Abstract
Meiotic recombination, including crossovers (COs) and gene conversions (GCs), impacts natural variation and is an important evolutionary force. COs increase genetic diversity by redistributing existing variation, whereas GCs can alter allelic frequency. Here, we sequenced Arabidopsis Landsberg erecta (Ler) and two sets of all four meiotic products from a Columbia (Col)/Ler hybrid to investigate genome-wide variation and meiotic recombination at nucleotide resolution. Comparing Ler and Col sequences uncovered 349,171 Single Nucleotide Polymorphisms (SNPs), 58,085 small and 2315 large insertions/deletions (indels), with highly correlated genome-wide distributions of SNPs, and small indels. A total of 443 genes have at least 10 nonsynonymous substitutions in protein-coding regions, with enrichment for disease-resistance genes. Another 316 genes are affected by large indels, including 130 genes with complete deletion of coding regions in Ler. Using the Arabidopsis qrt1 mutant, two sets of four meiotic products were generated and analyzed by sequencing for meiotic recombination, representing the first tetrad analysis with whole-genome sequencing in a nonfungal species. We detected 18 COs, six of which had an associated GC event, and four GCs without COs (NCOs), and revealed that Arabidopsis GCs are likely fewer and with shorter tracts than those in yeast. Meiotic recombination and chromosome assortment events dramatically redistributed genome variation in meiotic products, contributing to population diversity. In particular, meiosis provides a rapid mechanism to generate copy-number variation (CNV) of sequences that have different chromosomal positions in Col and Ler.
Natural genomic variations, including SNPs, insertions, deletions, and CNV, are prevalent in many species and can generate new alleles/genes, reshape gene structures, alter gene dosage, and change gene expression level (Long et al. 2003; Mitchell-Olds and Schmitt 2006). In human and animals, genome variations are associated with severe genetic diseases, such as Parkinson and Alzheimer (Stankiewicz and Lupski 2002; Hurles et al. 2008). In plants, genome variations contribute to adaptive fitness by affecting traits such as flowering time, disease resistance, and seed dormancy (Johanson et al. 2000; Michaels et al. 2003; Koornneef et al. 2004; Krieger et al. 2010). Recently, genome-wide studies using microarray or next-generation sequencing (NGS) indicate that natural variations in humans, mouse, and flies are more abundant than previously thought (Graubert et al. 2007; Emerson et al. 2008; Kidd et al. 2008).
Genome variations are shaped by meiotic recombination and chromosome assortment, which reshuffles the genome in every generation. Because chromosome assortment has limited possibilities, whereas meiotic recombination, either as crossover (CO) or noncrossover (NCO, or GC without exchange of flanking regions), can occur at many sites along the chromosome, recombination has a greater potential to increase genetic diversity. Furthermore, COs result in large-scale (megabases) reciprocal exchanges of genetic materials between homologous chromosomes, whereas GCs unidirectionally copy kilobase(s) or less of DNA sequences from one homolog to the other, thereby altering frequency of natural variations (Zickler and Kleckner 1999; Ma 2006). COs and NCOs can be inferred by analysis of haplotype markers in population studies (Hurst et al. 1972; Haubold et al. 2002), which can only detect fixed recombination events, not changes per meiosis.
In fungi such as the budding yeast Saccharomyces cerevisiae, meiotic products are kept together as spores in an ascus, forming a tetrad. This allows direct examination of the consequence of meiotic recombination in parallel cultures derived separately from the four spores, using “tetrad analysis” (Zickler and Kleckner 1999). Tetrad analysis has contributed significantly to the understanding of the molecular basis of meiotic recombination, including strong support for the steps in the double-strand break repair model (DSBR) (Zickler and Kleckner 1999; Keeney 2001; Ma 2006; Mezard et al. 2007) and used in yeast to examine the frequency and genome-wide distribution of meiotic recombination (Mancera et al. 2008; Qi et al. 2009). Here, we combined next-generation sequencing and tetrad analysis to investigate natural variations and meiotic recombination in the flowering plant Arabidopsis.
Arabidopsis thaliana is native to Europe and central Asia (Koornneef et al. 2004), with many genetic (geographical) variants (derivative lines are called accessions) adapted to different environments. The Columbia (Col) accession is widely used for molecular genetic studies and was sequenced by the Arabidopsis Genome Initiative as the genomic reference (Arabidopsis Genome Initiative 2000). Similar to Col, the Landsberg erecta (Ler) accession is also widely used for functional studies (Meyerowitz and Ma 1994). The Col and Ler lines have extensive DNA polymorphisms (Nordborg et al. 2005; Ziolkowski et al. 2009), even though they were both derived by George Redei at the University of Missouri, Columbia, from a heterogeneous population named Landsberg collected by Laibach: Col was selected from a group of nonirradiated Landsberg plants, whereas the Ler line was derived from X-ray-mutagenized Landsberg plants (http://arabidopsis.info/protocols/ler.html). Polymorphisms among accessions were analyzed by sequencing hundreds of short fragments or using oligonucleotide arrays (Nordborg et al. 2005; Clark et al. 2007). Moreover, genome variations among Col-0, Bur-0, and Tsu-1 were recently analyzed by Illumina sequencing (Ossowski et al. 2008). Because natural variations can have profound effects on gene function (Koornneef et al. 2004; Bentsink et al. 2010; Guyon-Debast et al. 2010; Todesco et al. 2010), a genome-wide examination of Arabidopsis natural variation, such as that between Col and Ler, will greatly facilitate the understanding of the effect of variation on gene functions, and the mapping of quantitative trait loci (QTL) using recombination inbred lines (RILs), each with a set of homozygoticized parental alleles.
Moreover, because the Arabidopsis qrt1 mutant meioses produce all four meiotic products as attached spores, which then develop into attached functional pollen grains, Arabidopsis offers a unique opportunity among plants and animals to carry out tetrad analysis, as was done using hundreds of DNA markers (Preuss et al. 1994; Francis et al. 2007). Here, we sequenced the Ler genome using high-throughput sequencing to uncover over 400,000 genome variations between Ler and Col, with functional implications. We then utilized SNP markers to examine meiotic recombination in two meioses by sequencing all products, allowing the detection of CO and NCO/GC events and the characterization of the GC tracts at nucleotide resolution. The sequencing data also displayed the redistribution of natural variations following a single meiotic generation.
Results
Sequencing of the Ler genome uncovered numerous SNPs with functional implications
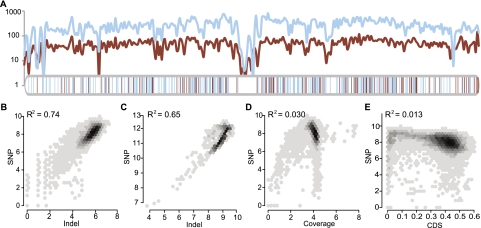
We sequenced Ler to obtain over 61.6 million reads, ∼62.7% of which were uniquely mapped to the Col reference genome, providing ∼18.7× coverage (Supplemental Table 1). We identified a total of 349,171 SNPs between the two genomes, offering a map of molecular markers with an average distance of 340 bp (median distance of 118 bp) between adjacent markers (Fig. 1A; Supplemental Fig. 1A,C). PCR amplification and Sanger sequencing of 23 randomly selected regions with 62 predicted SNPs on chromosome 1 confirmed all of them and also detected a few additional SNPs (data not shown), indicating that SNP identification was conservative and accurate, providing a valuable resource.
Figure 1.
The correlation of SNP and small indel densities. (A) Parallel change of SNP and small indel density on Chr1. The density was defined to be the number of SNPs/indels per 100 Kb. (Blue curve) SNP density; (red curve) small indels. Blue and red vertical bars below show the location of large deletions and insertions, respectively. (B,C) Linear regression of SNP and indel densities in 100-Kb (B) or 1-Mb (C) sliding windows. (D,E) Linear regression of SNP density with read coverage (D) or CDS fractions (E) in a 100-Kb sliding window. All values are log2 transformed before applying regression. A near zero R2 value suggests that read coverage or CDS fractions do not contribute much to the correlation.
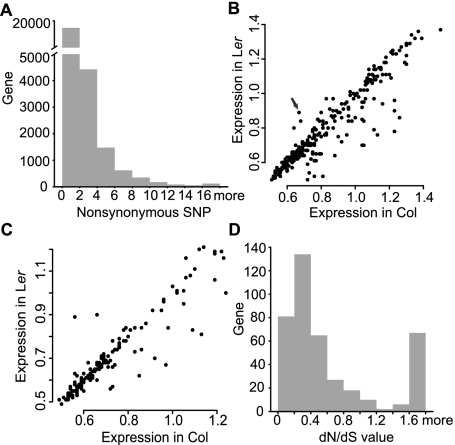
To assess the possible functional implications of genome variations, we analyzed their distribution and effect on protein sequences. Of the SNPs, 76,649 (∼22%) are in protein-coding sequences (CDS) and 194,911 (∼56%) in intergenic regions. Among the SNPs in CDS, nearly half are (35,798) caused by nonsynonymous changes, affecting 13,158 of the 27,169 annotated protein-coding genes. Most of the affected genes carry only a few nonsynonymous SNPs, but 443 genes have 10 or more nonsynonymous substitutions (Fig. 2A; referred to as the 443 genes hereafter). In addition, 357 SNPs lead to premature stop codons in 319 genes (referred to as the 319 genes) in Ler relative to Col, suggesting possible defects in the Ler alleles. In contrast, 89 genes had a sense codon in Ler corresponding to the stop codon in Col, suggesting longer coding regions in Ler (Supplemental Table 2) and possible defects in Col.
Figure 2.
Nonsynonymous SNPs and affected genes. (A) The number of nonsynonymous SNPs per gene. Although about half of the genes contain nonsynonymous SNPs, a much smaller set of genes has 10 or more nonsynonymous SNPs. (B) Expression levels in Col (x-axis) and Ler (y-axis) of genes with 10 or more nonsynonymous SNPs. The arrow points to AT5G58120, which has higher expression in Ler and encodes a disease resistance protein. (C) Expression levels in Col (x-axis) and Ler (y-axis) of genes with a Ler-premature stop codon. (D) Arabidopsis branch-specific dN/dS value of genes affected by 10 or more nonsynonymous SNPs with regard to A. lyrata. Only a few genes show neutral evolution, but most are under either positive or negative selection.
To obtain clues about the potentially functional effect of the coding differences, all genes were classified into three annotation types: “known,” “unknown” with EST or cDNA support, and “unknown” without expression information. Surprisingly, smaller fractions of the 443 genes were found in either of the “unknown” categories than in whole-genome frequencies (χ2 test, P = 0.0019; Supplemental Tables 2, 3), suggesting that the genes with amino acid differences are not more likely to be pseudogenes than the genome average. The 319 genes have more than expected unknown genes without expression information but, still, the majority had annotated functions (Supplemental Table 2). A total of 68.2% of the 443 genes are members of multigene families (χ2 test, P = 1.27 × 10−10), compared with 53% of all annotated protein-coding genes in multigene families, suggesting that nonsynonymous mutations might be more tolerated in multigene family members. In contrast, only 57.1% of the 319 genes belong to multigene families (χ2 test, P = 0.15). Strikingly, all members are altered (missense or nonsense) in Ler in six gene families, two of which are involved in resistance to biotrophic oomycetes and other pathogens (Rentel et al. 2008).
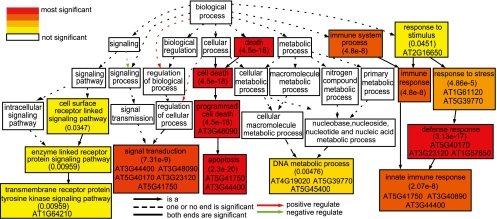
We further examined Gene Ontology (GO) for possible enrichment of specific categories among the genes affected by genome variations (Fig. 3; Supplemental Fig. 2). Among the 443 genes, those related to defense response, apoptosis, transmembrane receptor, and ATP binding are enriched, supporting the idea that Col and Ler differ in defense response. We also examined the expression profile according to the Plant Ontology (PO) database and found that most enriched PO groups are reproductive tissues and stages (Supplemental Table 4). According to microarray analyses (Schmid et al. 2003), most of mutated genes have similar or lower expression levels in Ler than that in Col (Fig. 2B,C). However, a few genes, such as the AT5G58120 gene encoding a disease-resistance protein, are expressed at higher levels in Ler than in Col (Fig. 2B). In short, changes in gene sequence and expression might impact functions, such as pathogen response.
Figure 3.
Gene Ontology groups enriched among genes with 10 or more nonsynonymous SNPs. Statistical significance is color coded, with Yekutieli FDR adjusted P-value shown in each significant group. Most enriched GO groups contain some genes with large dN/dS values (≥1.6), as shown by TAIR gene IDs in the box.
To investigate whether these genes have been under selection, we estimated the dN/dS ratio of the Arabidopsis thaliana branch after divergence from a close relative, Arabidopsis lyrata. dN is the rate (observed over possible changes) of nonsynonymous substitutions, whereas dS is the rate of synonymous substitutions. Neutrally evolved genes tend to have dN/dS values close to 1. In contrast, genes under negative or positive selection tend to have dN/dS values close to zero or larger than 1, respectively. Most of the 443 genes have dN/dS values of from 0 to ∼0.4 (negative selection indicative of highly conserved functions), but 50 genes have much larger dN/dS values (≥1.6) using all four amino acid frequency models, suggesting positive selection for new functions (Fig. 2D; Supplemental Table 5). Interestingly, a comparison of GO results with dN/dS analysis revealed that 21 of those 50 genes belong to enriched GO groups (Fig. 3), further supporting the idea of altered functions.
Numerous small indels with similar distribution patterns to those of SNPs
Small indels of several nucleotides were previously found to be more prevalent between Arabidopsis variants than among humans (Ossowski et al. 2008). We uncovered 58,085 indels of from 1 to 4 bp, with the median distance between indels being 919 bp, and noticed that distribution of SNPs and small indels showed parallel patterns across the entire genome (Fig. 1A; Supplemental Fig. 1B,C). To investigate the potential correlation between SNPs and small indel frequencies, linear regression with log transformation was applied in 10-Kb, 100-Kb, or 1-Mb sliding genomic windows. SNPs and indels are highly correlated in 100-Kb and 1-Mb windows, with R2 values of 0.74 and 0.65, respectively (Fig. 1B,C), more than that in 1-Kb windows (R2 = 0.52; Supplemental Fig. 1D). To exclude the possibility that more SNPs and indels might be detected in regions with high read coverage, the density of SNPs and indels were examined with regard to read coverage in each 100-Kb genomic window, but no correlation was observed (Fig. 1D; Supplemental Fig. 1E). Moreover, the fraction of CDS in each window cannot explain the change in frequencies of SNP/indel (Fig. 1E; Supplemental Fig. 1F). This strong correlation between frequencies of SNPs and small indels is similar to the mosaic pattern in mouse (Sakai et al. 2005; Tsang et al. 2005; Yang et al. 2011), which is probably due to variation in divergence time among different genomic regions. Since Col and Ler were derived from the same natural population, some genomic regions might have inherited the same haplotype, whereas in other regions Col and Ler might have had different haplotypes with longer divergence time and more polymorphisms.
Further, 1674 of the small indels are inside CDS, causing frameshift in 844 genes and nonframeshift changes in 461 genes (Supplemental Table 6). The number of affected genes is very large, considering that Col and Ler have diverged only ∼200,000 yr ago (Ziolkowski et al. 2009). Similar to the SNPs-impacted genes discussed in the previous section, most genes affected by indels have very low dN/dS values in comparison with A. lyrata, indicating that these genes have been under purifying selection during the 10 million years of divergence between A. thaliana and A. lyrata (Hu et al. 2011), but have changed more recently between Col and Ler (Supplemental Fig. 3A,B). Nevertheless, some of the genes affected by small indels have dN/dS ratios of 1.6 or higher (Supplemental Fig. 3A,B), suggesting that they might have been under positive selection. In addition, many genes with frameshift mutations have lower expression in Ler, but fewer nonframeshift genes do so (Supplemental Fig. 3D,E). GO categories of transmembrane receptors and ATP binding are over-represented among genes affected by frameshift mutations, whereas genes with nonframeshift mutations are enriched for transcription factors, suggesting functional differences in these categories (Supplemental Fig. 3G–J).
Detection of large indels and CNVs
To detect large genomic variations, we assembled paired-end reads from Ler into 30,217 contigs that ranged from 100 bp to 119 Kb (N50 = 11 Kb), representing ∼78% of the Col reference genome, and then aligned them with the Col genome. A total of 16,560 contigs were mapped to unique sites, 7503 had their segments mapped orderly, 994 had rearrangements for mapping positions, and the remaining were not mapped. From uniquely mapped contigs, we identified 1658 large deletions (median size, 730 bp) and 700 large insertions (median size, 266 bp) (Supplemental Table 7), spanning cumulatively 2841 and 372 Kb, respectively. To evaluate the indels, we compared them with the Monsanto Ler contigs (81,306) (downloaded from TAIR), which matched to ∼60% of the Col genome. About 78% of the deletions we detected were also uncovered by the Monsanto contigs, with ∼99% of them consistent in both data sets. Moreover, ∼71% of the insertions we detected were confirmed by the Monsanto contigs, with ∼96% of them in agreement. In addition, 28 of the indels we identified were tested by PCR, and 20 displayed different band sizes between Col and Ler (Supplemental Fig. 4A,B).
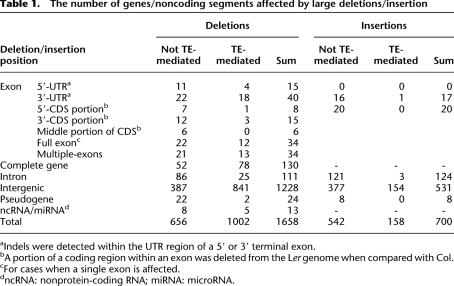
The 2315 large indels are widely dispersed along chromosomes (Fig. 1A; Supplemental Fig. 1C). A total of 1759 (75.9%) are located in intergenic regions, while the others contribute to the gain/loss of exons/introns/untranslated regions (UTRs) or even the entire genes (Table 1). One-hundred and thirty single-copy genes were absent in the Ler genome (Supplemental Table 8), with one example (At1g51430.1) confirmed by PCR (Supplemental Fig. 4C). Of these 130 genes, 25 were found to have an ortholog in A. lyrata. In addition, 107 putative genes were predicted from Ler-specific sequences; nine of the 107 were detected in A. lyrata. Furthermore, 186 genes with exons/UTRs affected by indels (Supplemental Table 9) could have changed/disrupted expression or functions. F-box genes encode subunits of E3 ubiquitin ligases that are involved in physiological and environmental responses and differ dramatically in gene number among A. thaliana, poplar, and rice (Xu et al. 2009). We found that eight F-box genes are absent from Ler (Supplemental Table 8) and 16 other F-box genes are partially affected (Supplemental Table 9), suggesting that these rapidly evolving genes are highly unstable even within the Arabidopsis species. In addition, Ler lacked four disease-resistance genes and part of six others (Supplemental Tables 8, 9). As large deletions affecting genes can cause phenotypic variations among different accessions (Kroymann et al. 2003), it was surprising to see that most genes affected by large indels show low dN/dS value compared with A. lyrata (Supplemental Fig. 3C), suggesting that they have been conserved and under purifying selection since the separation of the two species. However, many of these genes have lower expression levels in Ler than those in Col (Supplemental Fig. 3F), possibly due to nonsense-mediated decay. Furthermore, large indels also lead to reciprocal loss of genes in Col and Ler for 22 homologous gene pairs (Supplemental Table 10), providing possible examples of gene loss following duplication.
Table 1.
The number of genes/noncoding segments affected by large deletions/insertion
Large indels might include copy-number variations (CNVs) between Col and Ler. To test this, we used the sequences affected by these indels to search against the Col genome. We defined CNVs as indels of one or more copies of similar sequences. Using a criterion of 80% identity over 80% of the query, we identified 614 deletions (∼38%) and 20 insertions (∼3%) affecting sequences similar to other copies in the Col genome. A total of 85 of the CNVs affected genes (Supplemental Table 7), including some that occurred in tandemly duplicated genes, for example, in a cluster of genes encoding carbohydrate-binding X8 domain proteins with over 96% identity in amino acid sequence, Col has five copies (AT4G09462.1, AT4G09464.1, AT4G09465.1, AT4G09466.1, and AT4G09467.1), but Ler had a deletion of AT4G09467.1. A major type of CNVs affected transposable elements (TEs) or gain/loss of adjacent genes (Supplemental Table 7). For example, among the members of the ATREP1, ATREP2, and ATREP3 TE families present in Col, 13, 13, and 20, respectively, were absent in Ler (Supplemental Fig. 5). In our study, 997 of 1758 (56.7%) gain/loss of DNA segments in intergenic regions and 149 of 557 (26.8%) in genes contained a segment with high-sequence similarity to known TEs in Col, suggesting a role of TEs in generating CNVs.
Generating and sequencing “tetrads” of meiotic progeny plants
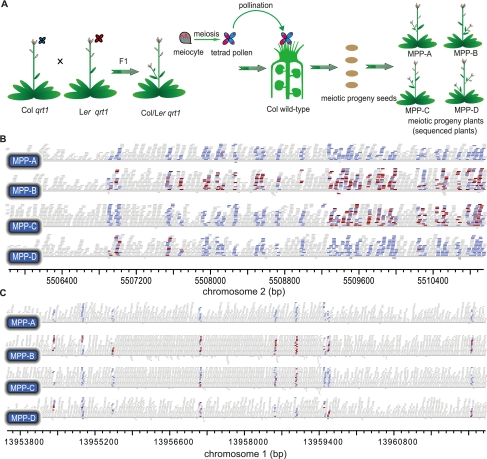
To observe meiotic recombination using tetrad analysis (Supplemental Fig. 6) with homologous chromosomes that have numerous polymorphic markers, we constructed a hybrid between Col and Ler, each mutated for the QRT1 gene (Fig. 4A). A tetrad of four attached pollen grains from the Col/Ler qrt1/qrt1 F1 hybrid was used to pollinate a single pistil of an emasculated Col flower, producing four seeds. Here we named the plants grown up from the four seeds as meiotic progeny plant (MPP)-A, (MPP)-B, (MPP)-C, and (MPP)-D (Fig. 4A), each containing the paternal genome with a mixture of Col and Ler DNA and the maternal genome of 100% Col DNA. We sequenced eight MPP genomes from two independent meioses, named as the first meiosis and the second meiosis, yielding sequence information for each plant with ∼8.2 to ∼16.6× coverage, matching 94% to ∼97.1% of the Col genome (Supplemental Table 1).
Figure 4.
Tetrad analysis detecting meiotic CO and NCO tracts using genomic sequencing in Arabidopsis. (A) A schematic illustration for generation of meiotic progeny plants (MPPs) to detect meiotic recombination using high-throughput sequencing. (Blue) Col genotype; (red) Ler genotype. (B) An example of detected CO on Chr2. MPP-A has a pure Col genotype, except one red bar, possibly due to sequencing error. MPP-B has equal representation of blue and red bars, carrying the Ler paternal genotype and the Col maternal genotype. Sequence exchange between MPP-C and MPP-D shows a CO event. One red bar in MPP-D to the right was likely due to sequencing error. The CO tract in the middle contains a 3:1 gene conversion, indicating repair of DSB in the Ler chromatid by using the homologous Col chromatid as a template. (C) An example of detected NCO in Chr1 from the first meiosis. Conversion occurred in the chromatid inherited by MPP-C from the Col to Ler genotype, leading to the 3:1 ratio in this region. (Blue horizontal bars) Mapped reads with a Col-specific SNP; (red bars) reads with a Ler-specific SNP; (gray bars) reads without a SNP. Each MPP plant contains one set of chromosomes from a Col/Col mother and another set of chromosomes from a Col/Ler hybrid.
Single-base resolution analysis of COs and NCOs/GCs
Meiotic CO events at single-base resolution were investigated by analyzing sequencing reads from different MPPs (Supplemental Fig. 7), such as the CO on Chr2 as revealed by mapped reads from MMP-C and MMP-D (Fig. 4B). We detected a total of 18 COs (Fig. 5), which were verified by PCR and conventional sequencing (data not shown). All COs were located in the intergenic regions and each chromosome experienced at least one CO (Fig. 5), consistent with its role in holding homologs together for accurate segregation. Nine COs were found in each meiosis (Fig. 6A; Supplemental Table 11), in remarkable agreement with a previous estimate of 9.24 COs from cytological and molecular genetic analyses (Sanchez Moran et al. 2001; Copenhaver et al. 2002).
Figure 5.
The distribution of COs and NCOs in the first and second meioses. Either meiosis has nine COs. (Cyan) Col genotype; (red) Ler genotype; (×) location of CO. A, B, C, and D represent four meiotic progeny plants, respectively. Green vertical bars show the detected NCOs positions. One or both of two closely spaced COs (*) in Chr3 from the second meiosis could be from an interference-insensitive pathway.
Figure 6.
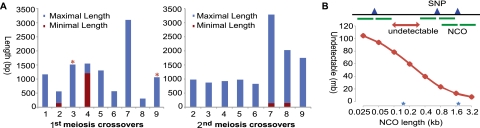
Properties of COs and NCOs in Arabidopsis. (A) The minimal (red) and maximal (blue) lengths of detected CO-associated conversion tracts from the first meiosis (left) and the second meiosis (right). The maximal length is the distance between two closest SNPs in unchanged regions flanking CO. The minimal length refers to the region having multiple 3:1 converted SNPs (one of the chromatids having a converted allele). In either the first or the second meiosis, there are two COs containing multiple converted SNPs, with maximal lengths ranging from ∼500 to ∼3000 bp. The red asterisks at the top of two bars indicate that the corresponding COs each had a 1-bp GC. In the second meiosis, CO-1 is not displayed here because it occurred in a large region without SNPs, making the position of CO-1 uncertain. (B) Predicted cumulative length of uncovered regions changes with possible length of NCO. NCO can only be detected if it covers at least one SNP, but is invisible between two adjacent SNPs. The predicted cumulative length of uncovered regions increases significantly when the length of NCO diminishes. Two blue stars show previously reported median NCO tract length from yeast (1.8 Kb) and human (156 bp).
Similar to the budding yeast, Arabidopsis uses the interference-sensitive pathway for the formation of a large majority of COs and the interference-insensitive pathways for a clearly detectable minority of COs (Copenhaver et al. 2002; de los Santos et al. 2003; Higgins et al. 2004; Hollingsworth and Brill 2004; Chen et al. 2005; Wijeratne et al. 2006). From physical distance between two COs, 16 of the 18 COs could be derived via the interference-sensitive pathway. In contrast, two COs were closely located on chromosome 3 in the second meiosis (Fig. 5, *), being only ∼244 Kb (<1 cM) apart, much smaller than the average interval (10.6 Mb) between other adjacent COs, suggesting that one or both of these COs could be generated by the interference-insensitive pathway.
Meiotic COs are known to distribute unevenly along the chromosomes, with recombination hot and cold spots (Copenhaver et al. 2002; Baudat et al. 2010). In Arabidopsis, putative hot spots have been reported in short regions (a few kilobases) on Chr4 (Drouaud et al. 2006). Here, one CO on Chr4 was in one of these regions (Supplemental Fig. 8). In addition, two COs, one in each meiosis analyzed here, around 25.6 Mb on Chr1, were located with a distance of only ∼2 Kb (Fig. 5), possibly representing a hot spot, which is within the size range of from 1 to 10 Kb for mammalian meiotic recombination hot spots (Kauppi et al. 2004).
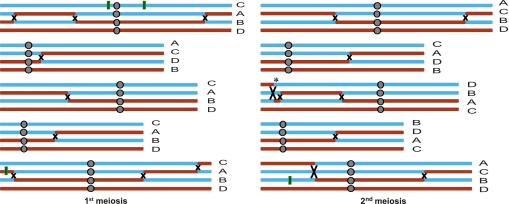
Tetrad analysis of all four meiotic products with single-base resolution allowed us to estimate the maximum (using flanking SNPs) and minimum (using converted SNPs) sizes of CO-associated conversion tracts (COCTs) for the first time in a multicellular organism (Supplemental Fig. 6; see Methods). One CO was located in a 129,507-bp region (Chr1: 8,733,517–8,863,024 bp) without SNPs, whereas the remaining 17 COs had maximum lengths of COCTs ranging from 306 to 3288 bp (Fig. 6A), with a median of maximum size of COCT tracts of 1115 bp, shorter than the median maximum estimate (2643 bp) of the budding yeast COCTs (Qi et al. 2009). Among the 17 COCTs described here, 47% had maximum lengths of <1 Kb and 35% had maximum lengths of 1–2 Kb. In another study of yeast CO utilizing microarrays (Mancera et al. 2008), the arithmetic average of minimal and maximal estimates of COCTs was defined as the midpoint length. The median of midpoint length of 18 COCTs detected here in Arabidopsis was 558 bp, significantly shorter than the 2-Kb value in yeast (Mancera et al. 2008) (Wilcoxon rank-sum test, P = 5.14 × 10−5).
According to the double-strand break repair model (DSBR) for meiotic recombination (Zickler and Kleckner 1999; Keeney 2001), the gap generated following the DSB is repaired using homologous sequences, leading to GC if there is sequence polymorphism. In the budding yeast, genome-wide analyses showed that most COCTs have a simple 3:1 GC pattern, but a small fraction of COCT regions had complex patterns (Mancera et al. 2008; Qi et al. 2009). Our analysis showed that all six COs with internal SNPs for GC detection were associated with 3:1 type GCs, including two with a single SNP, three with 140–150-bp COCTs, and a large one with a COCT of 1208 bp. It is possible that the size of the initial DSB gap in Arabidopsis could be ∼150 bp or shorter, whereas possible expansion of the dHJs could lead to longer conversion tracks. Interestingly, one CO (the CO on Chr2 in the first meiosis) spanned an 86-bp deletion in Ler, resulting in the removal of the deletion in one daughter cell via GC (Supplemental Fig. 9A,B).
In addition to COs, we also detected three and one NCO/GC events in the first and the second meioses, respectively, as confirmed by PCR and sequencing. All NCOs/GCs were located in intergenic regions. The longest NCO/GC tract, in the first meiosis, showed conversion of three SNPs spanning 1799 bp from Col to Ler in the MMP-C plant (Fig. 4C). The other NCO/GC tracts had minimum sizes (the region between the converted SNPs) of 1 bp. The estimated maximum NCO/GC tracts (the distance between the closest SNPs unaffected by the NCO) ranged from 3078 to 6696 bp (Supplemental Table 12). Because each MPP contains the maternal Col genome, we could only detect NCO/GC events with a sequence change from the Col allele to the Ler allele. Assuming equal frequency in both directions of conversion, Arabidopsis could have approximately six NCO/GC events per meiosis.
Redistribution of genome variations after meiosis
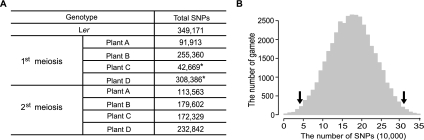
The sequences from the eight MPPs provided a unique opportunity to investigate the newly generated genetic architectures following meiosis. Figure 7A and Supplemental Table 13 show the patterns of redistributed SNPs and indels in the two sets of MPPs, compared with Col. For the first meiosis, the MPPs had 42,669–255,360 SNPs and 7632–53,149 indels. The MPPs from the second meiosis had 113,563–179,602 SNPs and 19,288–40,607 indels, quantitatively demonstrating the reshuffling of genetic variations due to meiotic recombination and chromosome assortment. To further investigate the redistribution of genetic variations, we simulated 10,000 meioses, producing 40,000 meiotic products, by comparing the genetic map and physical map and assigning COs according to genetic distance. The simulated COs were used to predict the number of SNPs and indels in meiotic products. Strikingly, two meiotic products had very small or large numbers of variations, respectively, in the tails of a simulated distribution of a number of SNPs and indels, with one highly similar to Col and the other to Ler (Fig. 7A,B). The generation of two extreme products might be due to two factors in this meiosis (first meiosis): (1) preferential occurrence of COs between the same two chromatids, such as the three COs for Chr1 and two COs for Chr5; (2) nonrecombinant chromatids tended to be assorted into the same two products (MPP-C or MPP-D), whereas recombinant chromosomes tended to be the other two products (MPP-A and MPP-B) (Fig. 5).
Figure 7.
The distribution of SNPs in meiotic products. (A) The number of SNPs in Ler and eight experimented meiotic products. As the Col sequence was used as reference, the pure Col region had zero SNPs and only bases different from Col were counted. Two extreme gametes are marked by black asterisks, with one highly similar to Col and the other very similar to Ler. (B) The distribution of SNPs in 40,000 simulated gametes. Simulation was performed according to genetic and physical maps of Arabidopsis. The unit of the x-axis is 10,000. Two black arrows indicate the location of two extreme gametes in the simulated distribution.
CNVs due to meiotic reshuffling of structural variants
CNVs can affect various biological processes (Zhang et al. 2009), thus estimating that the rate of CNV generation is critical to understanding their effects on genome evolution and gene functions. The rates of de novo CNV in human have been estimated; for example, one study found that the rate for total genome-wide new large CNVs (>100 Kb) is about 1.2 × 10−2 per genome per transmission (Itsara et al. 2010), and another study reported that most of over 4000 CNVs analyzed had individual rates of ∼10−5 per generation (Fu et al. 2010). Our limited analysis revealed that meiosis can rapidly generate CNVs among siblings, producing 21 and 32 CNVs in the two sets of four meiotic products, respectively (Supplemental Table 14). Further examination of Ler reads with PCR verification showed that these CNVs were due to reshuffling of existing highly similar sequences that map to different locations (Supplemental Fig. 10). These nonallelic similar sequences could be on the same chromosome, and a CO between them can lead to CNVs in the meiotic products (Supplemental Fig. 10B). When the similar sequences are on different chromosomes, only the assortment of the Col and Ler chromosomes is needed to cause CNVs in the meiotic products (Supplemental Fig. 10C). Arabidopsis can outcross 3% of the time in environments with natural populations (Platt et al. 2010), generating hybrids in which CNVs can be formed from reshuffling much more frequently compared with de novo mutation.
Discussion
Genetic variation and phenotypic variation
Natural variations are shaped by integrated forces of mutation, recombination, and selection, causing phenotypic differences and affecting individual adaptation to local environments. We have identified >400,000 SNPs, indels, and CNVs between Col and Ler, two accessions that are thought to have diverged ∼200,000 yr ago (Ziolkowski et al. 2009), even though there probably has been more recent gene flow between them (see Introduction). We have analyzed the Col/Ler genome variations and found evidence for functional divergence between alleles in Col and Ler, including multiple amino acid substitutions, premature stop codons or extension of reading frame in Ler, small indels causing frameshifts, and large indels of part or all of coding regions. Some of these genes have predicted functions that could influence the adaptive fitness of Col or Ler, potentially impacting traits such as disease resistance and flowering time for plant health and reproductive success, respectively.
In Arabidopsis, genetic variations from SNPs to CNVs could affect gene functions; for example, SNPs can change amino acids in phytochrome A (PHYA) and B (PHYB), affecting light responses and flowering time (Maloof et al. 2001; Filiault et al. 2008). A SNP also created a new splicing site that altered gene function (Guyon-Debast et al. 2010). Small and large indels in the RPS2 and MAM2 genes, respectively, caused pathogen sensitivity (Mindrinos et al. 1994; Kroymann et al. 2003). We found that genes responsible for biotic stress responses are enriched among genes specifically altered between Col and Ler, including those encoding F-box proteins, LRR (Leucine Rich Repeat)-RLK (Receptor-Like Kinase), RLP (Receptor-Like Protein), and NBS (Nucleotide Biding Site)-LRR proteins (Supplemental Table 15).
Possible relationship between frequency of CO, genome size, and length of synaptonemal complex
Human and mouse, as well as other animals and plants, have very different genome sizes, yet all have one to three COs per homolog pair (Baker et al. 1995; Barlow and Hulten 1998), similar to Arabidopsis, but unlike the two to 11 COs per chromosome in the budding yeast (Mancera et al. 2008; Qi et al. 2009) (Supplemental Table 16). Recent studies indicated that between individuals of the same species for human and others, the genetic distance (CO number) is positively correlated with the length of the synaptonemal complex (SC) but not the length of DNA (Lynn et al. 2002; Kleckner et al. 2003). However, this correlation does not seem to hold between species; for example, human, Arabidopsis, and the budding yeast have SC lengths per chromosomes of approximate 10–25, 2–3, and 1–2 microns, yet the CO numbers per chromosomes are 1–3, 1–3, and 2–11, respectively (Dresser and Giroux 1988; Barlow and Hulten 1998; Wijeratne et al. 2006). Strikingly, the ratios of genome size to SC length are very similar between human (∼10–12 Mb/micron) and Arabidopsis (∼12 Mb/micron), but much smaller in the budding yeast (∼0.5 Mb/micron). Because a similar number of chromatin loops are packed into the same SC length (Zickler and Kleckner 1999), the sizes of chromatin loops associated with SC are likely similar between human and Arabidopsis and are ∼20 times larger than that in yeast, providing a possible explanation for the difference in the number of COs per chromosome. Our results also suggest that Arabidopsis has shorter COCTs (CO-associated conversion tracts) than that in yeast (Mancera et al. 2008; Qi et al. 2009). The ability to conduct tetrad analysis in Arabidopsis offers great opportunities to gain further insights into the molecular control of meiosis in multicellular organisms.
The low frequency of detected NCOs and possible short GC tracts
We detected only ∼15 (nine COs + six NCOs) recombination events per meiosis; however, fluorescence immunolocalization studies in Arabidopsis detected about 120 AtRAD51 foci per meiotic cell (Sanchez-Moran et al. 2007), suggesting that there are over 100 DBS sites in a single meiosis. It is possible that there are many recombination events, but most are not detected because the sizes of DSB gaps and GC tracts are very small. An analysis of genomic regions relative to SNP distribution indicated that near 80% of the genome is at least 100 bp from any SNP (Fig. 6B). If the GC tract length is 100 bp or shorter, ∼80% or more of the NCO recombination events would be undetectable; therefore, our results could suggest that GC tracts in Arabidopsis are very short. Alternatively, in a fraction of the DBS repair events, the repair of meiotic heteroduplex DNA could also result in restoration of the parental genotypes in regions flanking the initial DSBs, as observed in yeast (Borts et al. 2000). A third possibility is that some DSBs might be repaired using the sister chromatids as templates, resulting in no GC. Recent studies showed that about one-third of the breaks could be repaired using the sister chromatid (Baarends and Mercier 2010); however, this could not fully explain the difference between the numbers of detected CO and NCO events and the observed AtRAD51 foci. Therefore, short GC tracts of ∼100 bp or less are at least one of the explanations for our results. On the other hand, if we assume the length of NCO in Arabidopsis is between those of yeast and human, the frequency of NCO is estimated to be four to approximately eight per meiosis (Fig. 6B; Supplemental Information). Another way to estimate the frequency of NCO per meiosis is to use the fraction of COs with detected GCs in COs, because all COs should have a conversion tract if there are SNPs. Among the 18 COs we observed, six had detectable Gcs, but 12 did not (Fig. 6). If the same fraction of NCO events were not detected due to the lack of SNPs, then there should be another 12 GCs/NCOs per meiosis, in addition to the six we estimated.
The redistribution of natural variations and generation of new CNVs
Although Arabidopsis is a predominantly self-pollinating plant, 3% outcrossing still allows gene transfer among different accessions (Platt et al. 2010). When the population faces the challenges of changes in the external environment, some hybrids that possess newly generated genotypes might confer better adaptation and out-compete others. Our data of four meiotic descendants from either of two meioses showed two interesting outcomes of outcrossing: (1) Meiosis indeed dramatically alters genetic variations, distributing the alleles from the two parents, creating new strains with new combinations of genes that are vastly different from either parent, and (2) reshuffling of existing structural variants can generate new CNVs in a rapid manner. These results provide a direct view of the landscapes of genetic variations at whole-genome scale, revealing how a single round of meiotic recombination and chromosome assortment can serve to reshape natural variations.
Methods
Plant material and growth conditions
Arabidopsis thaliana “Columbia-qrt1” and “Landsberg erecta-qrt1” from Dinesh Kumar's lab at Yale University were crossed to obtain F1 hybrids, whose tetrads of pollen were used to pollinate the Arabidopsis Columbia accession (Fig. 4A; see Supplemental Information for details). The resulting four seeds were named MPP-A, MPP-B, MPP-C, and MPP-D, and allowed to mature. All of the plants were grown under long-day conditions (16 h day and 8 h night) in a growth chamber at 18–22°C.
DNA isolation, genotyping, and genome resequencing
DNAs were extracted from each of MPPs and Ler leaves using the QIAGEN DNeasy Plant Mini kit (Cat#: 69104) and genotyped using SSLP markers, including NGA126, CIW4, NGA1126, and NGA63. Genomic DNA from Ler and meiosis progeny plants (MPPs) were subjected to Illumina sequencing. The genomic DNA of Ler were sequenced in six lanes of Genome Analyzer II (26,420,944 single-end reads of 36 bp) at FASTERIS SA, Switzerland and one lane of Genome Analyzer IIX (17,591,335 paired-end reads of 75 bp) at Beijing Genomics Institute (BGI), Shenzhen, China. Each MPP of the first meiosis was sequenced in two or three lanes of Genome Analyzer II (paired-end, 40 bp) at the National Center for Gene Research, Shanghai, China (see Supplemental Table 1 for read counts). Each MPP of the second meiosis was sequenced in one lane of Genome Analyzer I (single-end, 36 bp) and one lane of Genome Analyzer II (single-end, 35 bp) at FASTERIS SA and two lanes of Genome Analyzer IIX (single-end, 55 to ∼75 bp) at BGI (see Supplemental Table 1 for read counts).
Calling of small polymorphisms
The Tair9 assembly of Arabidopsis Columbia ecotype genome was downloaded from the TAIR (ftp://ftp.arabidopsis.org/home/tair/Sequences/whole_chromosomes) website. Sequenced reads of Ler were then mapped against the Col genome and SNPs were called using MAQ 0.7.1 (http://maq.sourceforge.net/) (Li et al. 2008), whereas small indels were predicted using inGAP (Qi et al. 2010). The mapping and SNP prediction procedure follows the online MAQ instruction from FASTQ format transformation to build consensus sequences. Only uniquely mapped reads with a mapping quality score equal to or greater than 20 were used in subsequent analyses. We further removed pseudo-SNPs due to repetitive sequences or amplification errors by Perl scripts written for the analysis here (available upon request). Finally, the sequencing reads of meiosis products were used to rule out errors in Col genome sequences and confirm predicted SNPs, which requires the first and second most abundant bases in SNP loci to be Col-specific base and Ler-specific bases or vice versa, otherwise the SNP loci were considered to be unreliable.
Identification of larger indels
All Ler paired-end Illumina reads were assembled using Velvet (Zerbino and Birney 2008) and the output contigs mapped to the Col genome by Mummer (Delcher et al. 1999), with redundant contigs removed before the prediction of large indels using custom scripts (available upon request). To minimize false positives, we implemented a step of read-mapping depth estimates in the pipeline, because indels affect the mapping pattern of paired ends in their flanking regions, and removed those predicted indels that lacked a “gapped region” in the landscape of read mapping. Finally, a homology search with coding regions in indels against Col genes was performed by a BLASTN search (identity>80%).
Very recently, Schneeberger et al. (2011) reported assembly of Ler sequences based on high-throughput sequencing reads. By using these contigs as a reference and importing more sequences from eight meiotic data sets and the Monsanto Ler contigs, we built a new assembly with the longest contig of ∼253 Kb and N50 of ∼26 Kb (more than twice as long as the ones from our reads only, respectively). The newly assembled contigs can be accessed along with the de novo assembly from reads in this study. When selecting Ler reads from eight meiotic data sets, we screened the alignment results of paired-end reads against the Col genome and collected 15,791,682 reads with at least one end unmapped. The insertion sizes and their standard deviation are estimated automatically by Velvet. The list of primers used for PCR-based verification of some predicted indels and the detailed information were provided in Supplemental Table 17.
Genes in Ler-inserted sequences were predicted using geneid. The reciprocal best BLAST hit and syntenic map between Col and A. lyarta from SynMap were used to identify the A. lyrata ortholog of Col-specific genes. Orthologs of Ler unique genes were identified by reciprocal best BLAST hit only.
Further bioinformatic analysis of SNP/indel affected genes
Gene Ontology analysis was performed in agriGO with default parameters (http://bioinfo.cau.edu.cn/agriGO/) (Du et al. 2010). dN/dS analysis was conducted using PAML with all four codon models (http://abacus.gene.ucl.ac.uk/software/paml.html) (Yang 2007). To calculate the Arabidopsis branch-specific dN/dS values, we used Poplar and A. lyrata orthologous genes downloaded from Phytozome (http://www.phytozome.net/) in tree-guided analysis. Since similar results were attained with all codon models, we reported results from the simplest codon model with the CodonFreq set to 0. Normalization of microarray data of affected genes was done in SNOMAD (http://pevsnerlab.kennedykrieger.org/ snomadinput.html) before comparing expression in Col and Ler (Colantuoni et al. 2002). The enrichment of SNP/indel-affected genes in multigene families was based on clustering of all Arabidopsis genes by MCL (Jiao et al. 2011).
Identification of crossovers and noncrossovers
Sequenced reads of meiosis progeny were mapped to the Col genome with MAQ 0.7.1. At each SNP locus, the read counts of all present bases were recorded. Crossovers were identified from the genome-wide distribution of the Ler allele at SNP loci. Ler-specific alleles flanked by Col markers were noted as potential GC events (Supplemental Fig. 7). To minimize noise from sequencing errors, we required high-quality calling of a Ler allele in at least three reads to support a converted SNP. Converted SNPs <1 Kb apart were grouped into one gene conversion event. To further reduce false GCs due to repetitive sequence, the 35-bp flanking sequences of each converted SNP were used as queries for BLAST searches against the Col genome. GCs with half or more converted SNPs in repetitive regions were ignored. The minimal length of CO/NCO was the length between the two farthest converted SNPs, and the maximal length was the length between the two closest unconverted SNPs. The midpoint length was the arithmetic average of minimal and maximal length.
To verify the detected COs and NCOs/GCs events, we used primers away from the SNPs supporting the recombination event to perform PCR (40 cycles of denaturing at 95°C for 30 sec, annealing at 54°C for 30 sec, and extension at 72°C for 1 min, and an additional step of 72°C for 10 min to allow complete extension). The PCR products were mixed with the corresponding primers and sequenced at the Nucleic Acid Facility at the Pennsylvania State University. The primers information is provided in the Supplemental Table 18.
Meiosis simulation and statistical analysis
Custom scripts (available upon requests) were used to generate 10,000 meioses by assigning crossovers to each chromosome according the probability of recombination estimated based on genetic and physical maps, with one crossover in each chromosome arm, so that the total number of crossovers per meiosis was 10. The integration of the genetic and physical map was according to a previous study (Meinke et al. 2009). The number of SNP/indel carried by each gamete was calculated based on the location of crossovers. Different iterations of simulations were tried, and because results from the 5000 meioses simulation were similar to 10,000 meioses, final results were based on the 10,000 meioses simulation.
Enrichment of GO groups was analyzed using Fisher's exact test with Yekutieli FDR multitest correction. The relation between multigene family members and genes with 10 or more nonsynonymous SNPs was analyzed by the χ2 test. Enrichment of unknown genes was also analyzed by the χ2 test (Zerbino and Birney 2008).
Data access
The high-throughput sequencing data from this study have been submitted to the NCBI Sequence Read Archive (SRA) (http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession numbers SRP007172 (http://www.ncbi.nlm.nih.gov/sra?term=SRP007172) and SRP008819 (http://www.ncbi.nlm.nih.gov/sra?term=SRP008819). SNPs, indels, large DNA polymorphisms, and Ler contigs are available at http://www.personal.psu.edu/hxm16/suppdatafile.zip.
Acknowledgments
We thank D. Kumar for providing the qrt1 mutant seeds, N. Altman for discussion on simulation of meiotic recombination, and X. Ma, L. Zhang, S. Schaeffer, M. Axtell, A. Nekrutenko, and anonymous reviewers for helpful comments and discussions. This work was supported by grants from the Chinese Ministry of Science and Technology (2011CB944600) and from the US Department of Energy (DE-FG02-02ER15332) to H.M.; Fudan University; and Rijk Zwaan. Additional support was provided by funds from the Institute of Hydrobiology to T.L. and the Biology Department and the Huck Institutes of the Life Sciences at the Pennsylvania State University.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.127522.111.
References
- Arabidopsis Genome Initiative 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Baarends WM, Mercier R 2010. Sisters dancing in meiosis. EMBO Rep 11: 76–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SM, Bronner CE, Zhang L, Plug AW, Robatzek M, Warren G, Elliott EA, Yu J, Ashley T, Arnheim N, et al. 1995. Male mice defective in the DNA mismatch repair gene PMS2 exhibit abnormal chromosome synapsis in meiosis. Cell 82: 309–319 [DOI] [PubMed] [Google Scholar]
- Barlow A, Hulten M 1998. Crossing over analysis at pachytene in man. Eur J Hum Genet 6: 350–358 [DOI] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327: 836–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Hanson J, Hanhart CJ, Blankestijn-de Vries H, Coltrane C, Keizer P, El-Lithy M, Alonso-Blanco C, de Andrs MT, Reymond M, et al. 2010. Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proc Natl Acad Sci 107: 4264–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borts RH, Chambers SR, Abdullah MF 2000. The many faces of mismatch repair in meiosis. Mutat Res 451: 129–150 [DOI] [PubMed] [Google Scholar]
- Chen C, Zhang W, Timofejeva L, Gerardin Y, Ma H 2005. The Arabidopsis ROCK-N-ROLLERS gene encodes a homolog of the yeast ATP-dependent DNA helicase MER3 and is required for normal meiotic crossover formation. Plant J 43: 321–334 [DOI] [PubMed] [Google Scholar]
- Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, Warthmann N, Hu TT, Fu G, Hinds DA, et al. 2007. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317: 338–342 [DOI] [PubMed] [Google Scholar]
- Colantuoni C, Henry G, Zeger S, Pevsner J 2002. SNOMAD (Standardization and NOrmalization of MicroArray Data): web-accessible gene expression data analysis. Bioinformatics 18: 1540–1541 [DOI] [PubMed] [Google Scholar]
- Copenhaver GP, Housworth EA, Stahl FW 2002. Crossover interference in Arabidopsis. Genetics 160: 1631–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM 2003. The Mus81/Mms81 endonuclease acts independently of double Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164: 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Kasif S, Fleischmann RD, Peterson J, White O, Salzberg SL 1999. Alignment of whole genomes. Nucleic Acids Res 27: 2369–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresser ME, Giroux CN 1988. Meiotic chromosome behavior in spread preparations of yeast. J Cell Biol 106: 567–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouaud J, Camilleri C, Bourguignon PY, Canaguier A, Berard A, Vezon D, Giancola S, Brunel D, Colot V, Prum B, et al. 2006. Variation in crossing-over rates across chromosome 4 of Arabidopsis thaliana reveals the presence of meiotic recombination “hot spots.” Genome Res 16: 106–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z 2010. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Cardoso-Moreira M, Borevitz JO, Long M 2008. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science 320: 1629–1631 [DOI] [PubMed] [Google Scholar]
- Filiault DL, Wessinger CA, Dinneny JR, Lutes J, Borevitz JO, Weigel D, Chory J, Maloof JN 2008. Amino acid polymorphisms in Arabidopsis phytochrome B cause differential responses to light. Proc Natl Acad Sci 105: 3157–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis KE, Lam SY, Harrison BD, Bey AL, Berchowitz LE, Copenhaver GP 2007. Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc Natl Acad Sci 104: 3913–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Zhang F, Wang Y, Gu X, Jin L 2010. Identification of copy number variation hotspots in human populations. Am J Hum Genet 87: 494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graubert TA, Cahan P, Edwin D, Selzer RR, Richmond TA, Eis PS, Shannon WD, Li X, McLeod HL, Cheverud JM, et al. 2007. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet 3: e3 doi: 10.1371/journal.pgen.0030003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon-Debast A, Lecureuil A, Bonhomme S, Guerche P, Gallois JL 2010. A SNP associated with alternative splicing of RPT5b causes unequal redundancy between RPT5a and RPT5b among Arabidopsis thaliana natural variation. BMC Plant Biol 10: 158 doi: 10.1186/1471-2229-10-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubold B, Kroymann J, Ratzka A, Mitchell-Olds T, Wiehe T 2002. Recombination and gene conversion in a 170-kb genomic region of Arabidopsis thaliana. Genetics 161: 1269–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Armstrong SJ, Franklin FC, Jones GH 2004. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: Evidence for two classes of recombination in Arabidopsis. Genes Dev 18: 2557–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N, Brill S 2004. The Mus81 solution to resolution: Generating meiotic crossovers without Holliday junctions. Genes Dev 18: 117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al. 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurles ME, Dermitzakis ET, Tyler-Smith C 2008. The functional impact of structural variation in humans. Trends Genet 24: 238–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst DD, Fogel S, Mortimer RK 1972. Conversion-associated recombination in yeast (hybrids-meiosis-tetrads-marker loci-models). Proc Natl Acad Sci 69: 101–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsara A, Wu H, Smith JD, Nickerson DA, Romieu I, London SJ, Eichler EE 2010. De novo rates and selection of large copy number variation. Genome Res 20: 1469–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100 [DOI] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C 2000. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Kauppi L, Jeffreys AJ, Keeney S 2004. Where the crossovers are: recombination distributions in mammals. Nat Rev Genet 5: 413–424 [DOI] [PubMed] [Google Scholar]
- Keeney S 2001. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol 52: 1–53 [DOI] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. 2008. Mapping and sequencing of structural variation from eight human genomes. Nature 453: 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N, Storlazzi A, Zickler D 2003. Coordinate variation in meiotic pachytene SC length and total crossover/chiasma frequency under conditions of constant DNA length. Trends Genet 19: 623–628 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D 2004. Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol 55: 141–172 [DOI] [PubMed] [Google Scholar]
- Krieger U, Lippman ZB, Zamir D 2010. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat Genet 42: 459–463 [DOI] [PubMed] [Google Scholar]
- Kroymann J, Donnerhacke S, Schnabelrauch D, Mitchell-Olds T 2003. Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proc Natl Acad Sci 100: 14587–14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Betran E, Thornton K, Wang W 2003. The origin of new genes: glimpses from the young and old. Nat Rev Genet 4: 865–875 [DOI] [PubMed] [Google Scholar]
- Lynn A, Koehler KE, Judis L, Chan ER, Cherry JP, Schwartz S, Seftel A, Hunt PA, Hassold TJ 2002. Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science 296: 2222–2225 [DOI] [PubMed] [Google Scholar]
- Ma H 2006. A molecular portrait of Arabidopsis meiosis. In The Arabidopsis book (ed. Somerville CR et al. ), pp. 1–39 American Society of Plant Biologists, Rockville, MD: doi: 10.1199/tab.0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloof JN, Borevitz JO, Dabi T, Lutes J, Nehring RB, Redfern JL, Trainer GT, Wilson JM, Asami T, Berry CC, et al. 2001. Natural variation in light sensitivity of Arabidopsis. Nat Genet 29: 441–446 [DOI] [PubMed] [Google Scholar]
- Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke D, Sweeney C, Muralla R 2009. Integrating the genetic and physical maps of Arabidopsis thaliana: identification of mapped alleles of cloned essential (EMB) genes. PLoS ONE 4: e7386 doi: 10.1371/journal.pone.0007386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz EM, Ma H 1994. Genetic variations of Arabidopsis thaliana. In The Arabidopsis book (ed. Meyerowitz EM and Somerville CR), pp. 1161–1268 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Mezard C, Vignard J, Drouaud J, Mercier R 2007. The road to crossovers: plants have their say. Trends Genet 23: 91–99 [DOI] [PubMed] [Google Scholar]
- Michaels SD, He Y, Scortecci KC, Amasino RM 2003. Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc Natl Acad Sci 100: 10102–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindrinos M, Katagiri F, Yu GL, Ausubel FM 1994. The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78: 1089–1099 [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T, Schmitt J 2006. Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature 441: 947–952 [DOI] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, et al. 2005. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3: e196 doi: 10.1371/journal.pbio.0030196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Clark RM, Lanz C, Warthmann N, Weigel D 2008. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res 18: 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt A, Horton M, Huang YS, Li Y, Anastasio AE, Mulyati NW, Agren J, Bossdorf O, Byers D, Donohue K, et al. 2010. The scale of population structure in Arabidopsis thaliana. PLoS Genet 6: e1000843 doi: 10.1371/journal.pgen.1000843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D, Rhee SY, Davis RW 1994. Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264: 1458–1460 [DOI] [PubMed] [Google Scholar]
- Qi J, Wijeratne A, Tomsho L, Hu Y, Schuster S, Ma H 2009. Characterization of meiotic crossovers and gene conversion by whole-genome sequencing in Saccharomyces cerevisiae. BMC Genomics 10: 475 doi: 10.1186/1471-2164-10-475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Zhao F, Buboltz A, Schuster SC 2010. inGAP: an integrated next-generation genome analysis pipeline. Bioinformatics 26: 127–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentel MC, Leonelli L, Dahlbeck D, Zhao B, Staskawicz BJ 2008. Recognition of the Hyaloperonospora parasitica effector ATR13 triggers resistance against oomycete, bacterial, and viral pathogens. Proc Natl Acad Sci 105: 1091–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Kikkawa Y, Miura I, Inoue T, Moriwaki K, Shiroishi T, Satta Y, Takahata N, Yonekawa H 2005. Origins of mouse inbred strains deduced from whole-genome scanning by polymorphic microsatellite loci. Mamm Genome 16: 11–19 [DOI] [PubMed] [Google Scholar]
- Sanchez Moran E, Armstrong SJ, Santos JL, Franklin FC, Jones GH 2001. Chiasma formation in Arabidopsis thaliana accession Wassileskija and in two meiotic mutants. Chromosome Res 9: 121–128 [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran E, Santos JL, Jones GH, Franklin FC 2007. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev 21: 2220–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU 2003. Dissection of floral induction pathways using global expression analysis. Development 130: 6001–6012 [DOI] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Ott F, Klein JD, Wang X, Lanz C, Smith LM, Cao J, Fitz J, Warthmann N, et al. 2011. Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc Natl Acad Sci 108: 10249–10254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR 2002. Genome architecture, rearrangements and genomic disorders. Trends Genet 18: 74–82 [DOI] [PubMed] [Google Scholar]
- Todesco M, Balasubramanian S, Hu TT, Traw MB, Horton M, Epple P, Kuhns C, Sureshkumar S, Schwartz C, Lanz C, et al. 2010. Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature 465: 632–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang S, Sun Z, Luke B, Stewart C, Lum N, Gregory M, Wu X, Subleski M, Jenkins NA, Copeland NG, et al. 2005. A comprehensive SNP-based genetic analysis of inbred mouse strains. Mamm Genome 16: 476–480 [DOI] [PubMed] [Google Scholar]
- Wijeratne AJ, Chen C, Zhang W, Timofejeva L, Ma H 2006. The Arabidopsis thaliana PARTING DANCERS gene encoding a novel protein is required for normal meiotic homologous recombination. Mol Biol Cell 17: 1331–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Ma H, Nei M, Kong H 2009. Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification. Proc Natl Acad Sci 106: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z 2007. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Yang H, Wang JR, Didion JP, Buus RJ, Bell TA, Welsh CE, Bonhomme F, Yu AH, Nachman MW, Pialek J, et al. 2011. Subspecific origin and haplotype diversity in the laboratory mouse. Nat Genet 43: 648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E 2008. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gu W, Hurles ME, Lupski JR 2009. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet 10: 451–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N 1999. Meiotic chromosomes: Integrating structure and function. Annu Rev Genet 33: 603–607 [DOI] [PubMed] [Google Scholar]
- Ziolkowski PA, Koczyk G, Galganski L, Sadowski J 2009. Genome sequence comparison of Col and Ler lines reveals the dynamic nature of Arabidopsis chromosomes. Nucleic Acids Res 37: 3189–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]