Abstract
Epigenetic modification can mediate environmental influences on gene expression and can modulate the disease risk associated with genetic variation. Epigenetic analysis therefore holds substantial promise for identifying mechanisms through which genetic and environmental factors jointly contribute to disease risk. The spatial and temporal variance in epigenetic profile is of particular relevance for developmental epidemiology and the study of aging, including the variable age at onset for many common diseases. This review serves as a general introduction to the topic by describing epigenetic mechanisms, with a focus on DNA methylation; genetic and environmental factors that influence DNA methylation; epigenetic influences on development, aging, and disease; and current methodology for measuring epigenetic profile. Methodological considerations for epidemiologic studies that seek to include epigenetic analysis are also discussed.
Keywords: DNA methylation; environment; epigenesis, genetic; folic acid
The association between environmental factors and disease risk has been investigated for a wide variety of disorders. There have been notable successes, such as the discovery of strong links between smoking, radon, and asbestos exposure and lung cancer (1). For many other common diseases that contribute substantially to the global health burden, however, environmental factors account for only a proportion of total disease risk and may interact with genes to exert their effect. Even the association between smoking and lung cancer may be moderated by genotype (2–4). Conversely, environmental modulation of genetic effects may act at many different levels and can influence the phenotype of even the most penetrant single-gene disorders.
Genetic epidemiology is now an integral component of the epidemiology paradigm (5–7). Some diseases, such as cystic fibrosis, are almost wholly caused by single genetic mutations (8). Most common diseases, however, cannot be explained by single genetic risk factors in isolation. Advances in the application of genetic methodologies will no doubt continue to identify new genetic variants with small independent effects, but a knowledge of both polygenic effects and complex interactions may be required to fully understand disease causation (2). In some cases, specific environmental exposures are associated with increasing mutation rate, with a dose-dependent increase in genetic damage and disease risk (9–12).
Environmental agents can also modify gene expression independently of the primary DNA sequence through a process known as epigenetics. Epigenetic modifications are mitotically heritable chemical/structural changes that regulate gene activity in the absence of underlying changes to DNA sequence. These modifications are the likely mediators of gene-environment interaction because genetic factors can modify the epigenetic response to the environment (13), and faulty epigenetic silencing can have downstream genetic consequences (14). The primary DNA sequence is generally fixed at conception, but epigenetic marks are dynamic and modifiable, probably throughout the life course. Recent work with human cell lines has shown evidence of dynamic reprogramming of epigenetic markings during the cell cycle (15–17). The expression of genetic risk is therefore likely to show varying penetrance over time in response to epigenetic profile. Such dynamism raises the possibility of novel preventive and/or therapeutic opportunities (18).
This review covers the following 8 topics:
Epigenetic mechanisms
Genetic factors that influence DNA methylation
Epigenetics and development
Aging and the epigenome
Epigenetics and disease
Environmental factors and the epigenome
Measuring epigenetic profile
Considerations for epidemiologic studies involving epigenetic analysis
EPIGENETIC MECHANISMS
Several epigenetic mechanisms regulate genes. The most robust and readily measured modification is DNA methylation. Its assessment requires only small amounts of genomic DNA with little specialized sample processing, and a wide range of fresh and archived tissues can be used. Other epigenetic marks include changes to histone proteins, around which DNA is packed, or involve functional noncoding RNAs. The interested reader is referred elsewhere for a detailed account of these mechanisms (19, 20). At present, analysis of histone proteins and noncoding RNAs is more technically challenging than analysis of DNA methylation and requires cells to be cryopreserved, snap-frozen in liquid nitrogen, or stored in RNA preservative.
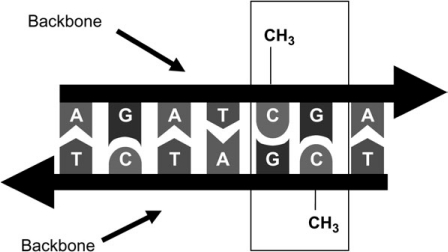
The DNA double helix is a ladder-like molecule in which each side, the backbone, is made up of phosphate groups and sugar molecules, and the rungs are composed of 4 bases: adenine (A), guanine (G), cytosine (C), and thymine (T). These bases are linked in pairs (known as base pairs) on opposite strands: C-G and A-T. In a CpG site, C and G are linked by a phosphate molecule in the “backbone” (Figure 1). CpG islands are regions of DNA that contain a high density of CpG sites (21). In many cases, these are located in the control region of genes or in association with repetitive DNA elements (22). In general, low levels of DNA methylation (hypomethylation) are associated with higher gene activity and high levels of methylation with gene silencing (23). Repeat associated CpG islands and nonregulatory CpG sites generally exist in a methylated state (22, 24).
Figure 1.
Diagrammatic representation of CpG methylation (where cytosine (C) and guanine (G) are linked by a phosphate molecule). Approximately 60% of genes have CpG islands in a region called the promoter. This segment of DNA acts to control the activity, or expression, of that gene. An important property of CpG sites is that the cytosine can be methylated, by adding a methyl (CH3) molecule, to form methyl-CpG. Note that, because DNA is double stranded, with strands running in opposite directions, and cytosine bases on one strand pair with guanine on the other, CpG sites are methylated on both strands.
GENETIC FACTORS THAT INFLUENCE DNA METHYLATION
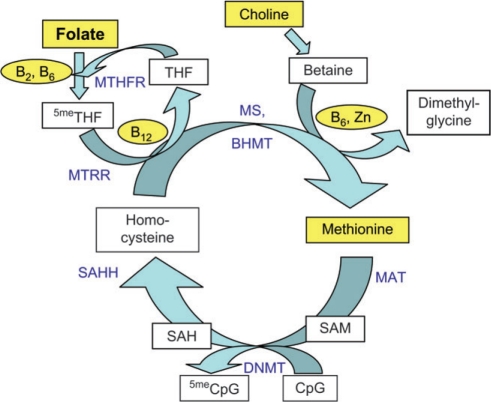
One-carbon metabolism is a process by which methyl groups are passed from one donor molecule to the next (25, 26) (Figure 2). This process produces S-adenosylmethionine, which donates its methyl group to cytosine in a reaction catalyzed by DNA methyltransferases (DNMTs; refer to the information below). The rate of passage through this cycle can be influenced by genetic polymorphisms that encode the enzymes involved (27). The C-to-T substitution at nucleotide 677 of the methylenetetrahydrofolate reductase gene, MTHFR (677C>T), for example, results in a more thermo-labile enzyme, and TT homozygous individuals, compared with CC homozygous individuals, have lower levels of DNA methylation (28, 29).
Figure 2.
One-carbon metabolism, which involves the transfer of a methyl group from one “donor” to the next, ending with the DNA methyltransferase (DNMT)–catalyzed transfer of a methyl group to DNA. Methyl donors are shown in boxes and cofactors in ellipses; dietary factors are shaded yellow. Note that, for simplicity, only part of the pathway is shown. CpG, cytosine (C) and guanine (G) linked by a phosphate molecule. Methyl donors: SAH, S-adenosylhomocysteine; SAM, S-adenosylmethionine; THF, tetrahydrofolate; 5meTHF, 5-methyl-THF. Enzymes: BHMT, betaine-homocysteine S-methyltransferase; B2, vitamin B2; B6, vitamin B6; B12, vitamin B12; MAT, methionine adenosyltransferase; MS, methionine synthase; MTHFR, 5,10-methylenetetrahydrofolate reductase; MTRR, 5-methyltetrahydrofolate-homocysteine methyltransferase reductase; SAHH, S-adenosylhomocysteine hydrolase; Zn, zinc.
The DNMT family regulates DNA methylation. DNMT1 maintains methylation levels following DNA replication, whereas DNMT3A and DNMT3B act de novo to add DNA methylation. These enzymes regulate the dynamic methylation of genes during the establishment of imprinting (parent of origin gene expression) or cell differentiation (30). Variants in DNMT1 have been identified as risk factors for disease, including, in a case-control study, systemic lupus erythematosus (31). A genetic deficiency of DNMT3B causes a recessive human disorder characterized by immunodeficiency, centromere instability, and facial anomalies (32). In case-control studies, variants in other DNMTs (i.e., DNMT3L, DNMT1) have been associated with cancers (33–36).
CpG sites themselves are subject to genetic variation that can alter the sequence of gene regulatory regions and potential methylation levels. The C allele of the 102 T>C variant of the serotonin receptor gene (5HT2A), for example, contains 2 additional CpG dinucleotides thought to facilitate greater methylation levels and lower gene expression (37) and has been associated with psychiatric phenotypes (reviewed by Serritti et al. (38)). Removal of CpG sites can potentially abolish binding sites for proteins involved in transcriptional regulation. In 2 prospective cohorts of colon cancer cases, a specific variant (C>T) in the O-6-methylguanine-DNA methyltransferase tumor suppressor gene (MGMT), for example, has been strongly associated with O-6-methylguanine-DNA methyltransferase promoter methylation and gene silencing (39).
DNA methylation levels may also be modified through genetic variation at non-CpG sites within, or in close proximity to, gene regulatory regions. Increasing methylation of a CpG island in the serotonin transporter gene (5HTT), for example, is associated with decreasing levels of gene expression, but this effect is evident only when the 5HTTLPR genotype (length of an upstream DNA repeat) is included (40).
EPIGENETICS AND DEVELOPMENT
DNA methylation levels and profile are very dynamic, especially during the epigenetic remodeling that takes place early in embryogenesis (Figure 3). The partially methylated egg and sperm genomes are globally demethylated soon after fertilization. Methylation is then reestablished progressively, starting in the early postconception period. Imprinted genes, however, retain the methylation profile of the parent of origin (41).
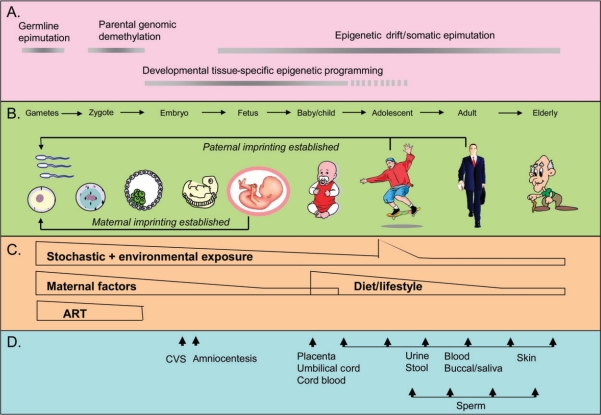
Figure 3.
Epigenetics, environment, and development. A. Dynamic epigenetic profile throughout the life course, with both genome-wide remodeling and tissue-specific programming events. Stochastic and environmentally induced epigenetic changes accumulated throughout the life course and may be passed through the germline to subsequent generations. Both paternal and maternal genomes are demethylated following fertilization. The exception is imprinted loci that are selectively marked during gametogenesis in either a paternal- or maternal-specific pattern. B. The process of cell differentiation in early development that produces different cell types involves distinct and specific epigenetic modifications. This process is largely completed prior to birth. C. Predicted periods of development likely to be sensitive to specific environmental exposures, including maternal diet and lifestyle, assisted reproductive technologies (ART), environmental toxins, or postnatal diet. D. Biospecimens available for epigenetic analysis included prenatal cerebrovascular system (CVS) and amniocentesis cells; full-term placenta, umbilical cord, and cord blood at birth; and several peripheral tissues thereafter, which can be resampled at multiple time points to track temporal epigenetic change.
Epigenetic marks can be stably passed from one cell to its descendants (42) and, in some cases, when such marks survive the epigenetic remodeling of gametogenesis and early embryogenesis, from parent to offspring (43–45). This process has been demonstrated in animal studies, but limited evidence for germline transmission in humans has also been reported and, in one case, has been linked to an increased risk of cancer (45).
Epimutation is estimated to be 100 times more frequent than genetic mutation (46, 47) and may occur randomly or in response to the environment. Periods of rapid cell division and epigenetic remodeling are likely to be most sensitive to stochastic or environmentally mediated epimutation.
AGING AND THE EPIGENOME
Both genome-wide and specific methylation profile/patterns change with age, and this may be genetically controlled (48). A generalized decrease in DNA methylation with age has been reported in mice and in cell lines (49–51), although this decrease may be tissue and/or gene specific (52–54). Decreased methylation may be accompanied by reactivation of previously silenced genes (46). Age-related methylation changes have been described in cancer (55) and in gene promoters associated with cancer risk (56, 57; reviewed by Issa (58)). Age-related epigenetic changes have also been demonstrated in sperm cells, but the direction of change over time appears gene specific (59). Within-pair differences in DNA methylation are greater in older than younger monozygotic twins (60). The contribution of genetic, environmental, and random factors to this cumulative discordance is unknown.
EPIGENETICS AND DISEASE
Disruption of epigenetic profile is a feature of most cancers (61–63) and is speculated to play a role in the etiology of other complex diseases (13, 64, 65), including asthma (66), allergy (67), obesity, type 2 diabetes, coronary heart disease, autism spectrum disorders (68), and bipolar disorder and schizophrenia (69–73). The potential to identify distinct epigenetic biomarkers associated with eating disorders has also been explored (74, 75). Disruption of epigenetic profile is also implicated in some adverse health outcomes for subjects conceived by means of assisted reproductive technologies (76).
The most striking disease-associated epigenetic change is seen in cancer. The increase in tumor suppressor gene CpG island methylation between cancerous and noncancerous tissue is often close to 100% (77) and is usually reversed at CpG sites associated with tandemly repetitive and interspersed repeat DNA in the same tumor cells (78–81). Specific profiles of methylation have also been associated with factors that predict prognosis (82).
To our knowledge, the dramatic difference in methylation levels observed in cancerous versus noncancerous tissue has not been found in other complex diseases, where methylation at any given CpG island or specific CpG sites in affected versus unaffected individuals may vary by less than 10% (83–85). Interpretation of functional consequences is therefore problematic. Results from small studies must also be interpreted with caution even when supported by in vitro functional demonstration of changes in gene activity with methylation change (86). However, for some genes, evidence exists that a small change in the level of DNA methylation, especially in the lower range, can dramatically alter gene expression (87, 88).
ENVIRONMENTAL FACTORS AND THE EPIGENOME
Diet
Diet is an important modifier of epigenetic profile (Figure 2; reviewed by Davis and Uthus (25), Cobiac (89), Lahiri et al. (90), and Muskiet (91)). Specific micronutrients involved in one-carbon metabolism include folate, an important primary methyl donor, whose availability is directly correlated with DNA methylation levels (91, 92). Low folate levels lead to hyperhomocysteinemia, which inhibits key metabolites of the one-carbon pathway (91, 93) and is associated with coronary heart disease and cancer (91, 94–96). To our knowledge, there is no evidence to date that these associations are mediated epigenetically. Diet can also change the epigenome via factors that alter the profile of histone modifications in cells, thereby altering gene expression levels (reviewed by Davis and Ross (97) and Herceg (98)). Given the demonstrated role of diet in regulating the epigenome, recommended dietary allowances of micronutrients have been proposed for maintenance of genome/epigenome stability (99).
The timing of nutritional insufficiency or other environmental exposures may also be critical (100). Epidemiologic evidence suggests transgenerational effects on health outcomes and mortality that are sensitive to the timing of environmental exposures. These effects may also be sex specific (101–105). Such associations have received a great deal of attention because of speculation about the underlying mechanisms. Pembrey et al. (105) identified several theoretical possibilities to explain the findings, including prions, viruses, RNA, responsive DNA sequences, or epigenetic changes. There is, however, no direct evidence that they are mediated via modification to epigenetic profile.
Animal studies have been more convincing. Maternal protein restriction in rats during pregnancy leads to a loss of methylation in the offspring at gene promoters associated with glucose metabolism in tissues such as liver, lung, and kidney (106, 107).
Other lifestyle factors
Human case-control studies have demonstrated that alcohol consumption increases methylation at gene promoters (108–111) and is associated with methylation-induced silencing of tumor suppressor genes in colorectal cancer (112) and hyperhomocysteinemia (113). Alcohol consumption is also associated with altered levels of methyltransferase activity and DNA methylation (71, 109). In both animal and in vitro studies, alcohol has been shown to impede the bioavailability of dietary folate and inhibit folate-dependent biochemical reactions (114, 115).
Cigarette smoking is associated with increased methylation at tumor suppressor genes in human case-control studies (116) and in mice (117) and with loss of methylation at oncogenes in human cancer cell lines (118). Disruption of DNMTs is implicated in the mechanism of induced change (118).
Endocrine disruptors
Endocrine disruptors are chemicals that interfere with the function of hormones by mimicking, blocking, or disrupting their synthesis, transport, or elimination. Hormones are chemical messengers that travel through the blood to target cells where they interact with receptors that, in turn, directly influence gene activity, usually via epigenetic mechanisms. Any endocrine disruption may therefore have epigenetic consequences. Diethylstilbestrol is a synthetic estrogen. Prenatal exposure to diethylstilbestrol has been shown to increase the risk of cervical and vaginal cancer and pregnancy-related problems in women and testicular abnormalities in men (119). Diethylstilbestrol exposure in animals decreases promoter DNA methylation in reproductive tissues and increases methylation (and hence decreases activity) of DNMT (120). Such exposure also decreases methylation of the cancer-causing oncogene c-fos (121) and the estrogen-responsive gene lactoferrin in mice (122). In animals, in utero or neonatal exposure to bisphenol A is associated with higher body mass, altered reproductive function, increasing cancer risk, and specific DNA methylation changes (123–125).
Genistein is an estrogen-like polyphenol found in soybeans that alters DNMT function and changes the DNA methylation status of several genes, including some tumor suppressors (126–128). Genistein may be protective against certain prostate and mammary cancers (127). Animal studies have also shown that transient exposure to vinclozolin (a fungicide) or methoxychlor (a pesticide) is associated with changes in DNA methylation of several genes (129) and decreased fertility in male offspring over several generations (130).
Other environmental agents
Heavy metals such as nickel, cadmium, and arsenic and ionizing and ultraviolet radiation (reviewed by Herceg (98)) have all been associated with an altered epigenetic profile. Chromium exposure in male mice, for example, causes hypomethylation of sperm genomic DNA and is associated with increasing risk of tumors and other abnormalities in progeny (131, 132).
Maternal behavior
One of the most interesting studies of environment-induced epigenetic change involved newborn rats exposed to differing degrees of maternal care (133, 134). Low-level maternal care was associated with decreased glucocorticoid receptor promoter methylation (increased gene activity) in the hippocampus and altered stress response in the young. This effect was reversible by either pharmacologic or dietary intervention in adulthood (135). Similar findings have been reported for the estrogen receptor alpha gene (136). No human version of these studies has yet been reported, but, given how widely stress is implicated in disease onset and relapse (137–141), it is difficult to overstate the potential importance of such findings.
Infective agents and the epigenome
Bacterial infection may directly modify the epigenetic profile of the host animal (142). Aberrant methylation of gastric mucosa genes is a common finding in humans infected with Helicobacter pylori and is an early event in gastric carcinogenesis (143–145). Insulin-like growth factor 2 methylation imprinting profile in the placenta is altered in mice infected with Campylobacter rectus during pregnancy (142). Similar alterations in DNA methylation have been reported in human cells and cell lines following viral or helminth infection (146–152). Methylation may be increased or decreased depending on the infectious agent (142, 153). Change in epigenetic profile in response to infection may play a role in the development of immune-related disorders and many cancers previously associated with infectious agents (e.g., gastric cancer).
MEASURING EPIGENETIC PROFILE
DNA methylation can be assessed by using very small amounts of genomic DNA obtained from fresh or archived tissue samples (154), including blood spots stored for many years (155). The “gold standard” for DNA methylation analysis involves sodium bisulfite treatment of genomic DNA for selective conversion of unmethylated cytosine residues to uracil, leaving methylated cytosine unchanged (156). This specific chemical change can be assessed by DNA sequencing, polymerase chain reaction amplification, or mass spectroscopy methods.
DNA methylation can also be studied on a genome-wide scale using a variety of enzyme-based or antibody affinity techniques that enrich for either methylated or unmethylated fractions of genomic DNA (157–162). These fractions can then be hybridized to DNA microarrays or sequenced en masse. Low-resolution array methodology has been used to identify genes consistently differentially methylated in a case-control study of psychosis (73) and has the potential to quickly (although not inexpensively) identify genes differentially methylated in a range of complex disorders in case-control studies.
As an alternative to mapping levels of DNA methylation at specific sites, the overall level of methylation in a tissue can be determined by directly measuring the amount of cytosine and methyl-cytosine (81, 163–165) or by a proxy using a combination of methylation-dependent DNA digestion and fluorescent tagging (166). Another alternative is to use other methods that assess levels of repeat-based methylation (167).
The development of ultra-high-throughput DNA sequencing technologies for the direct sequencing of enriched, methylated DNA fragments or of bisulfite-converted genomic DNA (168, 169) will ultimately permit measurement of comprehensive methylation profiles. The dynamic nature of epigenetic markings obviously warrants longitudinal biospecimen sampling where possible. In 2008, the National Institutes of Health made a significant commitment to research involving epigenomics (http://www.nih.gov/news/health/jan2008/od-22.htm) as part of an NIH Roadmap established in recognition of the importance of developing high-throughput and cost-effective methods for epigenetic analysis.
CONSIDERATIONS FOR EPIDEMIOLOGIC STUDIES INVOLVING EPIGENETIC ANALYSIS
Genetic-epidemiologic studies provide a framework for understanding the joint impact of genotype and environmental exposure on disease risk. Addition of epigenetic data will help clarify the functional basis underlying such joint effects by providing unbiased biologic measures arising from the combined effects of such interactions (and stochastic effects) that may lead to altered gene activity. The common disease genetic and epigenetic model provides a starting framework for including epigenetic data in genetic studies (13, 170).
Genetic and epigenetic analysis
Understanding the similarities and differences in genetic versus epigenetic data will help with planning the next wave of epidemiologic studies that prospectively incorporate both. Any peripheral tissue samples can be used for genotyping an individual. The level and pattern of epigenetic marks vary across different tissue and cell types, however, posing a formidable challenge for epigenetic analysis. Epigenetic marks in readily accessible cells (e.g., blood, buccal, skin), for example, may not reflect those in generally inaccessible tissues (e.g., brain). Analysis of postmortem tissue will provide valuable insight into the epigenetic profile of inaccessible tissues, but the identification and validation of peripheral epigenetic marker will be required for epidemiologic studies to benefit from incorporation of epigenetic data.
Genetic epidemiology involves the analysis of single nucleotide polymorphism alleles (usually binary) or variable numbers of simple repeats (usually tandem nucleotide elements), which can alter gene function via changes in coding sequence, RNA processing, or changing gene promoter sequence. A potentially methylatable CpG-rich region, in contrast, can comprise many dozens of individual CpG sites, each of which may have the potential to influence gene activity. CpG sites may be individually or coordinately methylated, but this is likely to vary from gene to gene and tissue to tissue. The relative biologic significance of methylation at a particular site versus average methylation across the entire region remains to be determined in most cases. A recent study of interindividual variation in methylation suggests that measurement of average methylation levels may suffice to characterize the methylation state of CpG-rich islands (171).
The genotype of an individual is generally fixed at conception, barring subsequent genetic mutations. The epigenotype, in contrast, is tissue and cell-type specific and may vary over time as a function of environmental exposure, aging, and random processes (48, 60, 172, 173). Analysis of epigenetic data, therefore, cannot rely upon the assumptions of Mendelian randomization (174, 175), which is predicated on the random assortment of genes transmitted from parents to offspring during gamete formation before disease onset. Associations between genotype and disease are therefore not usually biased because of reverse causation or confounding, unless linkage disequilibrium, pleiotropy, genetic heterogeneity, or population stratification are involved (174). Cross-sectional assessment of epigenetic profile does not permit any inference regarding direction of causation. Longitudinal data will be required to make causal inferences about observed associations between epigenetic marks and disease in cross-sectional data, and the timing of epigenetic sample collection will therefore be of paramount importance.
Modeling epigenetic data
Epigenetics is a relatively new field, and there is currently no consensus as to the most appropriate way to model methylation data. Very few epidemiologic studies currently integrate genetic (dichotomous variable), epigenetic (continuous variable), and environmental exposure data. Separate associations may exist between genotype and epigenotype, genotype and phenotype, and epigenotype and phenotype. Environmental exposures (e.g., cigarette smoking or alcohol consumption) may influence epigenotype and have independent effects upon the phenotype. The appropriate statistical model will thus depend on the question of interest and substantive knowledge concerning the relevant biologic pathways. If methylation is not allele specific, however, and that needs to be tested on a gene-by-gene basis, then there is no need to specify a model of inheritance (e.g., dominant, recessive, additive) for diseases caused by methylation changes.
Methods for modeling correlated outcomes include generalized estimating equations, mixed models, or the simpler approach of analyzing summary measures of the data such as means. Because potentially methylatable CpG sites often exist in clusters and may show coordinated methylation changes, analytic methods for summarizing correlated data may be required. Data reduction techniques such as principal components analysis have been used to model correlated CpG sites in studies in which DNA methylation is the exposure of interest (176).
Most analysis of epigenetic data summarizes methylation at individual CpG sites as proportions. The relation between methylation and gene expression may not be linear and may differ from site to site. Modern statistical software permits the modeling of proportional outcomes within the generalized linear framework (e.g., STaTa's GLM command (Stata Corporation, College Station, Texas), but the resulting estimates may be difficult for researchers to interpret. When epigenetic status or change in status over time is the outcome, then models for either threshold-based dichotomies or proportional data will be required. Threshold models, defined by a given level or pattern of methylation or a degree of change in methylation over time, will benefit from relevant functional data to identify meaningful thresholds.
Multiple testing
Similar to genome-wide association studies, a genome-wide search for epigenetic risk factors may investigate associations between a phenotype and thousands of biologic variables. As the number of tests performed increases, so does the probability of obtaining statistically significant results by chance. Tools such as the false discovery rate (177) permit identification of as many true associations as possible while minimizing the overall proportion of false-positive tests and can be applied to epigenetic analysis. Disentangling genetic and epigenetic effects in human studies is likely to prove challenging. Simple approaches might utilize standard epidemiologic analytic tools, such as examining associations between outcome and epigenotype within strata defined by different genotypes or examining interactions between different genetic and epigenetic factors, but these strategies are likely to require large sample sizes to achieve adequate power.
Studies of monozygotic twins provide an opportunity to examine the impact of unshared environmental exposures (e.g., discordance for cigarette smoking) on epigenetic profile while controlling for genotype and common familial environmental factors. An estimate of the association between changes in methylation and the outcome of interest not confounded by genetic background and shared environment can be obtained from the within-pairs coefficient of a suitable regression model. Carlin et al. (178) discuss models for estimating this coefficient and its interpretation in their review of regression modeling of twin data. Allelic effects on methylation against different genetic backgrounds can also be tested in dizygotic twins who share an allele in common versus dizygotic twins who differ at the allele of interest.
The dynamic nature of the epigenome may help explain the variable age at onset, progression, and outcomes associated with many common diseases and may provide new insights into the role of the environment in helping shape risk profiles. In this review, we have introduced epigenetics and discussed its role as a mediator of environmental effects on health and disease. Now that we are equipped with standard epidemiologic tools and statistical models, together with ever-advancing technology, the time is right to begin to answer important questions in this field.
Acknowledgments
Author affiliations: Orygen Youth Health Research Centre & Department of Psychiatry, University of Melbourne, Australia (Debra L. Foley); Developmental Epigenetics, Australia and Royal Children's Hospital & Department of Paediatrics, University of Melbourne, Australia (Jeffrey M. Craig, Ruth Morley, Richard Saffery); Centre for Adolescent Health, Australia and Royal Children's Hospital & Department of Paediatrics, University of Melbourne, Australia (Craig J. Olsson); Clinical Epidemiology and Biostatistics Unit, Australia and Royal Children's Hospital & Department of Paediatrics, University of Melbourne, Australia (Katherine Smith); and Murdoch Childrens Research Institute, Australia and Royal Children's Hospital & Department of Paediatrics, University of Melbourne, Australia (Jeffrey M. Craig, Ruth Morley, Craig J. Olsson, Terence Dwyer, Katherine Smith, Richard Saffery).
Conflict of interest: none declared.
Glossary
Abbreviation
- DNMT
DNA methyltransferases
References
- 1.Alberg AJ, Ford JG, Samet JM, et al. Epidemiology of lung cancer: ACCP evidence-based clinical practice guidelines. Chest. (2nd edition) 2007;132(3 suppl):29S–55S. doi: 10.1378/chest.07-1347. [DOI] [PubMed] [Google Scholar]
- 2.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40(5):616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 4.Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burke W, Press N. Genetics as a tool to improve cancer outcomes: ethics and policy. Nat Rev Cancer. 2006;6(6):476–482. doi: 10.1038/nrc1890. [DOI] [PubMed] [Google Scholar]
- 6.Khoury MJ. Genetic epidemiology and the future of disease prevention and public health. Epidemiol Rev. 1997;19(1):175–180. doi: 10.1093/oxfordjournals.epirev.a017940. [DOI] [PubMed] [Google Scholar]
- 7.Khoury MJ. Human Genome Epidemiology: A Scientific Foundation for Using Genetic Information to Improve Health and Prevent Disease. New York, NY: Oxford University Press; 2003. [Google Scholar]
- 8.Strausbaugh SD, Davis PB. Cystic fibrosis: a review of epidemiology and pathobiology. Clin Chest Med. 2007;28(2):279–288. doi: 10.1016/j.ccm.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 9.Ha M, Yoo KY, Cho SH. Glycophorin A mutant frequency in radiation workers at the nuclear power plants and a hospital. Mutat Res. 2002;501(1-2):45–56. doi: 10.1016/s0027-5107(02)00009-x. [DOI] [PubMed] [Google Scholar]
- 10.Meng Z, Qin G, Zhang B. DNA damage in mice treated with sulfur dioxide by inhalation. Environ Mol Mutagen. 2005;46(3):150–155. doi: 10.1002/em.20142. [DOI] [PubMed] [Google Scholar]
- 11.Jones IM, Galick H, Kato P, et al. Three somatic genetic biomarkers and covariates in radiation-exposed Russian cleanup workers of the Chernobyl nuclear reactor 6–13 years after exposure. Radiat Res. 2002;158(4):424–442. doi: 10.1667/0033-7587(2002)158[0424:tsgbac]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 12.Smith SJ, Li Y, Whitley R, et al. Molecular epidemiology of p53 protein mutations in workers exposed to vinyl chloride. Am J Epidemiol. 1998;147(3):302–308. doi: 10.1093/oxfordjournals.aje.a009450. [DOI] [PubMed] [Google Scholar]
- 13.Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20(8):350–358. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 14.Jacinto FV, Esteller M. MGMT hypermethylation: a prognostic foe, a predictive friend. DNA Repair (Amst) 2007;6(8):1155–1160. doi: 10.1016/j.dnarep.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 15.Brown SE, Fraga MF, Weaver IC, et al. Variations in DNA methylation patterns during the cell cycle of HeLa cells. Epigenetics. 2007;2(1):54–65. doi: 10.4161/epi.2.1.3880. [DOI] [PubMed] [Google Scholar]
- 16.Kangaspeska S, Stride B, Métivier R, et al. Transient cyclical methylation of promoter DNA. Nature. 2008;452(7183):112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 17.Métivier R, Gallais R, Tiffoche C, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452(7183):45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 18.Ptak C, Petronis A. Epigenetics and complex disease: from etiology to new therapeutics. Annu Rev Pharmacol Toxicol. 2008;48:257–276. doi: 10.1146/annurev.pharmtox.48.113006.094731. [DOI] [PubMed] [Google Scholar]
- 19.van Vliet J, Oates NA, Whitelaw E. Epigenetic mechanisms in the context of complex diseases. Cell Mol Life Sci. 2007;64(12):1531–1538. doi: 10.1007/s00018-007-6526-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saetrom P, Snøve O, Jr, Rossi JJ. Epigenetics and microRNAs. Pediatr Res. 2007;61(5 pt 2):17R–23R. doi: 10.1203/pdr.0b013e318045760e. [DOI] [PubMed] [Google Scholar]
- 21.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196(2):261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 22.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13(8):335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 23.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31(2):89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 25.Davis CD, Uthus EO. DNA methylation cancer susceptibility and nutrient interactions. Exp Biol Med (Maywood) 2004;229(10):988–995. doi: 10.1177/153537020422901002. [DOI] [PubMed] [Google Scholar]
- 26.Friso S, Choi SW. Gene-nutrient interactions in one-carbon metabolism. Curr Drug Metab. 2005;6(1):37–46. doi: 10.2174/1389200052997339. [DOI] [PubMed] [Google Scholar]
- 27.Friso S, Choi SW. Gene-nutrient interactions and DNA methylation. J Nutr. 2002;132(8 suppl):2382S–2387S. doi: 10.1093/jn/132.8.2382S. [DOI] [PubMed] [Google Scholar]
- 28.Castro R, Rivera I, Ravasco P, et al. 5,10-methylenetetrahydrofolate reductase (MTHFR) 677C→T and 1298A→C mutations are associated with DNA hypomethylation. J Med Genet. 2004;41(6):454–458. doi: 10.1136/jmg.2003.017244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friso S, Choi SW, Girelli D, et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci U S A. 2002;99(8):5606–5611. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brenner C, Fuks F. DNA methyltransferases: facts, clues, mysteries. Curr Top Microbiol Immunol. 2006;301:45–66. doi: 10.1007/3-540-31390-7_3. [DOI] [PubMed] [Google Scholar]
- 31.Park BL, Kim LH, Shin HD, et al. Association analyses of DNA methyltransferase-1 (DNMT1) polymorphisms with systemic lupus erythematosus. J Hum Genet. 2004;49(11):642–646. doi: 10.1007/s10038-004-0192-x. [DOI] [PubMed] [Google Scholar]
- 32.Hansen RS, Wijmenga C, Luo P, et al. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci U S A. 1999;96(25):14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montgomery KG, Liu MC, Eccles DM, et al. The DNMT3B C→T promoter polymorphism and risk of breast cancer in a British population: a case-control study. Breast Cancer Res. 2004;6(4):R390–R394. doi: 10.1186/bcr807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee SJ, Jeon HS, Jang JS, et al. DNMT3B polymorphisms and risk of primary lung cancer. Carcinogenesis. 2005;26(2):403–409. doi: 10.1093/carcin/bgh307. [DOI] [PubMed] [Google Scholar]
- 35.Kelemen LE, Sellers TA, Schildkraut JM, et al. Genetic variation in the one-carbon transfer pathway and ovarian cancer risk. Cancer Res. 2008;68(7):2498–2506. doi: 10.1158/0008-5472.CAN-07-5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cebrian A, Pharoah PD, Ahmed S, et al. Genetic variants in epigenetic genes and breast cancer risk. Carcinogenesis. 2006;27(8):1661–1669. doi: 10.1093/carcin/bgi375. [DOI] [PubMed] [Google Scholar]
- 37.Polesskaya OO, Aston C, Sokolov BP. Allele C-specific methylation of the 5-HT2A receptor gene: evidence for correlation with its expression and expression of DNA methylase DNMT1. J Neurosci Res. 2006;83(3):362–373. doi: 10.1002/jnr.20732. [DOI] [PubMed] [Google Scholar]
- 38.Serretti A, Drago A, De Ronchi D. HTR2A gene variants and psychiatric disorders: a review of current literature and selection of SNPs for future studies. Curr Med Chem. 2007;14(19):2053–2069. doi: 10.2174/092986707781368450. [DOI] [PubMed] [Google Scholar]
- 39.Ogino S, Hazra A, Tranah GJ, et al. MGMT germline polymorphism is associated with somatic MGMT promoter methylation and gene silencing in colorectal cancer. Carcinogenesis. 2007;28(9):1985–1990. doi: 10.1093/carcin/bgm160. [DOI] [PubMed] [Google Scholar]
- 40.Philibert R, Madan A, Andersen A, et al. Serotonin transporter mRNA levels are associated with the methylation of an upstream CpG island. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(1):101–105. doi: 10.1002/ajmg.b.30414. [DOI] [PubMed] [Google Scholar]
- 41.Kelsey G. Genomic imprinting—roles and regulation in development. Endocr Dev. 2007;12:99–112. doi: 10.1159/000109637. [DOI] [PubMed] [Google Scholar]
- 42.Bird A. Perceptions of epigenetics. Nature. 2007;447(7143):396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 43.Chong S, Whitelaw E. Epigenetic germline inheritance. Curr Opin Genet Dev. 2004;14(6):692–696. doi: 10.1016/j.gde.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 44.Rakyan VK, Chong S, Champ ME, et al. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc Natl Acad Sci U S A. 2003;100(5):2538–2543. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suter CM, Martin DI, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet. 2004;36(5):497–501. doi: 10.1038/ng1342. [DOI] [PubMed] [Google Scholar]
- 46.Bennett-Baker PE, Wilkowski J, Burke DT. Age-associated activation of epigenetically repressed genes in the mouse. Genetics. 2003;165(4):2055–2062. doi: 10.1093/genetics/165.4.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goyal R, Reinhardt R, Jeltsch A. Accuracy of DNA methylation pattern preservation by the Dnmt1 methyltransferase. Nucleic Acids Res. 2006;34(4):1182–1188. doi: 10.1093/nar/gkl002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bjornsson HT, Sigurdsson MI, Fallin MD, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299(24):2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singhal RP, Mays-Hoopes LL, Eichhorn GL. DNA methylation in aging of mice. Mech Ageing Dev. 1987;41(3):199–210. doi: 10.1016/0047-6374(87)90040-6. [DOI] [PubMed] [Google Scholar]
- 50.Wilson VL, Smith RA, Ma S, et al. Genomic 5-methyldeoxycytidine decreases with age. J Biol Chem. 1987;262(21):9948–9951. [PubMed] [Google Scholar]
- 51.Wilson VL, Jones PA. DNA methylation decreases in aging but not in immortal cells. Science. 1983;220(4601):1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- 52.Richardson BC. Role of DNA methylation in the regulation of cell function: autoimmunity, aging and cancer. J Nutr. 2002;132(8 suppl):2401S–2405S. doi: 10.1093/jn/132.8.2401S. [DOI] [PubMed] [Google Scholar]
- 53.Richardson B. Impact of aging on DNA methylation. Ageing Res Rev. 2003;2(3):245–261. doi: 10.1016/s1568-1637(03)00010-2. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Z, Deng C, Lu Q, et al. Age-dependent DNA methylation changes in the ITGAL (CD11a) promoter. Mech Ageing Dev. 2002;123(9):1257–1268. doi: 10.1016/s0047-6374(02)00014-3. [DOI] [PubMed] [Google Scholar]
- 55.Liu L, Wylie RC, Andrews LG, et al. Aging, cancer and nutrition: the DNA methylation connection. Mech Ageing Dev. 2003;124(10-12):989–998. doi: 10.1016/j.mad.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 56.Issa JP, Ottaviano YL, Celano P, et al. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7(4):536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 57.Ahuja N, Issa JP. Aging, methylation and cancer. Histol Histopathol. 2000;15(3):835–842. doi: 10.14670/HH-15.835. [DOI] [PubMed] [Google Scholar]
- 58.Issa JP. CpG-island methylation in aging and cancer. Curr Top Microbiol Immunol. 2000;249:101–118. doi: 10.1007/978-3-642-59696-4_7. [DOI] [PubMed] [Google Scholar]
- 59.Flanagan JM, Popendikyte V, Pozdniakovaite N, et al. Intra- and interindividual epigenetic variation in human germ cells. Am J Hum Genet. 2006;79(1):67–84. doi: 10.1086/504729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fraga MF, Ballestar E, Paz MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102(30):10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ballestar E, Esteller M. epigenetic gene regulation in cancer. Adv Genet. 2008;61:247–267. doi: 10.1016/S0065-2660(07)00009-0. [DOI] [PubMed] [Google Scholar]
- 62.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 63.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7(1):21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 64.Hatchwell E, Greally JM. The potential role of epigenomic dysregulation in complex human disease. Trends Genet. 2007;23(11):588–595. doi: 10.1016/j.tig.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 65.Schumacher A, Petronis A. Epigenetics of complex diseases: from general theory to laboratory experiments. Curr Top Microbiol Immunol. 2006;310:81–115. doi: 10.1007/3-540-31181-5_6. [DOI] [PubMed] [Google Scholar]
- 66.Miller RL, Ho SM. Environmental epigenetics and asthma: current concepts and call for studies. Am J Respir Crit Care Med. 2008;177(6):567–573. doi: 10.1164/rccm.200710-1511PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Steinke JW, Rich SS, Borish L. 5. Genetics of allergic disease. J Allergy Clin Immunol. 2008;121(2 suppl):S384–S387. doi: 10.1016/j.jaci.2007.07.029. quiz S416. [DOI] [PubMed] [Google Scholar]
- 68.Schanen NC. Epigenetics of autism spectrum disorders. Hum Mol Genet. 2006;15(spec no 2):R138–R150. doi: 10.1093/hmg/ddl213. [DOI] [PubMed] [Google Scholar]
- 69.Abdolmaleky HM, Smith CL, Zhou JR, et al. Epigenetic alterations of the dopaminergic system in major psychiatric disorders. Methods Mol Biol. 2008;448:187–212. doi: 10.1007/978-1-59745-205-2_9. [DOI] [PubMed] [Google Scholar]
- 70.Abdolmaleky HM, Cheng KH, Faraone SV, et al. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum Mol Genet. 2006;15(21):3132–3145. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abdolmaleky HM, Cheng KH, Russo A, et al. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;134B(1):60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- 72.Abdolmaleky HM, Smith CL, Faraone SV, et al. Methylomics in psychiatry: modulation of gene-environment interactions may be through DNA methylation. Am J Med Genet B Neuropsychiatr Genet. 2004;127B(1):51–59. doi: 10.1002/ajmg.b.20142. [DOI] [PubMed] [Google Scholar]
- 73.Mill J, Tang T, Kaminsky Z, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82(3):696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frieling H, Gozner A, Römer KD, et al. Global DNA hypomethylation and DNA hypermethylation of the alpha synuclein promoter in females with anorexia nervosa. Mol Psychiatry. 2007;12(3):229–230. doi: 10.1038/sj.mp.4001931. [DOI] [PubMed] [Google Scholar]
- 75.Frieling H, Bleich S, Otten J, et al. Epigenetic downregulation of atrial natriuretic peptide but not vasopressin mRNA expression in females with eating disorders is related to impulsivity. Neuropsychopharmacology. 2008;33(11):2605–2609. doi: 10.1038/sj.npp.1301662. [DOI] [PubMed] [Google Scholar]
- 76.Niemitz EL, Feinberg AP. Epigenetics and assisted reproductive technology: a call for investigation. Am J Hum Genet. 2004;74(4):599–609. doi: 10.1086/382897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007;16(spec no 1):R50–R59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- 78.Cadieux B, Ching TT, VandenBerg SR, et al. Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res. 2006;66(17):8469–8476. doi: 10.1158/0008-5472.CAN-06-1547. [DOI] [PubMed] [Google Scholar]
- 79.Schulz WA, Steinhoff C, Florl AR. Methylation of endogenous human retroelements in health and disease. Curr Top Microbiol Immunol. 2006;310:211–250. doi: 10.1007/3-540-31181-5_11. [DOI] [PubMed] [Google Scholar]
- 80.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775(1):138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 81.Ehrlich M. Cancer-linked DNA hypomethylation and its relationship to hypermethylation. Curr Top Microbiol Immunol. 2006;310:251–274. doi: 10.1007/3-540-31181-5_12. [DOI] [PubMed] [Google Scholar]
- 82.Cho NY, Kim BH, Choi M, et al. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J Pathol. 2007;211(3):269–277. doi: 10.1002/path.2106. [DOI] [PubMed] [Google Scholar]
- 83.Chan Y, Fish JE, D'Abreo C, et al. The cell-specific expression of endothelial nitric-oxide synthase: a role for DNA methylation. J Biol Chem. 2004;279(33):35087–350100. doi: 10.1074/jbc.M405063200. [DOI] [PubMed] [Google Scholar]
- 84.Huntriss J, Lorenzi R, Purewal A, et al. A methylation-dependent DNA-binding activity recognising the methylated promoter region of the mouse Xist gene. Biochem Biophys Res Commun. 1997;235(3):730–738. doi: 10.1006/bbrc.1997.6876. [DOI] [PubMed] [Google Scholar]
- 85.Kitazawa R, Kitazawa S. Methylation status of a single CpG locus 3 bases upstream of TATA-box of receptor activator of nuclear factor-kappaB ligand (RANKL) gene promoter modulates cell- and tissue-specific RANKL expression and osteoclastogenesis. Mol Endocrinol. 2007;21(1):148–158. doi: 10.1210/me.2006-0205. [DOI] [PubMed] [Google Scholar]
- 86.Oates NA, van Vliet J, Duffy DL, et al. Increased DNA methylation at the AXIN1 gene in a monozygotic twin from a pair discordant for a caudal duplication anomaly. Am J Hum Genet. 2006;79(1):155–162. doi: 10.1086/505031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boyes J, Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992;11(1):327–333. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hsieh CL. Dependence of transcriptional repression on CpG methylation density. Mol Cell Biol. 1994;14(8):5487–5494. doi: 10.1128/mcb.14.8.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cobiac L. Epigenomics and nutrition. Forum Nutr. 2007;60:31–41. doi: 10.1159/000107065. [DOI] [PubMed] [Google Scholar]
- 90.Lahiri DK, Maloney B, Basha MR, et al. How and when environmental agents and dietary factors affect the course of Alzheimer's disease: the “LEARn” model (latent early-life associated regulation) may explain the triggering of AD. Curr Alzheimer Res. 2007;4(2):219–228. doi: 10.2174/156720507780362164. [DOI] [PubMed] [Google Scholar]
- 91.Muskiet FA. The importance of (early) folate status to primary and secondary coronary artery disease prevention. Reprod Toxicol. 2005;20(3):403–410. doi: 10.1016/j.reprotox.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 92.Leopardi P, Marcon F, Caiola S, et al. Effects of folic acid deficiency and MTHFR C677T polymorphism on spontaneous and radiation-induced micronuclei in human lymphocytes. Mutagenesis. 2006;21(5):327–333. doi: 10.1093/mutage/gel031. [DOI] [PubMed] [Google Scholar]
- 93.Yi P, Melnyk S, Pogribna M, et al. Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation. J Biol Chem. 2000;275(38):29318–29323. doi: 10.1074/jbc.M002725200. [DOI] [PubMed] [Google Scholar]
- 94.Fanapour PC, Yug B, Kochar MS. Hyperhomocysteinemia: an additional cardiovascular risk factor. WMJ. 1999;98(8):51–54. [PubMed] [Google Scholar]
- 95.Herrmann W. The importance of hyperhomocysteinemia as a risk factor for diseases: an overview. Clin Chem Lab Med. 2001;39(8):666–674. doi: 10.1515/CCLM.2001.110. [DOI] [PubMed] [Google Scholar]
- 96.Wu LL, Wu JT. Hyperhomocysteinemia is a risk factor for cancer and a new potential tumor marker. Clin Chim Acta. 2002;322(1-2):21–28. doi: 10.1016/s0009-8981(02)00174-2. [DOI] [PubMed] [Google Scholar]
- 97.Davis CD, Ross SA. Dietary components impact histone modifications and cancer risk. Nutr Rev. 2007;65(2):88–94. doi: 10.1111/j.1753-4887.2007.tb00285.x. [DOI] [PubMed] [Google Scholar]
- 98.Herceg Z. Epigenetics and cancer: towards an evaluation of the impact of environmental and dietary factors. Mutagenesis. 2007;22(2):91–103. doi: 10.1093/mutage/gel068. [DOI] [PubMed] [Google Scholar]
- 99.Fenech M. The Genome Health Clinic and Genome Health Nutrigenomics concepts: diagnosis and nutritional treatment of genome and epigenome damage on an individual basis. Mutagenesis. 2005;20(4):255–269. doi: 10.1093/mutage/gei040. [DOI] [PubMed] [Google Scholar]
- 100.Morley R, Dwyer T. Fetal origins of adult disease? Clin Exp Pharmacol Physiol. 2001;28(11):962–966. doi: 10.1046/j.1440-1681.2001.03557.x. [DOI] [PubMed] [Google Scholar]
- 101.Bygren LO, Kaati G, Edvinsson S. Longevity determined by paternal ancestors’ nutrition during their slow growth period. Acta Biotheor. 2001;49(1):53–59. doi: 10.1023/a:1010241825519. [DOI] [PubMed] [Google Scholar]
- 102.Golding J, Pembrey M, Jones R, et al. ALSPAC—the Avon Longitudinal Study of Parents and Children. I. Study methodology. Paediatr Perinat Epidemiol. 2001;15(1):74–87. doi: 10.1046/j.1365-3016.2001.00325.x. [DOI] [PubMed] [Google Scholar]
- 103.Kaati G, Bygren LO, Edvinsson S. Cardiovascular and diabetes mortality determined by nutrition during parents’ and grandparents’ slow growth period. Eur J Hum Genet. 2002;10(11):682–688. doi: 10.1038/sj.ejhg.5200859. [DOI] [PubMed] [Google Scholar]
- 104.Kaati G, Bygren LO, Pembrey M, et al. Transgenerational response to nutrition, early life circumstances and longevity. Eur J Hum Genet. 2007;15(7):784–790. doi: 10.1038/sj.ejhg.5201832. [DOI] [PubMed] [Google Scholar]
- 105.Pembrey ME, Bygren LO, Kaati G, et al. Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet. 2006;14(2):159–166. doi: 10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]
- 106.Lillycrop KA, Phillips ES, Jackson AA, et al. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr. 2005;135(6):1382–1386. doi: 10.1093/jn/135.6.1382. [DOI] [PubMed] [Google Scholar]
- 107.Burdge GC, Slater-Jefferies J, Torrens C, et al. Dietary protein restriction of pregnant rats in the F0 generation induces altered methylation of hepatic gene promoters in the adult male offspring in the F1 and F2 generations. Br J Nutr. 2007;97(3):435–439. doi: 10.1017/S0007114507352392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bleich S, Lenz B, Ziegenbein M, et al. Epigenetic DNA hypermethylation of the HERP gene promoter induces down-regulation of its mRNA expression in patients with alcohol dependence. Alcohol Clin Exp Res. 2006;30(4):587–591. doi: 10.1111/j.1530-0277.2006.00068.x. [DOI] [PubMed] [Google Scholar]
- 109.Bönsch D, Lenz B, Fiszer R, et al. Lowered DNA methyltransferase (DNMT-3b) mRNA expression is associated with genomic DNA hypermethylation in patients with chronic alcoholism. J Neural Transm. 2006;113(9):1299–1304. doi: 10.1007/s00702-005-0413-2. [DOI] [PubMed] [Google Scholar]
- 110.Bönsch D, Lenz B, Kornhuber J, et al. DNA hypermethylation of the alpha synuclein promoter in patients with alcoholism. Neuroreport. 2005;16(2):167–170. doi: 10.1097/00001756-200502080-00020. [DOI] [PubMed] [Google Scholar]
- 111.Bönsch D, Lenz B, Reulbach U, et al. Homocysteine associated genomic DNA hypermethylation in patients with chronic alcoholism. J Neural Transm. 2004;111(12):1611–1616. doi: 10.1007/s00702-004-0232-x. [DOI] [PubMed] [Google Scholar]
- 112.van Engeland M, Weijenberg MP, Roemen GM, et al. Effects of dietary folate and alcohol intake on promoter methylation in sporadic colorectal cancer: the Netherlands cohort study on diet and cancer. Cancer Res. 2003;63(12):3133–3137. [PubMed] [Google Scholar]
- 113.Cravo ML, Camilo ME. Hyperhomocysteinemia in chronic alcoholism: relations to folic acid and vitamins B(6) and B(12) status. Nutrition. 2000;16(4):296–302. doi: 10.1016/s0899-9007(99)00297-x. [DOI] [PubMed] [Google Scholar]
- 114.Abdolmaleky HM, Thiagalingam S, Wilcox M. Genetics and epigenetics in major psychiatric disorders: dilemmas, achievements, applications, and future scope. Am J Pharmacogenomics. 2005;5(3):149–160. doi: 10.2165/00129785-200505030-00002. [DOI] [PubMed] [Google Scholar]
- 115.Mason JB, Choi SW. Effects of alcohol on folate metabolism: implications for carcinogenesis. Alcohol. 2005;35(3):235–241. doi: 10.1016/j.alcohol.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 116.Belinsky SA, Palmisano WA, Gilliland FD, et al. Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res. 2002;62(8):2370–2377. [PubMed] [Google Scholar]
- 117.Pulling LC, Vuillemenot BR, Hutt JA, et al. Aberrant promoter hypermethylation of the death-associated protein kinase gene is early and frequent in murine lung tumors induced by cigarette smoke and tobacco carcinogens. Cancer Res. 2004;64(11):3844–3848. doi: 10.1158/0008-5472.CAN-03-2119. [DOI] [PubMed] [Google Scholar]
- 118.Liu H, Zhou Y, Boggs SE, et al. Cigarette smoke induces demethylation of prometastatic oncogene synuclein-gamma in lung cancer cells by downregulation of DNMT3B. Oncogene. 2007;26(40):5900–5910. doi: 10.1038/sj.onc.1210400. [DOI] [PubMed] [Google Scholar]
- 119.Rubin MM. Antenatal exposure to DES: lessons learned…future concerns. Obstet Gynecol Surv. 2007;62(8):548–555. doi: 10.1097/01.ogx.0000271138.31234.d7. [DOI] [PubMed] [Google Scholar]
- 120.Sato K, Fukata H, Kogo Y, et al. Neonatal exposure to diethylstilbestrol alters the expression of DNA methyltransferases and methylation of genomic DNA in the epididymis of mice. Endocr J. 2006;53(3):331–337. doi: 10.1507/endocrj.k06-009. [DOI] [PubMed] [Google Scholar]
- 121.Li S, Hansman R, Newbold R, et al. Neonatal diethylstilbestrol exposure induces persistent elevation of c-fos expression and hypomethylation in its exon-4 in mouse uterus. Mol Carcinog. 2003;38(2):78–84. doi: 10.1002/mc.10147. [DOI] [PubMed] [Google Scholar]
- 122.Li S, Washburn KA, Moore R, et al. Developmental exposure to diethylstilbestrol elicits demethylation of estrogen-responsive lactoferrin gene in mouse uterus. Cancer Res. 1997;57(19):4356–4359. [PubMed] [Google Scholar]
- 123.Prins GS, Birch L, Tang WY, et al. Developmental estrogen exposures predispose to prostate carcinogenesis with aging. Reprod Toxicol. 2007;23(3):374–382. doi: 10.1016/j.reprotox.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ho SM, Tang WY, Belmonte de Frausto J, et al. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006;66(11):5624–5632. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 2007;104(32):13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fang MZ, Jin Z, Wang Y, et al. Promoter hypermethylation and inactivation of O(6)-methylguanine-DNA methyltransferase in esophageal squamous cell carcinomas and its reactivation in cell lines. Int J Oncol. 2005;26(3):615–622. [PubMed] [Google Scholar]
- 127.Day JK, Bauer AM, DesBordes C, et al. Genistein alters methylation patterns in mice. J Nutr. 2002;132(8 suppl):2419S–2423S. doi: 10.1093/jn/132.8.2419S. [DOI] [PubMed] [Google Scholar]
- 128.Dolinoy DC, Weidman JR, Waterland RA, et al. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114(4):567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chang HS, Anway MD, Rekow SS, et al. Transgenerational epigenetic imprinting of the male germline by endocrine disruptor exposure during gonadal sex determination. Endocrinology. 2006;147(12):5524–5541. doi: 10.1210/en.2006-0987. [DOI] [PubMed] [Google Scholar]
- 130.Anway MD, Cupp AS, Uzumcu M, et al. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308(5727):1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cheng RY, Hockman T, Crawford E, et al. Epigenetic and gene expression changes related to transgenerational carcinogenesis. Mol Carcinog. 2004;40(1):1–11. doi: 10.1002/mc.20022. [DOI] [PubMed] [Google Scholar]
- 132.Yu W, Sipowicz MA, Haines DC, et al. Preconception urethane or chromium(III) treatment of male mice: multiple neoplastic and non-neoplastic changes in offspring. Toxicol Appl Pharmacol. 1999;158(2):161–176. doi: 10.1006/taap.1999.8692. [DOI] [PubMed] [Google Scholar]
- 133.Szyf M, Weaver I, Meaney M. Maternal care, the epigenome and phenotypic differences in behavior. Reprod Toxicol. 2007;24(1):9–19. doi: 10.1016/j.reprotox.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 134.Weaver IC, Cervoni N, Champagne FA, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 135.Weaver IC, Champagne FA, Brown SE, et al. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25(47):11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Champagne FA, Weaver IC, Diorio J, et al. Maternal care associated with methylation of the estrogen receptor-alpha1b promoter and estrogen receptor-alpha expression in the medial preoptic area of female offspring. Endocrinology. 2006;147(6):2909–2915. doi: 10.1210/en.2005-1119. [DOI] [PubMed] [Google Scholar]
- 137.Hammen C. Stress and depression. Annu Rev Clin Psychol. 2005;1:293–319. doi: 10.1146/annurev.clinpsy.1.102803.143938. [DOI] [PubMed] [Google Scholar]
- 138.Mohr DC, Pelletier D. A temporal framework for understanding the effects of stressful life events on inflammation in patients with multiple sclerosis. Brain Behav Immun. 2006;20(1):27–36. doi: 10.1016/j.bbi.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 139.Post RM, Leverich GS. The role of psychosocial stress in the onset and progression of bipolar disorder and its comorbidities: the need for earlier and alternative modes of therapeutic intervention. Dev Psychopathol. 2006;18(4):1181–1211. doi: 10.1017/S0954579406060573. [DOI] [PubMed] [Google Scholar]
- 140.Stojanovich L, Marisavljevich D. Stress as a trigger of autoimmune disease. Autoimmun Rev. 2008;7(3):209–213. doi: 10.1016/j.autrev.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 141.Thrall G, Lane D, Carroll D, et al. A systematic review of the effects of acute psychological stress and physical activity on haemorheology, coagulation, fibrinolysis and platelet reactivity: implications for the pathogenesis of acute coronary syndromes. Thrombo Res. 2007;120(6):819–847. doi: 10.1016/j.thromres.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 142.Bobetsis YA, Barros SP, Lin DM, et al. Bacterial infection promotes DNA hypermethylation. J Dent Res. 2007;86(2):169–174. doi: 10.1177/154405910708600212. [DOI] [PubMed] [Google Scholar]
- 143.Miyazaki T, Murayama Y, Shinomura Y, et al. E-cadherin gene promoter hypermethylation in H. pylori-induced enlarged fold gastritis. Helicobacter. 2007;12(5):523–531. doi: 10.1111/j.1523-5378.2007.00519.x. [DOI] [PubMed] [Google Scholar]
- 144.Kitajima Y, Ohtaka K, Mitsuno M, et al. Helicobacter pylori infection is an independent risk factor for Runx3 methylation in gastric cancer. Oncol Rep. 2008;19(1):197–202. [PubMed] [Google Scholar]
- 145.Perri F, Cotugno R, Piepoli A, et al. Aberrant DNA methylation in non-neoplastic gastric mucosa of H. pylori infected patients and effect of eradication. Am J Gastroenterol. 2007;102(7):1361–1371. doi: 10.1111/j.1572-0241.2007.01284.x. [DOI] [PubMed] [Google Scholar]
- 146.Zhu R, Li BZ, Li H, et al. Association of p16INK4A hypermethylation with hepatitis B virus X protein expression in the early stage of HBV-associated hepatocarcinogenesis. Pathol Int. 2007;57(6):328–336. doi: 10.1111/j.1440-1827.2007.02104.x. [DOI] [PubMed] [Google Scholar]
- 147.Northrop JK, Thomas RM, Wells AD, et al. Epigenetic remodeling of the IL-2 and IFN-gamma loci in memory CD8 T cells is influenced by CD4 T cells. J Immunol. 2006;177(2):1062–1069. doi: 10.4049/jimmunol.177.2.1062. [DOI] [PubMed] [Google Scholar]
- 148.Limpaiboon T, Khaenam P, Chinnasri P, et al. Promoter hypermethylation is a major event of hMLH1 gene inactivation in liver fluke related cholangiocarcinoma. Cancer Lett. 2005;217(2):213–219. doi: 10.1016/j.canlet.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 149.Pang Y, Norihisa Y, Benjamin D, et al. Interferon-gamma gene expression in human B-cell lines: induction by interleukin-2, protein kinase C activators, and possible effect of hypomethylation on gene regulation. Blood. 1992;80(3):724–732. [PubMed] [Google Scholar]
- 150.Fang JY, Mikovits JA, Bagni R, et al. Infection of lymphoid cells by integration-defective human immunodeficiency virus type 1 increases de novo methylation. J Virol. 2001;75(20):9753–9761. doi: 10.1128/JVI.75.20.9753-9761.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mikovits JA, Young HA, Vertino P, et al. Infection with human immunodeficiency virus type 1 upregulates DNA methyltransferase, resulting in de novo methylation of the gamma interferon (IFN-gamma) promoter and subsequent downregulation of IFN-gamma production. Mol Cell Biol. 1998;18(9):5166–5177. doi: 10.1128/mcb.18.9.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gutiérrez MI, Siraj AK, Khaled H, et al. CpG island methylation in Schistosoma- and non-Schistosoma-associated bladder cancer. Mod Pathol. 2004;17(10):1268–1274. doi: 10.1038/modpathol.3800177. [DOI] [PubMed] [Google Scholar]
- 153.Habib M, Fares F, Bourgeois CA, et al. DNA global hypomethylation in EBV-transformed interphase nuclei. Exp Cell Res. 1999;249(1):46–53. doi: 10.1006/excr.1999.4434. [DOI] [PubMed] [Google Scholar]
- 154.Safar AM, Spencer H, Su X, et al. Promoter hypermethylation for molecular nodal staging in non-small cell lung cancer. Arch Pathol Lab Med. 2007;131(6):936–941. doi: 10.5858/2007-131-936-PHFMNS. [DOI] [PubMed] [Google Scholar]
- 155.Wong NC, Morley R, Saffery R, et al. Archived Guthrie blood spots as a novel source for quantitative DNA methylation analysis. Biotechniques. 2008;45(4):423–424. doi: 10.2144/000112945. [DOI] [PubMed] [Google Scholar]
- 156.Frommer M, McDonald LE, Millar DS, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89(5):1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Schumacher A, Kapranov P, Kaminsky Z, et al. Microarray-based DNA methylation profiling: technology and applications. Nucleic Acids Res. 2006;34(2):528–542. doi: 10.1093/nar/gkj461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128(4):669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 159.Wilson IM, Davies JJ, Weber M, et al. Epigenomics: mapping the methylome. Cell Cycle. 2006;5(2):155–158. doi: 10.4161/cc.5.2.2367. [DOI] [PubMed] [Google Scholar]
- 160.Beck S, Rakyan VK. The methylome: approaches for global DNA methylation profiling. Trends Genet. 2008;24(5):231–237. doi: 10.1016/j.tig.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 161.Rakyan V, Down T, Thorne N, et al. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs) Genome Res. 2008;18(9):1518–1529. doi: 10.1101/gr.077479.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Irizarry RA, Ladd-Acosta C, Carvalho B, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18(5):780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ehrlich M, Wang RY. 5-Methylcytosine in eukaryotic DNA. Science. 1981;212(4501):1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- 164.Gama-Sosa MA, Slagel VA, Trewyn RW, et al. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983;11(19):6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gama-Sosa MA, Wang RY, Kuo KC, et al. The 5-methylcytosine content of highly repeated sequences in human DNA. Nucleic Acids Res. 1983;11(10):3087–3095. doi: 10.1093/nar/11.10.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Karimi M, Johansson S, Ekström TJ. Using LUMA: a Luminometric-based assay for global DNA-methylation. Epigenetics. 2006;1(1):45–48. doi: 10.4161/epi.1.1.2587. [DOI] [PubMed] [Google Scholar]
- 167.Weisenberger DJ, Campan M, Long TI, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33(21):6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Taylor KH, Kramer RS, Davis JW, et al. Ultradeep bisulfite sequencing analysis of DNA methylation patterns in multiple gene promoters by 454 sequencing. Cancer Res. 2007;67(18):8511–8518. doi: 10.1158/0008-5472.CAN-07-1016. [DOI] [PubMed] [Google Scholar]
- 170.Feinberg AP. Epigenetics at the epicenter of modern medicine. JAMA. 2008;299(11):1345–1350. doi: 10.1001/jama.299.11.1345. [DOI] [PubMed] [Google Scholar]
- 171.Bock C, Walter J, Paulsen M, et al. Inter-individual variation of DNA methylation and its implications for large-scale epigenome mapping [electronic article] Nucleic Acids Res. 2008;36(10):e55. doi: 10.1093/nar/gkn122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23(8):413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 173.Fraga MF, Agrelo R, Esteller M. Cross-talk between aging and cancer: the epigenetic language. Ann N Y Acad Sci. 2007;1100:60–74. doi: 10.1196/annals.1395.005. [DOI] [PubMed] [Google Scholar]
- 174.Davey Smith G, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32(1):1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 175.Ebrahim S, Davey Smith G. Mendelian randomization: can genetic epidemiology help redress the failures of observational epidemiology? Hum Genet. 2008;123(1):15–33. doi: 10.1007/s00439-007-0448-6. [DOI] [PubMed] [Google Scholar]
- 176.Heijmans BT, Kremer D, Tobi EW, et al. Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum Mol Genet. 2007;16(5):547–554. doi: 10.1093/hmg/ddm010. [DOI] [PubMed] [Google Scholar]
- 177.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Carlin JB, Gurrin LC, Sterne JA, et al. Regression models for twin studies: a critical review. Int J Epidemiol. 2005;34(5):1089–1099. doi: 10.1093/ije/dyi153. [DOI] [PubMed] [Google Scholar]