Short abstract
Recent microarray analysis has revealed multiple levels of genomic sensitivity to glucose and highlighted the power of genome-wide analysis to detect cellular responses to minute environmental changes in the yeast Saccharomyces cerevisiae.
Abstract
The yeast Saccharomyces cerevisiae shows a great variety of cellular responses to glucose via several glucose-sensing and signaling pathways. Recent microarray analysis has revealed multiple levels of genomic sensitivity to glucose and highlighted the power of genome-wide analysis to detect cellular responses to minute environmental changes.
Over the years, the yeast Saccharomyces cerevisiae has consolidated its position as an excellent model for the analysis of signal transduction pathways and their control of metabolic pathways, stress responses, growth and differentiation. In particular, the molecular mechanisms underlying the metabolic responses to different nutrient conditions have been studied extensively. With the elucidation of the complete yeast genome and the introduction of genome-wide expression analysis, the impact of distinct nutrient conditions on transcriptional responses can be explored on a genome-wide scale. Because of the powerful set of genetic and genomic tools that are available, yeast is the perfect organism for combining genome-wide studies of gene expression with the experimental elucidation of signaling networks; such studies have, among other findings, detected changes in expression of many genes of unknown function. Recent work has extended genome-wide studies of nutrient regulation to include the yeast response to very low concentrations of glucose [1].
Glucose signaling pathways regulating transcription
S. cerevisiae has a strong preference for glucose as its carbon source. This is reflected in the preferential conversion of glucose into ethanol, even under aerobic conditions, and in the preferential utilization of glucose when cells are grown in mixtures of glucose and other sugars. In addition, growth on glucose is faster than on other carbon sources and a range of glucose-dependent effects can be observed on properties correlated with growth and stationary phase. During its evolution, S. cerevisiae has gained sophisticated regulatory mechanisms to sense fluctuating levels of glucose, and as a result has many different glucose-sensing and signaling pathways [2]. Three of these pathways are particularly well understood. The first is the main glucose repression pathway which controls, for example, respiration and the preferential utilization of glucose [3]; the second is the Snf3/Rgt2 pathway, which controls the expression of glucose carriers [4]; and the third is the Gpr1/Gpa2 pathway, which controls the synthesis of cAMP and activity of the protein kinase A (PKA) pathway [5] (Figure 1). In addition, many other glucose-induced regulatory effects have been described for which the glucose-sensing mechanism(s) and signaling pathway(s) are unclear. Glucose sensing via these pathways affects specific targets at the post-translational and/or transcriptional level, and a variety of transcription factors have been identified as targets of the glucose-signaling pathways (summarized in Table 1).
Figure 1.
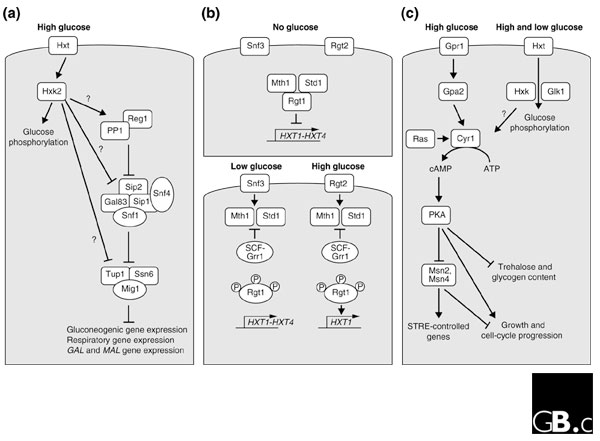
A simplified schematic representation of the three well-characterized glucose-response pathways in S. cerevisiae. (a) The main glucose repression pathway. In response to high glucose concentrations, the complex containing the Snf1 kinase inhibits the Mig1 repressor-containing complex and thus represses genes involved in respiration, gluconeogenesis and the metabolism of alternative carbon sources, such as galactose (GAL genes) and maltose (MAL genes). Protein phosphatase type 1 (PP1) acts in a complex with Reg1 to down-regulate Snf1 in low-glucose conditions. Glucose phosphorylation by Hxk2 is required for this pathway, but the step at which it acts is not known. (b) The Snf3/Rgt2 glucose-sensing pathway. In the absence of glucose, Rgt1 acts in a complex with Std1 and Mth1 as a transcriptional repressor of the HXT1-HXT4 genes. When glucose is present, the transcription factor Rgt1 is inactivated through SCF-Grr1-mediated inactivation and degradation of Mth1 and Std1, and hyperphosphorylation by an unknown kinase, resulting in dissociation of Rgt1 from the HXT promoters. Snf3 triggers the induction of HXT1-HXT4 in response to low glucose concentrations. High glucose concentrations further enhance HXT1 expression through Rgt2 in a process that involves conversion of Rgt1 into a transcriptional activator. (c) The Gpr1/Gpa2 glucose-sensing pathway. High glucose concentrations activate cAMP synthesis by the adenylate cyclase Cyr1 (which is dependent on Ras) through the Gpr1/Gpa2 G-protein-coupled receptor system in a glucose-phosphorylation-dependent manner. The resulting activation of protein kinase A (PKA) affects a wide variety of target genes involved in, for example, carbon metabolism and stress resistance. Some of these effects are mediated by the Msn2 and Msn4 transcription factors. STRE, stress-response element. See text for further details.
Table 1.
DNA sequences bound by glucose-regulated transcription factors
| Transcription factor | Binding site* | Main function | References |
| Cat8 | YCCRTYSRNCCG | Activator of gluconeogenic genes | [27] |
| Sip4 | YCCRTYSRNCCG | Activator of gluconeogenic genes | [28] |
| Mig1/Mig2 | T(C/G)(C/T)GGGG | Repressors of SUC, MAL and GAL genes | [29] |
| Gcr1 | CTTCC | Activator of glycolytic genes | [30] |
| Rgt1 | CGGANNA | Regulator of HXT gene expression | [31] |
| Nrg1/Nrg2 | Not determined | Repressors of SUC, GAL and FLO11 genes, interact with Snf1 | [32-34] |
| Azf1 | AAGAAAAA | Induction of CLN3 gene expression | [23] |
| Msn2/Msn4 | CCCCT | Induction of STRE-regulated genes | [20] |
| Gis1 | T(T/A)AGGGAT | Activator of post-diauxic shift (PDS) genes | [21] |
| Pop2-Ccr4-Not | Not determined | Global transcriptional regulator of growth-, metabolism- and stationary phase related proteins | [35] |
*Within DNA sequences, Y denotes a pyrimidine and R a purine. Abbreviation: STRE, stress-response element.
The first of the three well-characterized pathways, the main glucose repression pathway (Figure 1a) has been studied extensively for many years and is responsible for the repression - when glucose is available - of genes involved in respiration, gluconeogenesis and the metabolism of alternative carbon sources [3,6]. A central glucose-responsive repressor complex, made up of the Mig1, Ssn6 and Tup1 proteins, represses the transcription of gluconeogenic genes, respiratory genes and genes encoding enzymes involved in the utilization of galactose and maltose (Table 1). In the presence of glucose, the repressor complex is inhibited by a complex comprised of the protein kinase Snf1, the associated regulatory β subunits Sip1, Sip2 or Gal83, and the activating subunit Snf4 [7,8]. The Snf1 kinase was recently shown to be activated through phosphorylation of a conserved threonine residue in its activation loop by the Pak1, Tos3 and Elm1 kinases [9,10]. Down-regulation of Snf1 in low-glucose conditions is catalyzed by protein phosphatase type 1 (PP1), encoded by GLC7, which acts in a complex with the regulatory subunit Reg1/Hex2 [11]. Although several glucose effects on the Snf1 complex have been identified, the precise nature of the glucose-sensing mechanism involved in glucose repression still remains unclear. Glucose phosphorylation by Hxk2 is required for glucose repression, but whether this links to Snf1 function and/or to direct involvement of Hxk2 in transcriptional regulation in the nucleus remains to be elucidated [12].
The second of the well-studied glucose-response pathways, the Snf3/Rgt2 glucose-sensing pathway (Figure 1b), is involved in controlling the expression of glucose transporter genes [4]. High glucose concentrations induce the HXT1 gene through Rgt2, while low glucose concentrations induce the HXT1-HXT4 genes through Snf3. S. cerevisiae has seven functional glucose transporter genes (HXT1-HXT7) that are well characterized and many more for which the nature and physiological role is rather unclear [4,13]; the expression of many of these genes depends on the concentration of glucose in the medium. The Snf3 and Rgt2 proteins are glucose-carrier homologs but are apparently unable to carry out active glucose transport; instead, Snf3 and Rgt2 specifically mediate regulation of the active glucose carriers by low and high glucose concentrations, respectively [14,15]. In the absence of glucose, Mth1 and Std1 stabilize the Rgt1-Ssn6-Tupi repressor-corepressor complex. When glucose is present, the transcription factor Rgt1 is inactivated through SCF-Grr1-mediated inactivation and degradation of Mth1 and Std1, and hyperphosphorylation by an unknown kinase, resulting in dissociation of Rgt1 from the HXT promoters [16,17]. Remarkably, whereas Rgt1 represses HXT gene expression in the absence of glucose, at high glucose concentrations Rgt1 is converted to an activator that is required for maximal expression of the low-affinity glucose transporter HXT1 (Table 1) [18]. The HXT2, HXT4, HXT6 and SNF3 genes are repressed by Mig1 via the main glucose-repression pathway when glucose is abundant [13].
When cells that have been growing on nonfermentable carbon sources have glucose added to the medium, the third well-studied pathway, the Gpr1/Gpa2 glucose-sensing pathway (Figure 1c), is involved in the rapid increase in cAMP levels that is mediated by the adenylate cyclase Cyr1; the activity of Cyr1 in yeast depends on the Ras proteins. Activation of the Gpr1/Gpa2 pathway requires both extracellular glucose detection by the Gpr1-Gpa2 G-protein-coupled receptor system and intracellular glucose phosphorylation by one of the sugar kinases Hxk1, Hxk2 or Glk1 [2]. Subsequent activation of PKA affects the transcriptional regulation of genes involved in sugar metabolism, stress resistance and growth [5]. PKA mediates the repression of stress-resistance genes through the inactivation of the transcription factors Msn2 and Msn4. When activated, Msn2 and Msn4 are translocated from the cytoplasm to the nucleus [19], where they act through stress response elements (STREs) in the promoters of responsive genes (Table 1) [20]. The related post-diauxic shift element (Table 1) is affected in a similar way by the Gis1 transcription factor, which acts downstream in the Gpr1/Gpa2-PKA pathway [21].
Apart from these three well-characterized glucose-sensing pathways, other glucose-induced regulatory phenomena have been identified for which the signaling pathway(s) remain unclear. Examples include the glucose-induced upshift of ribosomal protein genes and CLN3 expression; ribosomal gene expression seems to involve, to some extent, the PKA pathway [22], while induction of the CLN3 gene requires the Azf1 transcription factor (Table 1) [23], but does not require active growth, is not blocked by mutations in the Ras/cAMP pathway or in the RGT2, SNF3, HXK2 or MIG1 genes and apparently depends on active glucose metabolism. The Yak1 kinase is also regulated by an unknown glucose-sensing pathway [24]; when glucose is exhausted, Yak1 accumulates in the nucleus and phosphorylates Pop2, a subunit of the Ccr4-Not complex that regulates the transcription of many genes involved in control of growth, metabolism and proper entry into stationary phase.
In addition to these complex transcriptional regulation pathways, glucose also controls mRNA stability, translation rate, protein turnover and enzyme activity by a range of different mechanisms. Most of these signaling pathways and mechanisms have been studied using classical methods, taking one or a few genes at a time, and glucose signaling in yeast was already recognised as being 'very complex' before the advent of genomic approaches.
Genome-wide analysis of responses to glucose
The introduction of genome-wide gene-expression analysis has extended the scope of glucose regulation to much larger parts of the genome than were previously implicated [25]. The recent work by the group of Al Brown [1] has added another dimension to glucose regulation by revealing different levels of genomic sensitivity to glucose in yeast. They have shown that very low (0.01%), low (0.1%) and high (1.0%) glucose concentrations elicit strikingly different responses in the genome-wide expression pattern. They confirmed the extreme sensitivity of yeast to glucose by showing that genes involved in carbon metabolism are strongly upregulated in the presence of only 0.01% glucose. In addition, the strong differential expression profiles of the different hexose transporter genes (HXT1-HXT7) was evident from the transcript-profiling data [1]. Remarkably, the expression of not only gluconeogenic genes, but also genes involved in the glyoxylate and tricarboxylic acid cycles, is very sensitive to these minute amounts of glucose. This shows that even at very low concentrations of glucose, yeast cells start to adapt to fermentative growth, although respiration is still active.
Glucose-controlled mRNA stability
Another novel aspect of the recent work from the Brown group is the influence of different glucose signals on ribosome biogenesis [1]. On the one hand, the microarray data confirmed that ribosomal gene expression is induced in response to high, but not low, glucose signals. On the other hand, mRNA stability assays revealed that glucose, even at concentrations as low as 0.01%, transiently stabilizes the mRNAs for ribosomal proteins. Ribosomal protein biosynthesis requires a major investment of cellular resources because ribosomal protein mRNAs are abundantly present in fast-growing cells and yet have short half-lives. The stabilization of the mRNAs for ribosomal proteins by a low-glucose signal, therefore, appears to be an 'economical' means by which to maintain proliferative capacity in a nutrient-limiting regime without synthesis of new ribosomal proteins. Typically for glucose regulation, this temporary stabilization of ribosomal protein mRNAs is not controlled by just one signaling pathway; rather, the mRNAs for different ribosomal subunits appear to be regulated by distinct glucose-signaling mechanisms.
The genes involved in gluconeogenesis are strongly repressed at glucose concentrations as low as 0.01%. This response to very low glucose concentrations is mediated by two regulatory phenomena: transcriptional repression and accelerated mRNA degradation [26]. Yin et al. [26] also showed that the degradation of gluconeogenic mRNA is dependent on the high-affinity glucose sensor Snf3, the phosphorylation of internalized sugars by hexokinases, the protein phosphatase-1 subunit Reg1 and the serine/threonine protein kinase Ume5.
In conclusion, genome-wide transcription analyses will broaden our understanding of the global response of a yeast cell to fluctuating nutrient levels. The glucose response in yeast is an excellent illustration of the complexity of transcriptional regulation by nutrients. Because of this complexity, microarray experiments will be very useful in identifying groups of genes regulated through the different signaling pathways. In addition, this technique will be indispensable for identifying the scope of transcriptional regulation and the extent of overlap between the different signaling pathways, in particular as a function of nutrient concentration. The remarkable sensitivity of the yeast transcriptome to very low glucose concentrations has revealed a new field of application for microarrays: identification of cellular responses to minute changes in the environmental conditions. Microarray analysis appears to provide unprecedented power to identify even the smallest cellular responses.
References
- Yin Z, Wilson S, Hauser NC, Tournu H, Hoheisel JD, Brown AJP. Glucose triggers different global responses in yeast, depending on the strength of the signal, and transiently stabilizes ribosomal protein mRNAs. Mol Microbiol. 2003;48:713–724. doi: 10.1046/j.1365-2958.2003.03478.x. [DOI] [PubMed] [Google Scholar]
- Rolland F, Winderickx J, Thevelein JM. Glucose-sensing and -signaling mechanisms in yeast. FEMS Yeast Res. 2002;2:183–201. doi: 10.1016/S1567-1356(02)00046-6. [DOI] [PubMed] [Google Scholar]
- Gancedo JM. Yeast carbon catabolite repression. Microbiol Mol Biol Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan S, Johnston M. Function and regulation of yeast hexose transporters. Microbiol Mol Biol Rev. 1999;63:554–569. doi: 10.1128/mmbr.63.3.554-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein JM, de Winde JH. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;33:904–918. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- Vincent O, Carlson M. Gal83 mediates the interaction of the Snf1 kinase complex with the transcription activator Sip4. EMBO J. 1999;18:6672–6681. doi: 10.1093/emboj/18.23.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MC, McCartney RR. Beta-subunits of Snf1 kinase are required for kinase function and substrate definition. EMBO J. 2000;19:4936–4943. doi: 10.1093/emboj/19.18.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SP, Leiper FC, Woods A, Carling D, Carlson M. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc Natl Acad Sci USA. 2003;100:8839–8843. doi: 10.1073/pnas.1533136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath N, McCartney RR, Schmidt MC. Yeast Pak1 kinase associates with and activates Snf1. Mol Cell Biol. 2003;23:3909–3917. doi: 10.1128/MCB.23.11.3909-3917.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz P, Alms GR, Haystead TA, Carlson M. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol Cell Biol. 2000;20:1321–1328. doi: 10.1128/MCB.20.4.1321-1328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno F, Herrero P. The hexokinase 2-dependent glucose signal transduction pathway of Saccharomyces cerevisiae. FEMS Microbiol Rev. 2002;26:83–90. doi: 10.1016/S0168-6445(01)00077-8. [DOI] [PubMed] [Google Scholar]
- Boles E, Hollenberg CP. The molecular genetics of hexose transport in yeasts. FEMS Microbiol Rev. 1997;21:85–111. doi: 10.1016/S0168-6445(97)00052-1. [DOI] [PubMed] [Google Scholar]
- Özcan S, Dover J, Rosenwald AG, Wolfl S, Johnston M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci USA. 1996;93:12428–12432. doi: 10.1073/pnas.93.22.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan S, Dover J, Johnston M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:2566–2573. doi: 10.1093/emboj/17.9.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FN, Johnston M. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J. 1997;16:5629–5638. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MC, McCartney RR, Zhang X, Tillman TS, Solimeo H, Wolfl S, Almonte C, Watkins SC. Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4561–4571. doi: 10.1128/mcb.19.7.4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley AL, Lakshmanan J, Aryal BK, Özcan S. Glucose-mediated phosphorylation converts the transcription factor Rgt1 from a repressor to an activator. J Biol Chem. 2003;278:10322–10327. doi: 10.1074/jbc.M212802200. [DOI] [PubMed] [Google Scholar]
- Görner W, Durchschlag E, Martinez-Pastor M, Estruch P, Ammerer G, Hamilton B, Ruis H, Schüller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Pastor MT, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- Pedruzzi I, Burckert N, Egger P, De Virgilio C. Saccharomyces cerevisiae Ras/cAMP pathway controls post-diauxic shift element-dependent transcription through the zinc finger protein Gis1. EMBO J. 2000;19:2569–2579. doi: 10.1093/emboj/19.11.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C, Struhl K. Protein kinase A mediates growth-regulated expression of yeast ribosomal protein genes by modulating RAP1 transcriptional activity. Mol Cell Biol. 1994;14:1920–1928. doi: 10.1128/mcb.14.3.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb LL, Hall DD, Heideman W. AZF1 is a glucose-dependent positive regulator of CLN3 transcription in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:1607–1614. doi: 10.1128/MCB.22.5.1607-1614.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya H, Shimizu-Yoshida Y, Omori A, Iwashita S, Katoh M, Sakai A. Yak1p, a DYRK family kinase, translocates to the nucleus and phosphorylates yeast Pop2p in response to a glucose signal. Genes Dev. 2001;15:1217–1228. doi: 10.1101/gad.884001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Yin ZK, Hatton L, Brown AJP. Differential post-transcriptional regulation of yeast mRNAs in response to high and low glucose concentrations. Mol Microbiol. 2000;35:553–565. doi: 10.1046/j.1365-2958.2000.01723.x. [DOI] [PubMed] [Google Scholar]
- Rahner A, Scholer A, Martens E, Gollwitzer B, Schuller HJ. Dual influence of the yeast Cat1p (Snf1p) protein kinase on carbon source-dependent transcriptional activation of gluconeogenic genes by the regulatory gene CAT8. Nucleic Acids Res. 1996;24:2331–2337. doi: 10.1093/nar/24.12.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage P, Yang X, Carlson M. Yeast SNF1 protein kinase interacts with SIP4, a C6 zinc cluster transcriptional activator: a new role for SNF1 in the glucose response. Mol Cell Biol. 1996;16:1921–1928. doi: 10.1128/mcb.16.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehlin JO, Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 1990;9:2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huie MA, Baker HV. DNA-binding properties of the yeast transcriptional activator, Gcr1p. Yeast. 1996;12:307–317. doi: 10.1002/(SICI)1097-0061(19960330)12:4<307::AID-YEA912>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Kim JH, Polish J, Johnston M. Specificity and regulation of DNA binding by the yeast glucose transporter gene repressor Rgt1. Mol Cell Biol. 2003;23:5208–5216. doi: 10.1128/MCB.23.15.5208-5216.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchin S, Vyas VK, Carlson M. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol Cell Biol. 2002;22:3994–4000. doi: 10.1128/MCB.22.12.3994-4000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Winston F. NRG1 is required for glucose repression of the SUC2 and GAL genes of Saccharomyces cerevisiae. BMC Genet. 2001;2:5. doi: 10.1186/1471-2156-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas VK, Kuchin S, Carlson M. Interaction of the repressors Nrg1 and Nrg2 with the Snf1 protein kinase in Saccharomyces cerevisiae. Genetics. 2001;158:563–572. doi: 10.1093/genetics/158.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Salvadore C, Chiang YC, Collart MA, Liu HY, Denis CL. The Ccr4 and Caf1 proteins of the Ccr4-not complex are physically and functionally separated from Not2, Not4, and Not5. Mol Cell Biol. 1999;19:6642–6651. doi: 10.1128/mcb.19.10.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]


