Abstract
The SecA molecular nanomachine in bacteria uses energy from ATP hydrolysis to drive posttranslational secretion of pre-proteins through the SecYEG translocon. Cytosolic SecA exists in a dimeric, ‘closed’ state with relatively low ATPase activity. After binding to the translocon, SecA undergoes major conformational rearrangement, leading to a state that is structurally more ‘open’, has elevated ATPase activity, and is active in translocation. The structural details underlying this conformational change in SecA remain incompletely defined. Most SecA crystal structures report on the cytosolic form; only one structure sheds light on a form of SecA that has engaged the translocon. We have used mild destabilization of SecA to trigger conformational changes that mimic those in translocation-active SecA and thus study its structural changes in a simplified, soluble system. Results from circular dichroism, tryptophan fluorescence, and limited proteolysis demonstrate that the SecA conformational reorganization involves disruption of several domain-domain interfaces, partial unfolding of the second nucleotide binding fold (NBF) II, partial dissociation of the helical scaffold domain (HSD) from NBF I and II, and restructuring of the 30 kDa C-terminal region. These changes account for the observed high translocation SecA ATPase activity because they lead to the release of an inhibitory C-terminal segment (called intramolecular regulator of ATPase 1, or IRA1), and of constraints on NBF II (or IRA2) that allow it to stimulate ATPase activity. The observed conformational changes thus position SecA for productive interaction with the SecYEG translocon and for transfer of segments of its passenger protein across the translocon.
Protein secretion is an essential process in all forms of life. In gram-negative bacteria, newly synthesized proteins destined for integration into membranes or secretion into the extracellular milieu predominantly traverse the secretory (Sec) pathway (1, 2). Most pre-proteins are targeted to the Sec pathway by a cleavable N-terminal signal sequence and by the peripheral membrane motor protein, SecA. E. coli SecA is a large, dynamic 102 kDa protein that forms homodimers and interacts with many different players during the translocation cycle, including pre-proteins, SecB, membrane, and the SecYEG translocon (3). The cytosolic chaperone SecB binds to a subset of pre-proteins, keeping them in a translocation-competent state. The pre-protein/SecB complex then interacts with SecA, and is localized to the translocon (2). The association of the pre-protein/SecB/SecA ternary complex with SecYEG induces a conformational change in SecA while ATP binding results in SecB release and initiation of translocation (1). The energy derived from ATP hydrolysis by SecA and the proton motive force subsequently drive the translocation of the pre-protein into the periplasmic space (4–6).
SecA is a multi-domain protein (Figure 1) with two tandem ATP-binding domains belonging to the DEAD-helicase superfamily, nucleotide-binding fold I (NBF I) and nucleotide-binding fold II (NBF II) (7). At the interface between NBF I and II is the nucleotide-binding cleft, and both NBF I and NBF II contain helicase motifs needed for ATP hydrolysis (7). NBF II, also known as IRA2 (8), undergoes a disorder-order transition during the ATP catalytic cycle (9) and has higher B factors than NBF I in a crystal structure of SecA from B. subtilis (7). A domain not found in other DEAD-helicases is the pre-protein cross-linking domain (PPXD), which interrupts NBF I (10). Two fragments of SecA can be individually expressed in E. coli or isolated by proteolytic cleavage: N68, a stable 68 kDa fragment of SecA comprised of NBF I, NBF II, and PPXD, and C34, formed by the α-helical scaffold domain (HSD), the α-helical wing domain (HWD), and the C-terminal linker (CTL) (11). The CTL region of the molecule, which includes a zinc-binding motif, contains the SecB-binding site and is also proposed to interact with anionic phospholipids (12). The ATPase activity of cytosolic SecA is suppressed by a helix-loop-helix motif in the HSD, called the IRA1 (11), and is positively regulated by NBF II (8). Therefore, C-terminally truncated constructs of SecA such as N68 (11) and SecA64 (13) possess elevated and unregulated ATPase activity.
Figure 1.
Domain organization and structural plasticity of the SecA PPXD. The overall domain organization of SecA in the three different crystal structures is similar, but the orientation of PPXD is different. (A) In the B. subtilis SecA structure (7) (PDB: 1M6N), the PPXD makes several intramolecular interactions with the HWD. The extreme C-terminal 40 residues are unresolved in this structure. The tryptophan residues corresponding to E. coli SecA Trps are shown in cyan spheres except for W519 and W541, which are located in a helical region of NBF II that is not present in B. subtilis SecA. Asterisks indicate the three tryptophan residues that contribute most to the overall SecA fluorescence (22). (B) In the structure of monomeric B. subtilis SecA (15) (PDB: 1TF5) the PPXD no longer interacts with the HWD. The last 61 residues are unresolved in this structure. (C) In the structure of the SecA-SecYEG complex (PDB: 3DIN, only SecA structure is shown) from T. maritime (16) the PPXD has rotated more extensively, making contact with NBF II. The last 55 residues are unresolved in this structure. (D) Linear representation of the E. coli SecA sequence illustrating the six different domains, with the color scheme that we use throughout this paper.
Various translocation components induce conformational changes in SecA during the pre-protein translocation cycle. SecA crystal structures show alterations in the positioning of the PPXD domain in relation to the HWD and NBF II (7, 14–16) (Figure 1). The recent 4.5 Å structure of SecYEG-bound SecA (16) shows the PPXD rotating away from HWD and making contact with NBF II (Figure 1C), thus forming a clamp region that is proposed to act as a channel for pre-protein translocation. Moreover, solution studies have suggested that even larger conformational changes may occur in SecA with at least two extreme conformational states: a compact, closed form in cytosolic SecA, and a more open state in translocation-active SecA. While ADP binding (17) and reduced temperature (18) favor the closed conformation, factors such as increased temperature (19), mutations (20), denaturants (21), association with model membranes (22, 23), and binding to SecYEG (24) push SecA into a more open conformation. A complete understanding of the complex mechanism of SecA-mediated protein translocation cycle requires identifying and characterizing the various conformational states of SecA and deducing their roles in the translocation cycle.
The most dramatic conformational change is believed to occur in ‘translocation-active SecA’. Generating this state requires the presence of all the components of translocation machinery making it challenging to study. We have used the strategy of mild perturbing the SecA native state in aqueous buffer and exploring how it shifts to populate a higher energy state on its energy landscape (25, 26). Associating properties of the newly populated state with functional characteristics of translocation-active SecA has allowed us to interrogate the conformational features of this elusive state. One of the hallmark features of translocation-active SecA is its enhanced ATPase activity (27), and such an activated state of SecA is reported to stably exist in low concentrations of denaturants such as guanidinium chloride or urea (21). In this study, we have characterized SecA in a low concentration of urea, and our findings provide a compelling model for the conformational transition in SecA that accompanies SecA-membrane/translocon binding and commitment of the pre-secretory complex to move the pre-protein across the membrane. The picture that emerges is that of a delicate balance of intradomain metastability and stabilizing interdomain interactions that are readily destabilized upon interaction with functional partners (membrane lipids, SecB, SecYEG, precursor protein, signal peptide, ATP).
EXPERIMENTAL PROCEDURES
Reagents
Unless otherwise mentioned, laboratory reagents were purchased from Sigma, VWR, or Fisher.
Construction of pET-17b SecA Plasmid
The gene was amplified by PCR from the pT7-SecA2 plasmid (D. Oliver, Wesleyan University) using Taq DNA polymerase (New England Biolabs, Ipswich, MA). The 2.7 kb PCR fragment was subcloned into the pGEM®-T vector (Promega, Madison, WI), digested with NdeI and XhoI restriction enzymes (New England Biolabs, Ipswich, MA) and ligated into the same sites in pET-17b (Novagen, Madison, WI) creating the pET-17b SecA plasmid. DNA sequencing (Davis Sequencing, Davis, CA) verified the correct sequence of the SecA gene.
Protein Expression and Purification
SecA protein was expressed in E. coli BL21(DE3) strain. Cells were grown in LB supplemented with LinA salts at 37 °C to an OD600 of 0.5, induced with 0.75 mM isopropylthiogalactoside, and grown for another 2.5 h at 37 °C. Cells were lysed using the Microfluidizer® (M-110L Microfluidics, Newton, MA), and soluble SecA protein was purified as described previously (19) with minor modifications. In brief, a three sequential step purification protocol was performed using a DEAE Sepharose CL-6B (Sigma, St. Louis, MO) column, a SP Sepharose Fast Flow (Sigma, St. Louis, MO) column, and a Superdex 200 prep grade (GE Healthcare, Piscataway, NJ) column. The fractions containing pure protein were pooled, concentrated and exchanged against buffer on a 50 kDa concentrator and stored at −80 °C. Protein concentration was determined from absorbance at 275 nm after dilution into 6 M guanidinium chloride, 20 mM sodium phosphate, pH 6.5 using an extinction coefficient of 0.73 OD per mg mL−1 (19). Far-UV CD and enzyme-coupled ATPase measurements were performed on each batch of purified protein to verify secondary structure and confirm protein activity.
ATPase Assay
Samples containing 1 μM SecA in 50 mM Tris pH 8.0, 30 mM KCl, 30 mM NH4Cl, 5 mM Mg(OAc)2, 1 mM dithiothreitol (DTT) (Buffer A) containing increasing urea concentrations were incubated at 22 °C for 12 – 14 h. ATP was added at a final concentration of 5 mM, and samples were incubated for an additional hour at 22 °C. The ATPase activity of SecA was measured using the malachite green-ammonium molybdate ATPase assay (28) with o-phosphoric acid (Sigma, St. Louis, MO) as a standard curve. Absorbance was measured at 660 nm on a Genesys 10 UV scanning spectrophotometer (Thermo Electron Corp, Waltham, MA).
Preparation of Urea-Activated SecA (u-SecA)
Low urea-treated, ATPase-hyperactive SecA, u-SecA, was generated by incubating c-SecA in 20 mM HEPES, pH 7.5, 50 mM KCl, 1 mM MgCl2, 1 mM DTT (Buffer B) containing 2.2 M urea for 4 h at 22 °C.
Gluteraldehyde Crosslinking
Gluteraldehyde was added to a final concentration of 0.1% (v/v) to various concentrations of c-SecA and u-SecA. The crosslinking reaction was performed at room temperature for 2 min followed by quenching with 100 mM Tris, pH 8.0. The samples were boiled for 5 min and analyzed on a 6% tricine-SDS-PAGE. The protein bands were visualized by Coomassie Blue staining.
Biotin-Labeled Signal Peptide-Binding Assay
Biotinylated-KRRLamB19C signal peptide (Bio-MMITLRKRRKLPLAVAVAAGVCSAQAMA, with Biotin attached to the N-terminal α-amine (GL Biochem Ltd., Shanghai)) was added to increasing concentrations of c-SecA and u-SecA (0.1 μM to 2 μM) to a final concentration of 1 μM signal peptide, and equilibrated for 2 h at 22 °C. Samples were cross-linked with 0.1% (v/v) gluteraldehyde at room temperature for 2 min and quenched with 100 mM Tris, pH 8.0. Each sample was run on two 6% tricine-SDS-PAGE gels. Samples analyzed on one of the gels were transferred to a PVDF membrane, and the other gel was stained with Coomassie Blue. The PVDF membranes were probed with a streptavidin-horseradish peroxidase conjugate (GE Healthcare) at a 1:1000 dilution in phosphate buffered saline, 0.05% Tween-20. Protein bands containing SecA crosslinked to the signal peptide were detected using the SuperSignal West Pico kit (Pierce, Rockford IL) following the manufacturer’s instructions, and chemiluminescence was detected using a G:Box gel documentation unit (Syngene, Frederick, MD).
Equilibrium Fluorescence Measurements
Samples containing 1 μM SecA were incubated at 22 °C for 12 – 14 h in various urea concentrations in Buffer A. The tryptophan fluorescence spectrum (300 – 380 nm) of SecA was measured on a QM-1 spectrofluorometer (Photon Technology International, Birmingham, NJ) at 22 °C with an excitation wavelength of 295 nm, and both bandwidths set at 2 nm.
Circular Dichroism (CD)
Far-UV CD measurements of 1 μM SecA in various concentrations of urea incubated at 22 °C overnight were recorded using a J715 spectropolarimeter (JASCO, Easton, MD) from 215 – 250 nm in a one-mm pathlength cell. Scan speed was set at 20 nm/min, and the data were averaged over five scans. The raw CD data were converted into [θ]MRE using the equation, [θ]MRE = (100 × θ)/(C × l), where θ is the ellipticity in degrees, C is the protein concentration (M), and l is the cuvette pathlength in cm. The α-helical content was calculated using the equation, % α-helix = (− [θ]MRE, 222 nm +3000)/39000 (29).
Limited Proteolysis
Samples of c-SecA and u-SecA at 2.5 μM were subjected to a time course of limited α-chymotrypsin (α-CT) (1:150 w/w) digestion from 0 to 15 min. At the end of each time point, the reaction was quenched with the addition of 5 mM 4-(2-aminoethyl) benzenesulfonyl fluoride, 3X SDS sample buffer, followed by flash freezing in liquid nitrogen. The samples were run in duplicate on two 8% or 10% tricine-SDS-PAGE gels. One gel was stained with Coomassie Blue, and the other was transferred to a PVDF membrane for further analysis.
Antibody Detection
Bands generated from the α-CT digestion of c-SecA and u-SecA were identified by sequential probing of the same blot with six different region-specific SecA antisera, A1 to A6 developed by Ramamurthy and Oliver (30). Each of the primary antibodies was used at a 1:5000 dilution in Tris-buffered saline with 0.05% Tween-20 and a goat anti-rabbit IgG alkaline phosphatase-conjugated secondary antibody (Sigma, St. Louis, MO) was used according to the manufacturer’s protocol. The blots were developed using AP Lumino (Pierce, Rockford, IL) reagent using the G:Box gel documentation unit (Syngene, Frederick, MD). Blots were stripped in 50 mM Tris, pH 6.8 containing 2% SDS, and 100 mM β-mercaptoethanol at 55 °C for an hour. The stripped blot was re-probed with the secondary antibody to ensure no background signal before proceeding with the next region-specific antibody.
Proteolytic Band Assignment
To determine the identity of the α-CT proteolytic bands, the ExPASy Peptide Cutter tool (http://web.expasy.org/peptide_cutter/) was used to estimate the probability of cleavage at each aromatic residue. Additionally, the molecular weights for each possible SecA fragment generated from α-CT digestion (from one cleavage to complete cleavage) were manually determined using ExPASy Compute pI/Mw tool (http://web.expasy.org/compute_pi/) and sorted by molecular weight. The Coomassie-stained, A1, A2, A3, A4, A5, and A6 antibody lanes were aligned using full-length SecA for each time point. From the aligned blots, bands under 40 kDa were selected for identification. Protein identification was performed by MALDI-TOF/TOF mass spectrometry (MS) on a few selected bands (Center for Advanced Proteomics Research, University of Medicine and Dentistry of New Jersey). The MS error was 50 ppm with most peptides having an error less than 20 ppm. The MS/MS sequencing confidence indicator was 99.7% or higher for all peptides. Although sequencing did not yield homogenous SecA fragments, the results from individually sequenced MS of peptides were used to determine the different SecA regions associated with a particular band. If an aromatic residue was found in a sequenced peptide more than twice, the residue was eliminated as a possible digestion site. Using the band assignments, we were also able to roughly determine the number of times a given site was cleaved, indicated on the cartoon diagrams (Figure 6B), and the size of the arrow indicates the frequency of cleavage after a given residue.
Figure 6.
Limited α-CT proteolysis of c-SecA and u-SecA. (A) Proteolytic fragments generated by α-CT digestion of c-SecA (C) and u-SecA (U) for times from 10 s to 15 min were probed with region-specific antibodies against SecA (30). The labels A1 through A6 represent the Western blots probed with each of the six region-specific antibodies. The 10 sec α-CT digest used for further analyses of accessibility in c-SecA and u-SecA is highlighted by a black box in each blot. Bands 1, 2, 3, and 4 are observed in both c-SecA and u-SecA. Bands marked by arrows in the blots probed by antibodies A3, and A4 are the peptide fragments < 20 kDa that are selectively found in c-SecA. Band 4 in the A5 and A6 blots corresponds to the 30 kDa reorganized C-terminal fragment that stably exist in u-SecA but not in c-SecA. (B) SecA domain organization showing the regions recognized by the different antibodies: A1–antibody against residues 1–209, A2–antibody against residues 211–350, A3–antibody against residues 351–509, A4–antibody against residues 519–664, A5–antibody against residues 665–820, A6–antibody against residues 822–901. The vertical arrows represent the deduced α-CT cleavage sites with the thickness of the arrows indicating the relative lability of sites, based on a 10 s treatment with enzyme. Cleavage sites for c-SecA are indicated above the domain diagram while sites for u-SecA are shown below.
Labeling Signal Peptide with 4-Maleimido-benzophenone (MBP)
The Bio-KRRLamB19C signal peptide at 1 mM concentration in 20 mM HEPES pH 7.5, 1.2 mM TCEP was alkylated at the Cys19 position by addition of 1.5 mM MBP. The reaction was stirred for 30 min at room temperature, followed by quenching with trifluoroacetic acid. The MBP-labeled peptide (Bio-KRRLamB19C-MBP) was purified by preparative reverse phase HPLC using a phenyl column (25x250 mm, 10 mm, 300 Å, Vydac). The identity and purity of the peptide was confirmed by mass spectrometry analysis using an Esquire-LC Ion trap mass spectrometer (Bruker Daltonics, Billerica, MA).
Photoactivated Crosslinking
In the absence of light, 2.5 μM Bio-KRRLamB19C-MBP was added to 2.5 μM c-SecA and u-SecA and incubated at room temperature for 15 min. Samples were illuminated with 365 nm light at a distance of ~5 cm for a minute. To obtain similar α-CT digestion patterns to u-SecA, 2.2 M urea was added to the crosslinked c-SecA, and the sample was incubated at 22 °C for 4 h. A 2 min α-CT digestion was performed on the Bio-KRRLamB19C-MBP crosslinked samples of c-SecA and u-SecA.
RESULTS
Urea-Induced Active Form of SecA
To understand the conformational rearrangement in SecA as it converts to the membrane/translocon-activated form, we generated a mild denaturant-destabilized form of SecA based on previous work that showed an enhancement in SecA ATPase activity in low concentrations of guanidinium chloride (21). We followed SecA ATPase activity at 22 °C in the presence of increasing urea concentration (Figure 2, open circles) and in agreement with the earlier report, the ATPase activity of SecA increased in low denaturant. We observed a maximum eightfold increase in activity at 2.2 M urea and termed this urea intermediate u-SecA. The extent of increase in the ATPase activity of SecA is similar to SecA activation during pre-protein translocation (27). The high enzymatic activity of SecA in 2.2 M urea also indicates that the core ATPase unit of the protein, NBF I, remains well folded in low urea. Importantly, based on fluorescence (Figure 2, filled circles) and CD (Figure 2, filled triangles) monitored over the same range of urea concentrations as the activity, SecA undergoes a structural transition to a state that is intermediate between native and unfolded states as its activity increases (see further discussion below).
Figure 2.
Low concentrations of urea activate the ATPase activity and alter the conformation of SecA. The ATPase activity of SecA (open circles) increases as the urea concentration is increased, and peaks at 2.2 M urea. The Trp fluorescence of SecA at 340 nm (filled circles) indicates that the structure of the protein has been partially perturbed at 2.2 M urea. The far-UV CD signal at 222 nm (filled triangles) indicates loss of helical secondary structure of SecA at the same point. The Trp and CD signal were normalized to the signal of SecA in the absence of urea (100%). The error bars in the ATPase activity data represent the standard deviation from three experiments.
U-SecA Is Predominantly Monomeric
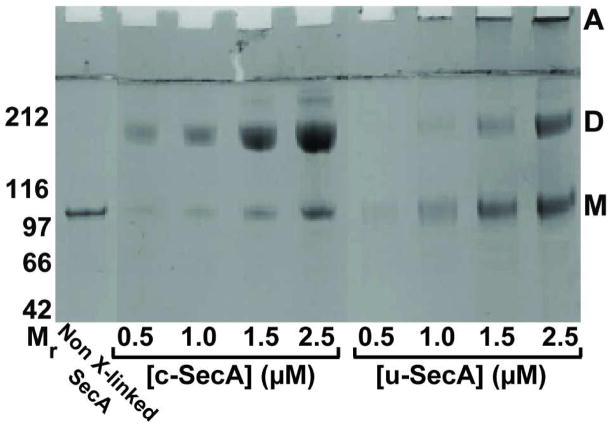
A major unresolved question relating to the molecular mechanism of SecA during the translocation cycle is whether SecA at the translocon functions as a monomer or a dimer (7, 16, 21–24, 31–38). We explored the oligomeric state of c-SecA and u-SecA using a qualitative assay based on gluteraldehyde cross-linking (Figure 3). As expected, c-SecA exists primarily as a dimer at all concentrations tested (39, 40), while u-SecA shows a preponderance of the monomeric state at low protein concentrations. Dimeric and higher order species are populated in u-SecA at higher protein concentrations (above 1.5 μM) suggesting a partitioning between the monomeric and dimeric state in u-SecA. The presence of aggregates of u-SecA is likely the result of non-specific association. The indication of a monomeric intermediate in our experiments contrasts with an earlier report of a dimeric intermediate during urea-unfolding of SecA (41). The observed discrepancy is likely due to the known effect of temperature, concentration, and buffer conditions (such as pH and salt) on the partitioning of SecA between monomer and dimer (40).
Figure 3.
The oligomeric state of the c-SecA and u-SecA monitored by gluteraldehyde cross-linking followed by SDS-PAGE analysis. The different oligomeric forms of SecA, M – monomer; D – dimer; A – aggregates, are indicated on the right side of the gel. The concentrations of c-SecA and u-SecA are shown below the gel. The diffuse nature of the crosslinked band arises from intramolecular crosslinking occurring along with the intersubunit crosslinking. The molecular weight standards, Mr, are shown in kDa in this and all other figures.
Monomeric u-SecA Binds Signal Peptide
The pre-protein binding activity of u-SecA was assessed by examining the binding of a biotin-labeled, LamB signal sequence (modified for water solubility (42)). Both c-SecA and u-SecA bound the signal peptide (Figure 4A and 4B), with the signal sequence associating primarily with dimeric c-SecA and monomeric u-SecA. Furthermore, unlabeled signal peptide efficiently competed with the biotin-labeled peptide in a dose-dependent manner indicating reversible binding of the signal sequence (Figure S1). Additionally, we measured the ATPase activity of both c-SecA and u-SecA in the presence of increasing concentrations of signal peptide. Similar to the results seen with SecA64 (13) signal peptide had little effect on c-SecA activity but decreased u-SecA activity by about 60% (data not shown). Thus, SecA in 2.2 M urea at 22 °C has enhanced ATPase activity that is inhibited by signal peptide binding, and is able to efficiently and reversibly bind signal sequences, supporting the use of u-SecA as a mimic of a translocation-active SecA.
Figure 4.
Signal peptide binding to SecA. Gluteraldehyde crosslinking was employed to assess signal peptide binding to c-SecA (A) and u-SecA (B) using a biotinylated model signal peptide. The presence of signal peptide was detected with streptavidin-horseradish peroxidase (bottom panels in A & B); signal peptide binds in a concentration dependent manner primarily to dimeric c-SecA and monomeric u-SecA. The top gel panel in both A and B shows Coomassie blue staining of the duplicate gel run in parallel.
Structural Comparison of u-SecA and c-SecA
Further insights into the conformational changes in u-SecA can be extracted from the observed tryptophan fluorescence and far-UV CD changes upon urea titration (Figure 2). The seven Trp residues in SecA (Figure 1A, cyan spheres) provide a tool to probe its tertiary structure. At 22 °C, SecA fluorescence at 340 nm begins to decrease at 1.4 M urea and by 2.2 M urea, when ATPase activity peaks, has dropped by 50% (Figure 2, filled circles). Through mutagenesis, Ding et al. demonstrated that Trps 701, 723, and 775 contribute most to the overall fluorescence signal of SecA (22). These three tryptophans are located in the C-terminal one-third of the molecule (within C34), with W701 and W723 in the HWD, and W775 in the IRA1 sub-domain. Therefore, we conclude that the C34 portion is largely responsible for the conformational change of SecA in low urea. We also monitored the change in the tryptophan fluorescence as c-SecA transitions to u-SecA by the ratio of the fluorescence intensity at 330 nm to 355 nm (Figure 5, filled circles). This ratio is very sensitive to shifts in the emission maximum and provides information about the environment of Trp residues (43). A decrease in the F330/F355 ratio indicates a red-shifted fluorescence. U-SecA exhibits a significantly red-shifted fluorescence relative to c-SecA, indicating that one or more of the three Trps becomes more solvent exposed upon treatment of SecA with urea.
Figure 5.
Changes in the secondary and tertiary structure of SecA in low urea concentrations. The ratio of fluorescence intensity at 330 and 355 nm (filled circles) and the α-helical content (filled triangles) are plotted at low urea concentrations to reveal the extent of loss in helicity and the nature of the changes in Trp environments in u-SecA.
The CD spectral changes in SecA upon urea titration (Figure 2, filled triangles) can be interpreted in terms of altered helical content based on ellipticity at 222 nm, and allow us to estimate that helical content decreases from 38% in c-SecA (close to an estimate of 44.6% for E. coli SecA built by homology on the B. subtilis SecA crystal structure) to 31.5% at 2.2 M urea (Figure 5), arguing for only modest change in overall secondary structure of SecA in low urea.
U-SecA Undergoes Reorganization at the Inter-Domain Interfaces
We compared limited proteolytic susceptibility of c-SecA and u-SecA and probed the resulting fragments with the six different region-specific SecA antibodies (A1 – A6) as a means to assess restructuring in specific SecA domains. Digestion with α-CT for a time course from 0 to 15 min showed that u-SecA is significantly more protease sensitive than c-SecA, consistent with its more open structure (Figures 6 & S2). Quantitation of undigested SecA at each digestion time point (Figure S2) shows that in c-SecA ~40% of full-length SecA remains after 15 min of digestion, while in u-SecA full-length SecA is completely digested within five min (Figure 6). Interestingly, while digestion is more rapid in u-SecA, the patterns of digestion for c-SecA and u-SecA are quite similar, with the notable exception that a 30 kDa protease-resistant species selectively accumulates in u-SecA (band 4, Figure 6).
Probing with region-specific antibodies provided greater insight into structural changes in u-SecA (Figure 6). The overall band pattern observed by A1 (against NBF I) and A2 (against PPXD) antibodies show minimal changes, suggesting a global integrity of the subdomains NBF I and PPXD in both c-SecA and u-SecA (Figure 6A). In contrast, the banding patterns observed with the remaining antibodies vary more widely between c-SecA and u-SecA, pointing to alterations in NBF II and the other C-terminal domains. Bands marked as 1, 2, 3, and 4 are found in both c-SecA and u-SecA at all-time points. Bands 1, 2, and 3, recognized by the A1 antibody, arise from the removal of a short segment from the N-terminus and cleavages within the PPXD leaving the bulk of NBF I with variable PPXD lengths. Band 4 arises from the C-terminal portion of SecA, as indicated by its recognition by the A5 and A6 antibodies. Note that band 4 is present in u-SecA from 10 sec to the 15 min time points, and not in c-SecA after 2 min. The persistence of band 4 suggests that upon cleavage, the C-terminal portion of u-SecA reorders into a more stable structure. The α–CT digestion of c-SecA shows significant cleavage in NBF II, leading to peptide fragments < 20 kDa in size that are recognized by the A3 and A4 antibodies (marked by arrows in Figure 6A), similar to previous reports (3).
We further identified the α-CT fragments in c-SecA and in u-SecA by a combination of mass spectrometry-based sequencing of several proteolytic bands ≤ 40 kDa, prediction of α-CT cleavage sites, band molecular weight, and antibody reactivity. We largely focused on the earliest points of the digestion time course, as they likely arise from primary cleavages, and thus report on flexible regions in both forms of SecA. All the identified cleavage sites (Figure 6B) fell into one of three classes: Class I sites were observed in both c-and u-SecA, Class II sites were preferentially cleaved in u-SecA, and Class III sites were cleaved only in u-SecA. Class I sites arise from cleavage at residues F10, F50 and F399 in NBF I; Y236, Y299 and F320 in the PPXD; and Y893 in the CTL (Figure 7). The finding that Class I cleavage sites, which are the same in c-SecA and u-SecA, map primarily to NBF I and PPXD domains argues that these domains retain similar domain architecture in u-SecA as in cytoplasmic SecA.
Figure 7.
Structural interpretation of conformationally diagnostic accessibility data for u-SecA from limited proteolysis, fluorescence, and literature results. Residue positions that were cut by α-CT in c-SecA and u-SecA based on the data shown in Figure 6B are mapped onto the NMR structure of E. coli SecA (PDB: 2VDA). Spacefill residues colored as brown and red represent class II and III cleavage sites, respectively, and W775 is shown in light pink spacefill. Class I sites are shown in spacefill in the same color as the domain to which they belong. SecA residues that are reported to undergo conformational change in the presence of its natural ligands are shown with their Cα atoms in spacefill: Motif IV (in cyan), buried in c-SecA, is reported to become accessible to make contact with SecYEG (56) in translocation-active SecA. Motif VI (in grey) is a highly mobile region in c-SecA by NMR studies (9, 47). And residues in black represent residues that become more mobile based on EPR labeling when SecA interacts with phospholipids (48). Interdomain interfaces that we propose to be disrupted in the transition from c-SecA to u-SecA are indicated by yellow hatched lines.
The Class II cleavage sites, including F70, W349, and F598, and Class III cleavages (Y134, Y384, Y428, and Y648) reveal conformational features specific to u-SecA (Figure 7). Overall, they map to domain-domain interfaces, arguing for significant domain dissociation in u-SecA. Interestingly, F598 is positioned at the interface between the N-terminal HSD and NBF II, and Y648, a buried residue in all SecA crystal structures, is at the interface of the HSD and IRA1. Cleavage at these residues in u-SecA illustrates that the domain-domain interactions of the HSD with NBF II and IRA1 are perturbed relative to c-SecA. Based on solvent-accessible surface areas (calculated from the B. subtilis crystal structure (7), PDB: 1M6N using Vadar (44)), Y428 should be inaccessible to protease in c-SecA, and therefore, cleavage at Y428 and its close spatial proximity to F598 suggest that this region of NBF II is altered in u-SecA. The remaining two cleavage sites, F79 and Y134, are relatively solvent-exposed in SecA crystal structures. We can speculate at this point that preferential cleavage in u-SecA for these sites that map to the NBF I/NBF II interface, reflects loosening of this domain-domain contact. Finally, the absence of cleavage sites between residues Y648 and Y893 in u-SecA, along with the persistent ~30 kDa band detected by both A5 and A6 antibodies, supports the remodeling of the C-terminal region in u-SecA.
Signal Peptide Binds Differently to c-SecA and u-SecA
We carried out limited proteolysis of c-SecA and u-SecA that had been photo-crosslinked with LamB signal peptide and probed proteolytic fragments with both region-specific SecA antibodies and streptavidin to identify regions that were involved in signal peptide binding or proximal to the signal peptide-binding site (Figure 8). Both crosslinked c-SecA and u-SecA were digested in 2.2 M urea to obtain a similar proteolytic pattern. The biotin detection (labeled B in Figure 8) shows a more intense signal for full length, undigested u-SecA than for c-SecA, suggesting qualitatively a greater affinity of signal peptide for u-SecA. Inspection of fragments that yield a signal upon probing with streptavidin indicate that the signal peptide crosslinks to the N-terminal domain (similar to N68 or SecA64) in both c-SecA and u-SecA, although the higher intensity for the 45 kDa fragment in c-SecA than in u-SecA suggests that it may be more labile in u-SecA. Provocatively, signal peptide crosslinks significantly to a 30 kDa band, recognized by C-terminal antibodies A5 and A6, in u-SecA, but not in c-SecA. These results argue compellingly that u-SecA presents a signal peptide-binding site that is at least in part distinct from that in c-SecA.
Figure 8.
Signal peptide photo-crosslinking to c-SecA (A) and u-SecA (B). Equimolar amounts of c-SecA or u-SecA and signal peptide were crosslinked using a photo-activatable crosslinker, as described in the text. S, B, and A1 – A6 denote, respectively, Coomassie blue visualization of the gel, enhanced chemiluminescence detection of SecA crosslinked to biotinylated signal peptide, and Western blotting with the six region-specific SecA antibodies. Note that signal peptide crosslinked significantly to a 30 kDa band in u-SecA that is also recognized by the two C-terminal SecA antibodies, A5 and A6.
DISCUSSION
SecA has been extensively studied since its key role in bacterial protein export was discovered (1, 3, 45, 46). Numerous findings have combined with structural data from crystallography and NMR to reveal the ability of SecA to interact with several partners, the delicate balance of domain-domain contacts in SecA that enable it to undergo a dramatic and functionally critical conformational change, and the capacity of SecA to act as the ‘motor’ in protein translocation—harnessing ATP energy and driving protein segments across the SecYEG translocon. Yet deducing the structural features of SecA when it is active in translocation has proven extremely difficult. The strategy employed here, gentle perturbation with denaturant, has yielded new insights, which can be coupled with previous findings to paint a picture of translocation-active SecA with enhanced structural detail.
It was previously known that the N-terminal domains of SecA were in the helicase family, and that NBF I was the minimal unit with ATPase catalytic activity (8). This activity was shown to be suppressed in cytosolic SecA by interactions between the C-terminal segment from residues 783 to 795, dubbed IRA1, and disruption of these interactions allowed the NBF II domain to activate ATP hydrolysis. This conformational disruption mirrors the activation of SecA ATPase upon proteolytic cleavage to remove the C-terminal third of the molecule (11,13), treatment with increased temperature (19), or binding of ATP (19) or model membranes (22,23). A similar conformational transition has been described when SecA docks onto the SecYEG translocon (24). The fact that gentle treatment with denaturant seemed to cause a very similar shift in conformation of SecA led us to explore the nature of the conformational change in denaturant-treated, yet soluble SecA, under conditions where its ATPase rate was comparable to that measured for translocation-active SecA.
U-SecA, populated at 2.2 M urea and 22 °C, displays an 8-fold elevated ATPase activity, favors a monomeric state, and binds signal peptides. All of these properties validate its use as a mimic of translocation-active SecA, putting aside for the purposes of this study the controversies that remain about whether SecA acts as a monomer or dimer during translocation. We found that u-SecA has undergone a major conformational change to a state with enhanced solvent exposure of tryptophan side chains and reduced helicity relative to c-SecA. We conclude that the fluorescence change likely arises because the C-terminal domain of u-SecA is displaced from contacts with the HSD and NBF I. In particular, we posit that the observed increase in Trp solvent exposure in u-SecA (Figures 2 and 5) can be accounted for largely by increased solvent accessibility of W775, which is on IRA1 and faces NBF 1 (shown in pink spacefill in Figure 7). This conclusion is supported by an earlier report that W775 becomes more solvent exposed both during the endothermic transition in SecA as well as when SecA interacts with signal peptide (22). In addition to the increased exposure of W775, we also saw enhanced proteolytic cleavage at residue Y428 in NBF II in u-SecA relative to c-SecA (shown in brown spacefill in Figure 7). Furthermore, we attribute the drop in helicity upon urea treatment of SecA to a combination of partial unraveling of the HSD and conformational perturbation of NBF II. The metastability of NBF II was first pointed out by Sianidis et al. (8), who concluded that a major fraction of the change in the helical content of SecA during thermal unfolding could be assigned to NBF II (or IRA2). Our own NMR study showed that residues 564–579 of NBF II (in grey spacefill in Figure 7) are highly mobile in c-SecA (47). Keramisanou et al. also confirmed the inherent flexibility of IRA2 using extensive NMR and biochemical approaches (9). More recently, an EPR study showed that helical residues 600–610 in NBF II and helical residues 636–638 in the N-terminal HSD (site examined are shown in black spacefill in Figure 7) are more mobile upon interaction of SecA with phospholipids than in soluble SecA (48). Previous structural studies had pointed to plasticity of the NBF I/NBF II interface and its importance in a progressive conformational shift of SecA from its initial encounter with pre-protein in the cytoplasm to its motor action driving pre-protein across the SecYEG channel (16).
In addition to these conformational changes we observed the intriguing result that a 30 kDa fragment recognized by antibodies specific to the C-terminal HSD, HWD and CTL region of SecA was persistently protease resistant in u-SecA. This observation is reminiscent of the earlier finding that a membrane-inserting 30 kDa fragment corresponding to the C-terminal one-third of SecA is protease resistant in membrane-bound SecA (49–51). Importantly, Price et al. reported that this 30 kDa fragment is more stable than the 30 kDa fragment generated by limited proteolysis of c-SecA (51). While it is not straightforward to interpret protease-resistance of a membrane-associated species in terms of the stability of the fragment, it is tempting to speculate that the conformational rearrangements in u-SecA that lead to a stable 30 kDa species involve a similar reordering within this domain to what occurs when SecA interacts with a membrane. We suspect that dissociation and reordering of the C-terminal domain may also account in part for the overall diminished helicity of u-SecA, as the C-terminal end of the HSD is likely to partially unravel as its contacts with the C-terminal domain are broken, just as we posit for the N-terminal end of the HSD upon disruption of contacts with NBF II.
How do these observations relate to SecA’s translocation function? Some features of u-SecA can be reconciled in light of the crystal structure of the SecA-SecYEG complex (16), which reflects the state of SecA when it first engages the translocation machinery. In this structure, SecA is monomeric and has experienced a striking conformational change leading to docking of the PPXD onto a single SecYEG trimer. Formation of an interface between NBF I and SecY involves the exposure of motif IV (residues 182, 185, 186, and 188, shown in blue spacefill in Figure 7). In c-SecA these residues are packed against the N-terminal portion of the HSD and the so-called stem region (10) of the PPXD. In contrast, the disruption of the interface between HSD and NBF I that we conclude occurs in u-SecA is fully consistent with the exposure of motif IV in SecA upon binding to SecYEG.
While some of our results on u-SecA can be reconciled with the structure of the SecA/SecYEG complex, others cannot. As noted by Zimmer et al. (16), the structure of the SecA/SecYEG complex reflects the initial encounter between the translocase and the translocon. From several lines of evidence, we are persuaded that u-SecA resembles a state of SecA subsequent to this initial encounter. Specifically, u-SecA displays enhanced proteolytic susceptibility at Y428 and F598, both of which are inaccessible in the SecA/SecYEG complex. These residues would become accessible as the interaction between NBF II and the N-terminal end of the HSD is weakened, which we postulate to be occurring in u-SecA, accompanied by perturbations to the interface between NBFs I and II, and consequent enhanced ATPase activity. We also conclude from our results on u-SecA that W775 is solvent-exposed in this state, and we see enhanced proteolytic cleavage at Y648. Neither of these residues would be solvent exposed in c-SecA or in SecA in complex with SecYEG, leading us to conclude that the interaction between the C-terminal end of the long HSD helix and IRA1 is disrupted in u-SecA, thus exposing W775 and Y648, which are intimately involved in the interhelix packing. Thus, the structural changes in u-SecA suggest that it represents a state downstream of the SecA/SecYEG initial encounter complex, potentially resembling SecA when it is engaged in moving pre-protein across the translocon.
How SecA recognizes signal peptides in order to ensure fidelity of targeting and to facilitate translocation has been the subject of numerous studies, with complex results that cannot readily be rationalized in terms of one signal peptide-binding site. Now having in u-SecA a model for a state of SecA populated at a later stage in translocation, we have compared its signal peptide interaction to that of c-SecA, which provides a model for the initial interaction of pre-proteins with SecA. Although preliminary, our finding that signal peptide crosslinked to u-SecA via a 30 kDa fragment recognized by antibodies A5 and A6 (Figure 8) suggests that the signal peptide-binding site on u-SecA involves the C-terminal domain of SecA. Some involvement of the C-terminal domain was suggested in the Gelis et al. NMR study (14), but other previous models do not implicate the C-terminal domain in signal peptide binding (10, 52). Interestingly, FRET based mapping of signal peptide binding to SecA (53) and a recent study characterizing the signal peptide binding-site employing genetic, biochemical and physiological also show that in addition to the PPXD, the C-domain of SecA interacts with the signal peptide (54).
Merging our results with the extensive past work on SecA allow us to propose a model for the series of structural changes in SecA during pre-protein translocation (Figure 9). In the cytosol, SecA is in a dimeric, auto-inhibited state with low ATPase activity (step 1). The interaction of IRA1 (light green) with the rest of the HSD (dark green) is a key lynchpin stabilizing the auto-inhibited state. Interaction with pre-protein (with or without SecB), ATP, membrane lipids, and the SecYEG translocon initiates architectural rearrangements accompanied by domain dissociation events (step 2). Interactions of the PPXD (yellow) with the HWD (orange) are disrupted, such that the PPXD moves closer to and forms new contacts with NBF I (blue) and NBF II (purple), promoting SecA binding to the translocon as observed in the crystal structure of SecA in the SecYEG-bound form (16). In the translocon-docked state (step 2), the two-helix finger is positioned to interact directly with the SecY channel (55), and SecA is primed for pre-protein translocation. Conversion to translocation-active SecA (step 3), as mimicked by u-SecA, involves further domain rearrangements: in particular, breaking contacts between the N-terminal end of the long helix of the HSD and NBF II and disruption of the interaction of IRA1 and the C-terminal end of the long helix of the HSD, which relieves the auto-inhibition of ATPase activity and results in the translocating ATPase rate. Concurrently, the 30 kDa C-terminal domain encompassing the HWD, IRA1, and the CTL (red) restructures as a proteolytically stable domain, which enables its productive interaction with the membrane, translocon, and pre-protein. In this state, the signal peptide on the pre-protein makes new contacts with the C-terminal region of SecA. This model for translocation-active SecA adds to our current understanding and offers a structural framework for the events that are at the heart of SecA’s roles in translocation. For example, the proposed role of IRA1 (also known as the ‘two-helix finger’) in escorting the polypeptide through the translocon may be initiated by the restructuring of the C-terminal region of SecA and creation of a new binding site for signal peptide.
Figure 9.
Proposed model for conformational changes in SecA during protein translocation. Cytosolic SecA is shown in a closed antiparallel dimeric form (1); upon interaction with membrane-associated SecYEG, the SecA dimer dissociates (2), and in a subsequent step SecA transitions to a domain-dissociated, translocation-active state (3). This new conformation (3), which we argue is modeled by u-SecA, represents SecA docked at the SecYEG translocon and engaged in productive translocation of pre-proteins. The different domains of SecA are color-coded as throughout this paper: blue, NBF I; yellow, PPXD; purple, NBF II; green, N-terminal HSD; orange, HWD; dark green, C-terminal HSD; and red, CTL.
Supplementary Material
Acknowledgments
Funding: This work was supported by a grant from the NIH to L.M.G. (Grant GM034962).
We gratefully thank Don Oliver at Wesleyan University for the generous gift of the E. coli region-specific SecA antibodies, and Eugenia Clerico for critical reading of the manuscript.
Abbreviations
- α-CT
α-chymotrypsin
- c-SecA
cytosolic SecA
- CD
circular dichroism
- CTL
C-terminal linker
- DTT
dithiothreitol
- HSD
helical scaffold domain
- HWD
α-helical wing domain
- IRA1
intramolecular regulator of ATPase 1
- IRA2
intramolecular regulator of ATPase 2
- MBP
4-maleimido-benzophenone
- NBF I
nucleotide-binding fold I
- NBF II
nucleotide-binding fold II
- PPXD
pre-protein cross-linking domain
- u-SecA
SecA in 2.2 M urea
Footnotes
Two figures showing a signal peptide competition binding assay and quantitation of limited proteolysis of SecA by Coomassie staining are provided as Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 2.du Plessis DJ, Berrelkamp G, Nouwen N, Driessen AJ. The lateral gate of SecYEG opens during protein translocation. J Biol Chem. 2009;284:15805–15814. doi: 10.1074/jbc.M901855200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vrontou E, Economou A. Structure and function of SecA, the preprotein translocase nanomotor. Biochim Biophys Acta. 2004;1694:67–80. doi: 10.1016/j.bbamcr.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Chen L, Tai PC. ATP is essential for protein translocation into Escherichia coli membrane vesicles. Proc Natl Acad Sci U S A. 1985;82:4384–4388. doi: 10.1073/pnas.82.13.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shiozuka K, Tani K, Mizushima S, Tokuda H. The proton motive force lowers the level of ATP required for the in vitro translocation of a secretory protein in Escherichia coli. J Biol Chem. 1990;265:18843–18847. [PubMed] [Google Scholar]
- 6.Yamane K, Ichihara S, Mizushima S. In vitro translocation of protein across Escherichia coli membrane vesicles requires both the proton motive force and ATP. J Biol Chem. 1987;262:2358–2362. [PubMed] [Google Scholar]
- 7.Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science. 2002;297:2018–2026. doi: 10.1126/science.1074424. [DOI] [PubMed] [Google Scholar]
- 8.Sianidis G, Karamanou S, Vrontou E, Boulias K, Repanas K, Kyrpides N, Politou AS, Economou A. Cross-talk between catalytic and regulatory elements in a DEAD motor domain is essential for SecA function. EMBO J. 2001;20:961– 970. doi: 10.1093/emboj/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keramisanou D, Biris N, Gelis I, Sianidis G, Karamanou S, Economou A, Kalodimos CG. Disorder-order folding transitions underlie catalysis in the helicase motor of SecA. Nat Struct Mol Biol. 2006;13:594–602. doi: 10.1038/nsmb1108. [DOI] [PubMed] [Google Scholar]
- 10.Papanikou E, Karamanou S, Baud C, Frank M, Sianidis G, Keramisanou D, Kalodimos CG, Kuhn A, Economou A. Identification of the preprotein binding domain of SecA. J Biol Chem. 2005;280:43209–43217. doi: 10.1074/jbc.M509990200. [DOI] [PubMed] [Google Scholar]
- 11.Karamanou S, Vrontou E, Sianidis G, Baud C, Roos T, Kuhn A, Politou AS, Economou A. A molecular switch in SecA protein couples ATP hydrolysis to protein translocation. Mol Microbiol. 1999;34:1133–1145. doi: 10.1046/j.1365-2958.1999.01686.x. [DOI] [PubMed] [Google Scholar]
- 12.Breukink E, Nouwen N, van Raalte A, Mizushima S, Tommassen J, de Kruijff B. The C terminus of SecA is involved in both lipid binding and SecB binding. J Biol Chem. 1995;270:7902–7907. doi: 10.1074/jbc.270.14.7902. [DOI] [PubMed] [Google Scholar]
- 13.Triplett TL, Sgrignoli AR, Gao FB, Yang YB, Tai PC, Gierasch LM. Functional signal peptides bind a soluble N-terminal fragment of SecA and inhibit its ATPase activity. J Biol Chem. 2001;276:19648–19655. doi: 10.1074/jbc.M100098200. [DOI] [PubMed] [Google Scholar]
- 14.Gelis I, Bonvin AM, Keramisanou D, Koukaki M, Gouridis G, Karamanou S, Economou A, Kalodimos CG. Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell. 2007;131:756–769. doi: 10.1016/j.cell.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osborne AR, Clemons WM, Jr, Rapoport TA. A large conformational change of the translocation ATPase SecA. Proc Natl Acad Sci U S A. 2004;101:10937–10942. doi: 10.1073/pnas.0401742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.den Blaauwen T, van der Wolk JP, van der Does C, van Wely KH, Driessen AJ. Thermodynamics of nucleotide binding to NBS-I of the Bacillus subtilis preprotein translocase subunit SecA. FEBS Lett. 1999;458:145–150. doi: 10.1016/s0014-5793(99)01139-4. [DOI] [PubMed] [Google Scholar]
- 18.Benach J, Chou YT, Fak JJ, Itkin A, Nicolae DD, Smith PC, Wittrock G, Floyd DL, Golsaz CM, Gierasch LM, Hunt JF. Phospholipid-induced monomerization and signal-peptide-induced oligomerization of SecA. J Biol Chem. 2003;278:3628–3638. doi: 10.1074/jbc.M205992200. [DOI] [PubMed] [Google Scholar]
- 19.Fak JJ, Itkin A, Ciobanu DD, Lin EC, Song XJ, Chou YT, Gierasch LM, Hunt JF. Nucleotide exchange from the high-affinity ATP-binding site in SecA is the rate-limiting step in the ATPase cycle of the soluble enzyme and occurs through a specialized conformational state. Biochemistry. 2004;43:7307–7327. doi: 10.1021/bi0357208. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt M, Ding H, Ramamurthy V, Mukerji I, Oliver D. Nucleotide binding activity of SecA homodimer is conformationally regulated by temperature and altered by prlD and azi mutations. J Biol Chem. 2000;275:15440–15448. doi: 10.1074/jbc.M000605200. [DOI] [PubMed] [Google Scholar]
- 21.Song M, Kim H. Stability and solvent accessibility of SecA protein of Escherichia coli. J Biochem (Tokyo) 1997;122:1010–1018. doi: 10.1093/oxfordjournals.jbchem.a021840. [DOI] [PubMed] [Google Scholar]
- 22.Ding H, Mukerji I, Oliver D. Lipid and signal peptide-induced conformational changes within the C-domain of Escherichia coli SecA protein. Biochemistry. 2001;40:1835–1843. doi: 10.1021/bi002058w. [DOI] [PubMed] [Google Scholar]
- 23.Natale P, den Blaauwen T, van der Does C, Driessen AJ. Conformational state of the SecYEG-bound SecA probed by single tryptophan fluorescence spectroscopy. Biochemistry. 2005;44:6426–6432. doi: 10.1021/bi047488r. [DOI] [PubMed] [Google Scholar]
- 24.Zimmer J, Li W, Rapoport TA. A novel dimer interface and conformational changes revealed by an X-ray structure of B. subtilis SecA. J Mol Biol. 2006;364:259–265. doi: 10.1016/j.jmb.2006.08.044. [DOI] [PubMed] [Google Scholar]
- 25.Englander SW. Protein folding intermediates and pathways studied by hydrogen exchange. Annu Rev Biophys Biomol Struct. 2000;29:213–238. doi: 10.1146/annurev.biophys.29.1.213. [DOI] [PubMed] [Google Scholar]
- 26.Smock RG, Gierasch LM. Sending signals dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papanikou E, Karamanou S, Baud C, Sianidis G, Frank M, Economou A. Helicase Motif III in SecA is essential for coupling preprotein binding to translocation ATPase. EMBO Rep. 2004;5:807–811. doi: 10.1038/sj.embor.7400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 29.Morrisett JD, David JS, Pownall HJ, Gotto AM., Jr Interaction of an apolipoprotein (apoLP-alanine) with phosphatidylcholine. Biochemistry. 1973;12:1290–1299. doi: 10.1021/bi00731a008. [DOI] [PubMed] [Google Scholar]
- 30.Ramamurthy V, Oliver D. Topology of the integral membrane form of Escherichia coli SecA protein reveals multiple periplasmically exposed regions and modulation by ATP binding. J Biol Chem. 1997;272:23239–23246. doi: 10.1074/jbc.272.37.23239. [DOI] [PubMed] [Google Scholar]
- 31.Kusters I, van den Bogaart G, Kedrov A, Krasnikov V, Fulyani F, Poolman B, Driessen AJ. Quaternary structure of SecA in solution and bound to SecYEG probed at the single molecule level. Structure. 2011;19:430–439. doi: 10.1016/j.str.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 32.Or E, Rapoport T. Cross-linked SecA dimers are not functional in protein translocation. FEBS Lett. 2007;581:2616–2620. doi: 10.1016/j.febslet.2007.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, Na B, Yang H, Tai PC. Additional in vitro and in vivo evidence for SecA functioning as dimers in the membrane: dissociation into monomers is not essential for protein translocation in Escherichia coli. J Bacteriol. 2008;190:1413–1418. doi: 10.1128/JB.01633-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Keyzer J, van der Sluis EO, Spelbrink RE, Nijstad N, de Kruijff B, Nouwen N, van der Does C, Driessen AJ. Covalently Dimerized SecA Is Functional in Protein Translocation. J Biol Chem. 2005;280:35255–35260. doi: 10.1074/jbc.M506157200. [DOI] [PubMed] [Google Scholar]
- 35.Jilaveanu LB, Zito CR, Oliver D. Dimeric SecA is essential for protein translocation. Proc Natl Acad Sci U S A. 2005;102:7511–7516. doi: 10.1073/pnas.0502774102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duong F. Binding, activation and dissociation of the dimeric SecA ATPase at the dimeric SecYEG translocase. EMBO J. 2003;22:4375–4384. doi: 10.1093/emboj/cdg418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Or E, Boyd D, Gon S, Beckwith J, Rapoport T. The bacterial ATPase SecA functions as a monomer in protein translocation. J Biol Chem. 2005;280:9097–9105. doi: 10.1074/jbc.M413947200. [DOI] [PubMed] [Google Scholar]
- 38.Das S, Stivison E, Folta-Stogniew E, Oliver D. Reexamination of the role of the amino terminus of SecA in promoting its dimerization and functional state. J Bacteriol. 2008;190:7302–7307. doi: 10.1128/JB.00593-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sardis MF, Economou A. SecA: a tale of two protomers. Mol Microbiol. 2010;76:1070–1081. doi: 10.1111/j.1365-2958.2010.07176.x. [DOI] [PubMed] [Google Scholar]
- 40.Woodbury RL, Hardy SJ, Randall LL. Complex behavior in solution of homodimeric SecA. Protein Sci. 2002;11:875–882. doi: 10.1110/ps.4090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doyle SM, Braswell EH, Teschke CM. SecA folds via a dimeric intermediate. Biochemistry. 2000;39:11667–11676. doi: 10.1021/bi000299y. [DOI] [PubMed] [Google Scholar]
- 42.Wang Z, Jones JD, Rizo J, Gierasch LM. Membrane-bound conformation of a signal peptide: a transferred nuclear Overhauser effect analysis. Biochemistry. 1993;32:13991–13999. doi: 10.1021/bi00213a032. [DOI] [PubMed] [Google Scholar]
- 43.Jiang JX, London E. Involvement of denaturation-like changes in Pseudomonas exotoxin a hydrophobicity and membrane penetration determined by characterization of pH and thermal transitions. Roles of two distinct conformationally altered states. J Biol Chem. 1990;265:8636–8641. [PubMed] [Google Scholar]
- 44.Willard L, Ranjan A, Zhang H, Monzavi H, Boyko RF, Sykes BD, Wishart DS. VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 2003;31:3316–3319. doi: 10.1093/nar/gkg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kusters I, Driessen AJ. SecA, a remarkable nanomachine. Cell Mol Life Sci. 2011;68:2053–2066. doi: 10.1007/s00018-011-0681-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Papanikou E, Karamanou S, Economou A. Bacterial protein secretion through the translocase nanomachine. Nat Rev Microbiol. 2007;5:839–851. doi: 10.1038/nrmicro1771. [DOI] [PubMed] [Google Scholar]
- 47.Chou YT, Swain JF, Gierasch LM. Functionally significant mobile regions of Escherichia coli SecA ATPase identified by NMR. J Biol Chem. 2002;277:50985–50990. doi: 10.1074/jbc.M209237200. [DOI] [PubMed] [Google Scholar]
- 48.Cooper DB, Smith VF, Crane JM, Roth HC, Lilly AA, Randall LL. SecA, the motor of the secretion machine, binds diverse partners on one interactive surface. J Mol Biol. 2008;382:74–87. doi: 10.1016/j.jmb.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Economou A, Wickner W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 50.Snyders S, Ramamurthy V, Oliver D. Identification of a region of interaction between Escherichia coli SecA and SecY proteins. J Biol Chem. 1997;272:11302–11306. doi: 10.1074/jbc.272.17.11302. [DOI] [PubMed] [Google Scholar]
- 51.Price A, Economou A, Duong F, Wickner W. Separable ATPase and membrane insertion domains of the SecA subunit of preprotein translocase. J Biol Chem. 1996;271:31580–31584. doi: 10.1074/jbc.271.49.31580. [DOI] [PubMed] [Google Scholar]
- 52.Musial-Siwek M, Rusch SL, Kendall DA. Selective photoaffinity labeling identifies the signal peptide binding domain on SecA. J Mol Biol. 2007;365:637–648. doi: 10.1016/j.jmb.2006.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Auclair SM, Moses JP, Musial-Siwek M, Kendall DA, Oliver DB, Mukerji I. Mapping of the signal peptide-binding domain of Escherichia coli SecA using Forster resonance energy transfer. Biochemistry. 2010;49:782–792. doi: 10.1021/bi901446r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grady LM, Michtavy J, Oliver DB. Characterization of the Escherichia coli SecA Signal Peptide-binding Site. J Bacteriol. doi: 10.1128/JB.06150-11. e-pub ahead of print Nov. 4, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Erlandson KJ, Miller SB, Nam Y, Osborne AR, Zimmer J, Rapoport TA. A role for the two-helix finger of the SecA ATPase in protein translocation. Nature. 2008;455:984–987. doi: 10.1038/nature07439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsukazaki T, Mori H, Fukai S, Ishitani R, Mori T, Dohmae N, Perederina A, Sugita Y, Vassylyev DG, Ito K, Nureki O. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature. 2008;455:988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.