Abstract
Objectives
Multiple linkage and association studies have suggested chromosome 8q24 as a promising candidate region for bipolar disorder (BP). We performed a detailed association analysis assessing the contribution of common genetic variation in this region to the risk of BP.
Methods
We analyzed 2,756 single nucleotide polymorphism (SNP) markers in the chromosome 8q24 region of 3,512 individuals from 737 families. In addition, we extended genotype imputation methods to family-based data and imputed 22,725 HapMap SNPs in the same region on 8q24. We applied a family-based method to test 15,552 high-quality genotyped or imputed SNPs for association with BP.
Results
Our association analysis identified the most significant marker (p = 4.80 × 10−5), near the gene encoding potassium voltage-gated channel KQT-like protein (KCNQ3). Other marginally significant markers were located near adenylate cyclase 8 (ADCY8) and ST3 beta-galactoside alpha-2,3-sialyltransferase 1 (ST3GAL1).
Conclusions
We developed an approach to apply MACH imputation to family-based data, which can increase the power to detect association signals. Our association results showed suggestive evidence of association of BP with loci near KCNQ3, ADCY8, and ST3GAL1. Consistent with genes identified by genome-wide association studies for BP, our results are consistent with the involvement of ion channelopathy in BP pathogenesis. However, common variants are insufficient to explain linkage findings in 8q24; other genetic variations should be explored.
Keywords: 8q24, bipolar disorder, imputation, ion channelopathy
Bipolar disorder (BP) is a common, complex psychiatric disease characterized by recurrent depression and manias, with an estimated lifetime prevalence of about 1% (1). Family and twin studies have reported a strong familial aggregation of BP, suggesting that genetic factors account for 60–85% of disease risk (2). While a large number of genetic variants were reported to be either linked or associated with BP, few have been replicated (3, 4). Only recent large, genome-wide association studies (GWAS) were able to identify the first BP genes. Ferreira et al. (5) analyzed a combined sample of 4,387 BP patients and 6,209 controls and reported genome-wide significant associations to BP with single nucleotide polymorphisms (SNPs) in Ankyrin 3 (ANK3) and in the alpha 1C subunit of the L-type voltage-gated calcium channel (CACNA1C), and the same SNPs in both ANK3 (6) and CACNA1C (7) were replicated by independent studies. However, these two variants account only for a small proportion of BP’s heritability; most heritable risk remains unexplained.
Some of this heritability may be explained by variants located in regions previously identified by linkage studies (4). Since the development and subsequent evolution of the human genome map and modern mapping methodologies, over 40 genome-wide linkage reports on BP and at least three meta-analyses (8, 9) were published [for a review see Barnett and Smoller (10)]. We first reported linkage to BP on the 8q24 region with a nonparametric linkage (NPL) score of 3.25 (11, 12). Cichon et al. (13) also reported a genome-wide significant two-point logarithm of odds (LOD) score (D8S514; LOD = 3.62) at 8q24 in a genome-wide linkage scan of 75 BP families. These results were included in a meta-analysis of 11 studies by McQueen et al. (9) which reported a genome-wide significant LOD score of 3.40 in a region on chromosome 8q24 under a broad model of BP [bipolar I disorder (BP-I) and bipolar II disorder (BP-II)]. Moreover, Macayran et al. (14) reported a child with BP carrying a duplication of 8q22.1– q24.1 caused by an unbalanced translocation.
To identify the genetic variants that account for the linkage signal in this region, we previously performed an association analysis with 249 candidate gene SNPs covering a 3.4 Mb region in a sample of 583 affected offspring from 258 nuclear families with evidence of linkage to BP. We detected a suggestive level of associations with SNPs three kb upstream of ST3GAL1 (15). We further typed an extended sample of 3,512 individuals from 737 multiplex families for 1,458 SNPs across a ~16 Mb region on 8q24. We tested each marker for association with BP and found suggestive but not experiment-wide significant associations with SNPs in several genes (16).
However, this SNP panel tagged (r2 > 0.8) only ~54% of known common polymorphisms in the 8q24 region (16). To fill the gaps, we designed a complementary panel of 1,536 additional SNPs in the same 8q24 region and typed the panel on the same sample (16). Here we present the joint analysis of all 3,072 SNPs. Furthermore, we developed an approach to apply the imputation method MACH (17) to family-based data. We imputed 22,725 HapMap SNPs in a ~18 Mb region on 8q24 flanking the linkage peak reported by McQueen et al. (9). After careful data cleaning, we tested all remaining variants for association with BP, and obtained evidence of a suggestive level of association between BP with loci near KCNQ3, ADCY8, and ST3GAL1. None of the observed associations is sufficient to account for the previously reported linkage signal.
Materials and methods
Samples
This study combined the Johns Hopkins sample of 65 families and the National Institute of Mental Health (NIMH) sample of 672 families; both samples have been described elsewhere [Hopkins sample (11); NIMH sample (11, 18; see also https://www.nimhgenetics.org/nimh_human_genetics_initiative/)]. Both samples collected multiplex families segregating BP, ascertained for a linkage study of BP. Family members were assessed using the Schedule for Affective Disorders–Lifetime Version (SADS-L) (19) or the Diagnostic Interview for Genetic Studies (DIGS) (20). Diagnoses of BP-I and schizoaffective disorder, bipolar type (SABP) were based on Research Diagnostic Criteria (RDC) in the first sample and DSM-III-R criteria in the second sample (criteria are essentially the same). BP-II diagnosis was based on RDC, with the additional requirement of recurrent major depression. The final best-estimate diagnostic procedure engaged two noninterviewing psychiatrists to review all the data for a consensus clinical diagnosis. In the case of disagreement, a third psychiatrist reviewed discordant diagnoses and adjudicated a final diagnosis.
Our sample comprised 3,525 genotyped individuals, including 1,391 males and 2,134 females from 737 families (16). As the initial linkage peak was obtained using a broad definition of affected/unaffected status, we defined individuals diagnosed with BP-I, schizoaffective disorder, SABP, or BP-II as affected (n = 1,958), and individuals who were determined to have never been diagnosed with BP, depression, schizophrenia, or any Axis I diagnosis as unaffected (n = 515). The remaining individuals were defined as ‘phenotype uncertain’ (n = 1,052); in this category were individuals with borderline or uncertain affective disorders such as single episodes of depression and nonaffective disorders such as substance use disorders, anxiety disorders, and other illness that could have potentially been an expression of overlapping vulnerability with mood disorders.
Genotype data
Genotype data were collected in two phases. We selected 1,536 SNPs in the region from 123.1 to 139.1 Mb (Build 35) on chromosome 8q24 using FESTA (21) for the first phase that was performed at the Center of Inherited Disease Research (CIDR) (16). We aimed to tag all the known common variants [minor allele frequency (MAF) > 0.05] with r2 ≥ 0.5 in region 123 to 131 Mb identified by Cichon et al. (13) and r2 ≥ 0.8 in region 131 to 139 Mb identified by McInnis et al. (11). After careful quality control (16), 1,461 SNPs were included in the final analysis.
To improve coverage, we selected and typed additional 1,536 SNPs conditional on the first marker set using FESTA (21). We designed this marker set to maximize the number of SNPs tagged using the same r2 and MAF criteria as in phase I. Moreover, we retyped 24 SNPs from phase I to estimate genotyping error rates. All markers were selected to have an Illumina (Illumina, Inc., San Diego, CA, USA) design cutoff score of 0.6, per manufacturer’s instructions, to generate a customized Illumina panel of 1,536 SNPs. These SNPs were genotyped using the University of Michigan’s Department of Psychiatry/Molecular and Behavioral Neuroscience Institute (Ann Arbor, MI, USA) microarray core facility on a local Illumina BeadStation system, following manufacturer’s instructions.
Quality control of the phase II data used PEDSTATS (22). We removed all SNPs that did not satisfy all of the following criteria: successful genotyping rate ≥ 90%; number of non-Mendelian inheritance (NMI) errors < 6; Hardy-Weinberg equilibrium (HWE) test using the entire sample with p-value ≥ 10−6; and MAF ≥ 5%. After applying these quality-control criteria, we retained 1,295 SNPs of the 1,536 for analysis for a combined dataset of 2,756 SNPs.
Statistical analyses
Single-marker association analysis
We performed single-marker association tests with the LAMP program (23), a maximum-likelihood method that jointly models linkage and association, to incorporate the large family sizes in our dataset (maximum family size: n = 23). For our main analysis, we assumed a multiplicative model with a population prevalence of 1%. In addition, we compared to the results obtained under dominant/recessive and a free model without any genetic model assumptions.
Imputation
We used the MACH program (17) to impute genotypes for all markers in the 8q24 region using the CEU population from HapMap (Build 35) database as references (24). MACH implements a hidden Markov model to impute unknown SNP genotypes, modeling samples as unrelated individuals. To extend the algorithm to related individuals, we performed MACH in two steps. First, we selected 200 independent individuals from our sample and analyzed them with MACH to calibrate imputation parameters such as the estimates of imputation error rates. Based on these estimates, we then imputed genotypes for the entire sample, treating individuals as independent. In total, we imputed 22,725 SNPs in an 18 Mb region by expanding one Mb at each end of our genotyped region.
We estimated imputation error rates by masking 2% of the original genotypes before imputation and then comparing the true genotypes with their imputed counterparts. To identify and remove poorly imputed markers, we evaluated three statistics: First, we removed all markers with MAF < 0.05 (n = 1,905). Second, we assessed the estimated squared correlation between imputed genotypes and true genotypes r̂2 calculated by MACH. We excluded markers with r̂2 < 0.3 (n = 4,225), which has been shown to remove ~70% of badly imputed SNPs (17). Moreover, the family structure in our dataset allowed us to estimate the imputation quality by counting the number of NMIs for each imputed SNP. We removed imputed SNPs with > 30 NMIs (n = 1,042). A total of 15,552 SNPs were included in the final association analysis.
Results
Genotyping quality and coverage
We estimated the genotyping error rate by comparing genotypes of 24 SNPs that were typed in both phases for all individuals. The estimated average mismatch rate was 0.26% per SNP. The combined marker set tagged 94.1% (78.3%) of the common HapMap SNPs (MAF > 0.05) in the 8q24 region with r2 ≥ 0.50 (r2 ≥ 0.80).
Single-marker association analysis
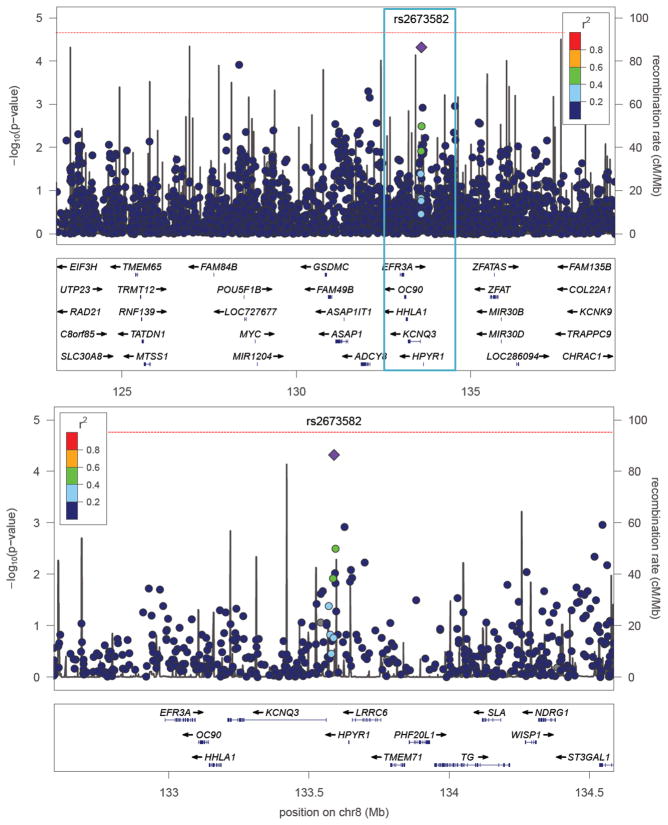
We tested each SNP for association with BP conditional on the underlying family structure using LAMP (23), assuming a multiplicative model with a disease prevalence of 1%. The most significantly associated marker was rs2673582 (p = 4.80 × 10−5), which is located 27 Kb upstream of KCNQ3 (Fig. 1). Applying Bonferroni correction for an experiment-wide type 1 error probability of 0.05 with 2,756 tests generates a single-marker critical value of 1.8 × 10−5. Hence, the observed p-value can be considered to be suggestively significant. Three other SNPs had p-values < 10−3, two of which (rs3750889 and rs1023096) are located within ADCY8 gene and are in high linkage disequilibrium (r2 = 0.86) (Table 1). A marker identified by Zandi et al. (16) in ST3GAL1 was ranked fifth in our analysis (p = 1.1 × 10−3). Results obtained under a dominant/recessive model or a free model were not fundamentally different (data not shown).
Fig. 1.
Panel A shows p-values (−log10p) from an association test for each genotyped SNP versus position (Mb) across linkage peak on 8q24 (9). Panel B magnifies one Mb surrounding the most significant marker rs2673582 (purple diamond). Below each plot, a subset of genes in this region is shown. Light gray lines display recombination rates as estimated from HapMap. The colors of the circles indicate the strength of linkage disequilibrium (LD) with rs2673582; gray circles indicate missing LD information (36). The vertical line indicates an experiment-wide significance threshold according to Bonferroni correction for multiple tests.
Table 1.
Top 10 results of single-marker association tests for genotyped markers using LAMP under a multiplicative model with a disease prevalence of 1%
| Marker | Position (Mb) | MAF | Gene | Location | p-value |
|---|---|---|---|---|---|
| rs2673582 | 133.59 | 0.425 | KCNQ3 | 27 Kb upstream | 4.80E-05 |
| rs4871780 | 128.36 | 0.421 | 1.20E-04 | ||
| rs3750889 | 132.07 | 0.406 | ADCY8 | Intron | 5.00E-04 |
| rs1023096 | 132.10 | 0.419 | ADCY8 | Intron | 7.00E-04 |
| rs6986303 | 134.55 | 0.289 | ST3GAL1 | Intron | 1.10E-03 |
| rs6984550 | 133.63 | 0.200 | KCNQ3 | 64 Kb upstream | 1.20E-03 |
| rs10095649 | 135.23 | 0.133 | 0.0026 | ||
| rs4523235 | 132.31 | 0.303 | 0.0027 | ||
| rs10094837 | 135.27 | 0.138 | 0.0028 | ||
| rs17602731 | 133.59 | 0.314 | KCNQ3 | 32 Kb upstream | 0.0032 |
MAF = minor allele frequency.
Imputation
To assess the performance of the imputation-method MACH on family-based data, we randomly masked 2% of genotypes and treated them as missing, then estimated the performance by comparing the imputed genotypes to the true genotypes. The estimated imputation error rate was 0.0577 per genotype and 0.035 per allele, respectively. We further assessed the quality of imputed genotypes for each marker using both the number of NMIs among imputed SNPs and the estimated r̂2 values generated by MACH. A total of 4,225 markers failed only the r̂2 criteria, 1,145 failed only the NMI criteria, and 103 markers failed both. While the number of NMIs and the imputation r̂2 were negatively correlated (coefficient −0.41), removing imputed SNPs by the number of observed NMIs provided an additional filter for identifying poorly imputed markers.
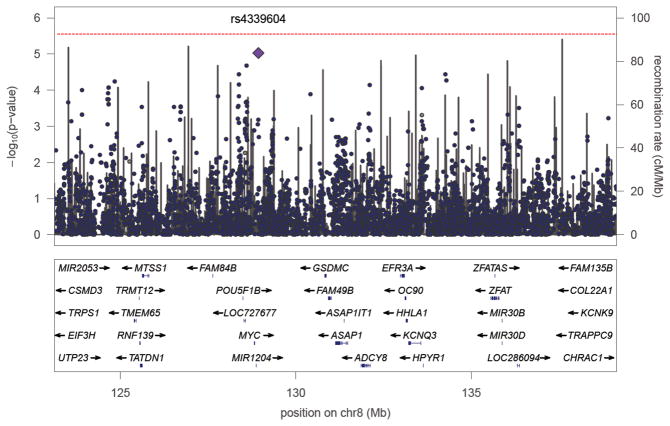
We tested the imputed genotypes of 15,552 SNPs for association with BP in a two-step procedure. First, we tested each marker using MQLS (25), a fast test that corrects for relatedness among individuals. However, MQLS loses power by not fully modeling the pedigree structure. Hence, we followed up each promising signal (p < 0.1) using LAMP (23). Our results showed 11 SNPs with p-values < 10−4, with the most significant being rs4339604 (p = 9.4 × 10−6, MAF = 0.057, physical position = 128.93 Mb), followed by rs7824868 (p = 2.1 × 10−5, MAF = 0.11, physical position = 128.59 Mb) (Fig. 2). Note that the most significant result near 128 Mb is located in a gene desert.
Fig. 2.
P-values (−log10p) for association of genotyped and imputed SNPs on 8q24. The horizontal axis shows position in Mb. The purple diamond indicates the most significant imputed SNP rs4339604. A subset of genes in this region is shown below the main plot. The gray lines indicate recombination rates as estimated from HapMap. The vertical line indicates an experiment-wide significance threshold according to Bonferroni correction for multiple tests.
Discussion
We analyzed a sample of 3,512 individuals in 737 families and tested 2,756 genotyped SNPs spanning ~16 Mb across the previously identified linkage peak in the 8q24 region (9). Furthermore, we imputed and tested all common HapMap SNPs in this region. Among the genotyped markers, the most significantly associated SNPs are located close to 133 Mb near KCNQ3, which is consistent with the linkage peak identified by genome-wide linkage analysis (9). Our results provided further suggestive evidence that genetic variants in ST3GAL1 or ADCY8 may be associated with BP (15, 16). Compared to our previous results (16), we identified the same candidate genes with somewhat more significant association p-values; all variants identified by Zandi et al. were also nominally significant (p < 0.05) in our analysis. However, appropriately evaluating these p-values is challenging. Adjusting the most significant result for the number of sequenced markers results in a Bonferroni-corrected p = 0.13 (uncorrected p = 4.8 × 10−5). However, Bonferroni correction assumes independent tests, and the SNPs in this region are highly correlated. Moreover, permutation analysis cannot be applied to assess significance because of the family structure in our dataset. Hence, it is difficult to assess experiment-wide statistical significance of our results. Including imputed SNPs added additional signals with suggestive evidence for association, although no SNPs were significant after stringent (Bonferroni) correction for multiple testing.
All genes implicated by our analysis have previously been implicated as candidates for BP and/or other psychiatric disorders. KCNQ3 has been shown to be expressed highly specifically to brain and coexpressed with KCNQ2 in most brain regions (26). KCNQ2 has been implicated to be associated with BP through the phosphatidyl-inositol phosphate pathway (27), and both KCNQ2 and KCNQ3 are key components to form a voltage-gated potassium channel that is important in the regulation of neuronal excitability (26). Although no peer-reviewed evidence has been forthcoming on KCNQ3 as a susceptibility gene for BP disorder, a recently published U.S. patent proposed using a single nucleotide mutation in KCNQ3 gene to assess the presence of or predisposition to schizophrenia, BP, or a related mental disorder in a subject (28). Furthermore, our findings have an intriguing connection to replicated GWAS results. ANK3 anchors voltage-gated sodium channels, and both ANK3 and subunits of the calcium channel are down-regulated in response to lithium treatment in mice (29). Hence, the results from both ANK3 and KCNQ3 are consistent with the involvement of an ion channelopathy in BP (30), which was also supported by a recent pathway-based analysis on GWAS data in BP (31).
The product of ADCY8 catalyzes the formation of cyclic AMP from ATP, where cyclic AMP may be involved in BP pathogenesis as a target for lithium and other mood stabilizing agents (32, 33). De Mooij-van Malsen et al. (34) showed that ADCY8 was differentially expressed in specific brain regions as a function of avoidance behavior in mice. The authors further explored the human homologous 8q24 region using a candidate gene approach to test association with BP with genotypes from a GWAS and reported nominally significant associations with ADCY8 (p = 0.0055) and KCNQ3 (p = 0.0029). The product of the ST3GAL1 gene is a type II membrane protein that catalyzes the transfer of sialic acid from CMP-sialic acid to galactose-containing substrates. A recent family-based association of candidate genes reported evidence of association of ST3GAL1 to BP (empirical p-value < 0.005) (35).
As none of the signals we observed can sufficiently explain the linkage signal in 8q24, it is likely that additional BP variants exist in this region. However, as testing 15,552 additional imputed SNPs did not generate additional interesting signals, our panel of 2,756 SNPs likely captured most of the common haplotype variation in the 8q24 region. Therefore, typing additional common variants in this region would not result in new findings. Our results clearly show that the common variants in the 8q24 region do not explain the previously observed linkage peak (9). This may be for one of two reasons: (i) the linkage peak may be a false positive, and the replications of the linkage peak are the result of publication bias; or (ii) the causal genetic variants in this region may be individually rare SNPs or copy number variants, which association tests of common SNP markers have low power to detect. To assess the contribution of rare variants in 8q24, it will be necessary to sequence a set of candidate genes or the entire 8q24 region in a sample of BP cases. Our results indicate at least two potential starting points.
In summary, we identified three biologically feasible signals for association with BP, but more research is required to understand the contribution of genes in the 8q24 region to BP.
Acknowledgments
We thank Ryan Welch for help in plotting the figures and Yun Li for help with MACH. This work was supported by NIMH grant R01-MH070775, the Prechter Bipolar Research Fund, and the University of Michigan Medical School Biomedical Research Program.
Footnotes
The authors of this paper do not have any commercial associations that might pose a conflict of interest in connection with this manuscript.
References
- 1.Merikangas KR, Akiskal HS, Angst J, et al. Lifetime and 12-month prevalence of bipolar spectrum disorder in the National Comorbidity Survey replication. Arch Gen Psychiatry. 2007;64:543–52. doi: 10.1001/archpsyc.64.5.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smoller JW, Finn CT. Family, twin, and adoption studies of bipolar disorder. Am J Med Genet C Semin Med Genet. 2003;123C:48–8. doi: 10.1002/ajmg.c.20013. [DOI] [PubMed] [Google Scholar]
- 3.Serretti A, Mandelli L. The genetics of bipolar disorder: genome ‘hot regions,’ genes, new potential candidates and future directions. Mol Psychiatry. 2008;13:742–71. doi: 10.1038/mp.2008.29. [DOI] [PubMed] [Google Scholar]
- 4.Burmeister M, McInnis MG, Zöllner S. Psychiatric genetics: progress amid controversy. Nat Rev Genet. 2008;9:527–540. doi: 10.1038/nrg2381. [DOI] [PubMed] [Google Scholar]
- 5.Ferreira MA, O’Donovan MC, Meng YA, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott LJ, Muglia P, Kong XQ, et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc Natl Acad Sci USA. 2009;106:7501–7506. doi: 10.1073/pnas.0813386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sklar P, Smoller JW, Fan J, et al. Whole-genome association study of bipolar disorder. Mol Psychiatry. 2008;13:558–569. doi: 10.1038/sj.mp.4002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badner JA, Gershon ES. Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry. 2002;7:405–411. doi: 10.1038/sj.mp.4001012. [DOI] [PubMed] [Google Scholar]
- 9.McQueen MB, Devlin B, Faraone SV, et al. Combined analysis from eleven linkage studies of bipolar disorder provides strong evidence of susceptibility loci on chromosomes 6q and 8q. Am J Hum Genet. 2005;77:582–595. doi: 10.1086/491603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnett JH, Smoller JW. The genetics of bipolar disorder. Neuroscience. 2009;164:331–343. doi: 10.1016/j.neuroscience.2009.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McInnis MG, Lan TH, Willour VL, et al. Genome-wide scan of bipolar disorder in 65 pedigrees: supportive evidence for linkage at 8q24, 18q22, 4q32, 2p12, and 13q12. Mol Psychiatry. 2003;8:288–298. doi: 10.1038/sj.mp.4001277. [DOI] [PubMed] [Google Scholar]
- 12.Avramopoulos D, Willour VL, Zandi PP, et al. Linkage of bipolar affective disorder on chromosome 8q24: follow-up and parametric analysis. Mol Psychiatry. 2004;9:191–196. doi: 10.1038/sj.mp.4001388. [DOI] [PubMed] [Google Scholar]
- 13.Cichon S, Schumacher J, Muller DJ, et al. A genome screen for genes predisposing to bipolar affective disorder detects a new susceptibility locus on 8q. Hum Mol Genet. 2001;10:2933–2944. doi: 10.1093/hmg/10.25.2933. [DOI] [PubMed] [Google Scholar]
- 14.Macayran JF, Brodie SG, Rao PN, et al. Duplication 8q22.1-q24.1 associated with bipolar disorder and speech delay. Bipolar Disord. 2006;8:294–298. doi: 10.1111/j.1399-5618.2006.00306.x. [DOI] [PubMed] [Google Scholar]
- 15.Zandi PP, Avramopoulos D, Willour VL, et al. SNP fine mapping of chromosome 8q24 in bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:625–630. doi: 10.1002/ajmg.b.30486. [DOI] [PubMed] [Google Scholar]
- 16.Zandi PP, Zöllner S, Avramopoulos D, et al. Family-based SNP association study on 8q24 in bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:612–618. doi: 10.1002/ajmg.b.30651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dick DM, Foroud T, Flury L, et al. Genome-wide linkage analyses of bipolar disorder: a new sample of 250 pedigrees from the National Institute of Mental Health Genetics Initiative. Am J Hum Genet. 2003;73:107–114. doi: 10.1086/376562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Endicott J, Spitzer RL. A diagnostic interview: the schedule for affective disorders and schizophrenia. Arch Gen Psychiatry. 1978;35:837–844. doi: 10.1001/archpsyc.1978.01770310043002. [DOI] [PubMed] [Google Scholar]
- 20.Nurnberger JI, Jr, Blehar MC, Kaufmann CA, et al. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry. 1994;51:849–859. doi: 10.1001/archpsyc.1994.03950110009002. [DOI] [PubMed] [Google Scholar]
- 21.Qin ZS, Gopalakrishnan S, Abecasis GR. An efficient comprehensive search algorithm for tagSNP selection using linkage disequilibrium criteria. Bioinformatics. 2006;22:220–225. doi: 10.1093/bioinformatics/bti762. [DOI] [PubMed] [Google Scholar]
- 22.Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21:3445–3447. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- 23.Li M, Boehnke M, Abecasis GR. Joint modeling of linkage and association: identifying SNPs responsible for a linkage signal. Am J Hum Genet. 2005;76:934–949. doi: 10.1086/430277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Ding J, Abecasis GR. Mach 1.0: Rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet. 2006;S79:2290. [Google Scholar]
- 25.Thornton T, McPeek MS. Case-control association testing with related individuals: a more powerful quasi-likelihood score test. Am J Hum Genet. 2007;81:321–337. doi: 10.1086/519497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schroeder BC, Kubisch C, Stein V, Jentsch TJ. Moderate loss of function of cyclic-AMP-modulated KCNQ2/KCNQ3 K+ channels causes epilepsy. Nature. 1998;396:687–690. doi: 10.1038/25367. [DOI] [PubMed] [Google Scholar]
- 27.Carter CJ. Multiple genes and factors associated with bipolar disorder converge on growth factor and stress activated kinase pathways controlling translation initiation: implications for oligodendrocyte viability. Neurochem Int. 2007;50:461–490. doi: 10.1016/j.neuint.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 28.Chumakov I, Cohen D, Macciardi F. Compositions and methods for treating mental disorders. WO/2006/067056. [Last accessed November 1, 2010];U S Patent. http://www.wipo.int/pctdb/en/wo.jsp?WO=2006067056.
- 29.McQuillin A, Rizig M, Gurling HM. A microarray gene expression study of the molecular pharmacology of lithium carbonate on mouse brain mRNA to understand the neurobiology of mood stabilization and treatment of bipolar affective disorder. Pharmacogenet Genomics. 2007;17:605–617. doi: 10.1097/FPC.0b013e328011b5b2. [DOI] [PubMed] [Google Scholar]
- 30.Gargus JJ. Ion channel functional candidate genes in multigenic neuropsychiatric disease. Biol Psychiatry. 2006;60:177–185. doi: 10.1016/j.biopsych.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 31.Askland K, Read C, JM Pathways-based analyses of whole-genome association study data in bipolar disorder reveal genes mediating ion channel activity and synaptic neurotransmission. Hum Genet. 2009;125:63–79. doi: 10.1007/s00439-008-0600-y. [DOI] [PubMed] [Google Scholar]
- 32.Stewart RJ, Chen B, Dowlatshahi D, MacQueen GM, Young LT. Abnormalities in the cAMP signaling pathway in post-mortem brain tissue from the Stanley Neuropathology Consortium. Brain Res Bull. 2001;55:625–629. doi: 10.1016/s0361-9230(01)00524-x. [DOI] [PubMed] [Google Scholar]
- 33.Perez J, Tardito D, Mori S, Racagni G, Smeraldi E, Zanardi R. Abnormalities of cAMP signaling in affective disorders: implication for pathophysiology and treatment. Bipolar Disord. 2000;2:27–36. doi: 10.1034/j.1399-5618.2000.020104.x. [DOI] [PubMed] [Google Scholar]
- 34.de Mooij-van Malsen AJ, van Lith HA, Oppelaar H, et al. Interspecies trait genetics reveals association of Adcy8 with mouse avoidance behavior and a human mood disorder. Biol Psychiatry. 2009;66:1123–1130. doi: 10.1016/j.biopsych.2009.06.016. [DOI] [PubMed] [Google Scholar]
- 35.Perlis RH, Purcell S, Fagerness J, et al. Family-based association study of lithium-related and other candidate genes in bipolar disorder. Arch Gen Psychiatry. 2008;65:53–61. doi: 10.1001/archgenpsychiatry.2007.15. [DOI] [PubMed] [Google Scholar]
- 36.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]