Abstract
Pigmentation of skin, eye, and hair reflects some of the most evident common phenotypes in humans. Several candidate genes for human pigmentation are identified. The SNP rs12913832 has strong statistical association with human pigmentation. It is located within an intron of the nonpigment gene HERC2, 21 kb upstream of the pigment gene OCA2, and the region surrounding rs12913832 is highly conserved among animal species. However, the exact functional role of HERC2 rs12913832 in human pigmentation is unknown. Here we demonstrate that the HERC2 rs12913832 region functions as an enhancer regulating OCA2 transcription. In darkly pigmented human melanocytes carrying the rs12913832 T-allele, we detected binding of the transcription factors HLTF, LEF1, and MITF to the HERC2 rs12913832 enhancer, and a long-range chromatin loop between this enhancer and the OCA2 promoter that leads to elevated OCA2 expression. In contrast, in lightly pigmented melanocytes carrying the rs12913832 C-allele, chromatin-loop formation, transcription factor recruitment, and OCA2 expression are all reduced. Hence, we demonstrate that allelic variation of a common noncoding SNP located in a distal regulatory element not only disrupts the regulatory potential of this element but also affects its interaction with the relevant promoter. We provide the key mechanistic insight that allele-dependent differences in chromatin-loop formation (i.e., structural differences in the folding of gene loci) result in differences in allelic gene expression that affects common phenotypic traits. This concept is highly relevant for future studies aiming to unveil the functional basis of genetically determined phenotypes, including diseases.
Genome-wide association studies (GWAS) have successfully identified numerous genetic variants that are significantly associated with complex diseases or other phenotypic traits (Manolio 2010). Intriguingly, many of the identified disease or other phenotype-associated DNA variants are located in noncoding regions of the human genome. It remains largely unknown if and how these noncoding DNA variants influence phenotypic traits at the molecular level. Although linkage with unknown causal DNA variants is often assumed as a major factor to explain association of noncoding DNA variants, it has been suggested that some of these noncoding DNA variants constitute potential regulatory elements for distal genes (Coetzee et al. 2010; Freedman et al. 2011). However, so far, this has only been demonstrated for some low-penetrance disease susceptibility loci (Jia et al. 2009; Pomerantz et al. 2009; Ahmadiyeh et al. 2010; Harismendy et al. 2010; Wright et al. 2010).
Pigmentation of skin, eye, and hair reflects one of the clearest common phenotypes in humans, but is in fact a complex polygenetic trait. Several genes are involved in the human pigmentation pathway, and many DNA polymorphisms are associated with human pigmentation variation (Sturm 2009). The SNP rs12913832 is highly associated with human eye (Eiberg et al. 2008; Sturm et al. 2008; Liu et al. 2009, 2010), hair (Han et al. 2008; Branicki et al. 2009, 2011), and skin (Branicki et al. 2009; Valenzuela et al. 2010; Spichenok et al. 2011) color variation. This SNP is the key determinant of human eye color variation and has strong predictive power for human pigmentation (Liu et al. 2009, 2010; Valenzuela et al. 2010; Branicki et al. 2011; Spichenok et al. 2011). Interestingly, rs12913832 is not located in a pigment gene, but in intron 86 of the HERC2 gene, 21 kb upstream of the promoter of OCA2 (Fig. 1A). The OCA2 gene is involved in human pigmentation since it regulates melanosomal pH, and mutations in OCA2 cause oculocutaneous albinism type II (Brilliant 2001). The region directly surrounding HERC2 rs12913832 is highly conserved among animal species (Fig. 1A; Supplemental Table S1; Sturm et al. 2008), and this SNP strongly associates with OCA2 expression levels (Cook et al. 2009). However, if and how HERC2 rs12913832 directly regulates human pigmentation is currently unknown. It was previously hypothesized that this SNP acts as a distal regulatory region that either silences or enhances (Eiberg et al. 2008; Sturm et al. 2008; Sturm and Larsson 2009) OCA2 expression, but convincing experimental data that would confirm or reject either one of these hypotheses remain to be presented.
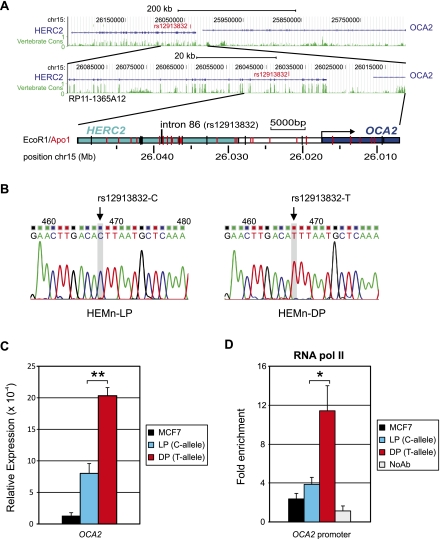
Figure 1.
Characterization and suitability of the HEMn cell system: The OCA2 gene is differentially expressed in HEMn-LP and HEMn-DP cells. (A) UCSC Browser NCBI36/hg18 assembly (http://genome.ucsc.edu/cgi-bin/hgGateway?db=hg18) overview of the OCA2-HERC2 locus (top panel). (Middle panel) The region covered on BAC RP11-1365A12. Vertebrate conservation (green); the position of rs12913832 (red). (Lower panel) A schematic overview of the region investigated in this study. Restriction enzyme digestion sites are indicated. (B) Sequence analysis of the region around HERC2 rs12913832 in HEMn-LP (left) and HEMn-DP (right). The genotypes of rs12913832 were determined by direct sequencing of PCR fragments containing rs12913832. (C) RT-qPCR analysis of OCA2 primary transcripts in MCF7 and HEMn cells demonstrates differential OCA2 expression between HEMn-LP and HEMn-DP cells. Each gene expression analysis is carried out in triplicate and normalized to an endogenous reference gene (ACTB). (D) ChIP-qPCR of RNA Pol II binding at the OCA2 promoter in MCF7, HEMn-LP, and HEMn-DP cells. Enrichment is calculated relative to necdin (NDN), and values are normalized to input measurements. All ChIP analyses are performed in triplicate. Data are represented as mean ± SEM; (*) p < 0.05; (**) p < 0.01.
In this study, we used various molecular approaches to investigate the functional role of HERC2 rs12913832 in human pigmentation variation. In darkly pigmented and lightly pigmented melanocyte cell lines with different genotypes for HERC2 rs12913832, we investigated the relation between the HERC2 rs12913832 genotype and OCA2 expression, surveyed the epigenomic landscape of the region around HERC2 rs12913832, and mapped the chromatin folding of the OCA2-HERC2 locus. Our study provides crucial mechanistic insight into how a noncoding DNA polymorphism can affect a common human phenotypic trait by modulating the chromatin folding of a gene locus.
Results
Characterization and suitability of the melanocyte cell system
To investigate the potential functional role of HERC2 rs12913832, we used commercially available primary Human Epidermal Melanocytes of neonatal origin derived from a lightly pigmented (HEMn-LP) and a darkly pigmented donor (HEMn-DP) (Cascade Biologics, Invitrogen). The MCF7 breast-cancer cell line was used for control purposes. Genotyping, either by iPLEX (Sequenom) (Liu et al. 2009) or direct DNA sequencing of PCR products, of SNPs statistically associated with human pigmentation revealed that HEMn-LP cells are homozygous for the rs12913832 C-allele, while HEMn-DP cells are homozygous for the rs12913832 T-allele (Fig. 1B). structure analysis (Pritchard et al. 2000) of DNA samples from HEMn-LP and HEMn-DP cells using 24 ancestry-informative autosomal SNPs together with a worldwide reference data set (HGDP-CEPH) (Lao et al. 2010) revealed a probable European and a probable Sub-Saharan African genetic origin of the respective cell donors (Supplemental Fig. S1). Both data sets are in agreement with the light and dark skin-color phenotype information of the HEMn-LP and HEMn-DP cell donors, as provided by Cascade Biologics.
We investigated the transcription status of the OCA2 gene in our skin melanocyte system by measuring primary OCA2 transcripts using quantitative RT-PCR (qRT-PCR). We observed higher expression of OCA2 in HEMn-DP cells carrying the HERC2 rs12913832 T-allele, than in HEMn-LP cells carrying the C-allele (Fig. 1C; p < 0.01), confirming previous observations (Cook et al. 2009). We also tested for the presence of RNA polymerase II (RNA pol II) on the OCA2 promoter using chromatin immunoprecipitation (ChIP) using an antibody against the POLR2A subunit of RNA polymerase II followed by qPCR (ChIP-qPCR). ChIP is a technology that identifies chromatin regions bound by a specific protein (i.e., a transcription factor) using an antibody specific to the protein of interest to enrich cross-linked fragmented chromatin bound by such a protein from the bulk of the chromatin. We observed a higher RNA Pol II recruitment to the OCA2 promoter in HEMn-DP cells than in HEMn-LP cells (Fig. 1D; p < 0.05). Both OCA2 gene expression and RNA pol II recruitment were very low in MCF7 control cells (Fig. 1C,D). From these results, we conclude that the HEMn-LP/HEMn-DP melanocyte system constitutes an appropriate system to investigate the functional effect of HERC2 rs12913832 on human pigmentation.
Identification of HERC2 rs12913832 as a melanocyte-specific enhancer
HERC2 rs12913832 was previously hypothesized to function as a regulatory region (Sturm et al. 2008; Cook et al. 2009). Gene regulatory regions are characterized by open chromatin and active chromatin marks (Giresi et al. 2007). Hence we used formaldehyde-assisted identification of regulatory elements (FAIRE) and ChIP-qPCR of histone modifications, to investigate the chromatin status of the HERC2 rs12913832 region. FAIRE identifies regulatory DNA regions based on differences in cross-link ability between chromatin of regulatory DNA regions and the bulk of the chromatin (Giresi et al. 2007). FAIRE analysis revealed an open chromatin structure in the region around HERC2 rs12913832 in both HEMn-LP and HEMn-DP cells, while this region was shown to be closed in MCF7 control cells (Fig. 2A). In addition, the level of enrichment identified by this assay was higher for HEMn-DP cells than for HEMn-LP cells (Fig. 2A; p < 0.05), suggesting that the chromatin of the HERC2 rs12913832 region is more open when the rs12913832 T-allele is present relative to when the C-allele is present. ChIP-qPCR analysis detected elevated levels of acetylated histone H3, an active chromatin mark, at HERC2 rs12913832 in HEMn-LP and HEMn-DP cells, but not in the MCF7 control cells (Fig. 2B). Both observations are consistent with the hypothesis that HERC2 rs12913832 acts as a human melanocyte-specific gene regulatory region.
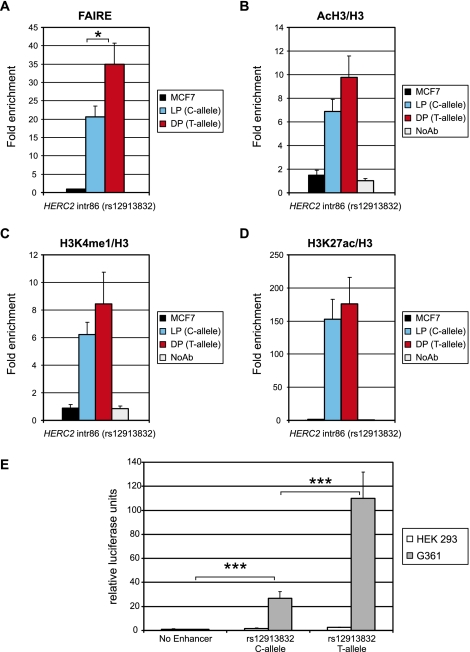
Figure 2.
The region directly surrounding HERC2 rs12913832 acts as a melanocyte-specific enhancer. (A) Formaldehyde assisted identification of regulatory elements (FAIRE) demonstrates low nucleosome occupancy at the rs12913832 region in HEMn-LP and HEMn-DP cells. (B) ChIP-qPCR of acetylated histone H3 demonstrates active chromatin marks at the rs12913832 region in HEMn-LP and HEMn-DP cells. (C) ChIP-qPCR of histone H3 mono methylated on lysine 4 demonstrates that the rs12913832 region in HEMn-LP and HEMn-DP cells has enhancer potential. (D) ChIP-qPCR of histone H3 acetylated on lysine 27 demonstrates that the enhancer at the rs12913832 region in HEMn-LP and HEMn-DP cells is active. The enrichments displayed are relative to NDN. ChIP values for histone H3 marks are normalized to histone H3 occupancy. (E) Luciferase reporter assay demonstrates differential melanocyte enhancer activity for the rs12913832 region. The rs12913832 region from HEMn-LP (C-allele) and HEMn-DP (T-allele) was inserted into a luciferase reporter plasmid and transfected into HEK293 or G361 melanoma cells. Luciferase expression is normalized to Renilla luciferase expression. Data are represented as mean ± SEM; (*) p < 0.05; (***) p < 0.005.
Enhancers are distal gene regulatory elements that act positively on gene expression and are enriched for mono-methylation of lysine 4 on histone H3 (Heintzman et al. 2007). Recently it was shown that active enhancers additionally accumulate acetylation of lysine 27 on histone H3 (Creyghton et al. 2010; Rada-Iglesias et al. 2010). ChIP-qPCR analysis of these chromatin marks revealed the presence of both enhancer marks on rs12913832 in both HEMn cell types, but not in MCF7 control cells (Fig. 2C,D). Similar to the differences observed with the FAIRE assay, higher enrichments for the chromatin marks were found on HERC2 rs12913832 in HEMn-DP cells than in HEMn-LP cells, although these differences do not reach statistical significance (Fig. 2B–D). The above observations are consistent with the HERC2 rs12913832 region operating as an enhancer in skin melanocytes. Interestingly, the level of the enhancer marks in the HERC2 rs12913832 alleles also positively correlates with OCA2 expression and the level of melanocyte pigmentation.
We further investigated the enhancer potential of the region encompassing HERC2 rs12913832 by cloning this region from HEMn-LP and HEMn-DP cells into a luciferase reporter vector. Upon transfection of these constructs into human embryonic kidney cells (HEK293), no substantial increase in luciferase expression was observed for the vectors containing the putative enhancer regions, as compared with the empty vector (Fig. 2E). However, robust luciferase expression was observed when these constructs were transfected into G361 melanoma cells, with an approximately fivefold higher activity for the rs12913832 T-allele than for the rs12913832 C-allele (Fig. 2E; p < 0.005). Sequencing of the cloned fragment revealed the presence of another SNP, rs6497271 (Supplemental Fig. S2A), which could theoretically modulate the observed enhancer activity. Analysis of chimeras of our reporter constructs containing different combinations of rs12913832 alleles and rs6497271 alleles, however, demonstrates that the enhancer activity of the HERC2 rs12913832 region is not modulated by rs6497271 (Supplemental Fig. S2B). These findings also confirmed the previously reported suggestion of Sturm et al. (2008) that this SNP could not explain association to the region because of very low minor allele frequency. Together, these independent experiments demonstrate that the region surrounding HERC2 rs12913832 acts as a melanocyte-specific enhancer whose activity depends on the allelic status of rs12913832.
Detection of a differential chromatin loop between the HERC2 rs12913832 enhancer and the OCA2 promoter
Distal enhancers increase the expression of their target genes by directly contacting the gene promoters via chromatin loops, which can be detected using chromosome conformation capture (3C) technology (Palstra 2009). In 3C, formaldehyde is used to trap interactions between chromatin segments that are in close proximity to each other. The cross-linked chromatin is subsequently digested using a restriction enzyme followed by intramolecular ligation under dilute conditions. The relative abundance of ligation products is determined by qPCR and is proportional to the frequency with which the various restriction fragments interact. We inferred that if the region surrounding HERC2 rs12913832 acts as a melanocyte-specific enhancer for the OCA2 gene, as indicated by our data, we should be able to detect a chromatin loop between the HERC2 rs12913832 region and the OCA2 gene. Indeed, locus-wide 3C analysis of an EcoRI restriction fragment containing rs12913832 showed, in HEMn-DP cells, an elevated relative cross-linking frequency for an OCA2 promoter–containing restriction fragment, indicating a chromatin loop between the rs12913832 T-allele and the OCA2 promoter (Fig. 3A). HEMn-LP cells, however, showed a lower cross-linking frequency of this fragment (Fig. 3A), demonstrating a decreased interaction efficiency between the rs12913832 C-allele and the OCA2 promoter. In contrast, MCF7 control cells showed a linear chromatin conformation (Fig. 3A).
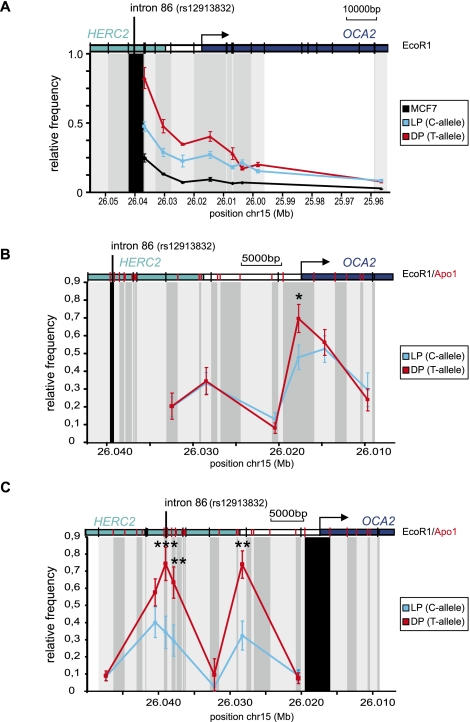
Figure 3.
A chromatin loop is formed between the HERC2 rs12913832 enhancer region and the OCA2 promoter. (A–C) Locus-wide cross-linking frequencies observed in MCF7 (black), HEMn-LP (cyan), and HEMn-DP (red) cells. The analyzed region of the human OCA2-HERC2 locus is depicted on the top of each graph. The x-axis shows the approximate position on chromosome 15 (UCSC Browser NCBI36/hg18 assembly; see also Fig. 1A). (Black shading) The position and size of the “fixed” restriction fragment; (gray shading) position and size of other restriction fragments analyzed. (Black vertical bars in the locus graph) EcoRI sites; (red vertical bars) ApoI sites. The cross-linking frequencies are normalized to the highest interaction within an experiment. (A) Cross-linking frequencies for an EcoRI restriction fragment containing rs12913832 in MCF7 and HEMn cells. In HEMn cells, high cross-linking frequencies are observed for a restriction fragment containing the OCA2 promoter. (B) Cross-linking frequencies for an ApoI restriction fragment containing rs12913832 in HEMn cells. In HEMn cells, high cross-linking frequencies are observed for a restriction fragment containing the OCA2 promoter. Cross-linking frequencies between the restriction fragment containing rs12913832 and the restriction fragment containing the OCA2 promoter are higher for the T-allele (red) than for the C-allele (cyan). (C) Cross-linking frequencies for an ApoI restriction fragment containing the OCA2 promoter in HEMn cells. High cross-linking frequencies with restriction fragments surrounding the rs12913832 enhancer region are observed in HEMn cells. Cross-linking frequencies between the restriction fragment containing rs12913832 and the restriction fragment containing the OCA2 promoter are higher for the T-allele (red) than for the C-allele (cyan). Data are represented as mean ± SEM; (*) p < 0.05; (**) p < 0.01; (***) p < 0.005.
To further investigate the observed differences in chromatin looping, we aimed for a higher resolution of the 3C experiments. We therefore used a more frequently digesting restriction enzyme (ApoI vs. EcoRI) that generates smaller restriction fragments. Analysis of locus-wide cross-linking frequencies of the ∼500-bp ApoI fragment containing HERC2 rs12913832 demonstrates a chromatin loop between rs12913832 and the OCA2 promoter that is more prominent in HEMn-DP cells (T-allele) than in HEMn-LP cells (C-allele) (Fig. 3B; p < 0.05). The difference in chromatin looping between the two HERC2 rs12913832 alleles and the OCA2 promoter becomes even clearer in the reciprocal analysis of the ApoI fragment that contains the OCA2 promoter (Fig. 3C; p < 0.005). Locus-wide 3C analysis in MCF7 control cells revealed a more linear chromatin organization (Supplemental Fig. S3A,B), and an interaction between HERC2 rs12913832 and the OCA2 promoter is absent (Supplemental Fig. S3C; p < 0.005). These data demonstrate that the formation of a chromatin loop between HERC2 rs12913832 and the OCA2 promoter is melanocyte-specific and is more efficient when the rs12913832 T-allele is present; this is in line with the higher OCA2 expression observed in HEMn-DP cells (T-allele) (Fig. 1C).
Interestingly, in both experiments, an additional peak in cross-linking frequencies was detected with a restriction fragment downstream from the HERC2 gene, suggesting the presence of an additional regulatory element in this region (Fig. 3B,C). This peak is also detected in MCF7 cells although at a lower frequency (Supplemental Figs. S3A,B, S4A) and therefore probably reflects a constitutive interaction not involved in melanocyte-specific OCA2 regulation. ENCODE data (The ENCODE Project Consortium et al. 2007) obtained from multiple cell lines and our own ChIP-qPCR experiments, indeed, demonstrate the presence of chromatin signatures associated with gene-regulatory elements in the OCA2–HERC2 intergenic region, thereby confirming this notion (Supplemental Fig. S4B,C).
Detection of an allele-specific chromatin loop between the HERC2 rs12913832 enhancer and the OCA2 promoter in combination with the previously observed link between rs12913832 alleles and OCA2 expression (Cook et al. 2009) demonstrates that the HERC2 rs12913832 enhancer directly regulates OCA2 expression.
Analysis of transcription factor binding at the HERC2 rs12913832 enhancer
Sturm et al. (2008) performed an in silico analysis of transcription factor binding sites present in the HERC2 region surrounding rs12913832. Potential binding sites for HLTF (also known as RUSH or SMARCA3), MITF, and LEF1 were identified (Supplemental Fig. S2A), and it was hypothesized that the C-allele of rs12913832 would disrupt the HLTF binding site (Sturm et al. 2008; Sturm and Larsson 2009). Both HLTF and LEF1 have previously been implicated in enhancer function and chromatin looping (Hewetson and Chilton 2008; Yun et al. 2009), whereas MITF is a key transcription factor in melanocyte development and pigment gene expression (Levy et al. 2006). To experimentally verify whether the HERC2 rs12913832 alleles are differentially bound by HLTF, we performed ChIP-qPCR experiments. HLTF binding was enriched above background for the rs12913832 T-allele, but not for the C-allele (Fig. 4A; p < 0.01), providing experimental evidence that the T-to-C mutation reduces HLTF binding to the rs12913832 region. To further investigate the role of HLTF in OCA2 expression, we overexpressed HLTF in HEMn-LP and HEMn-DP cells. In HEMn-LP cells, overexpression of HLTF resulted in reduced OCA2 expression (Fig. 4B). However, HERC2 expression was also reduced in HEMn-LP and in HEMn-DP cells, but to a similar extent, which therefore suggests that the reduced OCA2 expression in HEMn-LP cells is a systematic response rather than a functional response to HLTF overexpression (data not shown). In contrast, overexpression of HLTF in HEMn-DP cells resulted in elevated expression levels of OCA2 (Fig. 4B; p < 0.05), demonstrating a positive role of HLTF in the regulation of OCA2 expression, and this regulatory role depends on the T-allele of HERC2 rs12913832.
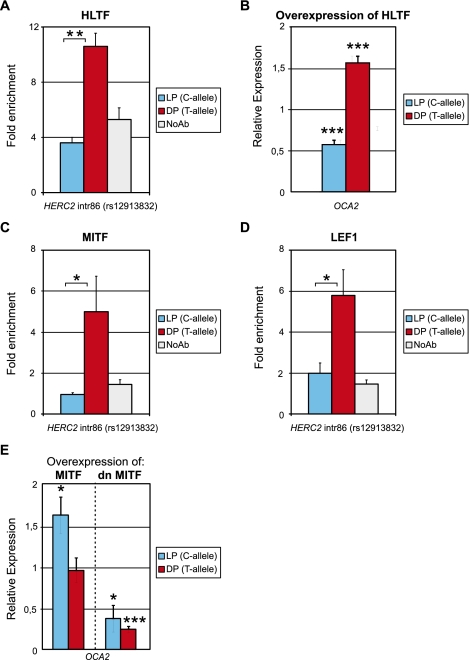
Figure 4.
The HERC2 rs12913832 enhancer is regulated by the transcription factors HLTF, MITF, and LEF1. (A) ChIP-qPCR of HLTF at the rs12913832 region in HEMn-LP (C-allele) and HEMn-DP (T-allele) cells. HLTF binding is only observed for the T-allele. (B) Overexpression of HLTF in HEMn-DP cells results in increased OCA2 expression but not in HEMn-LP cells. (C) ChIP-qPCR of MITF at the HERC2 rs12913832 region in HEMn-LP (C-allele) and HEMn-DP (T-allele) cells. MITF binding is only observed for the T-allele. (D) ChIP-qPCR of LEF1 at the HERC2 rs12913832 region in HEMn-LP (C-allele) and HEMn-DP (T-allele) cells. LEF1 binding is only observed for the T-allele. ChIP enrichments displayed are relative to NDN. (E) Overexpression of MITF in HEMn-LP cells results in increased OCA2 expression. This is not observed in HEMn-DP cells. Overexpression of a dominant negative MITF (dnMITF) results in decreased OCA2 expression in both HEMn-LP and HEMn-DP cells. Expression is relative to ACTB expression and nontransfected control cells. Data are represented as mean ± SEM; (*) p < 0.05; (**) p < 0.01; (***) p < 0.005.
Reduced recruitment of HLTF, a chromatin remodeler of the SWI-SNF family (MacKay et al. 2009), to the rs12913832 C-allele may lead to a more closed chromatin conformation that could affect the recruitment of other transcription factors such as MITF and LEF1. Indeed, we found increased binding of MITF and LEF1 to the region encompassing rs12913832 containing the T-allele, in comparison to this region containing the C-allele (Fig. 4C,D; p < 0.05). This is in line with a recent study demonstrating that MITF binds near HERC2 rs12913832 in a melanoma cell line (Strub et al. 2010). Furthermore, in melanoma cells, OCA2 expression is up-regulated upon overexpression of MITF (Hoek et al. 2008). In agreement with this, we found that overexpression of MITF in HEMn-LP cells resulted in increased OCA2 expression (Fig. 4E; p < 0.05), while overexpression of a dominant negative form of MITF (dnMITF) (Levy et al. 2010) resulted in reduced OCA2 expression (Fig. 4E; p < 0.05). Interestingly, overexpression of MITF in HEMn-DP cells failed to increase OCA2 expression, suggesting that binding of MITF to the rs12913832 enhancer is not limiting in HEMn-DP cells (Fig. 4E). However, overexpression of the dnMITF in HEMn-DP cells resulted in decreased OCA2 expression (Fig. 4E; p < 0.005). Together these data demonstrate that MITF indeed regulates OCA2 expression. It suggests that MITF preferentially binds to the HERC2 intron 86 region when it contains the rs12923832 T-allele, which is facilitated by the interaction of HLTF with the rs12913832 region.
Analysis of pigmentation-associated SNPs other than HERC2 rs12913832
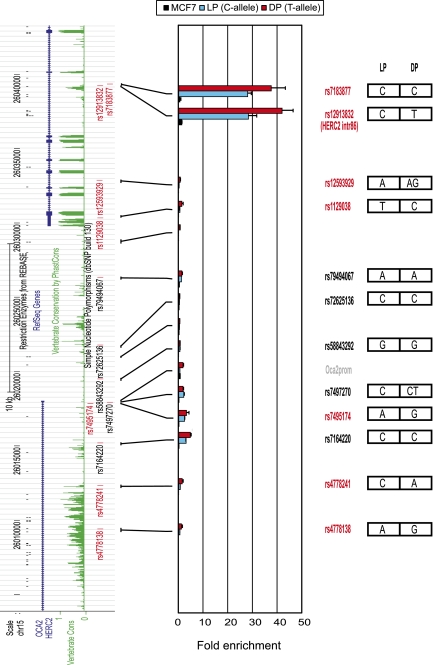
It remains possible that linkage with another DNA variant may be responsible for the regulatory effect of a noncoding SNP. To exclude the possibility that pigmentation-linked sequence variants other than HERC2 rs12913832 might explain our observations, we genotyped all SNPs within the analyzed 3′HERC2/5′OCA2 region that have been linked to pigmentation, including those in linkage disequilibrium (LD; r2 > 0.8) (Supplemental Table S1). This region of ∼35 kb contains 12 pigmentation-linked SNPs in total. Besides HERC2 rs12913832, six SNPs have been directly linked to pigmentation traits (Liu et al. 2009), while five additional SNPs are in high LD (r2 > 0.8) with these six SNPs (Supplemental Table S1). The HEMn-LP and HEMn-DP cells share the same genotype of only one pigmentation-linked SNP, namely, rs7183877, and have different alleles for the other five SNPs, whereas for the linked SNPs, the allelic status for all but one SNP is the same in HEMn-LP and HEMn-DP cells. The LD-SNP rs7497270 is homozygous in the HEMn-LP cells (C) and heterozygous in the HEMn-DP cells (CT). To assess the possible regulatory potential of these additional 11 pigmentation-linked SNPs, we interrogated these sites as well as several control sites using FAIRE analysis in HEMn-LP and HEMn-DP cells, and in the MCF7 control cell line (Fig. 5). As we showed before (Fig. 2A), HERC2 rs12913832 is highly enriched in HEMn cells as compared with MCF7 cells. The only other site with such a high enrichment is rs7183877. This site shares the C-allele in HEMn-LP and HEMn-DP cells and is located only 115 bp away from HERC2 rs12913832. This close physical location explains its high enrichment in the FAIRE assay, because FAIRE analysis has a resolution in the range of 0.5–1.5 kb. From the remaining investigated sites, only those in the promoter region and in the first intron of OCA2 have an appreciable although small enrichment in the FAIRE assay (Fig. 5). Irrespective of their genotypes, this enrichment appears to be higher in the HEMn-DP cells than in the HEMN-LP cells, probably reflecting the higher transcriptional status of OCA2 in the HEMN-DP cells (Fig. 1C,D). We therefore conclude that none of the SNPs that can be linked to pigmentation within the ∼35-kb HERC2/OCA2 region analyzed other than rs12913832 has regulatory potential.
Figure 5.
FAIRE analysis of pigmentation-associated SNPs other than HERC2 rs12913832 present within the 3′HERC2/5′OCA2 region does not reveal additional regulatory elements. (Left side) Tracks from the UCSC Browser (NCBI36/hg18 assembly; http://genome.ucsc.edu/cgi-bin/hgGateway?db=hg18) of the investigated 3′HERC2/5′OCA2 region. Pigmentation-associated SNPs (red); linked SNPs (r2 > 0.8) (black). The approximate location of the analyzed PCR amplicons is indicated. (Right side) The genotype of each SNP for HEMn-LP and HEMn-DP. The enrichments displayed are relative to NDN. Data are represented as mean ± SEM.
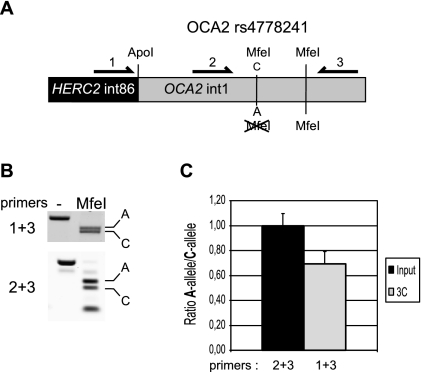
Furthermore, of the 12 pigmentation-linked SNPs, only three—rs12913832, rs7164220, and rs4778241—are significantly conserved among vertebrate species, which might suggest functionality (Supplemental Table S1). Both HEMn-LP and HEMn-DP cells share the C-allele for rs7164220. However, HEMn-LP and HEMn-DP cells differ in alleles for rs4778241 (C and A), and therefore it remains possible that the observed allele-specific chromatin loop between HERC2 rs12913832 and the OCA2 promoter is caused by this SNP. To investigate this, we made use of a commercially available cell line derived from an intermediate pigmented donor (HEMn-MP) (Cascade Biologics, Invitrogen). This cell line is homozygous for the C-allele of HERC2 rs12913832 but heterozygous for rs4778241 (C and A) (Supplemental Table S1). The presence of the rs4778241 C-allele generates an MfeI restriction site, which allowed us to design an allele-specific 3C assay (Fig. 6A). We used this assay to test the hypothesis that rs4778241, instead of HERC2 rs12913832, is responsible for the observed allele-specific chromatin loop between HERC2 rs12913832 and the OCA2 promoter. If this hypothesis holds true, we would expect to find an increased interaction of the ApoI restriction fragment containing HERC2 rs12913832 with the ApoI restriction fragment containing the A-allele of rs4778241 (also present in HEMn-DP cells). Instead, however, we detected an interaction between the rs12913832 ApoI fragment and both the ApoI fragments containing either the A-allele or C-allele of rs4778241 (Fig. 6B,C), and there seemed to be a preference (although not statistically significant) for the rs4778241 C-allele, which is also present in HEMn-LP cells. The ApoI restriction fragment containing HERC2 rs12913832 does not contain any sequence variation except for HERC2 rs12913832 (Supplemental Fig. 2A), enforcing the notion that the observed differential chromatin looping is solely dependent on the HERC2 rs12913832 allele.
Figure 6.
The chromatin loop between the HERC2 rs12913832 enhancer region and the OCA2 promoter is not caused by allelic differences in the 5′ region of OCA2. (A) Schematic overview of the allele-specific 3C assay. The allele-specific interaction between an ApoI fragment containing HERC2 rs12913832 and an ApoI fragment containing OCA2 rs4778241 is investigated. The presence of the C-allele of rs4778241 generates an additional MfeI restriction site. Primers 1 and 3 are used to detect the 3C product, while primers 2 and 3 are used to normalize the ratio of the A-allele over the C-allele. Ratios are determined by MfeI digestion of PCR products. The additional MfeI site is used to monitor the completeness of digestion. (B) Example of gel images of a representative MfeI digestion of PCR products. In the digested lane, the top band represents the A-allele and the bottom band the C-allele of rs4778241. (C) Quantification of multiple gel images as shown in B. The ratio of the bands generated by primer pair 2 + 3 after MfeI digestion is set to 1. Data are represented as mean ± SEM.
Together these observations make it highly improbable that the observed allele-specific chromatin loop between HERC2 rs12913832 and the OCA2 promoter is caused by a SNP other than HERC2 rs12913832 located in the 3′HERC2/5′OCA2 region.
Discussion
Overall, our study provides extensive experimental evidence that the genomic region directly surrounding HERC2 rs12913832 functions as a human melanocyte-specific enhancer element that influences OCA2 gene expression. In particular, we demonstrate that the HERC2 rs12913832 enhancer communicates with the OCA2 promoter via a long-range chromatin loop, and enhancer activity is mediated by the transcription factors HLTF, LEF1, and MITF. The HERC2 rs12913832 T-allele robustly recruits these transcription factors, which, in combination with increased looping to the OCA2 promoter, facilitates OCA2 expression. We propose that this leads to enhanced melanin production and the dark pigmentation phenotype. In contrast, the HERC2 rs12913832 C-allele mutates an HLTF binding site and diminishes HLTF recruitment, which correlates with decreased MITF and LEF1 recruitment and reduced chromatin-loop formation. This results in reduced OCA2 expression and the light pigmentation phenotype.
Linkage with an unknown causal DNA variant may be indicated as a plausible explanation for the regulatory effect of a noncoding SNP. We consider this hypothesis, however, unlikely for the following reasons: Our data clearly demonstrate that the region around HERC2 rs12923832 displays chromatin features associated with enhancers; it acts as a melanocyte-specific enhancer in luciferase reporter assays; it communicates with the OCA2 promoter region via a chromatin loop; and the transcription factors HLTF, MITF, and LEF interact with the HERC2 rs12913832 region in an allele-specific manner. Together, this demonstrates that the HERC2 rs12913832 region constitutes a genuine regulatory element for the OCA2 gene. Moreover, other pigmentation SNPs within the investigated region do not display chromatin features consistent with enhancer function and do not influence chromatin looping between HERC2 rs12923832 and the OCA2 promoter region. Considering our data in combination with previously published indirect evidence such as (1) the gene function of OCA2 being one of the major pigmentation genes that affects the amount and quality of melanin in melanocytes (Brilliant 2001); (2) several OCA2 mutations resulting in albinism (Rinchik et al. 1993; Brilliant 2001); and (3) a strong correlation between HERC2 rs12913832, OCA2 expression levels, and melanocytic melanin content (Cook et al. 2009), it is highly likely that HERC2 rs12913832 is the major causal variant for the regulation of OCA2 transcription and hence pigmentation. Our data, however, do not exclude the possibility that additional regulatory regions for the OCA2 gene are present within or even outside the HERC2/OCA2 region, but if existing at all, their effect is expected to be rather minor.
Previously, two different models proposed that the region around HERC2 rs12913832 acts as a regulatory element that influences OCA2 expression. Eiberg et al. (2008) suggested that the HERC2 rs12913832 region functions as a silencer for OCA2 expression, while Sturm and colleagues (Sturm et al. 2008; Sturm and Larsson 2009) suggested that the HERC2 rs12913832 region functions as an enhancer. Clearly, our study provides solid experimental evidence to support the model as proposed by Sturm and colleagues (Sturm et al. 2008; Sturm and Larsson 2009) and further extends it, since we demonstrate that the region around HERC2 rs12913832 displays multiple features of an enhancer element and forms an allele-specific chromatin loop with the OCA2 promoter. Since both HLTF and LEF1 have previously been shown to be involved in chromatin looping, we find it plausible that the increased binding of these factors to the HERC2 rs12913832 T-allele is responsible for the observed increased looping of the HERC2 rs12913832 region to the OCA2 promoter (Hewetson and Chilton 2008; Yun et al. 2009). The model proposed by Sturm and colleagues suggests that binding of MITF and LEF1 to the HERC2 intron 86 region fully depends on HLTF interaction with HERC2 rs12913832. However, based on our data, this part of the model requires some refinement. Using ChIP assays, we only detected MITF and LEF1 binding when the T-allele of HERC2 rs12913832 is present, but due to the inefficiency of ChIP assays, it is plausible that we missed low-level binding of these factors to the rs12913832 C-allele. Our overexpression studies demonstrate that the locus containing the C-allele of rs12913832 is responsive to MITF levels but irresponsive to HLTF levels. In contrast, the locus containing the rs12913832 T-allele is responsive to HLTF overexpression and to expression of the dnMITF but not to increased MITF levels. This suggests that MITF binding to the HERC2 rs12913832 region in HEMn cells is in an equilibrium that can be modulated by varying MITF levels and HLTF binding. Alternatively, other MITF-responsive OCA2 regulatory elements might be present.
For some disease-associated SNPs such as the cancer risk variants rs6983267 (Pomerantz et al. 2009; Ahmadiyeh et al. 2010; Wright et al. 2010) and rs11986220 (Jia et al. 2009), or DNA variants associated with coronary artery disease (Harismendy et al. 2010), it has been reported that they constitute transcription regulatory elements for distal genes, and the presence of a chromatin loop between the putative regulatory element and the promoter of the regulated gene has been demonstrated. However, none of these studies reports allelic differences in chromatin-loop formation. One study (Wright et al. 2010) that specifically investigated potential allelic differences in chromatin-loop formation between rs6983267 and the MYC gene in colon cancer cells failed to demonstrate such an allelic difference. In contrast, our study provides the key mechanistic insight that allele-dependent differences in chromatin-loop formation (i.e., structural differences in the folding of gene loci) functionally contribute to differences in allelic gene expression. Moreover, the predicted risk associated with above-mentioned rare disease phenotypes is modest, whereas HERC2 rs12913832 has strong predictive power for human pigmentation phenotypes (Liu et al. 2009, 2010; Valenzuela et al. 2010; Branicki et al. 2011; Spichenok et al. 2011). Therefore, our study demonstrates that genetic variation in gene regulatory elements can have a strong influence on common human phenotypic traits, which extends previous knowledge from rare phenotypes and puts it into a new and more general perspective.
We anticipate that many phenotypic traits, pathological and nonpathological, are modulated by DNA variants in distally located regulatory regions. In this study, we demonstrate that it is feasible to annotate regulatory function to a noncoding SNP and identify its target gene, using an approach that combines FAIRE, ChIP, and 3C. All of these techniques can be used at a genome-wide scale to study the epigenome landscape. Such a genome-wide approach combined with GWAS data allows us to assign function to many more noncoding SNPs in future studies that aim to unveil the functional basis of genetically determined phenotypic variation.
Methods
Cell culture
HEMn-LP (C-0025C; Lot #200708522; Cascade Biologics, Invitrogen), HEMn-MP (C-1025C; Lot# 070417901; Cascade Biologics, Invitrogen), and HEMn-DP (C-2025C, Lot# 6C0474; Cascade Biologics, Invitrogen) were grown in Medium 254 supplemented with HMGS according to the manufacturer's instructions (Cascade Biologics, Invitrogen). G361, HEK293, and MCF7 cells were cultured in DMEM/10% FCS at 37°C/5% CO2. HLTF was overexpressed using a pCDNA5 FRT/TO/FLAG SMARCA3 (MacKay et al. 2009). Transfection was performed using Lipofectamine LTX transfection reagent (Invitrogen) according to the manufacturer's instructions. MITF and dnMITF were overexpressed using adeno viral constructs (a kind gift from Dr. D. Fisher).
Genotyping
Pigmentation SNPs including HERC2 rs12913832 were genotyped with the iPLEX (Sequenom) multiplex reactions as described by Liu et al. (2009) using genomic DNA samples derived from the different cell lines. Genotypes for rs12913832 were confirmed by direct sequencing of PCR fragments containing rs12913832. Genotypes of rs7164220, rs7497270, rs58843292, rs72625136, and rs79494067 were determined by restriction digestion of PCR fragments. To infer the geographic origin/genetic ancestry of the cell-line donors, 24 autosomal SNPs sensitive to detect continental ancestry were genotyped via two SNaPshot (Applied Biosystems) multiplex reactions as described elsewhere (Lao et al. 2010). The continental ancestry of the samples was recovered by performing a structure analysis (Pritchard et al. 2000) in which data from the HGDP-CEPH panel served as reference, as described elsewhere (Lao et al. 2010).
Chromatin immunoprecipitation (ChIP)
ChIP was performed as described in the Millipore protocol (http://www.millipore.com/userguides/tech1/mcproto407), except that samples were cross-linked with 2% formaldehyde for 10 min at room temperature. To remove melanin, DNA was column purified before PCR (OneStep PCR Inhibitor Removal Kit, Zymo Research). Quantitative real-time PCR (CFX96 Real Time System, Bio-Rad) was performed using SYBR Green (Sigma-Aldrich) and Platinum Taq DNA Polymerase (Invitrogen) under the following cycling conditions: 10 min at 50°C, 5 min at 95°C, 45 cycles of 10 sec at 95°C, 30 sec at 60°C, and followed by a melting curve analysis. Enrichment was calculated relative to necdin (NDN), and values were normalized to input measurements. The antibodies used were RNA polymerase II (POLR2A, N-20; sc-899), LEF1 (H-70; sc-28687X), MITF (C-17; sc-11002X) from Santa Cruz Biotechnology, Ac-H3 (#06-599) from Millipore, PanH3 (#ab1791), acetylated Histone H3 K27 (#4729ab), and mono-methyl histone H3 K4 (#ab8895-50) from Abcam, HLTF (PAB12415) from Abnova. Primer sequences are listed in Supplemental Table S2.
Formaldehyde-assisted identification of regulatory elements (FAIRE)
FAIRE was performed as described before (Giresi et al. 2007), except that selected genomic sites were analyzed by quantitative real-time PCR (see ChIP). To remove melanin, DNA was column purified before PCR (OneStep PCR Inhibitor Removal Kit, Zymo research). Primer sequences are listed in Supplemental Table S2.
Transcription analysis
Total cellular RNA was isolated from the different cell lines with TriPure Isolation Reagent according to the manufacturer's instructions (Roche Diagnostics) or with the RNeasy Plus Mini kit (QIAGEN) following the manufacturer's instructions. To remove PCR-inhibiting substances such as melanin, RNA samples were column purified (OneStep PCR Inhibitor Removal Kit, Zymo Research Corporation) following the manufacturer's instructions. Subsequent DNase I digestion was performed with Ambion's Turbo DNA-free kit (Applied Biosystems) according to the manufacturer's protocol. The reverse-transcriptase (RT) reaction was performed using RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas GmbH) according to the manufacturer's instructions. Quantitative real-time PCR reactions for gene expression analysis were performed on a LightCycler 480 Realtime PCR System (Roche Diagnostics Nederland B.V.) with the Lightcycler 480 SYBR Green I master mix using the following parameters: initial denaturation for 10 min at 95°C, followed by 45 cycles for 10 sec at 95°C, for 30 sec at 60°C, and for 1 sec at 72°C, followed by a melting curve analysis and a final cooling step to 40°C. LightCycler 480 System Software v1.5 (Roche) was used to analyze the qPCR data. The reference gene ACTB was used to normalize the amplification signal between the samples of different cell lines, differences in treatment, and amount of input cDNA. The primer sequences are listed in Supplemental Table S2.
Luciferase assays
A 1450-bp fragment surrounding rs12913832 was PCR-amplified from genomic DNA obtained from HEMn-LP (C-allele) or HEMn-DP (T-allele) using the Expand Long Template PCR Kit (Roche). The PCR fragment was digested with HindIII and PstI, generating a 750-bp fragment that was subcloned in pBluescript. Sequencing verified the presence of the correct rs12913832 allele for each construct. Correct clones were digested with BamHI and Asp718I and cloned into the BglII/Asp718I sites of a modified pGL3-promoter vector (in which the SV40 promoter and the SV40 3′ UTR were replaced by an HSP promoter and an HSP 3′ UTR; Promega). Constructs were transfected into HEK293 or G361 melanoma cells using Lipofectamine 2000, and luciferase expression was normalized to Renilla luciferase expression. Chimerical luciferase reporter constructs containing different combinations of rs12913832 and rs6497271 alleles were generated by digesting HEMn-LP (C-allele) and HEMn-DP (T-allele) containing luciferase reporter constructs with XmnI and religating after swapping the fragments. Constructs were sequence-verified.
Chromosome conformation capture (3C) analysis
Chromosome conformation capture (3C) analysis was performed essentially as described (Palstra et al. 2003; Hagege et al. 2007) using EcoRI or ApoI as the restriction enzyme. To remove melanin, DNA was column purified before PCR (OneStep PCR Inhibitor Removal Kit, Zymo Research). Quantitative real-time PCR (CFX96 Real Time System, Bio-Rad) was performed using iTaq SYBR Green Supermix with ROX (Bio-Rad), under the following cycling conditions: 2 min at 50°C, 10 min at 95°C, 45 cycles of 15 sec at 95°C, 1 min at 60°C, followed by a melting curve analysis. Cross-linking frequencies between samples were normalized using primers in the nonexpressed human beta hemoglobin LCR and HBE1 (Palstra et al. 2003). A random template was generated as described previously (Palstra et al. 2003) using BAC RP11-1365A12 for the OCA2/HERC2 locus. For the human beta hemoglobin locus, we used a 185-kb PAC (Palstra et al. 2003). The primer sequences are listed in Supplemental Table S2.
Statistical analysis
Statistical calculations were performed in Excel, or using R software (for the data obtained in the ChIP assays AcH3/H3, H3K4Me1/H3, and H3K27Ac/H3). All P-values were calculated with t-tests.
Acknowledgments
We thank R.A. Poot for support in initiating this project and for useful comments on the manuscript. We also thank F. Grosveld for providing infrastructural support and for useful comments on the manuscript. R. Stadhouders and E. Soler are acknowledged for discussions, J. van Haren for providing the G361 melanoma cell line, D. Fisher (Dana-Farber Cancer Institute) for generously providing MITF constructs, K. van Duijn, M. Vermeulen, and Y. Choi for help with genotyping, K. van Duijn and O. Lao for help in genetic ancestry inferences, O. Lao for help with statistical analyses, and S. Walsh for valuable comments on the manuscript. R.J.P. was supported by the Netherlands Organisation for Scientific Research (NWO), grant number 700.57.408, and the Erasmus MC. M.K. was supported in part by the Netherlands Forensic Institute (NFI), the Erasmus MC, and a grant from the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO) within the framework of the Forensic Genomics Consortium Netherlands (FGCN).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.128652.111.
References
- Ahmadiyeh N, Pomerantz MM, Grisanzio C, Herman P, Jia L, Almendro V, He HH, Brown M, Liu XS, Davis M, et al. 2010. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc Natl Acad Sci 107: 9742–9746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branicki W, Brudnik U, Wojas-Pelc A 2009. Interactions between HERC2, OCA2 and MC1R may influence human pigmentation phenotype. Ann Hum Genet 73: 160–170 [DOI] [PubMed] [Google Scholar]
- Branicki W, Liu F, van Duijn K, Draus-Barini J, Pospiech E, Walsh S, Kupiec T, Wojas-Pelc A, Kayser M 2011. Model-based prediction of human hair color using DNA variants. Hum Genet 129: 443–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilliant MH 2001. The mouse p (pink-eyed dilution) and human P genes, oculocutaneous albinism type 2 (OCA2), and melanosomal pH. Pigment Cell Res 14: 86–93 [DOI] [PubMed] [Google Scholar]
- Coetzee GA, Jia L, Frenkel B, Henderson BE, Tanay A, Haiman CA, Freedman ML 2010. A systematic approach to understand the functional consequences of non-protein coding risk regions. Cell Cycle 9: 256–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook AL, Chen W, Thurber AE, Smit DJ, Smith AG, Bladen TG, Brown DL, Duffy DL, Pastorino L, Bianchi-Scarra G, et al. 2009. Analysis of cultured human melanocytes based on polymorphisms within the SLC45A2/MATP, SLC24A5/NCKX5, and OCA2/P loci. J Invest Dermatol 129: 392–405 [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiberg H, Troelsen J, Nielsen M, Mikkelsen A, Mengel-From J, Kjaer KW, Hansen L 2008. Blue eye color in humans may be caused by a perfectly associated founder mutation in a regulatory element located within the HERC2 gene inhibiting OCA2 expression. Hum Genet 123: 177–187 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman ML, Monteiro AN, Gayther SA, Coetzee GA, Risch A, Plass C, Casey G, De Biasi M, Carlson C, Duggan D, et al. 2011. Principles for the post-GWAS functional characterization of cancer risk loci. Nat Genet 43: 513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD 2007. FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res 17: 877–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T 2007. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat Protoc 2: 1722–1733 [DOI] [PubMed] [Google Scholar]
- Han J, Kraft P, Nan H, Guo Q, Chen C, Qureshi A, Hankinson SE, Hu FB, Duffy DL, Zhao ZZ, et al. 2008. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet 4: e1000074 doi: 10.1371/journal.pgen.1000074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, Ren B, Fu XD, Topol EJ, Rosenfeld MG, et al. 2010. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature 470: 264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Hewetson A, Chilton BS 2008. Progesterone-dependent deoxyribonucleic acid looping between RUSH/SMARCA3 and Egr-1 mediates repression by c-Rel. Mol Endocrinol 22: 813–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek KS, Schlegel NC, Eichhoff OM, Widmer DS, Praetorius C, Einarsson SO, Valgeirsdottir S, Bergsteinsdottir K, Schepsky A, Dummer R, et al. 2008. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res 21: 665–676 [DOI] [PubMed] [Google Scholar]
- Jia L, Landan G, Pomerantz M, Jaschek R, Herman P, Reich D, Yan C, Khalid O, Kantoff P, Oh W, et al. 2009. Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet 5: e1000597 doi: 10.1371/journal.pgen.1000597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao O, Vallone PM, Coble MD, Diegoli TM, van Oven M, van der Gaag KJ, Pijpe J, de Knijff P, Kayser M 2010. Evaluating self-declared ancestry of U.S. Americans with autosomal, Y-chromosomal and mitochondrial DNA. Hum Mutat 31: E1875–E1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy C, Khaled M, Fisher DE 2006. MITF: Master regulator of melanocyte development and melanoma oncogene. Trends Mol Med 12: 406–414 [DOI] [PubMed] [Google Scholar]
- Levy C, Khaled M, Robinson KC, Veguilla RA, Chen PH, Yokoyama S, Makino E, Lu J, Larue L, Beermann F, et al. 2010. Lineage-specific transcriptional regulation of DICER by MITF in melanocytes. Cell 141: 994–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, van Duijn K, Vingerling JR, Hofman A, Uitterlinden AG, Janssens AC, Kayser M 2009. Eye color and the prediction of complex phenotypes from genotypes. Curr Biol 19: R192–R193 [DOI] [PubMed] [Google Scholar]
- Liu F, Wollstein A, Hysi PG, Ankra-Badu GA, Spector TD, Park D, Zhu G, Larsson M, Duffy DL, Montgomery GW, et al. 2010. Digital quantification of human eye color highlights genetic association of three new loci. PLoS Genet 6: e1000934 doi: 10.1371/journal.pgen.1000934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay C, Toth R, Rouse J 2009. Biochemical characterisation of the SWI/SNF family member HLTF. Biochem Biophys Res Commun 390: 187–191 [DOI] [PubMed] [Google Scholar]
- Manolio TA 2010. Genomewide association studies and assessment of the risk of disease. N Engl J Med 363: 166–176 [DOI] [PubMed] [Google Scholar]
- Palstra R 2009. Close encounters of the 3C kind: Long-range chromatin interactions and transcriptional regulation. Brief Funct Genomics Proteomics 8: 297–309 [DOI] [PubMed] [Google Scholar]
- Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W 2003. The β-globin nuclear compartment in development and erythroid differentiation. Nat Genet 35: 190–194 [DOI] [PubMed] [Google Scholar]
- Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M, et al. 2009. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet 41: 882–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J 2010. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinchik EM, Bultman SJ, Horsthemke B, Lee ST, Strunk KM, Spritz RA, Avidano KM, Jong MT, Nicholls RD 1993. A gene for the mouse pink-eyed dilution locus and for human type II oculocutaneous albinism. Nature 361: 72–76 [DOI] [PubMed] [Google Scholar]
- Spichenok O, Budimlija ZM, Mitchell AA, Jenny A, Kovacevic L, Marjanovic D, Caragine T, Prinz M, Wurmbach E 2011. Prediction of eye and skin color in diverse populations using seven SNPs. Forensic Sci Int Genet 5: 472–478 [DOI] [PubMed] [Google Scholar]
- Strub T, Giuliano S, Ye T, Bonet C, Keime C, Kobi D, Le Gras S, Cormont M, Ballotti R, Bertolotto C, et al. 2010. Essential role of microphthalmia transcription factor for DNA replication, mitosis and genomic stability in melanoma. Oncogene 30: 2319–2332 [DOI] [PubMed] [Google Scholar]
- Sturm RA 2009. Molecular genetics of human pigmentation diversity. Hum Mol Genet 18: R9–R17 [DOI] [PubMed] [Google Scholar]
- Sturm RA, Larsson M 2009. Genetics of human iris colour and patterns. Pigment Cell Melanoma Res 22: 544–562 [DOI] [PubMed] [Google Scholar]
- Sturm RA, Duffy DL, Zhao ZZ, Leite FP, Stark MS, Hayward NK, Martin NG, Montgomery GW 2008. A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am J Hum Genet 82: 424–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela RK, Henderson MS, Walsh MH, Garrison NA, Kelch JT, Cohen-Barak O, Erickson DT, John Meaney F, Bruce Walsh J, Cheng KC, et al. 2010. Predicting phenotype from genotype: Normal pigmentation. J Forensic Sci 55: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JB, Brown SJ, Cole MD 2010. Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol Cell Biol 30: 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun K, So JS, Jash A, Im SH 2009. Lymphoid enhancer binding factor 1 regulates transcription through gene looping. J Immunol 183: 5129–5137 [DOI] [PubMed] [Google Scholar]