Abstract
We describe the first case of Japanese spotted fever and the first isolate of spotted fever group rickettsia from a patient in South Korea. The isolated rickettsia from the patient was identified as Rickettsia japonica by analysis of the nucleotide sequences of 16S rRNA, gltA, ompA, ompB, and sca4 genes.
Keywords: Rickettsia japonica, Japanese spotted fever, isolation, Korea
The Rickettsiaceae family comprises obligate intracellular bacteria and contains 2 genera: Rickettsia (typhus group and spotted fever group[SFG]) and Orientia. Scrub typhus caused by O. tsutsugamushi is the most prevalent rickettsiosis in South Korea (1). SFG rickettsiae were first demonstrated to exist in South Korea by the isolation of Rickettsia akari from the Korean vole in 1957 (2). However, not a single case of rickettsialpox or other SFG rickettsiosis has been documented in Korea. Recently, evidence for the existence of SFG rickettsiosis has been provided by serologic survey and DNA detection in South Korea (3–5). Moreover, SFG rickettsiae displaying homology with R. japonica and R. rickettsii were detected in Haemaphysalis ticks by polymerase chain reaction analysis of the citrate synthase (gltA) gene, 16S rRNA, and ompA genes (6). However, no human cases of SFG rickettsiosis have been reported, and no SFG strain has been isolated from a person so far.
In this report, we present the first documentation of Japanese spotted fever in South Korea and isolation of R. japonica. To our knowledge, this is the first report of an SFG rickettsia isolated from a patient in South Korea.
Case Report
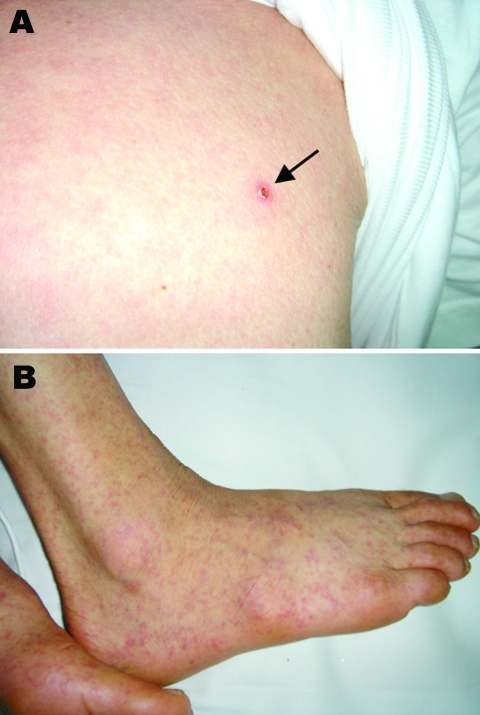
A 65-year-old farmer was admitted to a hospital in Incheon, South Korea, on July 9, 2004; he had experienced fever, back pain, and myalgia for 5 days before admission. He lived in Mueui Island, ≈20 km east of Incheon. On physical examination, he had fever of 38.6°C, cervical and axillary lymphadenopathies, and a maculopapular rash. An eschar, which was smaller and more shallow than those of scrub typhus, was noticed on the chest wall (Figure 1). Laboratory studies showed a hemoglobin level of 7.7 mmol/L, a leukocyte count of 8 × 109/L, and a platelet count of 87 × 109/L. The patient was treated with oral doxycycline (200 mg/day), but the fever persisted during the treatment. On the third hospital day, petechiae developed on the trunk and extremities, including palms and soles (Figure 1). The leukocyte count increased to 11.2 × 109/L, and the platelet count decreased further to 32 × 109/L. He also showed confusion, irritability, and radiographic evidence of interstitial pneumonitis. The patient was then given azithromycin (500 mg/day intravenously) instead of oral doxycycline because of the possibility that he was infected with doxycycline-resistant O. tsutsugamushi. His fever resolved during next 5 days and he was discharged.
Figure 1.
Skin findings of the patient. A small eschar (arrow) on the chest with erythematous rash (A) and petechiae (B) were observed.
The serum samples were tested for antibody against O. tsutsugamushi (Boryong), R. typhi (Wilmington), and the isolated strain (Inha1) by using the indirect fluorescent-antibody (IFA) test. The serum specimen taken on the day of admission was negative for antibodies against all Rickettsia spp. by the IFA test. The serum sample taken after 18 days was sent to the national reference laboratory (Division of Rickettsial Diseases, Department of Bacteriology, National Institute of Health, Seoul, Republic of Korea) and was reported to be positive against R. japonica with a titer of 1,024. The convalescent-phase serum sample, taken on the 30th day after admission, was positive for antibodies against the isolated strain (Inha1) with a titer of 5,120.
To isolate the pathogen, a few drops of blood taken on the day of admission were added directly to the monolayer cultures of ECV304 cells, a spontaneously transformed cell line derived from human umbilical vein endothelial cells (obtained from S.Y. Kim) (7). After incubation for 24 h, monolayers of ECV304 were maintained in M199 medium containing 10% fetal calf serum and observed daily with an inverted microscope. The infected cells exhibited few cytopathologic changes, displaying only a few rounded cells. On day 28, IFA staining was done by using the convalescent-phase serum of the patient to visualize the rickettsiae. Many intracellular bacteria were observed inside the cells; they were not seen in the staining with control serum. From these results, we tentatively identified our isolated bacterium (strain Inha1), as a member of SFG rickettsiae.
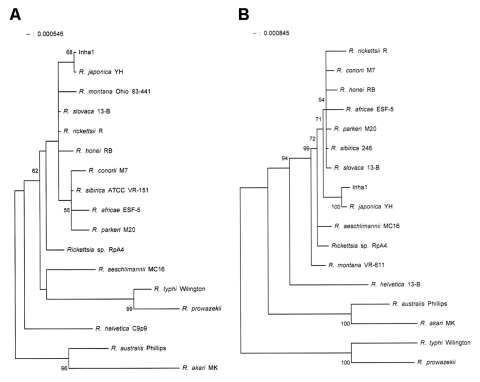
Amplification and sequencing of the 16S rRNA, gltA, ompA, ompB, and sca4 genes were as described by Lee et al. (6). Sequences were aligned by using the multiple-alignment algorithm in the MegAlign software package (Windows version 3.12e; DNASTAR, Madison, WI, USA), and phylogenetic trees were constructed by the neighbor-joining method with the MEGA program (8). Sequences were compared to the sequences with those of 16 reference strains of SFG rickettsiae (9–11). The nucleotide sequence (AY743328) of the 16S rRNA gene was identical to that of R. japonica YH. Inha1 demonstrated 16S rRNA sequence similarities of 95.9%–99.7% to the other strains of SFG rickettsiae. In the phylogenetic tree, Inha1 formed a cluster with R. japonica YH, separate from the other strains of SFG rickettsiae (Figure 2). The nucleotide sequence (AY743327) of the gltA gene of Inha1 showed a high similarity (99.8%) with that of R. japonica YH. The sequence similarities of Inha1 to the other strains of SFG rickettsiae were 93.6%–99.1%.
Figure 2.
Phylogenetic tree based on 16S rRNA gene sequences (A) and gltA gene sequences (B) of the isolated rickettsial strain (Inha1). The phylogenetic tree was constructed by the neighbor-joining method with MEGA software. Bootstrap analysis was performed with 100 replicates. The GenBank accession numbers for the 16S rRNA gene sequences included are as follows: Rickettsia japonica, L36213; R. rickettsii, L36217; Rickettsia sp. RpA4, AF120026; R. aeschlimannii, U74757; R. africae, L36098; R. conorii, AE008647; R. honei, AF060705; R. montana, U11016; R. parkeri, U12461; R. sibirica, D38628; R. slovaca, L36224; R. helvetica, L36212; R. australis, U12459; R. akari, L36099; R. typhi, U12463; and R. prowazekii, M21789. The GenBank accession numbers for the gltA gene sequences of these bacteria are U59724, U59729, AF120029, U59722, U59733, U59730, U59726, U74756, U59732, U59734, U59725, U59723, U59718, U59717, U59714, and M17149, respectively.
The nucleotide sequence of the ompA, ompB, and sca4 of Inha1 strain also showed a high similarity to R. japonica YH and the sequence similarities to R. japonica YH were 100, 99.9, and 99.9%, respectively.
Conclusions
To identify the isolate at species level, we determined the sequence of five genes that have been used for the phylogenetic classification of rickettsiae (9–14). The sequences are identical or highly homologous to those of R. japonica. Although the sequence similarity of the gltA gene of Inha1 strain to R. japonica YH was 99.8%, the other 4 genes show sufficient similarity to fulfill the criteria suggested by Fournier et al. (9).
R. japonica was first isolated in Japan from human patients and ticks (15). The isolation of R. japonica in South Korea is not surprising because of the geographic proximity of South Korea to Japan. Furthermore, among 4 SFG rickettsiae detected in Korean ticks, 3 strains were highly homologous to R. japonica (6). Therefore, R. japonica may be the most dominant SFG rickettsiae distributed in South Korea, and the geographic distribution of R. japonica may be more widespread than previously known. However, other SFG rickettsiae, including R. sibirica, may be present in northeastern Asia. To clarify this issue, more strains of SFG rickettsiae must be isolated from other locations within SouthKorea.
Acknowledgments
This work was supported by a grant from Korea Center for Disease Control and Prevention in 2005.
Biography
Dr Chung is a member of the Korean Society of Infectious Diseases. His primary research interests are infections by intracellular organisms, especially Rickettsia and Plasmodium falciparum.
Footnotes
Suggested citation for this article: Chung M-H, Lee S-H, Kim M-J, Lee J-H, Kim E-S, Lee J-S, et al. Japanese spotted fever, Korea. Emerg Infect Dis [serial on the Internet]. 2006 Jul [date cited]. http://dx.doi.org/10.3201/eid1207.051372
References
- 1.Seong SY, Choi MS, Kim IS. Orientia tsutsugamushi infection: overview and immune responses. Microbes Infect. 2001;3:11–21. 10.1016/S1286-4579(00)01352-6 [DOI] [PubMed] [Google Scholar]
- 2.Jackson EB, Danauskas JX, Coale MC, Smadel JE. Recovery of Rickettsia akari from the Korean vole Microtus fortis pelliceus. Am J Hyg. 1957;66:301–8. [DOI] [PubMed] [Google Scholar]
- 3.Lee KR, Baek LJ, Song KJ, Woo KD, Lee YJ, Lee HW. Seroepidemiologic study of hemorrhagic fever with renal syndrome, scrub typhus, murine typhus, spotted fever and leptospirosis in Korea, 1991. Korean University Medical Journal. 1994;31:73–88. [Google Scholar]
- 4.Jang WJ, Kim JH, Choi YJ, Jung KD, Kim YG, Lee SH, et al. First serologic evidence of human spotted fever group rickettsiosis in Korea. J Clin Microbiol. 2004;42:2310–3. 10.1128/JCM.42.5.2310-2313.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi YJ, Jang WJ, Ryu JS, Lee SH, Park KH, Pail HS, et al. Spotted fever group and typhus group rickettsioses in humans, South Korea. Emerg Infect Dis. 2005;11:237–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JH, Park H, Jung KD, Jang WJ, Koh ES, Kang SS, et al. Identification of the spotted fever group rickettsiae detected from Haemaphysalis longicornis in Korea. Microbiol Immunol. 2003;47:301–4. [DOI] [PubMed] [Google Scholar]
- 7.Shin EY, Lee JY, Park MK, Chin YH, Jeong GB, Kim SY, et al. Overexpressed alpha3beta1 and constitutively activated extracellular signal-regulated kinase modulate the angiogenic properties of ECV304 cells. Mol Cells. 1999;9:138–45. [PubMed] [Google Scholar]
- 8.Kumar S, Tamura K, Masatoshi N. MEGA: molecular evolutionary genetics analysis, version 1.01. University Park (PA): Pennsylvania State University; 1993. [Google Scholar]
- 9.Fournier PE, Dumler JS, Greub G, Zhang J, Wu Y, Raoult D. Gene sequence-based criteria for identification of new rickettsia isolates and description of Rickettsia heilongjiangensis sp. nov. J Clin Microbiol. 2003;41:5456–65. 10.1128/JCM.41.12.5456-5465.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roux V, Raoult D. Phylogenetic analysis of the genus Rickettsia by 16S rDNA sequencing. Res Microbiol. 1995;146:385–96. 10.1016/0923-2508(96)80284-1 [DOI] [PubMed] [Google Scholar]
- 11.Roux V, Rydkina E, Eremeeva M, Raoult D. Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the rickettsiae. Int J Syst Bacteriol. 1997;47:252–61. 10.1099/00207713-47-2-252 [DOI] [PubMed] [Google Scholar]
- 12.Roux V, Raoult D. Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer-membrane protein rOmpB (ompB). Int J Syst Evol Microbiol. 2000;50:1449–55. 10.1099/00207713-50-4-1449 [DOI] [PubMed] [Google Scholar]
- 13.Sekeyova Z, Roux V, Raoult D. Phylogeny of Rickettsia spp. inferred by comparing sequences of gene D, which encodes an intracytoplasmic protein. Int J Syst Evol Microbiol. 2001;51:1353–60. [DOI] [PubMed] [Google Scholar]
- 14.Fournier PE, Roux V, Raoult D. Phylogenetic analysis of spotted fever group rickettsiae by study of the outer surface protein rOmpA. Int J Syst Bacteriol. 1998;48:839–49. 10.1099/00207713-48-3-839 [DOI] [PubMed] [Google Scholar]
- 15.Mahara F. Japanese spotted fever: report of 31 cases and review of the literature. Emerg Infect Dis. 1997;3:105–11. 10.3201/eid0302.970203 [DOI] [PMC free article] [PubMed] [Google Scholar]