Abstract
The identification of the molecular events that drive cancer transformation is essential to the development of targeted agents that improve the clinical outcome of lung cancer. Many studies have reported genomic driver mutations in non-small-cell lung cancers (NSCLCs) over the past decade; however, the molecular pathogenesis of >40% of NSCLCs is still unknown. To identify new molecular targets in NSCLCs, we performed the combined analysis of massively parallel whole-genome and transcriptome sequencing for cancer and paired normal tissue of a 33-yr-old lung adenocarcinoma patient, who is a never-smoker and has no familial cancer history. The cancer showed no known driver mutation in EGFR or KRAS and no EML4-ALK fusion. Here we report a novel fusion gene between KIF5B and the RET proto-oncogene caused by a pericentric inversion of 10p11.22–q11.21. This fusion gene overexpresses chimeric RET receptor tyrosine kinase, which could spontaneously induce cellular transformation. We identified the KIF5B-RET fusion in two more cases out of 20 primary lung adenocarcinomas in the replication study. Our data demonstrate that a subset of NSCLCs could be caused by a fusion of KIF5B and RET, and suggest the chimeric oncogene as a promising molecular target for the personalized diagnosis and treatment of lung cancer.
Lung cancer remains a leading cause of mortality in cancer, with around 1.38 million deaths worldwide annually (Ferlay et al. 2010). With a conventional chemotherapeutic regimen, the median survival time for lung cancer patients in advanced stages is <1 yr from diagnosis (Schiller et al. 2002). Tobacco smoking is known to be a major risk factor of lung cancer in Western countries, where 85%–90% of all lung cancers were attributed to smoking (Toh et al. 2006). However, ∼25% of lung cancer patients worldwide are “never-smokers” (Lee et al. 2011). Data from many Asian countries have shown that never-smokers constitute 30%–40% of non-small-cell lung cancer (NSCLC). NSCLC accounts for ∼80% of lung cancer cases (Subramanian and Govindan 2007), and the dominant histological type is adenocarcinoma (>50%) (Pao and Girard 2011).
Lung cancer of never-smokers tends to be driven by single somatic mutation events, rather than global genetic and epigenetic changes (Lee et al. 2011). A subset of somatic mutations has been reported in NSCLCs in the past few years, such as EGFR, KRAS, and EML4-ALK genes (which are conventionally called the triple-markers) (Pao and Girard 2011). Mutations in the tyrosine kinase domain of EGFR, which are associated preferentially with NSCLCs of nonsmokers and Asians, are sensitive to EGFR-targeted therapy, such as gefitinib (Paez et al. 2004). Missense mutations in KRAS are common in the lung adenocarcinomas of smokers and induce resistance to EGFR inhibitors (Riely et al. 2008). More recently, the EML4-ALK fusion gene was identified in NSCLC (Soda et al. 2007), which is generated by inversion in chromosome 2. This fusion gene, formed by chromosomal rearrangement, is more frequently detected in the lung adenocarcinoma of young patients, regardless of ethnicity, with no or little history of cigarette smoking (Wong et al. 2009). ALK-positive lung cancer constitutes ∼5% of NSCLCs and is highly sensitive to ALK inhibitors, such as crizotinib (Pao and Girard 2011).
Although several genetic mutations have been reported previously, a large proportion of lung cancer patients have been observed to have none of them in their cancer genome. More than 40% of NSCLCs appear to be driven by unknown genetic events (Harris 2010; Pao and Girard 2011).
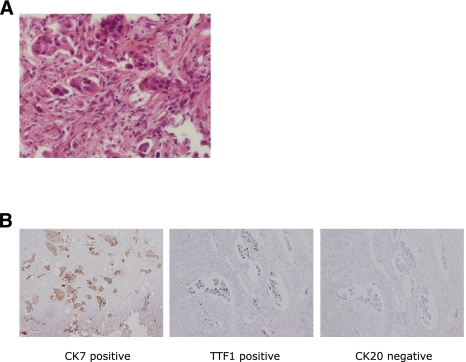
Here we report a novel fusion gene generated by a chromosomal inversion event in a young, never-smoker lung adenocarcinoma patient, whose cancer was negative for the triple-markers, using massively parallel DNA and RNA sequencing. The patient, known as AK55, was healthy until he was 33 yr of age, when a poorly differentiated adenocarcinoma developed in the right upper lobe of a lung (Fig. 1A). He had no known family history of cancers from his grandparents, and he was a never-smoker. Metastases in liver and multiple bones were also detected in positron emission tomography (PET) studies. For a pathological diagnosis, he underwent computed tomography (CT) guided biopsy of the primary lung cancer, as well as ultrasound-guided biopsy of the liver metastasis.
Figure 1.
Pathology of lung adenocarcinoma analyzed in this study. (A) A paraffin section stained by hematoxylin and eosin from a primary lung cancer tissue obtained by CT-guided biopsy (400×). In the cancer tissue, poorly differentiated tumor cell nests were present in the desmoplastic stroma. In addition, the cancer cells had plump cytoplasm and large pleomorphic nuclei. (B) Immunohistochemical analyses of the cancer (from metastatic tumor in the cervical spine). From left to right, CK7 (positive), TTF1 (positive), and CK20 (negative). The results highly suggest that the origin of this cancer is lung adenocarcinoma.
The immunohistochemical analyses, such as CK7, CK20, and TTF1, were consistent with lung adenocarcinoma (Fig. 1B; positive for CK7 and TTF1, negative for CK20). In pathologic studies, his lung adenocarcinoma was negative for known EGFR, KRAS, and ALK mutations (EML4-ALK fusion gene). The specimen from AK55 was referred to the Genomic Medicine Institute at Seoul National University (GMI-SNU) for the identification of the driver mutations of the cancer by high-throughput analysis of whole-genome and transcriptome sequencing.
Results
Whole-genome analysis
From whole-genome deep sequencing of liver metastatic lung cancer tissue and normal tissue (blood) of AK55, we obtained 47.77× and 28.27× average read-depth, respectively (Table 1). The whole-genome coverage of the liver metastatic lung cancer tissue was evenly distributed (excepting normal “spikes” [Kim et al. 2009] of repetitive sequences in the centromeric or telomeric regions), suggesting no evidence of aneuploidy in the cancer tissue (Fig. 2A). The bimodal distribution of read-allele frequency of single nucleotide variants (SNVs) on 0.5 and 1.0 also supports the euploidy of the genome of liver metastasis (Supplemental Fig. 1). The whole-genome sequence of blood DNA demonstrated that AK55 did not have any remarkable cancer-related SNVs archived in OMIM (Online Mendelian Inheritance in Man) and SNPedia (http://www.snpedia.org), suggesting his lung cancer was unlikely to be driven by germline mutations. We identified 10,390 nonsynonymous SNVs, 334 coding sequence (CDS) indels (insertions and deletions), and 70 candidates of large deletion on CDS from the whole-genome sequences of liver metastasis (Supplemental Tables 1–3). Comparison of the whole-genome sequences between liver metastatic cancer and normal tissue identified 10 nonsynonymous somatic mutations (eight SNVs and two indels) (Supplemental Tables 1–3; Supplemental Fig. 2). These 10 somatic mutations did not occur in genes with known driver mutations, such as EGFR, KRAS, BRAF, PIK3CA, AKT1, MAP2K1, and MET (Pao and Girard 2011). Given the known functions of the genes affected by the somatic mutations and functional annotation of the eight SNVs using the SIFT algorithm (Kumar et al. 2009), those somatic mutations are not thought to have a significant impact on lung cancer transformation. The somatic mutations may be present in the primary lung cancer or may have occurred during metastasis. In any case, we suggest they are unlikely to be driver mutations.
Table 1.
Summary statistics of sequencing analysis of the lung cancer patient AK55
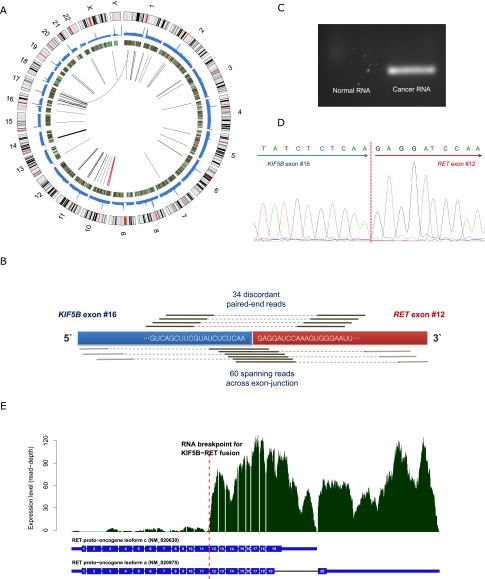
Figure 2.
Discovery of novel transforming KIF5B-RET fusion gene in lung adenocarcinoma. (A) Graphical representation of whole-genome and transcriptome sequencing data from the liver metastatic lung cancer tissue. Chromosome ideograms are shown in the outer layer. Coverage of cancer whole-genome sequencing is shown in the first middle layer. Expression level of genes is shown in the second middle layer using heatmap. Intra- and interchromosomal fusion genes are shown in the central layer. The thickness of lines shows the amount of evidence (number of spanning reads). The KIF5B-RET fusion gene is shown in red. (B) Detection of KIF5B-RET fusion gene from transcriptome sequencing. We identified 34 “discordant paired-end reads” and 60 “spanning reads” across the exon-junction. A discordant paired-end read is defined as a read whose end-sequences are aligned to each of the fusion partner genes. A spanning read is a read, one of whose end-sequences is aligned across the junction of the predicted fusion transcript. In this analysis, the fusion occurred between the 16th exon of KIF5B and 12th exon of RET. (C) Validation of KIF5B-RET fusion transcript in RNA (cDNA) from liver metastatic cancer tissue by PCR amplification and electrophoresis. The fusion gene is only detected in the liver metastatic lung cancer tissue of AK55. The negative control cDNA (normal) were extracted from the blood of a healthy Korean individual (AK1) (Kim et al. 2009). (D) Validation of the fusion gene breakpoint using Sanger sequencing in cDNA. (E) RNA expression level of each RET exon. Active expression is observed from the 12th exon, downstream from the junction of the predicted KIF5B-RET fusion gene. This suggests that the RET oncogene is expressed exclusively from the fusion gene, rather than the natural RET gene.
By comparison of DNA and RNA sequences, we examined A-to-I RNA editing in the liver metastatic cancer tissues (Supplemental Table 4; Shah et al. 2009; Ju et al. 2011). We found 10 RNA editing candidate sites in total; however, their functional impacts were not sufficient to be fundamental driver mutations of cancer.
Fusion gene analysis
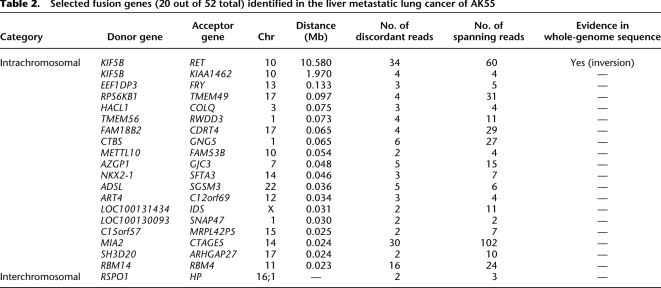
Next, we analyzed transcriptome sequencing data from the liver metastatic lung cancer. We focused on detecting fusion genes since not only hematologic but also solid cancers are known to be driven by fusion genes resulting from pathogenic chromosomal translocation or inversion (Tomlins et al. 2005; Wong et al. 2009; Tao et al. 2011; Welch et al. 2011). Our approaches identified 52 fusion genes (Fig. 2A; Table 2; Supplemental Table 5; Supplemental Methods). Of these, 94.2% (n = 49) were intrachromosomal fusions between adjacent genes (<135 kb), which may not have any functional roles in oncogenesis (Table 2; Nacu et al. 2011). In addition, one (1.9%) was an interchromosomal fusion, but this was generated by haptoglobin (HP), which is highly expressed in liver. Although the existence of this fusion gene is interesting biologically, given the molecular function of the gene, it is not believed to have an impact on cellular transformation. The remaining two (3.8%) were KIF5B-RET and KIAA1462-KIF5B fusion genes, which were intrachromosomal fusions between remote genes (more than ∼2 Mb). Of these, KIAA1462-KIF5B was excluded, since its expression level is low and KIAA1462 is a hypothetical protein of which the molecular function is not known. Out of the 52 fusion genes, we could detect a corresponding chromosomal rearrangement (e.g., large deletion, inversion, or translocation) only in KIF5B-RET from the whole-genome sequence of the liver metastatic lung cancer tissue (described later). To our knowledge, this fusion gene has not been reported in human cancer previously.
Table 2.
Selected fusion genes (20 out of 52 total) identified in the liver metastatic lung cancer of AK55
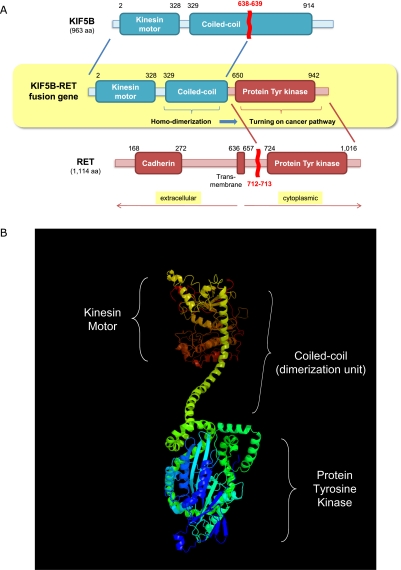
The final gene fusion, KIF5B-RET, is interesting in particular since RET is a well-known tyrosine-kinase proto-oncogene (Takahashi et al. 1985). The name of RET originated from “REarranged during Transfection,” as the DNA sequence of this gene was discovered rearranged when discovered. Although RET is essential for the development of the enteric nervous system and the kidney in embryogenesis (Durbec et al. 1996), its expression level in normal lung tissue is generally very low (Su et al. 2002). However, chimeric forms of the RET oncogene with many kinds of partner genes are well known as driver mutations in papillary thyroid carcinoma (PTC) (Alberti et al. 2003). The molecular activation mechanism of chimeric RET tyrosine kinase with dimerization units of diverse partner genes is well understood in PTC. The dimerization unit stimulates autophosphorylation of the tyrosine kinase unit in the chimeric oncogene (Alberti et al. 2003). Interestingly, KIF5B, the fusion partner gene of RET in the cancer tissue of AK55, contains a dimerization unit (coiled-coil domain) which induces homo-dimerization (Score et al. 2006; Takeuchi et al. 2009; Daire and Pous 2011).
Therefore, we further confirmed the characteristics of this gene fusion event using the transcriptome sequencing data. The fusion transcript was highly expressed, as evidenced by 34 discordant paired-end reads and 60 spanning reads across the fusion-junction (Table 2; Fig. 2B). These data showed that the end of the 16th exon of KIF5B and the start of the 12th exon of the RET proto-oncogene (which is a rearrangement hotspot in RET fusion gene in PTC) (Alberti et al. 2003) were integrated. The fusion transcripts were validated using PCR amplification and Sanger sequencing of cDNA from the liver metastatic lung cancer tissue (Fig. 2C,D). In addition, the expression profile of RET showed that the exons after fusion breakpoints (from 12th to 20th exon) are exclusively expressed (Fig. 2E) in the liver metastatic lung cancer tissue, suggesting most of the RET expression in the cancer took place from the fusion gene rather than from the natural RET gene. The expression levels of these exons (exons 12–20) are ∼10 times higher than those of lung cancers without RET rearrangement (Supplemental Table 6). In addition, given the genetic sequence, the fusion protein would contain both a dimerization unit (coiled-coil domain of KIF5B) and a tyrosine kinase unit (from RET) (Fig. 3A,B).
Figure 3.
Molecular characteristics of KIF5B-RET fusion kinase. (A) Functional domains of KIF5B-RET fusion kinase. The fusion kinase consists of 638 N-terminal residues of KIF5B and 402 C-terminal residues of RET kinase. As a result, the fusion protein consists of a protein kinase domain together with a coiled-coil domain. The coiled-coil domain induces dimerization of the fusion kinase, which activates the oncogenic protein tyrosine kinase domain by autophosphorylation. (B) The three-dimensional structure of the KIF5B-RET chimeric oncogene, as predicted by the PHYRE2 algorithm (Kelley and Sternberg 2009). The N- and C-terminal of the fusion protein are colored in red and blue, respectively.
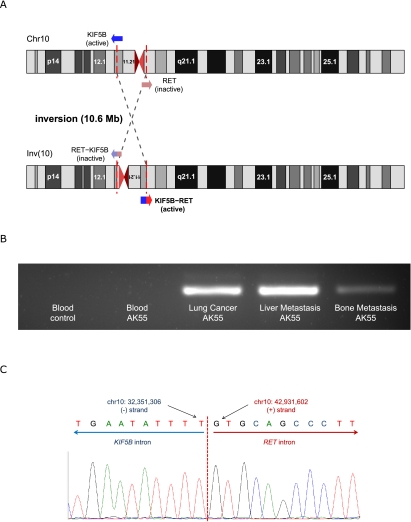
KIF5B and RET are 10.6 Mb away from each other, located at 10p11.22 and 10q11.21, respectively. Because the coding strands for the two genes are different, a 10.6-Mb-long inversion event is necessary for generating the fusion gene (Fig. 4A). We confirmed this genomic inversion event in the liver metastatic lung cancer tissue by detecting highly confident reads supporting the inversion (eight reads). By considering the count of the reads supporting normal chromosome structure (six reads), we conclude that the KIF5B-RET fusion was present in the major subpopulation of liver metastatic lung cancer tissue (Supplemental Methods). In blood, however, there was no corresponding chromosomal rearrangement in the whole-genome sequencing. We found a single read, suggesting the inversion in the whole-genome sequence from primary lung cancer, which alone is not sufficient to confirm the origin of this inversion. Hence, we validated the chromosomal inversion using PCR amplification and Sanger sequencing. DNA samples from three cancer tissues of AK55 (primary lung cancer, bone, and liver metastatic lung cancer tissues), but not normal blood, showed PCR products resulting from the inversion event (Fig. 4B). This confirms that the fusion gene also exists in the primary lung cancer as well as in bone metastatic cancer tissue. Sanger sequencing identified the breakpoints of the inversion with nucleotide resolution (chr10: 32,351,306–42,931,601) (Fig. 4C). Interestingly, a single-base-pair deletion was identified 2 bp adjacent to the breakpoint (chr10: 42,931,604), suggesting an error-prone DNA repair mechanism, such as nonhomologous end joining (NHEJ), fork stalling and template switching (FoSTeS), or microhomology-mediated break-induced replication (MMBIR), might have contributed to this inversion event after double-strand DNA breaks (Hastings et al. 2009; Zhang et al. 2009; Kee and D'Andrea 2010). Furthermore, the G-quadruplex (a non-B DNA) structure is predicted in the ∼100 bp upstream of the rearrangement hotspot of RET, which is known to be fragile and a source of chromosomal rearrangements (Nambiar et al. 2011).
Figure 4.
A chromosomal rearrangement for generating KIF5B-RET fusion in the lung cancer tissue of AK55. (A) Detection of a 10.6-Mb-long inversion event in chromosome 10 from the whole-genome sequencing of the liver metastatic lung cancer. KIF5B is generally expressed with its universal promoter. By the inversion event, this promoter activates global expression of the KIF5B-RET fusion gene. (B) Validation of the KIF5B-RET fusion gene DNA by inversion-specific PCR amplification and electrophoresis. The fusion gene is only detected in the cancer tissues of AK55 (primary lung cancer, liver and bone metastatic lung cancer tissues). The negative control DNA sample was extracted from a healthy Korean individual (AK1) (Kim et al. 2009). (C) Identification of the fusion gene and inversion breakpoint using Sanger sequencing. The inversion breakpoints were located in the introns of KIF5B and RET as predicted. Two bases downstream from the breakpoint (chr10: 42,931,604, hg18), a 1-bp deletion was identified, suggesting error-prone DNA repair mechanisms might contribute to this inversion event after double-strand DNA breaks.
Recurrence of KIF5B-RET in primary lung adenocarcinomas
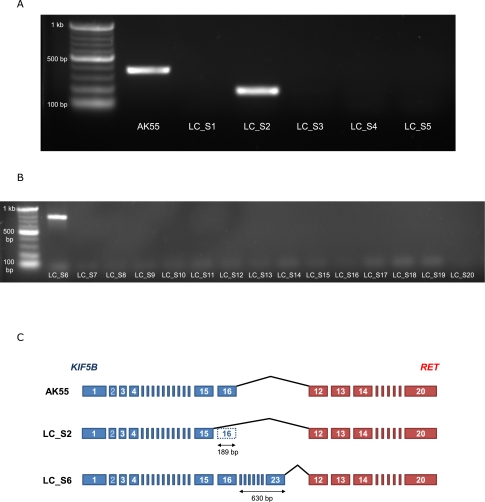
In order to show that the KIF5B-RET fusion gene also exists in other primary lung adenocarcinomas, we analyzed transcriptomes of five additional triple-negative (EGFR, KRAS, and EML4-ALK) primary lung adenocarcinomas using massively parallel sequencing (here we call them LC_S1–LC_S5) (Supplemental Table 7). KIF5B-RET fusion transcripts were found in LC_S2. As in AK55, RET was highly expressed from 12th exon (Supplemental Table 6). Because KIF5B is generally expressed in differentiated tissue (Su et al. 2002), the KIF5B-RET fusion gene could be expressed by the active promoter of KIF5B in those lung cancer tissues (AK55 and LC_S2). We validated this fusion transcript in LC_S2 using cDNA PCR (Fig. 5A).
Figure 5.
Replication studies of the KIF5B-RET fusion gene in an additional five triple-negative lung adenocarcinomas. (A) cDNA PCR targeting KIF5B-RET fusion transcripts and gel electrophoresis in the liver metastatic lung cancer of AK55 and five additional triple-negative lung adenocarcinomas. cDNA from AK55 and LC_S2 shows clear evidence of the fusion transcript. Because the fusion transcript in AK55 contains one more exon of KIF5B (exon 16) compared with that in LC_S2 (exon 15), the size of the PCR product in AK55 is longer than that in LC_S2. (B) cDNA PCR targeting KIF5B-RET fusion transcripts and gel electrophoresis in 15 double-negative lung adenocarcinomas. LC_S6 shows clear evidence of the fusion transcript. The fusion transcript in LC_S6 contains seven more exons of KIF5B (exons 17–23) compared with that in AK55. (C) Comparison of schematic KIF5B-RET fusion transcripts between AK55, LC_S2, and LC_S6. Each rectangle indicates an exon of KIF5B (blue) and RET (red) gene.
In addition, we further assessed the KIF5B-RET fusion gene using cDNA PCR of 15 more double-negative (EGFR and EML4-ALK were negative in pathologic studies; KRAS mutation status was unknown) primary lung adenocarcinomas (LC_S6–LC_S20). LC_S6 showed the KIF5B-RET fusion gene (Fig. 5B). The breakpoint of the fusion gene in LC_S6 was identified using Sanger sequencing (Supplemental Fig. 3). Overall, we identified two cases of the KIF5B-RET fusion gene (LC_S2 and LC_S6) in 20 primary lung adenocarcinomas in the replication study. These results clearly show that KIF5B-RET fusion is not rare and that the fusion transcript exists in the primary lung adenocarcinomas. In addition, because it would be very unlikely to find identical nonfunctional fusion genes in different cancer tissues, these results also provide indirect evidence that the expression of the KIF5B-RET fusion gene has an important functional impact in lung cancer.
Interestingly in LC_S2 and LC_S6, exon 12 of RET was joined to exon 15 (LC_S2) and exon 23 (LC_S6) instead of to exon 16 of KIF5B as in AK55 (Fig. 5C). These suggest that the double-strand breaks of DNA in KIF5B may not be consistent among primary lung cancers. However, because their coiled-coil domains are well preserved in the KIF5B-RET chimeric oncogene in both the samples (the length of coiled-coil domain in the fusion gene was 247 and 520 amino acids in LC_S2 and LC_S6, respectively), the dimerization activity is probably not very different compared with that of AK55 (310 amino acids).
Discussion
AK55 was referred for genetic study because of atypical features of his lung cancer: never-smoker, young age, multiple metastases, no family history, and no known somatic mutations in the conventional cancer marker tests in the clinic. Massively parallel DNA and RNA sequencing of the cancer and normal tissues successfully identified the genetic cause of his lung adenocarcinoma. As sequencing technologies develop, whole-genome and transcriptome sequencing can be achieved within the time necessary for making therapeutic decisions. In addition, the cost of sequencing has been reduced dramatically in recent years. Now, human whole-genome deep sequencing analysis together with transcriptome analysis can be completed for under $10,000 within 3 wk from the tissue sampling. Given the fact that each cancer is transformed by its unique mutation events and this information is essential for medical therapeutic decisions, it is clear that cancer genome and transcriptome sequencing should be performed as a crucial cancer diagnostic procedure in the near future. This will especially benefit those patients whose cancer does not have any known cancer driver mutations.
Integration of genome and transcriptome sequencing has several remarkable advantages for identifying somatic cancer mutation. First, cross-checking of the results between DNA and RNA sequencing enables the extensive removal of false positives. Because the human genome consists of ∼3 Gb, whole-genome sequencing generally includes thousands of false-positive calls even though its accuracy is >99.9999%. Second, transcriptome sequencing allows us to concentrate on specific genomic variants that are related to the genes under active transcription in the cancer. Cancer genomes may include hundreds of somatic passenger mutations. To isolate important driver mutations among them, we can use the information of expression levels in the cancer. Third, integration of DNA and RNA sequencing provides an opportunity to find variations generated during the gene transcription process, such as RNA editing. Lastly, transcriptome sequencing can provide information of fusion genes generated by chromosomal inversion or translocation more easily than whole-genome sequencing. Ab initio detection of chromosomal rearrangements, such as translocation and inversion, is still challenging in whole-genome sequencing alone without specific targets. These difficulties can clearly be overcome by combining whole-genome and transcriptome sequencing.
The oncogenic effect of RET was first identified in PTC, where diverse kinds of chromosomal translocations and inversions led to the formation of PTC/RET fusion genes (Alberti et al. 2003). Specific point mutations have also been reported as drivers in multiple endocrine neoplasia (MEN) types 2A and 2B (Alberti et al. 2003). In addition, activated RET has been observed in prostate cancer (Dawson et al. 1998), pancreatic cancer (Zeng et al. 2008), and melanoma (Ohshima et al. 2010). The direct transforming impact of RET as a driver is also supported by RET transgenic mice studies, which generated a variety of malignancies (Portella et al. 1996; Kawai et al. 2000). However, this gene has not been highlighted in lung cancer previously.
By using the integrated sequencing technologies, we demonstrated a novel KIF5B-RET fusion gene in a lung adenocarcinoma for the first time. Although we found the fusion gene in a small number of samples, the evidence for its transforming role is convincing for the following reasons: (1) The fusion stands out based on direct data analysis from lung cancers not containing any known driver mutation; (2) RET is an established oncogene in other tumors; (3) the tyrosine kinase domain of RET is highly expressed exclusively in lung cancer samples containing the fusion gene; and (4) the partner gene (KIF5B) has the coiled-coil domain, which is a well-known dimerization unit and is necessary for activation of the fusion oncogene. Given the frequencies of known somatic mutations in lung adenocarcinoma, e.g., EGFR (∼15%), KRAS (∼25%) mutations, the EML4-ALK fusion gene (∼5%), and others (∼15%) (Harris 2010; Pao and Girard 2011), a considerable number of unexplored mutations still remain to be identified (∼40% of lung adenocarcinoma). Because we found two KIF5B-RET fusions out of 20 primary lung adenocarcinomas (one of the five triple-negative [EGFR, KRAS and EML4-ALK] and one of the 15 double-negative [EGFR and EML4-ALK]), we may estimate that the frequency of the fusion gene would be ∼6% in lung adenocarcinoma (4.3%–8%; 1/5 × 40% = 8% from triple-negative cancers; 1/15 × 65% = 4.3% from double negative cancers). Interestingly, The Cancer Genome Atlas (TCGA) data set showed RET overexpression in three out of 32 samples (9.4%) (Supplemental Fig. 4). Because the sample size of our study is small (three cases of the KIF5B-RET fusion gene in a total of 21 samples) and there may be additional molecular mechanisms for RET overexpression (in TCGA data set), such as negative mutants in the regulating domains of RET, genomic amplification, and RET fusion with other partner genes, further epidemiological studies are highly necessary to understand the frequency of the KIF5B-RET fusion with more accuracy.
In summary, we reported a novel KIF5B-RET fusion oncogene as a driver mutation of lung adenocarcinoma. This fusion gene may be a good therapeutic molecular target for treatments of lung cancer. Developments of specific agents targeting KIF5B-RET will provide more advanced therapeutic strategies for lung adenocarcinoma.
Methods
All protocols of this study were approved by the Institutional Review Board of Seoul St. Mary's Hospital (approval no. KC11OISI0603). In pathologic studies in the hospital (Seoul National University Hospital), mutations of EGFR and KRAS and of the EML4-ALK fusion gene were examined from primary lung adenocarcinoma tissues. The results were all negative. Regarding EGFR and KRAS, nucleotide variations of exon 18–21 (EGFR) and exon 2 (KRAS) were studied using PCR and DNA sequencing as previously reported (Lynch et al. 2004; Eberhard et al. 2005). For EML4-ALK, fluorescence in situ hybridization study was performed as previously reported (Kwak et al. 2010).
We obtained paraffin-embedded tissues from primary lung cancer and bone metastasis. A frozen tissue from biopsy of liver metastasis was also available to use. In addition, we extracted venous blood from AK55. Genomic DNA was extracted from the lung cancer, bone metastasis, liver metastasis, and blood. Furthermore, we extracted RNA from the frozen liver metastasis. Then cDNA was synthesized from total RNA as described previously (Ju et al. 2011). Sequencing libraries were generated according to the standard protocol of Illumina Inc. for high-throughput sequencing. Excluding the genomic DNA from the paraffin-embedded bone metastasis (the DNA concentration of which was too low for it to qualify under our criteria for generating the sequencing library), samples were sequenced using Illumina HiSeq2000 and Genome Analyzer IIx (Table 1). Because the DNA of the primary lung cancer was extracted from a small amount of DNA in the paraffin-embedded tissue, the short-read redundancy was too high for analysis. Hence, our primary comparisons were done between the sequences from the liver metastasis and blood. The sequencing experiments were performed using the standard methods of Illumina and our previous reports (Kim et al. 2009; Ju et al. 2011).
For the replication study, we obtained five more frozen primary lung adenocarcinoma tissues (LC_S1–LC_S5), which were determined to be EGFR-, KRAS-, and EML4-ALK–negative in the pathologic studies mentioned above. We sequenced the transcriptome (cDNA from whole mRNA) using Illumina Hiseq2000 as described above. In addition, we also obtained 15 frozen primary lung adenocarcinoma tissues (LC_S6–LC_S20) from patients negative for EGFR and EML4-ALK but whose KRAS status was unknown (these 15 samples were assessed using PCR and Sanger sequencing as described below).
Short reads were aligned to the NCBI reference human genome assembly (build 36.3, hg18) using the GSNAP (Wu and Nacu 2010) alignment program, with allowance for 5% mismatches. We called genomic variants of each sample, e.g., SNVs, short indels, and large deletions, using modified criteria from our previous publications in whole-genome sequencing (Kim et al. 2009; Ju et al. 2011). Briefly, SNVs and indels were defined based on satisfaction of the following three conditions: (1) The number of uniquely mapped reads at the position should be two or more; (2) the average base quality (phred Q score) for the position should be eho; and (3) the read-allele frequency at the position should be eoul. For detection of the somatic mutations (SNVs and indels) in the liver metastatic lung cancer tissues, we used the following conditions: (1) nonsynonymous SNVs or indels in the liver metastatic lung cancer tissues; (2) the SNV allele count should be zero in whole-genome sequence of blood; (3) the wild-type allele count should be 10 or more in the whole-genome sequence of blood; and (4) the candidate positions should not be detected in Korean genomes (Ju et al. 2011) and in the 1000 Genomes Project (Durbin et al. 2010). Large deletion candidates on CDS were identified using modified criteria from our previous publications in whole-genome sequencing (Ju et al. 2011). Briefly, (1) there should be two or more long-insert paired-end reads; (2) the sequencing read-depth of the large deletion candidate region should be <90% of that of its flanking regions; (3) the size of large deletions should be ≥200 bp. To detect large deletions caused by somatic mutation, we visually compared the read-depth between whole-genome sequence of the liver metastatic lung cancer and the blood tissue (Supplemental Figure 2). For identifying RNA editing in liver metastatic lung cancer tissue, we used more conservative criteria than those used previously (Ju et al. 2011), since we have only a pair of DNA and RNA sequencing data of lung cancer (liver metastasis) and, hence, comparison of many samples was impossible. Our criteria were (1) clear nonsynonymous A-to-G SNVs (in coding strand) in transcriptome sequencing with ≥10 high-quality (phred Q score ≥25) mismatches, read-allele frequency ≥20%; (2) clear wild type in genome sequencing (count for SNVs allele = 0, count for wild type ≥10; (3) not located on repetitive genomic regions.
RNA sequencing data were also analyzed as described previously (Ju et al. 2011). By using the GSNAP alignment tool (Wu and Nacu 2010), we aligned short reads from transcriptome sequencing to a set of constructed mRNA sequences instead of the reference human genome to avoid mapping errors resulting from mRNA splicing (Ju et al. 2011). We allowed 5% mismatches for the alignment. The expression profiles were calculated using the RPKM unit as described (Mortazavi et al. 2008). For SNV detection in transcriptome sequencing (for RNA editing), 15 bp of both ends of a read were trimmed and not used, since end sequences are easily misaligned (mostly due to alternative splicing of transcripts).
For detection of fusion genes using transcriptome sequencing, we used discordant read pairs, where the reads of a pair were aligned to different genes, and exon-spanning reads across the fusion breakpoint of chimeric transcripts. Because there can be a lot of false positives in the massively parallel sequencing reads, we further applied three filter criteria as follows: (1) the homology filter, (2) the fusion-spanning read filter, and (3) the fusion point filter. We described the details of methods for identifying fusion genes in the Supplemental Methods. Regarding the final fusion gene candidates, we assessed corresponding genomic rearrangements, such as inversions, translocations, and large deletions in the whole-genome sequencing data.
We validated our findings using PCR amplification and Sanger sequencing of genomic DNA and cDNA. The PCR reactions were 10 min at 95°C; 30 cycles of 30 sec at 95°C, 10 sec at 62°C, and 10 sec at 72°C; and, finally, 10 min at 72°C. PCR and Sanger sequencing primers for genomic inversion of AK55 were 5′-CAGAATTTCACAAGGAGGGAAG-3′ (KIF5B) and 5′-CAGGACCTCTGACTACAGTGGA-3′ (RET). The primers for the fusion transcripts were 5′-GTGAAACGTTGCAAGCAGTTAG-3′ (KIF5B) and 5′-CCTTGACCACTTTTCCAAATTC-3′ (RET). For cDNA PCR in replication studies, we used different KIF5B primers (5′-TAAGGAAATGACCAACCACCAG-3′) since the KIF5B fusion breakpoint in LC_S2 was different to that in AK55. All the Sanger sequencing experiments were performed at Macrogen Inc. (http://www.macrogen.com).
Data access
Sequencing reads are uploaded in EBI-SRA under accession number ERP001071 (whole-genome sequencing; http://www.ebi.ac.uk/ena/data/view/ERP001071) and ERP001058 (transcriptome sequencing; http://www.ebi.ac.uk/ena/data/view/ERP001058). The short-read data are also available from our website (ftp://ftp.gmi.ac.kr/asianGenome/AK55/, http://tiara.gmi.ac.kr/). The program developed and used for detecting fusion genes in this study (GFP; Gene Fusion Program) is also available from ftp://ftp.gmi.ac.kr/pub/GFP/.
Acknowledgments
We thank the lung cancer patients who participated in this study and provided their cancer tissues for medical research. We thank the scientists and medical doctors who contributed to this work but were not included in the author list. We thank The Cancer Genome Atlas (TCGA) Project Team and their specimen donors for providing expression profile data. This work has been supported by Macrogen Inc. (MG2011009).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.133645.111.
References
- Alberti L, Carniti C, Miranda C, Roccato E, Pierotti MA 2003. RET and NTRK1 proto-oncogenes in human diseases. J Cell Physiol 195: 168–186 [DOI] [PubMed] [Google Scholar]
- Daire V, Pous C 2011. Kinesins and protein kinases: key players in the regulation of microtubule dynamics and organization. Arch Biochem Biophys 510: 83–92 [DOI] [PubMed] [Google Scholar]
- Dawson DM, Lawrence EG, MacLennan GT, Amini SB, Kung HJ, Robinson D, Resnick MI, Kursh ED, Pretlow TP, Pretlow TG 1998. Altered expression of RET proto-oncogene product in prostatic intraepithelial neoplasia and prostate cancer. J Natl Cancer Inst 90: 519–523 [DOI] [PubMed] [Google Scholar]
- Durbec P, Marcos-Gutierrez CV, Kilkenny C, Grigoriou M, Wartiowaara K, Suvanto P, Smith D, Ponder B, Costantini F, Saarma M, et al. 1996. GDNF signalling through the Ret receptor tyrosine kinase. Nature 381: 789–793 [DOI] [PubMed] [Google Scholar]
- Durbin R, Altshuler D, Abecasis G, Bentley D, Chakravarti A, Clark A, Collins FS, De La Vaga F, Donnelly P, Egholm M, et al. 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS, Ince WL, Janne PA, Januario T, Johnson DH, et al. 2005. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol 23: 5900–5909 [DOI] [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM 2010. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127: 2893–2917 [DOI] [PubMed] [Google Scholar]
- Harris T 2010. Does large scale DNA sequencing of patient and tumor DNA yet provide clinically actionable information? Discov Med 10: 144–150 [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR 2009. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 5: e1000327 doi: 10.1371/journal.pgen.1000327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju YS, Kim JI, Kim S, Hong D, Park H, Shin JY, Lee S, Lee WC, Yu SB, Park SS, et al. 2011. Extensive genomic and transcriptional diversity identified through massively parallel DNA and RNA sequencing of eighteen Korean individuals. Nat Genet 43: 745–752 [DOI] [PubMed] [Google Scholar]
- Kawai K, Iwashita T, Murakami H, Hiraiwa N, Yoshiki A, Kusakabe M, Ono K, Iida K, Nakayama A, Takahashi M 2000. Tissue-specific carcinogenesis in transgenic mice expressing the RET proto-oncogene with a multiple endocrine neoplasia type 2A mutation. Cancer Res 60: 5254–5260 [PubMed] [Google Scholar]
- Kee Y, D'Andrea AD 2010. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev 24: 1680–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ 2009. Protein structure prediction on the web: a case study using the Phyre server. Nat Protoc 4: 363–371 [DOI] [PubMed] [Google Scholar]
- Kim JI, Ju YS, Park H, Kim S, Lee S, Yi JH, Mudge J, Miller NA, Hong D, Bell CJ, et al. 2009. A highly annotated whole-genome sequence of a Korean individual. Nature 460: 1011–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4: 1073–1081 [DOI] [PubMed] [Google Scholar]
- Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Janne PA, Costa DB, et al. 2010. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 363: 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Kim JH, Kim SK, Ha SJ, Mok TS, Mitsudomi T, Cho BC 2011. Lung cancer in never smokers: change of a mindset in the molecular era. Lung Cancer 72: 9–15 [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. 2004. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350: 2129–2139 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nacu S, Yuan W, Kan Z, Bhatt D, Rivers CS, Stinson J, Peters BA, Modrusan Z, Jung K, Seshagiri S, et al. 2011. Deep RNA sequencing analysis of readthrough gene fusions in human prostate adenocarcinoma and reference samples. BMC Med Genomics 4: 11 doi: 10.1186/1755-8794-4-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar M, Goldsmith G, Moorthy BT, Lieber MR, Joshi MV, Choudhary B, Hosur RV, Raghavan SC 2011. Formation of a G-quadruplex at the BCL2 major breakpoint region of the t(14;18) translocation in follicular lymphoma. Nucleic Acids Res 39: 936–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima Y, Yajima I, Takeda K, Iida M, Kumasaka M, Matsumoto Y, Kato M 2010. c-RET molecule in malignant melanoma from oncogenic RET-carrying transgenic mice and human cell lines. PLoS ONE 5: e10279 doi: 10.1371/journal.pone.0010279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. 2004. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304: 1497–1500 [DOI] [PubMed] [Google Scholar]
- Pao W, Girard N 2011. New driver mutations in non-small-cell lung cancer. Lancet Oncol 12: 175–180 [DOI] [PubMed] [Google Scholar]
- Portella G, Salvatore D, Botti G, Cerrato A, Zhang L, Mineo A, Chiappetta G, Santelli G, Pozzi L, Vecchio G, et al. 1996. Development of mammary and cutaneous gland tumors in transgenic mice carrying the RET/PTC1 oncogene. Oncogene 13: 2021–2026 [PubMed] [Google Scholar]
- Riely GJ, Kris MG, Rosenbaum D, Marks J, Li A, Chitale DA, Nafa K, Riedel ER, Hsu M, Pao W, et al. 2008. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res 14: 5731–5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, Zhu J, Johnson DH 2002. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med 346: 92–98 [DOI] [PubMed] [Google Scholar]
- Score J, Curtis C, Waghorn K, Stalder M, Jotterand M, Grand FH, Cross NC 2006. Identification of a novel imatinib responsive KIF5B-PDGFRA fusion gene following screening for PDGFRA overexpression in patients with hypereosinophilia. Leukemia 20: 827–832 [DOI] [PubMed] [Google Scholar]
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, et al. 2009. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461: 809–813 [DOI] [PubMed] [Google Scholar]
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al. 2007. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448: 561–566 [DOI] [PubMed] [Google Scholar]
- Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, et al. 2002. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci 99: 4465–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian J, Govindan R 2007. Lung cancer in never smokers: a review. J Clin Oncol 25: 561–570 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Ritz J, Cooper GM 1985. Activation of a novel human transforming gene, ret, by DNA rearrangement. Cell 42: 581–588 [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Choi YL, Togashi Y, Soda M, Hatano S, Inamura K, Takada S, Ueno T, Yamashita Y, Satoh Y, et al. 2009. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res 15: 3143–3149 [DOI] [PubMed] [Google Scholar]
- Tao J, Deng NT, Ramnarayanan K, Huang B, Oh HK, Leong SH, Lim SS, Tan IB, Ooi CH, Wu J et al. 2011. CD44-SLC1A2 gene fusions in gastric cancer. Sci Transl Med 3: 77ra30 doi: 10.1126/scitranslmed.3001423 [DOI] [PubMed] [Google Scholar]
- Toh CK, Gao F, Lim WT, Leong SS, Fong KW, Yap SP, Hsu AA, Eng P, Koong HN, Thirugnanam A, et al. 2006. Never-smokers with lung cancer: epidemiologic evidence of a distinct disease entity. J Clin Oncol 24: 2245–2251 [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. 2005. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310: 644–648 [DOI] [PubMed] [Google Scholar]
- Welch JS, Westervelt P, Ding L, Larson DE, Klco JM, Kulkarni S, Wallis J, Chen K, Payton JE, Fulton RS, et al. 2011. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. JAMA 305: 1577–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DW, Leung EL, So KK, Tam IY, Sihoe AD, Cheng LC, Ho KK, Au JS, Chung LP, Pik Wong M 2009. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer 115: 1723–1733 [DOI] [PubMed] [Google Scholar]
- Wu TD, Nacu S 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26: 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q, Cheng Y, Zhu Q, Yu Z, Wu X, Huang K, Zhou M, Han S, Zhang Q 2008. The relationship between overexpression of glial cell-derived neurotrophic factor and its RET receptor with progression and prognosis of human pancreatic cancer. J Int Med Res 36: 656–664 [DOI] [PubMed] [Google Scholar]
- Zhang F, Carvalho CM, Lupski JR 2009. Complex human chromosomal and genomic rearrangements. Trends Genet 25: 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]