Abstract
Despite the recent discoveries of and interest in numerous structural variations (SVs)—which include duplications and inversions—in the human and other higher eukaryotic genomes, little is known about the etiology and biology of these SVs, partly due to the lack of molecular tools with which to create individual SVs in cultured cells and model organisms. Here, we present a novel method of inducing duplications and inversions in a targeted manner without pre-manipulation of the genome. We found that zinc finger nucleases (ZFNs) designed to target two different sites in a human chromosome could introduce two concurrent double-strand breaks, whose repair via non-homologous end-joining (NHEJ) gives rise to targeted duplications and inversions of the genomic segments of up to a mega base pair (bp) in length between the two sites. Furthermore, we demonstrated that a ZFN pair could induce the inversion of a 140-kbp chromosomal segment that contains a portion of the blood coagulation factor VIII gene to mimic the inversion genotype that is associated with some cases of severe hemophilia A. This same ZFN pair could be used, in theory, to revert the inverted region to restore genomic integrity in these hemophilia A patients. We propose that ZFNs can be employed as molecular tools to study mechanisms of chromosomal rearrangements and to create SVs in a predetermined manner so as to study their biological roles. In addition, our method raises the possibility of correcting genetic defects caused by chromosomal rearrangements and holds new promise in gene and cell therapy.
Many genetic diseases and cancer are associated with chromosomal rearrangements such as deletions, insertions, duplications, translocations, and inversions (Stephens et al. 2009; Stankiewicz and Lupski 2010). Even among healthy individuals, thousands of different structural variations (SVs) or copy number variations resulting from different chromosomal rearrangements are observed (Iafrate et al. 2004; Redon et al. 2006). Thus, genome SVs contribute to both genetic diversity and the onset of diseases. Despite the rapid progress in the identification of genome-wide SVs and growing interest in these variations among geneticists (Korbel et al. 2007; Park et al. 2010), experimental systems to create individual SVs, especially inversions and duplications, systematically in an otherwise genetically identical background are lacking. Genome-wide association studies could reveal associations of a set of SVs with a certain phenotype or disease but cannot provide a causal link between the two. To study individual SVs in cultured cells or model organisms, one must be able to isolate or construct isogenic clones that harbor such variations. We reasoned that mechanistic insights about their formation could help us develop the means to do so.
DNA sequence analyses of SVs suggest that both non-allelic homologous recombination (NAHR) and non-homologous end-joining (NHEJ) are involved in the generation of chromosomal rearrangements (Hastings et al. 2009; Conrad et al. 2010). Because DNA double-strand breaks (DSBs) greatly enhance the rate of both NAHR and NHEJ, it is assumed that most, if not all, chromosomal rearrangements arise from the repair of DSBs, through NAHR or NHEJ, that occur accidentally in the genome (van Gent et al. 2001). In an attempt to mimic chromosomal rearrangements experimentally, DNA-cleaving chemicals and irradiation have been used, which introduce DSBs at random sites (Rothkamm et al. 2001; Bueno et al. 2009). These studies showed that two DSBs occurring in different chromosomes could give rise to chromosomal translocations. Other chromosomal rearrangements, including deletions, duplications, and inversions, are more difficult than translocations to study, partly because these aberrations, unlike translocations, cannot easily be detected using standard karyotyping analysis.
We had shown that two concurrent DSBs induced by engineered ZFNs—artificial endonucleases composed of tailor-made zinc finger DNA-binding arrays and the FokI nuclease domain (Kim et al. 1996)—are sufficient to promote deletions in the human genome. But, unlike deletions that involve only a single intra-chromosomal ligation of DSB endpoints, duplications involve inter-chromosomal (or inter-chromatidal) ligations, and inversions involve two separate ligations. To our knowledge, it has never been investigated whether two concurrent DSBs could give rise to duplications and inversions, and, if they do, how frequently the events would occur.
To address these questions, we used ZFNs to introduce DSBs at pre-determined endogenous sites in the human genome. Our results revealed that two concurrent DSBs are sufficient to promote frequent genomic inversions and duplications as well as deletions. In addition, we constructed a ZFN pair that induces the inversion of a chromosomal segment that contains the promoter and exon 1 of the blood coagulation factor VIII (F8) gene. This ZFN might be used to restore genomic integrity in patients with severe hemophilia A. Thus, we present a novel, ZFN-based method for inducing SVs without pre-manipulations, such as the insertion of loxP elements, in the human genome.
Results
Isolation and analysis of clonal populations of cells with large chromosomal deletions
To isolate clonal populations of cells in which a cluster of homologous genes is deleted in the genome, we used previously characterized ZFN pairs, each of which targets one of two sites, separated by 230 kbp in length, on human chromosome 3 (Perez et al. 2008; Lee et al. 2010). A ZFN pair termed K230 can induce a DSB at the site 230 kbp upstream of the gene that encodes chemokine receptor 5 (CCR5) and can delete a cluster of homologous chemokine receptor genes, including CCR1, CCR2, CCR3, and CCR5, when co-expressed with ZFN-224 (this ZFN was termed S162 in Lee et al. [2010]), the ZFN pair that targets the CCR5 locus, in human cells (Fig. 1A). These chemokine receptors may have redundant roles (Choe et al. 1996; Doranz et al. 1996), and the isolation of single clones in which these genes are deleted could be useful for their functional analysis.
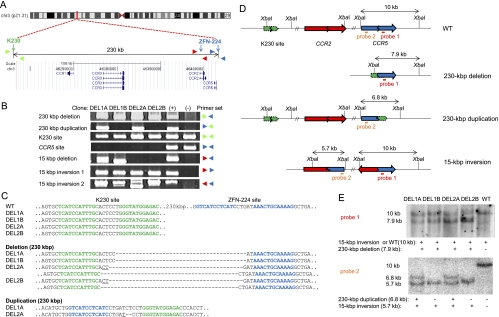
Figure 1.
Various chromosomal rearrangements induced by ZFNs. (A) Schematic of ZFN target sites and chemokine receptor genes on the human chromosome 3 (http://genome.ucsc.edu). Note that ZFN-224 has two target sites, one at the CCR5 locus and the other at the CCR2 locus. Colored triangles indicate approximate positions of PCR primers. (B) PCR products validating various chromosomal rearrangements. (+) indicates a positive control (cells transfected with plasmids encoding two ZFN pairs) and (−) indicates a negative control (cells transfected with empty plasmid). (C) DNA sequences of PCR products. Each ZFN target site is shown in boldface letters. K230 and CCR5 target sites are shown in green and blue letters, respectively. Note the absence of the intact CCR5 (ZFN-224) site in these clones. Microhomologies are underlined and inserted bases are shown in italics. Dashes indicate deleted bases. (D) Schematic of ZFN target sites in wild-type cells and in various clones with chromosomal rearrangements. Two probes (red and orange bars) and the site recognized by the restriction enzyme, XbaI, used for Southern blot analysis are indicated. (E) Southern blot analysis of chromosomal rearrangements. Genomic DNA digested with XbaI and hybridized with probe 1 (upper) or with probe 2 (lower). A direct evidence of the 15-kbp inversion was provided by DNA sequencing (Supplemental Fig. 1).
To this end, we transfected human embryonic kidney (HEK) 293T cells with plasmids that encode these two ZFN pairs and isolated single clones in two steps. First, transfected cells were grown in 96-well plates at a density of 50 cells per well for 10 d, and then genomic DNA was isolated from a portion of the cells and subjected to PCR analysis to detect chromosomal deletions. Next, cells from deletion positive pools were cloned by limiting dilution. Several deletion clones, in which the chromosomal DNA segment between the two ZFN target sites was deleted, were obtained from two different pools (Fig. 1B). As expected, clones isolated from different pools had their own distinctive junction sequences (Compare DEL1A and DEL1B clones obtained from a pool with DEL2A and DEL2B clones from the other pool in Fig. 1C), and clones from the same pools had the same junction sequences. (However, sibling clones from the same pools turned out to be not identical with each other. See below.) Interestingly, clones from one pool (DEL2A and DEL2B) had two different deletion junction sequences, suggesting that ZFNs induced large deletions in two homologous chromosomes in these clones.
Chromosomal rearrangements in deletion clones
To analyze deletion clones in further detail, we investigated whether they had any local mutations other than the 230-kbp deletions. To this end, we performed PCR to amplify wild-type DNA segments encompassing each of the two ZFN target sites and determined their DNA sequences. Unexpectedly, the DNA segment that encompasses the ZFN site at the CCR5 locus was not amplified from any of the four clones, although the DNA segment that encompasses the K230 site was successfully amplified and found to have no local mutations (Fig. 1B,C). HEK 293 is a multiploid cell line that has at least three copies of chromosome 3 (Bylund et al. 2004). Our results suggest that there must be additional chromosomal rearrangements that involve the CCR5 site in the deletion clones on the other two copies of chromosome 3. Because ZFN-224 induces an off-target DNA cleavage at the CCR2 locus, which is located 15 kbp upstream of the CCR5 locus, and can efficiently induce targeted deletion of the 15-kbp chromosomal segment between the two sites in human cells (Lee et al. 2010), we suspected that, in addition to the 230-kbp deletions, these clones might have 15-kbp deletions on other copies of chromosome 3. We analyzed these clones by PCR and found that two clones from one pool (DEL1A and DEL1B), indeed, had a 15-kbp deletion (Fig. 1B; Supplemental Fig. 1) but that the other two clones from the other pool (DEL2A and DEL2B) did not. At first, we were puzzled by these findings because we could not account for the lack of the ZFN-224 site on all copies of chromosome 3. We then hypothesized that genomic rearrangements other than deletions, such as duplications and inversions, might have occurred via NHEJ-mediated repair of DSBs induced by ZFNs in these clones.
To investigate whether ZFNs induced duplications or inversions, we performed PCR analysis using genomic DNA isolated from the four clones. As shown in Supplemental Figure 2A, when two DSB endpoints, each occurring at one of the two different sites in each sister chromatid (or homologous chromosome), are joined, the genomic DNA segment between the two DSBs would be deleted in one chromatid (or homologous chromosome) and duplicated in the other chromatid (or homologous chromosome). Duplications can be detected by PCR using appropriate primers. Likewise, PCR analysis using primers pointing the same direction relative to the wild-type chromosome can yield amplified products only when the middle DNA region is inverted (Supplemental Fig. 2B). These analyses showed that deletion clones, indeed, had additional genomic rearrangements other than 230-kbp or 15-kbp deletions (Fig. 1B). First, we found that all four clones had 15-kbp inversions; two breakpoint junctions corresponding to the inversion event were detected by PCR. As expected, the DNA sequences of two inversion junctions in clones from the same pool were identical with those of corresponding junctions in their sibling clones but were different from those in clones from the other pool (Supplemental Fig. 1). Furthermore, we were able to detect PCR products corresponding to 230-kbp duplications in two independent clones, DEL1A and DEL2A, whose breakpoint junction sequences were different from each other (Fig. 1B,C). Interestingly, the other two clones, DEL1B and DEL2B, lacked corresponding duplications (Fig. 1B). It appears that these two clones lost chromosomal segments corresponding to the 230-kbp duplications that are present in their sibling clones.
To confirm the presence of various genomic rearrangements, genomic DNA from these clones was further analyzed by Southern blot using two different probes (Fig. 1D,E). When probe 1 (which hybridizes downstream from the ZFN-224 site in the CCR5 locus) was used, a 7.9-kb DNA band that corresponds to the 230-kbp deletion was detected in all four clones but not in wild-type cells. (The relative intensity of this DNA band in DEL2A and DEL2B clones indicates that there are two different 230-kbp deletions in these clones, which is in line with the presence of two different deletion breakpoint junctions [Fig. 1C].) When probe 2 (which hybridizes upstream of the ZFN-224 site in the CCR5 locus) was used, a 6.8-kb band that corresponds to the 230-kbp duplication was detected in DEL1A and DEL2A clones but not in the other two clones or in wild-type cells. Whereas probe 2 hybridized to a 10-kb band in DNA from wild-type cells, indicating the presence of an intact CCR5 locus, this probe did not detect a 10-kb band for any of the clones, which is in line with the PCR analysis showing that all of them lacked the CCR5 site (Fig. 1B,E). In addition to duplications and deletions, all four clones showed inversion-specific bands (Fig. 1E, the 10-kb band with probe 1 and the 5.7-kb band with probe 2). (Loss of the CCR5 site in these clones suggests that the 10-kb band detected with probe 1 should be assigned to the 15-kbp inversion-specific band, not to the wild-type band.) Taken together, PCR and Southern blot analyses showed that DEL1A and DEL1B clones had a 230-kbp deletion, a 15-kbp deletion, and a 15-kbp inversion and that DEL2A and DEL2B clones had two 230-kbp deletions and a 15-kbp inversion. In addition, DEL1A and DEL2A clones, but not the other two clones, had a 230-kbp duplication in chromosome 3.
Duplications induced by ZFNs
To investigate whether duplications can be induced at any site and are not limited to sites recognized by certain combinations of ZFNs, we used previously characterized ZFNs that target the CCR5 and CCR2 loci. These two genes are adjacent to each other on chromosome 3 and are highly homologous. Thus, the target sites of some of these ZFNs are conserved between the two loci. Duplications were detected using PCR primers whose sequences correspond to the CCR2 region or to the CCR5 region, as shown in Figure 2A. We were able to obtain duplication-specific PCR products from cells treated with ZFNs that target two conserved sites at CCR2 and CCR5 loci but not from cells treated with Z30 and Z266, ZFN pairs that show genome editing activity at the CCR5 locus but not at the CCR2 locus because their recognition sites are not conserved at the CCR2 locus (Fig. 2B). This result suggests that two DSBs are necessary to induce duplications. The DNA sequences of the PCR amplicons showed that the 5′ portion of the CCR5 coding region was directly linked to the 3′ portion of the CCR2 coding region (Fig. 2C; Supplemental Fig. 3A). These rearrangements can be formed only when the 15-kbp genomic DNA segment between the two conserved ZFN target sites is duplicated.
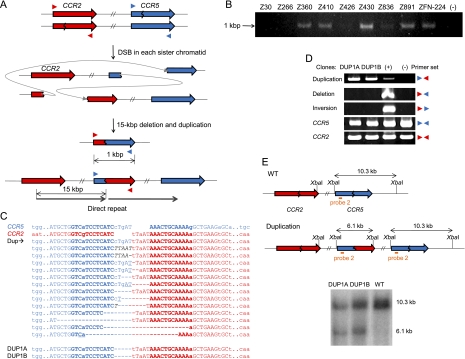
Figure 2.
Duplications induced by ZFNs. (A) Schematic representation of ZFN-mediated duplications. Zigzag lines indicate ZFN target sites. Colored triangles indicate approximate positions of PCR primers. (B) PCR products corresponding to the 15-kbp duplications in cells treated with various ZFNs. (−) indicates a negative control (cells transfected with empty plasmid). (C) DNA sequences of breakpoint junctions of the duplications. Nucleotide sequences of CCR5 and CCR2 sites are shown in blue and red colors, respectively. Dup indicates various duplication junctions induced by ZFN-224. Duplication junction sequences induced by other ZFNs are shown in Supplemental Figure 3. DNA sequences of breakpoint junctions in two clones, DUP1A and DUP1B, are also shown. Nonconserved bases at the CCR2 and CCR5 loci are shown in lowercase letters. Symbols are as in Figure 1. (D) PCR products validating duplications in two clones. No other mutations or rearrangements were detected in these clones. (+) is a positive control (cells treated with ZFN-224) and (−) is a negative control (cells transfected with empty plasmid). (E) Schematic of CCR2 and CCR5 loci in the wild-type and duplication alleles and Southern blot analysis. Symbols are as in Figure 1.
We also tested various combinations of two ZFNs to investigate whether two DSBs occurring at diverse sites could give rise to duplications of the chromosomal segment between the two DSBs. In every combination of two ZFNs we tested, we detected PCR products commensurate with duplications of genomic DNA between the two cleavage sites whose length ranged from 230 to 835 kbp (Supplemental Fig. 3B). No PCR product was obtained from cells treated with only one ZFN pair or from control cells transfected with empty vector. As seen with deletion events, small indels and microhomologies often were observed at the breakpoint junctions of duplications. These results suggest the involvement of NHEJ in the generation of ZFN-mediated duplications.
Next, we screened for clonal populations of cells in which the 15-kbp chromosomal segment between the CCR2 and CCR5 loci is duplicated after expression of ZFN-224. Several clones were isolated by limiting dilution and two of them, termed DUP1A and DUP1B, were further characterized by PCR and Southern blot (Fig. 2C–E), which confirmed the presence of the 15-kbp duplication. In these clones, no other genomic rearrangements, such as deletions or inversions, were detected (Fig. 2D). According to our model described above, two DSBs, each occurring in one of the two sister chromatids (or homologous chromosomes), could give rise to deletions in one chromatid and duplications in the other chromatid via inter-molecular ligation. It is interesting that duplication was not accompanied with corresponding deletion in DUP1A and DUP1B, suggesting that the deletion was lost in these clones. We also characterized previously isolated deletion clones (Lee et al. 2010), in which the 15-kbp segment between the CCR2 and CCR5 loci is deleted after expression of ZFN-224, to see whether these clones contain duplications in addition to deletions, and found no duplications. Taken together, these results show that it is possible to isolate distinctive clones, by limiting dilution, using ZFNs that generate two DSBs in a chromosome. Such clones can contain either a deletion alone or a duplication alone or both a duplication and the corresponding deletion.
Inversions induced by ZFNs
Genomic inversions often are associated with cancer and genetic diseases. For example, a 500-kbp DNA segment on chromosome 10 is inverted in thyroid cancer cells from patients exposed to irradiation associated with the Chernobyl nuclear power accident (Nikiforov et al. 1999; Nikiforova et al. 2000). Understanding the mechanisms of genomic inversion formation is of great interest and is a prerequisite for prevention and potential therapeutic intervention to thwart diseases caused by these genomic aberrations.
To study the etiology of inversions and to explore the potential for flip-flopping inverted DNA segments back to the normal state, we transiently expressed ZFNs in HEK 293 cells and then analyzed the cells' genomic DNA. Unlike deletions or duplications, which have only one breakpoint junction, genomic inversions generate two breakpoint junctions, which can be detected by PCR using appropriate primers (Fig. 3A). We obtained PCR products containing inversion breakpoint junctions from cells treated with various combinations of ZFNs that cleave genomic DNA at two sites but not from cells treated with a single ZFN pair that cleaves DNA only at one site or from control cells transfected with empty plasmid (Fig. 3B). We cloned these PCR products and determined their DNA sequences, which unequivocally confirmed genomic inversions in each case (Fig. 3C; Supplemental Fig. 4). We detected inversions of 15-kbp, 230-kbp, 243-kbp, 276-kbp, 781-kbp, 835-kbp, and 15-Mbp DNA segments at chromosome 3p21, which were induced with appropriate combinations of ZFNs. Again, DNA sequences at breakpoint junctions often showed indels and microhomologies, suggesting the involvement of NHEJ in ZFN-mediated genomic inversions.
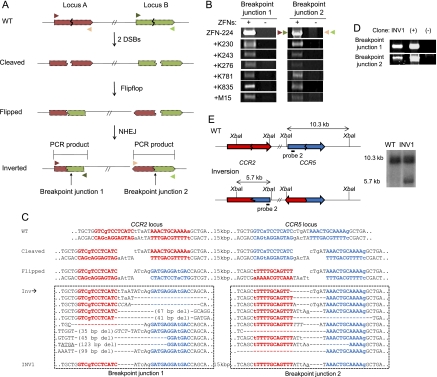
Figure 3.
Inversions induced by ZFNs. (A) Schematic representation of ZFN-mediated inversions. Zigzag lines indicate ZFN target sites. PCR primers (triangles) used for the detection of two breakpoint junctions that result from genomic inversions are shown. (B) PCR products corresponding to inversion events in cells treated with various combinations of two ZFN pairs. (C) DNA sequences of breakpoint junctions of the inversion events. Nucleotide sequences of CCR5 and CCR2 sites are shown in blue and red, respectively. (Inv) Various inversion junctions induced by ZFN-224. Inversion junction sequences induced by other ZFNs are shown in Supplemental Figure 4. The two breakpoint junction sequences in the INV1 clone are also shown. Nonconserved bases at the CCR2 and CCR5 loci are shown in lowercase letters. Symbols are as in Figure 1. (D) PCR products validating inversions in the INV1 clone. (+) and (−) are as in Figure 2D. (E) Schematic of CCR2 and CCR5 loci in the wild-type and inversion alleles and Southern blot analysis. Symbols are as in Figure 1.
Using ZFN-224, we also generated and isolated clonal populations of cells in which the 15-kbp chromosomal segment between the CCR2 and CCR5 loci is inverted. As expected, the inversion clone termed INV1 had two breakpoint junctions, which were detected by inversion-specific PCR (Fig. 3D). DNA sequence analysis of breakpoint junctions and Southern blot analysis confirmed the inversion event in this clone (Fig. 3E). Importantly, no other genomic rearrangements, such as deletions or duplications, were detected in this clone. These results suggest that two DSBs occurring in a chromosome can give rise to frequent inversions of DNA segments between the breaks and raise the possibility of using ZFNs for correcting genetic mutations caused by inversions.
Frequencies of genomic rearrangements
To estimate the frequencies of various genomic rearrangements induced by ZFNs, we performed digital PCR analysis using serially diluted genomic DNA samples isolated from cells transfected with ZFNs (Kim et al. 2010b). The Extreme Limiting Dilution Analysis program (Hu and Smyth 2009) was used for statistical analysis of this PCR-based method. The frequencies of duplications and inversions induced by various ZFNs ranged from 0.01% to 5% (Supplemental Table 1). Large genomic segments of up to hundreds of kbp in length could be duplicated or inverted efficiently, although these frequencies appeared to be somewhat lower than those of deletions that were generated using the same ZFNs, which range from 0.03% to 10% (Lee et al. 2010). In Supplemental Table 2, we compared the frequencies of duplications and inversions with those of corresponding deletions induced using the same ZFNs.
These high frequencies suggest that one should be able to isolate clones harboring intended genomic rearrangements by dilution cloning, as demonstrated above. For example, we identified 11 clones that contain 15-kbp duplications after screening 9000 cells (0.1%). This frequency is somewhat lower than that measured by the digital PCR analysis (0.3%). It appears that not all single cells that contain the duplications could survive to form colonies due to the cytotoxicity of ZFNs as observed previously (Kim et al. 2009).
Toward gene therapy of severe hemophilia A caused by DNA inversion
Hemophilia A is caused by various mutations in the F8 gene (Graw et al. 2005). Almost half of all severe hemophilia A cases result from inversions of portions of the F8 gene (Lakich et al. 1993; Bagnall et al. 2002). Such inversions are caused by NAHR between far-upstream sequences and their corresponding highly homologous sequences that are located in intron 1 or intron 22 of the F8 gene. Here, we focused on the inversion that involves the intron 1 homolog (Fig. 4A). Unfortunately, no cell lines are currently available that harbor either of these inversion genotypes. We used HEK 293 cells as a surrogate system, because both wild-type and mutant cells contain the same target sequences that are associated with the inversion genotypes.
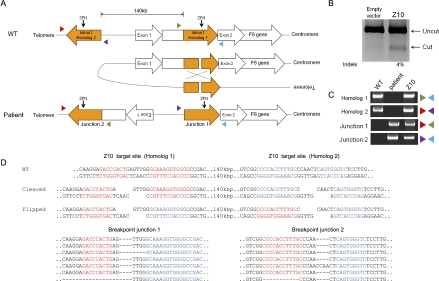
Figure 4.
Targeted inversion of the F8 gene using ZFNs. (A) Schematic representation of a chromosomal inversion that causes severe hemophilia A. NAHR between two homologous regions, one in intron 1 of the F8 gene (here named homolog 1) and the other located 140 kbp upstream (homolog 2), gives rise to an inversion found in severe hemophilia A. The two homologous regions are oriented in opposite directions. PCR primers (colored triangles) used to detect the inversion are shown. Z10 target sites are indicated by arrows. (B) Site-specific mutations at the Z10 target site in the F8 intron 1 revealed by the T7 endonuclease assay. (C) PCR products corresponding to wild-type and inversion genotypes. Genomic DNA isolated from a hemophilia A patient was used as a positive control for the inversion-specific PCR. (D) DNA sequences of breakpoint junctions of the inversion events. Z10 target sites are shown in red (homolog 1) or blue (homolog 2).
We constructed a ZFN pair that targets the intron 1 homolog and expressed this ZFN (termed Z10) in HEK 293 cells to test its genome-editing activity. We found that Z10 was highly active, inducing site-specific mutations at the target site (Fig. 4B). Next, we used PCR to test whether this ZFN could induce the inversion of the 140-kbp DNA segment that contains the promoter and exon 1 of the F8 gene. PCR amplicons specific to the DNA inversion found in severe hemophilia A patients were produced from HEK 293 cells transfected with Z10 but not from untransfected, control cells (Fig. 4C). DNA sequence analysis of these PCR products showed that indels were often, but not always, induced at the ZFN target site (Fig. 4D). These indels are characteristic of NHEJ-mediated DSB repair. But we cannot rule out the possibility that the ZFN-induced inversions were mediated first by NAHR and that indels were formed later by NHEJ repair of DSBs generated by persistent ZFN activity on the target site.
We then determined the inversion frequency of Z10 using digital PCR analysis (Kim et al. 2010b). The frequency of the 140-kbp inversion induced by Z10 ranged from 0.2% to 0.4% (Supplemental Table 1). Next, we screened for clones that contained the inverted genotype via limiting dilution. We obtained three independent clones from 3000 single cells (0.1%) and confirmed the inversion genotype using PCR (Supplemental Fig. 5A) and DNA sequencing (Supplemental Fig. 5B). HEK 293 cells contain three copies of the X chromosome and thus three copies of the X-linked F8 gene (Bylund et al. 2004). Each of these three clones was heterozygous, containing both the wild-type allele and an inverted allele. We determined the DNA sequence of the ZFN target site in these clones and found that only one clone contained an indel at the inversion junction (Supplemental Fig. 5B). No other local mutations were observed at the ZFN target site in other clones. Furthermore, no other genome rearrangements, such as deletions or duplications that involve the ZFN target site, were observed in these clones. We propose that this same ZFN could be used to revert the inverted chromosomal segment in mutant cells that carry the inversion genotype back to the wild-type orientation because the DNA sequence of the target site is the same in wild-type and mutant cells.
Discussion
Thanks to the rapid progress in DNA sequencing and microarray technologies, SVs in the human and other genomes are being identified at an accelerated rate. However, little is known about their etiology and biology. DNA sequence analyses of SVs suggest that three different mechanisms are involved in the generation of SVs: NAHR, NHEJ, and replication-based mechanisms (Hastings et al. 2009; Conrad et al. 2010). Experimental tests of each mechanism in mammalian and other higher eukaryotic cells have been limited due to the lack of appropriate tools. In this report, we used custom-designed ZFNs to demonstrate that two concurrent DSBs occurring in human chromosomes give rise to duplications, inversions, and deletions of pre-defined chromosomal segments in human cells, thus providing strong experimental evidence that supports NHEJ-mediated generation of SVs. (Because most of our ZFNs, except for the F8 gene-targeting Z10, were designed to target non-homologous regions, we did not observe targeted genome rearrangements via NAHR in this study. The limited homology between CCR2 and CCR5 appears not sufficient to promote rearrangements via NAHR.) DSBs are a frequently occurring form of DNA damage caused by chemical, physical, and biological stresses. Apparently, the repair of DSBs can give rise to genomic rearrangements, which contribute to SVs and genetic diversity.
Our results suggest that targeted genome rearrangements using ZFNs could provide a powerful platform for inducing individual SVs in cultured cells and model organisms. It has been shown that SVs are associated with various phenotypes and diseases such as susceptibility to viral infection (Gonzalez et al. 2005), obesity (Bochukova et al. 2010), mental disorders (International Schizophrenia Consortium 2008; Pinto et al. 2010), etc. The biological roles and phenotypic effects of thousands of newly discovered SVs in human and other organisms are largely unexplored and are likely to become the subjects of intense research. To study the biology of an SV of interest, researchers ultimately need to obtain a clone that harbors the variation and compare it with an isogenic wild-type clone. This approach has been hampered by a lack of specialized molecular methods to induce SVs in an isogenic background. As demonstrated in this report, ZFNs can be custom-designed to target any genomic locus and could allow creation of SVs in cultured cells and organisms.
In our study, the frequencies of ZFN-mediated genome rearrangements ranged from 10−4 to 10−1 and were largely independent on the length of rearranged genomic segments, at least up to hundreds of kbp in length (Supplemental Table 1). These high frequencies allowed us to isolate clonal populations of cells, by limiting dilution, in which pre-defined genomic segments were rearranged. The use of improved nuclease domains in the ZFNs (Guo et al. 2010; Doyon et al. 2011) or altered cell culture conditions (Doyon et al. 2010) might improve the frequencies of targeted genome rearrangements further. It remains to be seen whether these high frequencies of genome rearrangements can be achieved in non-transformed primary cells that are difficult to transfect. Surrogate reporters might be used for the enrichment of cells that contain nuclease-induced genome rearrangements, as demonstrated for the enrichment of cells that contain local mutations (Kim et al. 2011a).
Unfortunately, however, ZFNs that generate two concurrent DSBs in a chromosome often, but not always, give rise to two or more rearrangement events in a single cell, as we have shown in our analysis of clonal populations of cells. (Note that the complex rearrangements we observed in DEL1A/B and DEL2A/B clones were caused by induction of three rather than two DSBs in a cell.) Thus, deletions can be accompanied with duplications and even with inversions, or vice versa, in single cells. Because these events appear to occur randomly, a sufficient number of cells must be screened to isolate clones that harbor only the SV of interest. Indeed, we were able to isolate clones that contain only the intended inversion or duplication but no other rearrangements, as demonstrated above.
To induce chromosomal deletions, duplications, and inversions, one needs to use two pairs of ZFNs in general, unless a single ZFN pair targets two sites with identical or highly homologous sequences. Functional ZFNs can be formed not only between orthogonal pairs but also between unorthogonal pairs, which gives rise to off-target mutations. In this regard, we recommend use of two different autonomous ZFN pairs to prevent the formation of crossed dimers (Sollu et al. 2010).
We also propose that ZFNs can be used to correct genetic aberrations caused by chromosomal rearrangements. Many genetic diseases and cancer are known to result from recurrent chromosomal rearrangements. An example shown here is the inversion of a large genomic segment that disrupts the blood coagulation factor VIII gene, which is responsible for almost half of all cases of severe hemophilia A (Lakich et al. 1993). Both deletions and duplications of dosage-sensitive genes such as PLP1 result in genetic disorders (Lee et al. 2007). As demonstrated in this study, ZFNs may provide the means to correct these chromosomal aberrations. Of note is the possibility of implementing ZFN technology with induced pluripotent stem (iPS) cells (Takahashi et al. 2007). iPS cells derived from patients with genetic diseases contain the very genetic defects that cause the diseases and, thus, cannot be used per se in cell therapy. Tailor-made ZFNs could be used to correct the genetic aberrations in iPS or somatic cells, which are subsequently differentiated or de-differentiated, respectively, into appropriate cells and then delivered to patients. In principle, clones of gene-corrected iPS cells can be isolated by limiting dilution and then expanded. This approach is not limited by the efficiency of ZFN delivery into host cells as is conventional gene therapy using viral or nonviral vectors to deliver ZFNs directly to cells in the patient. Furthermore, clones of gene-corrected iPS cells can be carefully monitored, before use in cell therapy, to examine whether they contain any other undesired rearrangements or off-target mutations in the genome, a critical advantage of using gene-corrected iPS cells over direct delivery of ZFNs into patients.
Our findings that ZFNs can induce various chromosomal rearrangements raise a concern about ZFN-mediated gene therapy. A derivative of ZFN-224 used in this study is now under clinical investigation in the U.S. to treat patients with AIDS (Holt et al. 2010). The rationale behind this clinical study is that ZFN-224-treated T cells would be devoid of CCR5, an essential co-receptor of HIV, and become resistant to the viral infection. As shown in this and our previous study (Lee et al. 2010), ZFN-224 can induce deletion, inversion, and/or duplication of the 15-kbp chromosomal segment between the CCR2 and CCR5 loci in human cells. (Note, however, that Sangamo Biosciences Inc. claims that its CCR5-targeting ZFN has been optimized for clinical use [doi: 10.1038/news.2011.461].) Recently, two groups independently reported on genome-wide off-target effects of ZFNs, including ZFN-224 (Gabriel et al. 2011; Pattanayak et al. 2011). They carefully measured local mutation frequencies at potential off-target sites. Our results suggest that gross chromosomal rearrangements between off-target sites and the on-target site should also be monitored, because these events would be more detrimental than are local mutations.
ZFNs have traditionally been used to introduce local genomic modifications in higher eukaryotic cells and organisms by targeting a single site in the genome (Kim et al. 2010a, 2011b), and the possible outcomes of two concurrent ZFN-induced DSBs have only recently been explored (Kim et al. 2010a). ZFNs inducing two concurrent DSBs, each occurring in a different chromosome, have been shown to promote translocations in human cells (Brunet et al. 2009). We reported on the isolation and characterization of single clones that harbor 15-kbp deletions of the genomic segment between the CCR2 and CCR5 genes in human cells (Lee et al. 2010). However, these deletions generate CCR2–CCR5 hybrid genes and may not be appropriate for functional studies of these genes. In this report, we extended the scale of DNA excision to isolate clones that harbor 230-kbp deletions, which remove the entire cluster of four chemokine receptor genes, CCR1, CCR2, CCR3, and CCR5. (These homologous genes may have arisen from gene duplications, possibly mediated by DSB repair.) These genes may have redundant roles, and ZFNs used for making the 230-kbp genomic excision could provide valuable resources to study the functions of these genes.
Duplications and inversions constitute a large fraction of SVs in the genome and are responsible for many genetic diseases. To our knowledge, it has never been demonstrated that these important classes of genomic rearrangements can be induced in eukaryotic cells in a targeted manner. In principle, it may be possible to create duplications and inversions using recombinase systems such as Cre/loxP (Ramirez-Solis et al. 1995) or meganucleases such as I-SceI (Simsek and Jasin 2010). But these methods are not only laborious and time-consuming but also impractical for use in gene or cell therapy because they require pre-insertion of the target sequence into the genome. This report is the first demonstration that duplications and inversions can be induced in a targeted manner without pre-manipulation of the genome.
In summary, we have developed a novel method of targeted genome rearrangements using engineered ZFNs. We propose that ZFNs can be employed as molecular tools to study the mechanisms that cause chromosomal rearrangements and to create SVs in cultured cells and model organisms so as to study their biological roles. In addition, our method raises the possibility of correcting genetic defects caused by chromosomal rearrangements and holds new promise in gene and cell therapy.
Methods
Plasmids
Plasmids that encode ZFNs targeting the CCR5 gene are described (Kim et al. 2009), as are plasmids that encode ZFNs targeting sites far upstream of the CCR5 locus (Lee et al. 2010). Z10 was obtained from ToolGen Inc. Z10 and ZFN-224 have an obligatory heterodimeric nuclease domain (Miller et al. 2007) and all the other ZFNs have the wild-type nuclease domain.
Cell culture and transfection
HEK 293T/17 (ATCC, CRL-11268) cells were maintained in Dulbecco's modified Eagle medium (Welgene Biotech) supplemented with fetal bovine serum (10%), penicillin (100 units/mL), and streptomycin (100 μg/mL) at 37°C and 5% CO2. Cells were seeded at 1 × 106 cells/well in a six-well plate, incubated for 24 h, and then transfected with ZFN-encoding plasmids (total 4 μg) using polyethylenimine (Aldrich, cat#40872). At 3 d post-transfection, genomic DNA was isolated from cells using the G-DEX IIc Genomic DNA Extraction Kit (iNtRON, cat#17231) as described by the manufacturer's protocol and used for PCR analysis.
Isolation of clonal populations of cells
Clonal populations of cells that harbor various genomic rearrangements were isolated in a two-step dilution cloning process as described (Kim et al. 2010b). First, HEK 293T/17 cells transfected with plasmids encoding ZFNs were incubated for 6 h and seeded in three 96-well plates at a density of 50 or 30 cells per well on average. When grown to confluency, cell populations were split into two halves, one of which was used for PCR analysis to screen for pools of cells that contained clones with desired genomic rearrangements. Cells in positive wells were seeded in three 96-well plates at limiting dilution (0.4 cell per well on average). Each clone was cultured for two to three weeks, and a portion of cells in each well was used for PCR analysis to detect chromosomal rearrangements.
PCR analysis and DNA sequencing of breakpoint junctions
To detect chromosomal rearrangements, cells in 96-well plates were lysed in 50 μL lysis buffer (400 ng/μL proteinase K, 17 μM SDS), and genomic DNA (100 ng per reaction) was subjected to PCR analysis using appropriate primers (Supplemental Table 3). For sequencing analysis, PCR products corresponding to chromosomal rearrangements were purified using QIAquick Gel Extraction Kit (QIAGEN) and cloned into the T-Blunt vector using the T-Blunt PCR Cloning Kit (SolGent). Cloned PCR products were sequenced using M13 primers or primers used for PCR amplification.
Southern blot analysis
DNA probes were amplified from a suitable template with Phusion High-Fidelity DNA Polymerases (Finnzymes, cat#530) using appropriate primers (Supplemental Table 3), and PCR products were cloned into the T-Blunt vector and sequenced. DNA probes were prepared by PCR amplification of cloned DNA templates and labeling with [α-32P]-dCTP using the Megaprime DNA Labeling System (Amersham). Genomic DNA (30 μg) was treated with XbaI (New England Biolabs), electrophoresed on agarose gels (0.8%), and transferred to Hybond-N+membrane (Amersham). After hybridization at 65°C for 24 h, the membrane was washed and exposed to an imaging plate (Fuji Photo Film Co.), which was then scanned using the BAS-2500 system (FUJIFILM Life Science) and analyzed using Multi Gauge Ver3.0 (Fuji Photo Film Co.) software.
Frequencies of chromosomal rearrangements
The frequencies of chromosomal rearrangements were estimated by digital PCR analysis as described (Kim et al. 2010b). Genomic DNA samples isolated from cells transfected with plasmids encoding ZFNs were serially diluted in distilled water and diluted samples were then subjected to nested PCR using appropriate primers (Supplemental Table 3). Critical dilution points that support the amplification of breakpoint junctions were determined. The results were analyzed using the Extreme Limiting Dilution Analysis program (Hu and Smyth 2009) (http://bioinf.wehi.edu.au/software/elda/).
Genomic DNA from hemophilia A patients
Seoul National University Institutional Review Board (SNUIRB) approval was obtained for the analysis performed in this study. Blood cells of a hemophilia A patient were obtained at Korea Hemophilia Foundation Clinic, and genomic DNA was isolated using QIAamp DNA blood Maxi kit.
Acknowledgments
J.-S.K. is supported in part by the National Research Foundation of Korea (2011-0000402). We thank Dr. Ki-Young Yoo for providing the blood sample of a hemophilia patient.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.129635.111.
References
- Bagnall RD, Waseem N, Green PM, Giannelli F 2002. Recurrent inversion breaking intron 1 of the factor VIII gene is a frequent cause of severe hemophilia A. Blood 99: 168–174 [DOI] [PubMed] [Google Scholar]
- Bochukova EG, Huang N, Keogh J, Henning E, Purmann C, Blaszczyk K, Saeed S, Hamilton-Shield J, Clayton-Smith J, O'Rahilly S, et al. 2010. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature 463: 666–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet E, Simsek D, Tomishima M, DeKelver R, Choi VM, Gregory P, Urnov F, Weinstock DM, Jasin M 2009. Chromosomal translocations induced at specified loci in human stem cells. Proc Natl Acad Sci 106: 10620–10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno C, Catalina P, Melen GJ, Montes R, Sanchez L, Ligero G, Garcia-Perez JL, Menendez P 2009. Etoposide induces MLL rearrangements and other chromosomal abnormalities in human embryonic stem cells. Carcinogenesis 30: 1628–1637 [DOI] [PubMed] [Google Scholar]
- Bylund L, Kytola S, Lui WO, Larsson C, Weber G 2004. Analysis of the cytogenetic stability of the human embryonal kidney cell line 293 by cytogenetic and STR profiling approaches. Cytogenet Genome Res 106: 28–32 [DOI] [PubMed] [Google Scholar]
- Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, et al. 1996. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85: 1135–1148 [DOI] [PubMed] [Google Scholar]
- Conrad DF, Bird C, Blackburne B, Lindsay S, Mamanova L, Lee C, Turner DJ, Hurles ME 2010. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet 42: 385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doranz BJ, Rucker J, Yi Y, Smyth RJ, Samson M, Peiper SC, Parmentier M, Collman RG, Doms RW 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85: 1149–1158 [DOI] [PubMed] [Google Scholar]
- Doyon Y, Choi VM, Xia DF, Vo TD, Gregory PD, Holmes MC 2010. Transient cold shock enhances zinc-finger nuclease-mediated gene disruption. Nat Methods 7: 459–460 [DOI] [PubMed] [Google Scholar]
- Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC 2011. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods 8: 74–79 [DOI] [PubMed] [Google Scholar]
- Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C, Nowrouzi A, Bartholomae CC, Wang J, Friedman G, et al. 2011. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol 29: 816–823 [DOI] [PubMed] [Google Scholar]
- Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ, et al. 2005. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 307: 1434–1440 [DOI] [PubMed] [Google Scholar]
- Graw J, Brackmann HH, Oldenburg J, Schneppenheim R, Spannagl M, Schwaab R 2005. Haemophilia A: From mutation analysis to new therapies. Nat Rev Genet 6: 488–501 [DOI] [PubMed] [Google Scholar]
- Guo J, Gaj T, Barbas CF III 2010. Directed evolution of an enhanced and highly efficient FokI cleavage domain for zinc finger nucleases. J Mol Biol 400: 96–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G 2009. Mechanisms of change in gene copy number. Nat Rev Genet 10: 551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, Crooks GM, Kohn DB, Gregory PD, Holmes MC, et al. 2010. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol 28: 839–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Smyth GK 2009. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 347: 70–78 [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C 2004. Detection of large-scale variation in the human genome. Nat Genet 36: 949–951 [DOI] [PubMed] [Google Scholar]
- International Schizophrenia Consortium 2008. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 455: 237–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S 1996. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci 93: 1156–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS 2009. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res 19: 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Lee HJ, Carroll D 2010a. Genome editing with modularly assembled zinc-finger nucleases. Nat Methods 7: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Lee HJ, Kim E, Kim J-S 2010b. Analysis of targeted chromosomal deletions induced by zinc finger nucleases. Cold Spring Harb Protoc. doi: 10.1101/pdb.prot5477 [DOI] [PubMed] [Google Scholar]
- Kim H, Um E, Cho SR, Jung C, Kim JS 2011a. Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nat Methods 8: 941–943 [DOI] [PubMed] [Google Scholar]
- Kim S, Lee MJ, Kim H, Kang M, Kim JS 2011b. Preassembled zinc-finger arrays for rapid construction of ZFNs. Nat Methods 8: 7. [DOI] [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. 2007. Paired-end mapping reveals extensive structural variation in the human genome. Science 318: 420–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakich D, Kazazian HH Jr, Antonarakis SE, Gitschier J 1993. Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat Genet 5: 236–241 [DOI] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR 2007. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131: 1235–1247 [DOI] [PubMed] [Google Scholar]
- Lee HJ, Kim E, Kim JS 2010. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res 20: 81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. 2007. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol 25: 778–785 [DOI] [PubMed] [Google Scholar]
- Nikiforov YE, Koshoffer A, Nikiforova M, Stringer J, Fagin JA 1999. Chromosomal breakpoint positions suggest a direct role for radiation in inducing illegitimate recombination between the ELE1 and RET genes in radiation-induced thyroid carcinomas. Oncogene 18: 6330–6334 [DOI] [PubMed] [Google Scholar]
- Nikiforova MN, Stringer JR, Blough R, Medvedovic M, Fagin JA, Nikiforov YE 2000. Proximity of chromosomal loci that participate in radiation-induced rearrangements in human cells. Science 290: 138–141 [DOI] [PubMed] [Google Scholar]
- Park H, Kim JI, Ju YS, Gokcumen O, Mills RE, Kim S, Lee S, Suh D, Hong D, Kang HP, et al. 2010. Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing. Nat Genet 42: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V, Ramirez CL, Joung JK, Liu DR 2011. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods 8: 765–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, et al. 2008. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol 26: 808–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. 2010. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466: 368–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Solis R, Liu P, Bradley A 1995. Chromosome engineering in mice. Nature 378: 720–724 [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. 2006. Global variation in copy number in the human genome. Nature 444: 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothkamm K, Kuhne M, Jeggo PA, Lobrich M 2001. Radiation-induced genomic rearrangements formed by nonhomologous end-joining of DNA double-strand breaks. Cancer Res 61: 3886–3893 [PubMed] [Google Scholar]
- Simsek D, Jasin M 2010. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol 17: 410–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollu C, Pars K, Cornu TI, Thibodeau-Beganny S, Maeder ML, Joung JK, Heilbronn R, Cathomen T 2010. Autonomous zinc-finger nuclease pairs for targeted chromosomal deletion. Nucleic Acids Res 38: 8269–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR 2010. Structural variation in the human genome and its role in disease. Annu Rev Med 61: 437–455 [DOI] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. 2009. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462: 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872 [DOI] [PubMed] [Google Scholar]
- van Gent DC, Hoeijmakers JH, Kanaar R 2001. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet 2: 196–206 [DOI] [PubMed] [Google Scholar]