J, Suchy S, Senderowicz L, Victorsen A, Bild NA, Grundstad AJ, Hanley D, MacAlpine DM, Mannervik M, Venken K, Bellen H, White R, Gerstein M, Russell S, Grossman RL, Ren B, Posakony JW, Kellis M, White KP. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Perlmann T, Wrange O. Specific glucocorticoid receptor binding to DNA reconstituted in a nucleosome. EMBO J. 1988;7:3073–3079. doi: 10.1002/j.1460-2075.1988.tb03172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina B, Brüggemeier U, Beato M. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell. 1990;60:719–731. doi: 10.1016/0092-8674(90)90087-u. [DOI] [PubMed] [Google Scholar]
- Prufer K, Racz A, Lin GC, Barsony J. Dimerization with retinoid X receptors promotes nuclear localization and subnuclear targeting of vitamin D receptors. J Biol Chem. 2000;275:41114–41123. doi: 10.1074/jbc.M003791200. [DOI] [PubMed] [Google Scholar]
- Rayasam GV, Elbi C, Walker DA, Wolford RG, Fletcher JM, Edwards DP, Hager GL. Ligand specific dynamics of the progesterone receptor in living cells and during chromatin remodeling in vitro. Mol Cell Biol. 2005;25:2406–2418. doi: 10.1128/MCB.25.6.2406-2418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard-Foy H, Hager GL. Sequence specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987;6:2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishi V, Bhattacharya P, Chatterjee R, Rozenberg J, Zhao J, Glass K, Fitzgerald P, Vinson C. CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes. Proc Natl Acad Sci U S A. 2010;107:20311–20316. doi: 10.1073/pnas.1008688107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D. Epigenomics: Methylation matters. Nature. 2009;462:296–297. doi: 10.1038/462296a. [DOI] [PubMed] [Google Scholar]
- Serandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, Barloy-Hubler F, Brown M, Lupien M, Metivier R, Salbert G, Eeckhoute J. Epigenetic switch involved in activation of pioneer factor FOXAl-dependent enhancers. Genome Res. 2011 doi: 10.1101/gr.111534.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp ZD, Mancini MG, Hinojos CA, Dai F, Berno V, Szafran AT, Smith KP, Lele TT, Ingber DE, Mancini MA. Estrogen-receptor-alpha exchange and chromatin dynamics are ligand- and domain-dependent. J Cell Sci. 2006;119:4101–4116. doi: 10.1242/jcs.03161. [DOI] [PubMed] [Google Scholar]
- Siersbaek R, Nielsen R, John S, Sung MH, Baek S, Loft A, Hager GL, Mandrup S. Adipogenic development is associated with extensive early remodeling of the chromatin landscape and establishment of transcription factor ‘hotspots’. EMBO J. 2011;30:1459–1472. doi: 10.1038/emboj.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Kooren J, de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nat Methods. 2007;4:895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavreva DA, Muller WG, Hager GL, Smith CL, McNally JG. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol Cell Biol. 2004;24:2682–2697. doi: 10.1128/MCB.24.7.2682-2697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T. Green fluorescent protein. The green revolution. Curr Biol. 1995;5:262–264. doi: 10.1016/s0960-9822(95)00056-x. [DOI] [PubMed] [Google Scholar]
- Steger DJ, Grant GR, Schupp M, Tomaru T, Lefterova MI, Schug J, Manduchi E, Stoeckert CJ, Jr, Lazar MA. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev. 2010;24:1035–1044. doi: 10.1101/gad.1907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtil-Rodriguez A, Millan-Arino L, Quiles I, Ballare C, Beato M, Jordan A. Progesterone induction of the llbeta-hydroxysteroid dehydrogenase type 2 promoter in breast cancer cells involves coordinated recruitment of STAT5A and progesterone receptor to a distal enhancer and polymerase tracking. Mol Cell Biol. 2008;28:3830–3849. doi: 10.1128/MCB.01217-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LH, Pfendner EG, Senior MB, Frankel FR. Progesterone, glucocorticoid and estradiol receptors in MCF-7 cells bind to chromatin. Mol Cell Endocrinol. 1983;30:267–278. doi: 10.1016/0303-7207(83)90063-1. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. Histone variats - ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010 doi: 10.1038/nrm2861. (in press) [DOI] [PubMed] [Google Scholar]
- Tomura A, Goto K, Morinaga H, Nomura M, Okabe T, Yanase T, Takayanagi R, Nawata H. The subnuclear three-dimensional image analysis of androgen receptor fused to green fluorescence protein. J Biol Chem. 2001;276:28395–28401. doi: 10.1074/jbc.M101755200. [DOI] [PubMed] [Google Scholar]
- Venditti P, Di Croce L, Kauer M, Blank J, Becker PB, Beato M. Assembly of MMTV promoter minichromosomes with positioned nucleosomes precludes NF1 access but not restriction enzyme cleavage. Nucleic Acids Res. 1998;26:3657–3666. doi: 10.1093/nar/26.16.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicent GP, Ballare C, Nacht AS, Clausell J, Subtil-Rodriguez A, Quiles I, Jordan A, Beato M. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol Cell. 2006;24:367–381. doi: 10.1016/j.molcel.2006.10.011. [DOI] [PubMed] [Google Scholar]
- Vicent GP, Nacht AS, Smith CL, Peterson CL, Dimitrov S, Beato M. DNA instructed displacement of histones H2A and H2B at an inducible promoter. Mol Cell. 2004;16:439–452. doi: 10.1016/j.molcel.2004.10.025. [DOI] [PubMed] [Google Scholar]
- Vicent GP, Zaurin R, Nacht AS, Font-Mateu J, Le Dily F, Beato M. Nuclear factor 1 synergizes with progesterone receptor on the mouse mammary tumor virus promoter wrapped around a histone H3/H4 tetramer by facilitating access to the central hormone-responsive elements. J Biol Chem. 2010;285:2622–2631. doi: 10.1074/jbc.M109.060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicent GP, Zaurin R, Nacht AS, Li A, Font-Mateu J, Le Dily F, Vermeulen M, Mann M, Beato M. Two chromatin remodeling activities cooperate during activation of hormone responsive promoters. PLoS Genet. 2009;5:el000567. doi: 10.1371/journal.pgen.1000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss TC, John S, Hager GL. Single cell analysis of glucocorticoid receptor action reveals that stochastic post-chromatin association mechanisms regulate ligand-specific transcription. Mol Endocrinol. 2006;20:2641–2655. doi: 10.1210/me.2006-0091. [DOI] [PubMed] [Google Scholar]
- Voss TC, Schiltz RL, Sung MH, Johnson JA, John S, Hager GL. Combinatorial probabilistic chromatin interactions produce transcriptional heterogeneity. J Cell Sci. 2009;122:345–356. doi: 10.1242/jcs.035865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li W, Liu XS, Carroll JS, Janne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]