Abstract
Aims
Circulating microRNAs (miRNAs) have attracted major interest as biomarkers for cardiovascular diseases. Since RNases are abundant in circulating blood, there needs to be a mechanism protecting miRNAs from degradation. We hypothesized that microparticles (MP) represent protective transport vehicles for miRNAs and that these are specifically packaged by their maternal cells.
Methods and results
Conventional plasma preparations, such as the ones used for biomarker detection, are shown to contain substantial numbers of platelet-, leucocyte-, and endothelial cell-derived MP. To analyse the widest spectrum of miRNAs, Next Generation Sequencing was used to assess miRNA profiles of MP and their corresponding stimulated and non-stimulated cells of origin. THP-1 (monocytic origin) and human umbilical vein endothelial cell (HUVEC) MP were used for representing circulating MP at a high purity. miRNA profiles of MP differed significantly from those of stimulated and non-stimulated maternal THP-1 cells and HUVECs, respectively. Quantitative reverse transcription–polymerase chain reaction of miRNAs which have been associated with cardiovascular diseases also demonstrated significant differences in miRNA profiles between platelets and their MP. Notably, the main fraction of miRNA in plasma was localized in MP. Furthermore, miRNA profiles of MP differed significantly between patients with stable and unstable coronary artery disease.
Conclusion
Circulating MP represent transport vehicles for large numbers of specific miRNAs, which have been associated with cardiovascular diseases. miRNA profiles of MP are significantly different from their maternal cells, indicating an active mechanism of selective ‘packaging’ from cells into MP. These findings describe an interesting mechanism for transferring gene-regulatory function from MP-releasing cells to target cells via MP circulating in blood.
Keywords: MicroRNA, Microparticles, Vascular inflammation
1. Introduction
Microparticles (MP) are small (<1.1 µm), pro-inflammatory vesicles released by various cell types (e.g. leucocytes, endothelial cells, platelets) in a tightly regulated process. They contain cytoplasm and surface markers of their cells of origin.1,2 Once released into the circulation, MP bind and fuse with their target cells through receptor–ligand interactions, thereby acting as biological vectors mediating vascular inflammation and coagulation.3,4 Therefore, MP have been shown to play a pivotal role in several cardiovascular diseases.5,6 Increasing evidence indicates that inflammatory and pro-coagulatory effects of MP on their target cells are caused by a specific lipid composition (e.g. by phosphatidylserine) as well as by the transfer of inflammatory cell components from their cells of origin.1 However, it was also shown that MP transport mRNA thereby affecting protein expression of their target cells.7 Recent progress in the understanding of microRNA (miRNA) has prompted the questions of whether MP also affect their target cells via transferring endogenous miRNAs and whether these MP have different miRNA patterns than their maternal cells.
miRNAs are a class of small (∼22 nucleotides long), non-coding RNA that bind to messenger RNAs (mRNAs), thereby acting as endogenous post-translational gene regulators.8 Since more then 1000 different human miRNAs have already been discovered, the interaction between miRNAs and mRNAs is highly complex and currently not completely understood. However, approximately one-third of human protein-encoding genes are miRNA regulated, underlining the extraordinary impact of miRNA on protein expression.9,10 Recent data indicate that miRNAs play a vital role in many cardiovascular diseases and can be found in cardiac tissue as well as in circulating blood, opening the possibility to use them as diagnostic surrogate markers.11–15 Many pioneering studies describing specific miRNA patterns in cardiovascular diseases used plasma as their source of miRNAs.16,17 However, as RNase, which is abundant in circulating blood, rapidly degrades RNA in plasma, it is a central question how plasma miRNAs are protected from degradation.
We used Next Generation Sequencing (NGS) and quantitative reverse transcription–polymerase chain reaction (qRT–PCR) to systematically compare miRNA profiles of MP with the profiles of their maternal cells in their non-stimulated and stimulated states. Furthermore, we provide evidence for the central role of MP as transfer vehicles of miRNAs in circulation and for the proof of concept that miRNA profiles in circulating MP can be disease-specific.
2. Methods
2.1. MP attachment to endothelial cells
To show in proof-of-principle experiments that MP attach and fuse with target cells, such as human umbilical vein endothelial cells (HUVECs), a static adhesion assay was performed (Figure 1). THP-1 cells were stained (celltracker™, Invitrogen™, USA) before THP-1 MP release was induced [lipopolysaccharide (LPS), 15 µg/mL]. MP were isolated from cell culture supernatant, washed, and added on HUVECs (0.5–24 h, 37°C) before non-adherent MP were removed. Attachment of fluorescent THP-1 MP on HUVECs was quantified microscopically (Zeiss-Axio-Observer, Germany; ImageJ1.45I, USA).
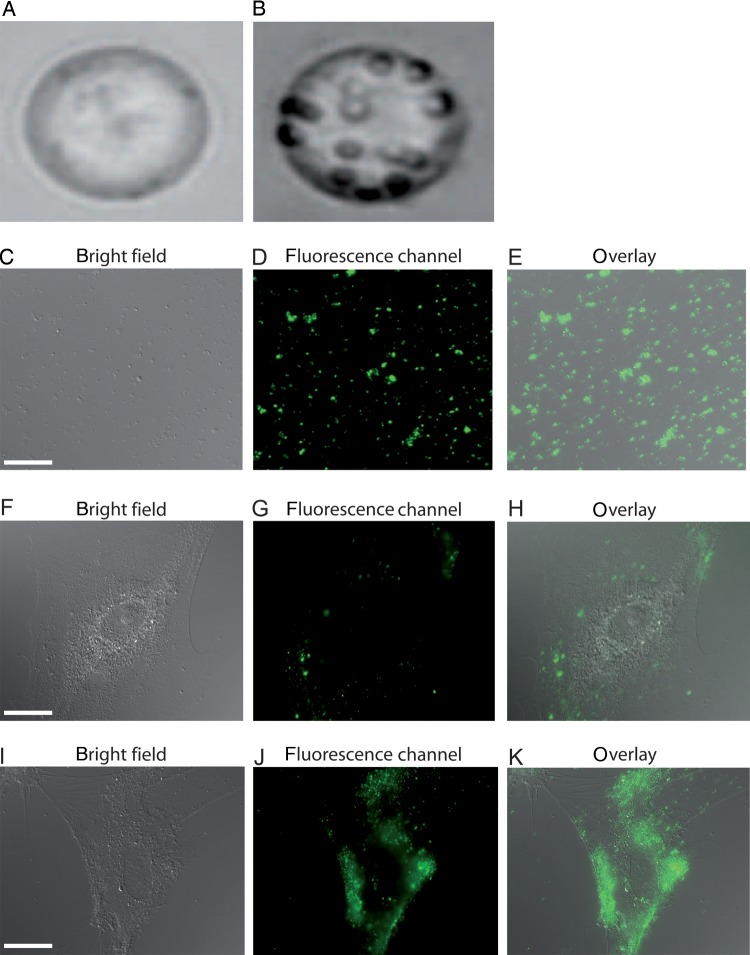
Figure 1.
MP derived from stimulated THP-1 cells bind to and fuse with HUVECs. In comparison to non-stimulated THP-1 cells (A), stimulated THP-1 cells (B) release more MP. Purity and sufficient staining of THP-1 MP were assessed with microscopy (C–E). After 30 min incubation, fluorescent MP began to attach on the HUVEC surface and were detectable as distinct fluorescence spots (F–H). Twenty hours after THP-1 MP application, HUVECs have internalized the majority of fluorescent MP exhibiting diffuse fluorescence (I–K). Scale bar indicates 10 μm.
2.2. Flow cytometry of MP
Investigating whether miRNAs detected in plasma of clinical studies are associated with MP, the amount of circulating MP was counted with FACS and MP subpopulations were specifically labelled. Therefore, plasma MP were labelled with AnnexinV-APC (Invitrogen™, USA) and co-stained with one of the following antibodies: anti-CD41-PE, anti-CD61-FITC, anti-CD18-FITC (all Beckman Coulter, USA), and anti-CD62E-FITC (R&D-Systems®, USA). MP were defined as particles <1.1 µm (1.1 µm beads, Sigma-Aldrich, Germany) expressing their characteristic surface receptors. All measurements were performed with FACSCanto (BD Bioscience, USA).
2.3. miRNAs of coronary artery disease patients
To investigate whether blood miRNAs are predominantly associated with MP, RNA of MP in plasma and plasma without MP of patients with coronary artery disease (CAD) was isolated (Trizol®, Invitrogen™, USA) and qRT–PCR was performed. Plasma of citrate anti-coagulated blood was prepared, MP were isolated (16 000 g, 90 min), and RNA of MP and MP-free plasma was isolated before cDNA synthesis was done (Qiagen, Germany). miRNA profiles of circulating MP of patients with stable CAD (n= 5) were compared with matched patients with acute coronary syndrome (ACS, n= 5) using qRT–PCR (Table 1). All patients gave their written informed consent. The study was approved by the local Ethics Committee.
Table 1.
Clinical characteristics of patients with stable CAD and ACS
| CAD (n= 5) | ACS (n= 5) | P-value | |
|---|---|---|---|
| Age | 76 ± 2 | 66 ± 4 | n.s. |
| Sex | |||
| Male | 3 | 5 | n.s. |
| Female | 2 | 0 | |
| Laboratory parameters | |||
| Leucocytes (103/μL) | 6 | 11 | n.s. |
| Platelets (103/μL) | 194 | 158 | n.s. |
| Troponin (ng/mL) | 0.05 | 36 | 0.04 |
| CK (U/L) | 20 | 901 | 0.01 |
| Cardiovascular risk factors | |||
| Smoking (%) | 20 | 40 | n.s. |
| Hypercholesterolaemia (%) | 40 | 100 | n.s. |
| Arterial hypertension (%) | 100 | 60 | n.s. |
| Diabetes mellitus (%) | 40 | 40 | n.s. |
| Fam history (%) | 20 | 40 | n.s. |
| Medication | |||
| Anti-platelet drugs (%) | 80 | 100 | n.s. |
| ACE/AT1 inhibitors (%) | 100 | 60 | n.s. |
| β-Blocker (%) | 20 | 80 | n.s. |
| Diuretics (%) | 60 | 40 | n.s. |
| Statins (%) | 40 | 80 | n.s. |
| Coronary artery diseases | |||
| 1-vessel disease | 1 | — | |
| 2-vessel diseases | 2 | 4 | |
| 3-vessel diseases | 2 | 1 | |
With the exception of cardiac enzymes, baseline characteristics of both groups were not significantly different. n.s., not significant.
2.4. Platelet MP preparation and qRT–PCR
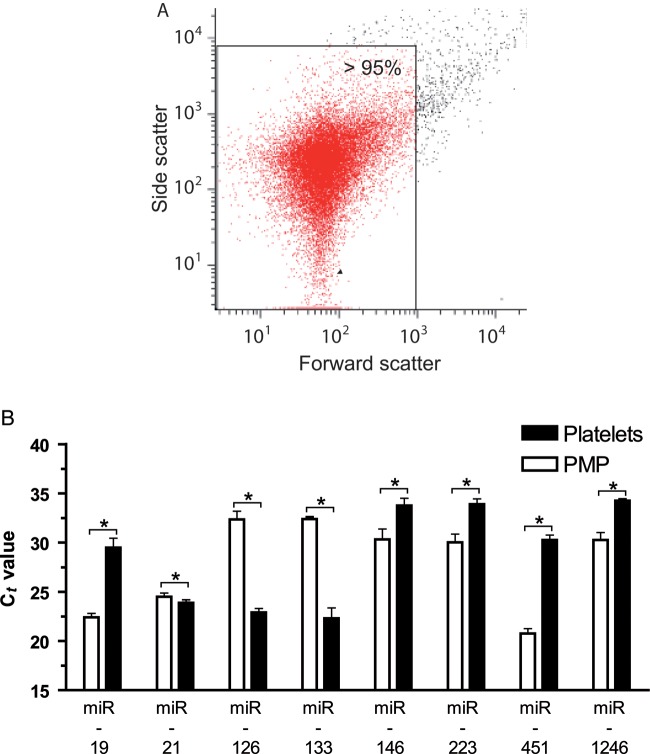
Platelet MP were prepared by stimulating platelets from platelet concentrates with 20 µM ADP, followed by centrifugation (16 000 g, 2 min) to separate platelets from platelet MP. The supernatant, which contained platelet MP, was again centrifuged (16 000 g, 90 min) and RNA was extracted of the pellet. Purity of MP was controlled with FACS (Figure 2A). Expression of miRNAs that have been described in plasma of cardiovascular disease patients or that are known to be expressed in platelets (miRNA-19, miRNA-21, miRNA-126, miRNA-133, miRNA-146, miRNA-223, miRNA-451, and miRNA-1246) were analysed in platelet and platelet MP samples with qRT–PCR (Qiagen). As the expression of housekeeping genes is often unevenly distributed between cells and MP, miRNA expression of platelets and platelet MP was calculated with the quantile-distribution method and is illustrated as ct values. The study was performed in accordance to the Helsinki declaration and was approved by the local Ethics Committee (Alfred-Medical-Research-Centre, Australia).
Figure 2.
Expression of miRNA associated with cardiovascular diseases differs between platelet MP and platelets. (A) Prepared platelet MP (PMP) samples that were used for RNA isolation were highly pure. (B) Several miRNAs, which are involved in cardiovascular diseases, were detected in PMP. Interestingly, many miRNAs had different expression levels in MP and platelets suggesting a selective miRNA packaging into PMP. *P< 0.05.
2.5. THP-1 cell stimulation and MP preparation
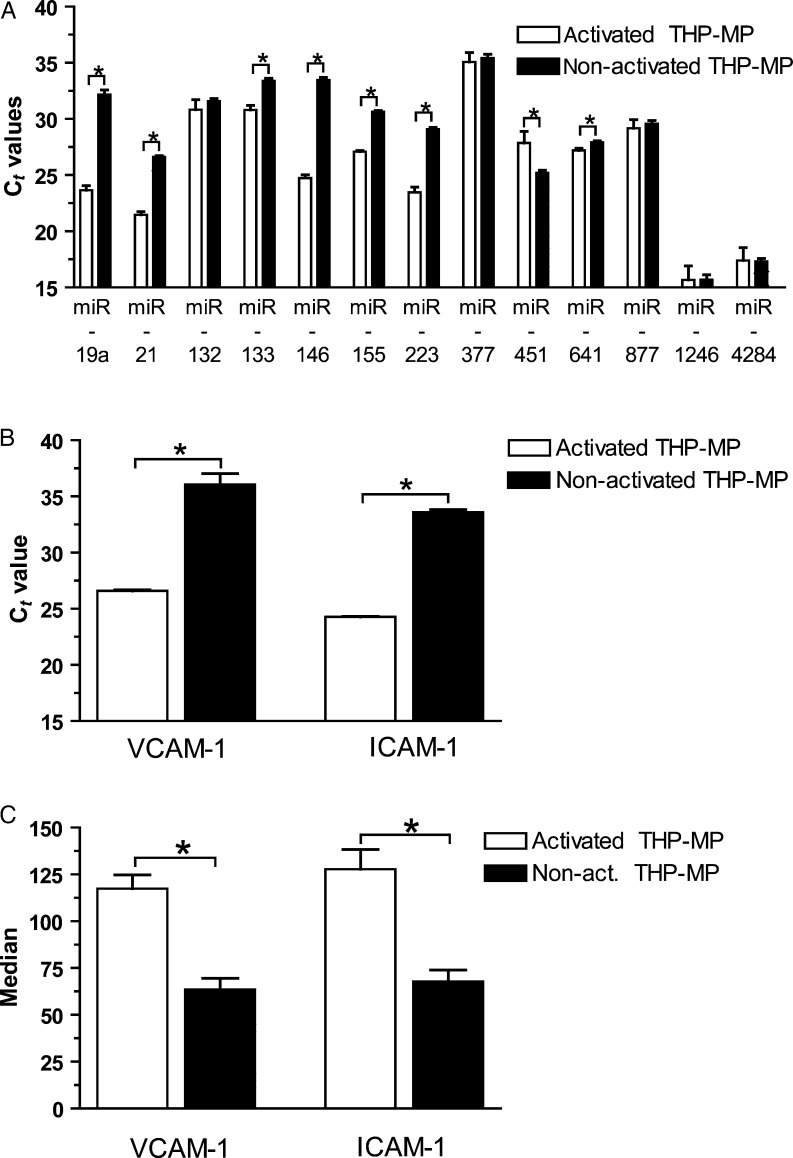
THP-1 cells (ATCC, USA) were stimulated with LPS (Fc 15 μg/mL, 24 h) to obtain stimulated THP-1 cells and THP-1 MP. After stimulation, the cells were centrifuged (120 g, 20 min) and RNA of the pellet containing stimulated THP-1 cells was purified. The corresponding supernatant was again centrifuged (16 000 g, 90 min) before RNA was extracted from the pellet which contained MP. To investigate whether MP of stimulated and non-stimulated THP-1 cells have different miRNA profiles, qRT–PCR of several inflammatory miRNAs was performed for both MP types (commercial primers, Qiagen). Furthermore, to investigate whether stimulated MP induce more endothelial inflammation than non-stimulated MP, HUVECs were incubated with both MP types and vascular cell adhesion molecule 1 (VCAM-1) and intercellular cell adhesion molecule 1 (ICAM-1) expression was quantified by qRT–PCR (commercial primers, Qiagen and Geneworks, Australia) and FACS (R&D-Systems®, Australia, and Chemicon, USA).
2.6. HUVECs stimulation and MP preparation
RNA extraction was done from HUVEC MP as well as from non-stimulated and stimulated HUVECs. To receive stimulated HUVECs and HUVEC MP, cells were stimulated with phorbol 12-myristate 13-acetate (Fc 100 ng/mL, 24 h) and RNA was isolated as described for THP-1.
2.7. NGS of RNA samples
In comparison to microarrays or qRT–PCR, NGS has the great advantage of being able to detect all known miRNAs in the analysed sample and not only a pool of pre-selected miRNAs. Therefore, to analyse the largest spectrum of miRNAs, NGS was used for THP-1 cells, HUVECs, and their MP. One microgram of total RNA was used in a library preparation according to the Illumina Small RNA Sequencing protocol (v1.5, Illumina, USA) using Illumina compatible adapters supplied in the AIR Bar-coded Adapter Set1 (BiooScientific, USA). Multiplexed libraries were sequenced at a concentration of 10 pM for 36 cycles. Fastx Toolkit was used to remove the 3′-adapter sequence, demultiplex, and remove the 6-mer barcode sequence. A custom script was used to identify all unique reads (contigs) and generate a counts matrix detailing how many instances a particular sequence appeared in each bar-coded sample. Each contig was aligned with BWA against the miRNA-base hairpin.18
2.8. Statistics
For qRT–PCR analysis, the housekeeping genes were significantly different between platelets and platelet MP. Therefore, quantile normalization was used (Bioconductor, USA) to test whether miRNA was differentially expressed.19 Quantile normalization was applied to raw data and analysis of variance (ANOVA) methods were used to analyse differences in the expression of each miRNA between both groups. For data analysis of NGS, read counts were normalized and imported into microarray data analysis package (Partek®Genomics Suite™, USA). Replicate data was averaged within the treatment groups and compared with control or reference groups. ANOVA was used to identify genes that were altered by more than two-fold. Statistics for categorical variables were performed with the Fisher exact test (graphpad version 5, USA). Alpha was set in a two-sided test as 0.05 and data are expressed as mean ± SEM.
3. Results
As the aim of the study was to investigate whether MP contain distinct miRNA patterns compared with their maternal cells, miRNA profiles of the most abundant blood MP (e.g. leucocytes, endothelial cells, platelets) and their corresponding stimulated and non-stimulated parent cells were analysed. To avoid contamination by other cell types, THP-1 cells and HUVECs were used as models for monocytes and endothelial cells.
3.1. MP generation and function as transport vehicle
Highly pure, not contaminated THP-1 MP were obtained from THP-1 cells. To provide evidence for the function as transport vehicles, THP-1 MP were incubated with HUVECs resulting in adhesion (>30 min) and fusion (>24 h) to HUVECs (Figure 1), suggesting that THP-1 MP can function as transport vesicles, therefore representing a model by which the role of MP in miRNA transport can be studied.
3.2. Quantification of plasma MP
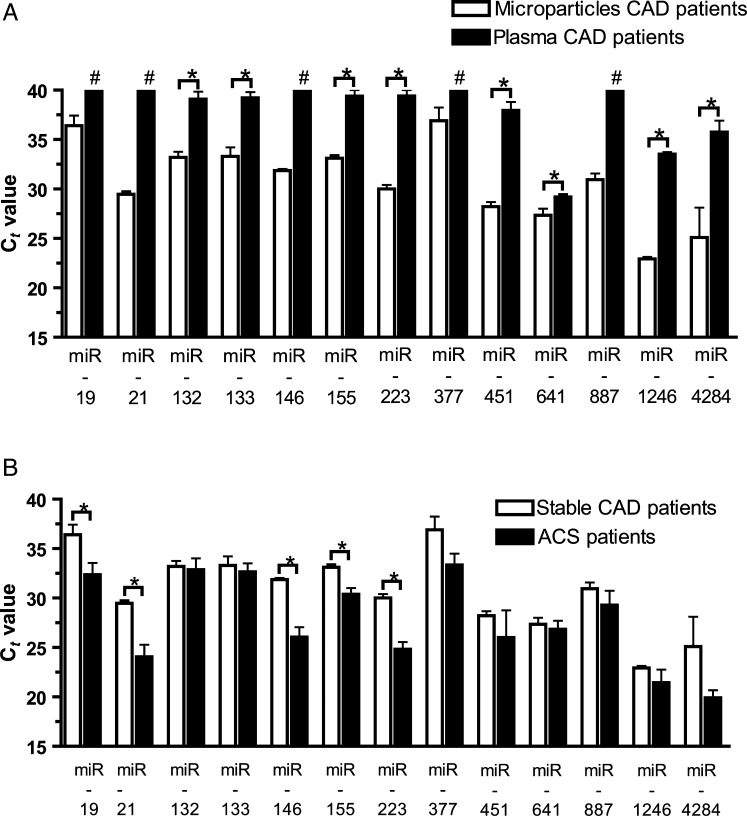
Pioneering clinical studies have found miRNAs in plasma of cardiovascular diseases patients.16,17 Hypothesizing that these miRNAs are mainly transported inside MP, we investigated whether plasma contains MP in which miRNAs can be detected. Plasma preparations contained ∼5200 MP/µL (5200 ± 1340 MP/µL), a number that is in accordance with previous observations.20 Forty-one to 45% of circulating MP were of platelet, 28% of leucocyte, and 8% of endothelial origin. To confirm that MP are the main miRNA compartment in plasma, qRT–PCRs of different miRNAs were performed analysing plasma MP compared with MP-free plasma. The predominant amount of plasma miRNAs was found to be associated with MP and only small amounts were detected in MP-free plasma, underlining the importance of MP as miRNA transport vesicles in circulation (Figure 3A).
Figure 3.
Circulating miRNAs are mainly associated with MP. The majority of plasma miRNAs are associated with MP and only small amounts can be found in MP-free plasma of patients with CAD (A). In comparison to patients with stable CAD, larger amounts of different pro-inflammatory miRNAs were found in patients with ACS, underlining the importance of MP-associated miRNAs in patients with acute vs. stable CAD (B). *P< 0.05; #specific miRNA is not detectable.
3.3. Comparison of miRNAs in patients with acute vs. stable CAD
To provide proof-of-concept data that differing patterns of MP-associated miRNAs characterize different diseases, patients with acute and stable CAD as determined in coronary angiography were compared with each other (Table 1). Even in a small study population, patients with stable CAD (n= 5) had significantly less MP-associated pro-inflammatory miRNAs than matched patients with ACS (n= 5) (Figure 3B). These data emphasize the importance of MP as transport vehicles for inflammatory miRNAs in cardiovascular disease patients.
3.4. miRNAs in platelets and their MP
As in many cardiovascular diseases, a co-incidence between increased platelet MP and circulating miRNAs can be found, we investigated whether platelet MP contain miRNAs and whether these miRNA profiles differ from that of platelets.16,21 As shown in Figure 2, miRNA-19, miRNA-21, miRNA-126, miRNA-133, miRNA-146, miRNA-223, miRNA-451, and miRNA-1246 were identified in platelet MP. Interestingly, miRNA-19, miRNA-146, miRNA-223, miRNA-451, and miRNA-1246 were significantly increased in platelet MP, whereas the expression of miRNA-126 and miRNA-133 were higher in platelets. In summary, platelet MP contain several miRNAs that have been detected in plasma of patients with cardiovascular diseases. Furthermore, differences in the miRNA profile of platelets and platelet MP suggest that platelets may have a mechanism to selectively transport miRNAs into MP.
3.5. NGS of miRNAs in THP-1 cells and their MP
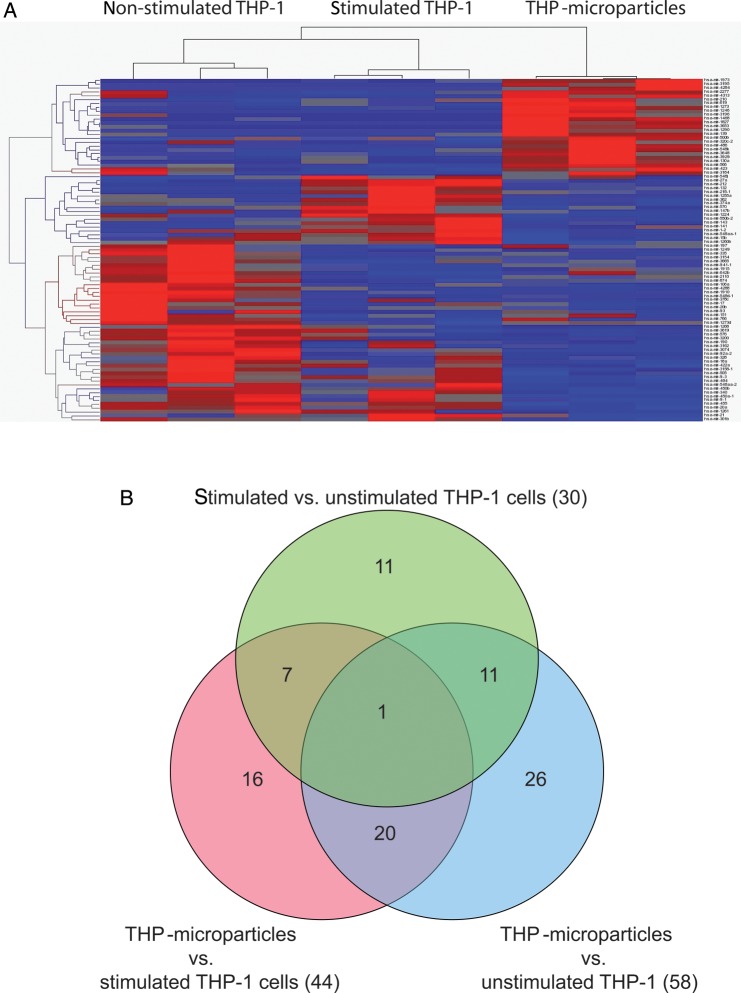
In all THP-1 samples, a total of 786 different miRNAs were detected. Forty-four miRNAs significantly discriminated between THP-1 MP and stimulated THP-1 cells (19 miRNAs up-regulated in THP-1 MP vs. 25 miRNAs up-regulated in stimulated THP-1; Figure 4 and Table 2). Fifty-eight miRNAs were significantly different in THP-1 MP in comparison to non-stimulated THP-1 cells (23 miRNAs up-regulated in THP-1 MP vs. 35 miRNAs up-regulated in non-stimulated THP-1) and 30 miRNAs differed between stimulated and non-stimulated THP-1 (22 miRNAs up-regulated in non-stimulated THP-1 vs. eight miRNAs up-regulated in stimulated THP-1). Hence, the miRNA profile of THP-1 cells changes under stimulation and stimulated THP-1 cells seem to package a different miRNA pool into MP compared with non-stimulated THP-1 cells.
Figure 4.
miRNA profiles of stimulated and non-stimulated THP-1 cells as well as THP-1 MP. (A) A heat map diagram shows miRNA clustering of RNA samples from stimulated and non-stimulated THP-1 cells and THP-1 MP as assessed by NGS. (B) As illustrated in a Venn diagram, miRNA was not randomly distributed between the groups suggesting the existence of a specific packaging mechanism of miRNAs into THP-1 MP.
Table 2.
Differences in miRNA abundance of THP-1 MP in comparison to stimulated THP-1 cells
| miRNA | Fold change | P-value |
|---|---|---|
| miRNA-1290 | 230↑ | 0.045 |
| miRNA-4284 | 191↑ | 0.043 |
| miRNA-1246 | 54↑ | 0.006 |
| miRNA-486 | 11↑ | 0.005 |
| miRNA-642b | 11↑ | 0.039 |
| miRNA-1973 | 10↑ | 0.013 |
| miRNA-130a | 7↑ | 0.022 |
| miRNA-3929 | 6↑ | 0.018 |
| miRNA-320 | 6↑ | 0.028 |
| miRNA-139 | 5↑ | 0.016 |
| miRNA-548k | 5↑ | 0.008 |
| miRNA-566 | 5↑ | <0.001 |
| miRNA-619 | 5↑ | 0.016 |
| miRNA-3648 | 5↑ | 0.021 |
| miRNA-423 | 3↑ | 0.021 |
| miRNA-500b | 3↑ | 0.046 |
| miRNA-3184 | 3↑ | 0.047 |
| miRNA-1273 | 3↑ | <0.001 |
| miRNA-2277 | 2↑ | 0.022 |
| miRNA-1255a | −28↓ | 0.039 |
| miRNA-450a-1 | −14↓ | 0.030 |
| miRNA-1224 | −12↓ | 0.001 |
| miRNA-143 | −8↓ | 0.011 |
| miRNA-1–2 | −7↓ | 0.008 |
| miRNA-340 | −6↓ | 0.011 |
| miRNA-212 | −5↓ | <0.001 |
| miRNA-132 | −5↓ | 0.026 |
| miRNA-15b | −4↓ | 0.008 |
| miRNA-9-3 | −4↓ | 0.005 |
| miRNA-301b | −4↓ | 0.011 |
| miRNA-550b-2 | −4↓ | 0.015 |
| miRNA-455 | −4↓ | <0.001 |
| miRNA-374a | −4↓ | 0.007 |
| miRNA-147b | −3↓ | 0.041 |
| miRNA-20a | −3↓ | 0.006 |
| miRNA-1260b | −3↓ | 0.029 |
| miRNA-570 | −3↓ | 0.030 |
| miRNA-484 | −3↓ | 0.010 |
| miRNA-548aa-1 | −3↓ | 0.018 |
| miRNA-21 | −3↓ | 0.042 |
| miRNA-218-1 | −3↓ | 0.041 |
| miRNA-362 | −3↓ | 0.003 |
| miRNA-450b | −2↓ | 0.029 |
| miRNA-27a | −2↓ | <0.001 |
3.6. Stimulated THP-1 MP contain increased inflammatory miRNAs and induce endothelial inflammation
Investigating whether MP from stimulated THP-1 cells contain different miRNA patterns than those of non-stimulated THP-1 cells, we found that several pro-inflammatory miRNAs are significantly up-regulated in stimulated vs. non-stimulated MP, indicating that the stimulus inducing MP release can influence miRNA transport into MP (Figure 5A). Consecutively, endothelial inflammation markers (VCAM-1 and ICAM-1) were up-regulated after incubation with stimulated THP-1 MP in comparison to non-stimulated THP-1 MP as determined in qRT–PCR (Figure 5B) and flow cytometry (Figure 5C).
Figure 5.
MP from stimulated THP-1 cells contain increased pro-inflammatory miRNA than from non-stimulated THP-1 cells and induce more vascular inflammation. (A) MP released from stimulated THP-1 cells have higher expression levels of several inflammatory miRNAs than those from non-stimulated THP-1 cells. Consecutively, they induce more endothelial inflammation as quantified by qRT–PCR (B) and flow cytometry (C) than MP from non-stimulated THP-1 cells. *P < 0.05.
3.7. NGS of miRNAs in HUVECs and their MP
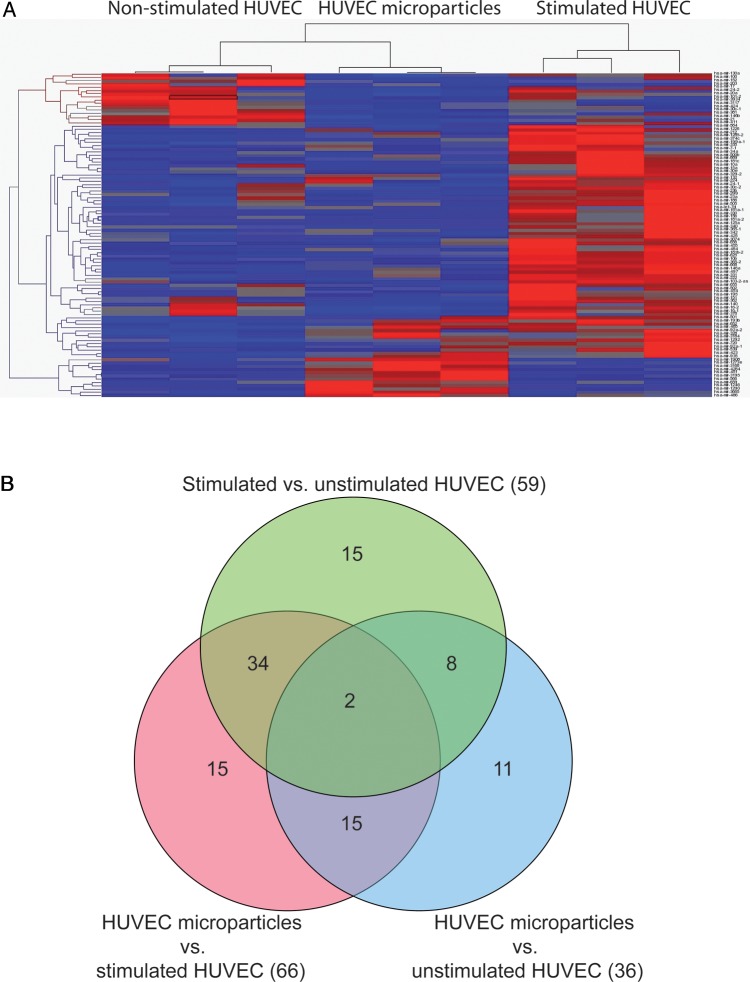
Analysing miRNA patterns of endothelial cells, a total of 470 different miRNAs were detected in HUVEC samples. Fifty-nine miRNAs significantly discriminated between stimulated and non-stimulated HUVECs (53 miRNAs up-regulated in stimulated HUVECs vs. six miRNAs up-regulated in non-stimulated HUVECs) (Figure 6 and Table 3). Sixty-six miRNAs were significantly different in HUVEC MP in comparison to stimulated cells (11 miRNAs up-regulated in HUVEC MP vs. 55 miRNAs up-regulated in stimulated HUVECs) and 36 miRNAs differed between HUVEC MP and non-stimulated HUVECs (18 miRNAs up-regulated in HUVEC MP vs. 18 miRNAs up-regulated in non-stimulated HUVECs). These data demonstrate that stimulated endothelial cells possess significantly different miRNA patterns compared with the non-stimulated endothelium. Furthermore, HUVECs seem to selectively pack their MP with distinct miRNAs.
Figure 6.
miRNA profiles of stimulated and non-stimulated HUVECs as well as HUVEC MP. (A) As illustrated in a heat map diagram, multiple miRNAs were differentially expressed in HUVEC MP and HUVECs. (B) As illustrated in a Venn diagram, miRNAs were not randomly distributed between the groups suggesting the existence of a specific packaging mechanism of miRNAs into HUVEC MP.
Table 3.
Differences in miRNA abundance of HUVEC MP in comparison to stimulated HUVECs
| miRNA | Fold change | P-value |
|---|---|---|
| miRNA-451 | 124↑ | 0.006 |
| miRNA-1908 | 15↑ | <0.001 |
| miRNA-4284 | 14↑ | 0.002 |
| miRNA-1246 | 12↑ | 0.02 |
| miRNA-3168 | 12↑ | 0.018 |
| miRNA-1290 | 11↑ | 0.011 |
| miRNA-3195 | 10↑ | 0.011 |
| miRNA-659 | 9↑ | 0.035 |
| miRNA-1273e | 6↑ | 0.026 |
| miRNA-3665 | 5↑ | 0.001 |
| miRNA-486 | 3↑ | 0.014 |
| miRNA-15a | 13↓ | 0.012 |
| miRNA-30e | 12↓ | 0.022 |
| miRNA-299 | 10↓ | 0.004 |
| miRNA-15b | 9↓ | 0.027 |
| miRNA-378 | 8↓ | 0.036 |
| miRNA-454 | 8↓ | 0.049 |
| miRNA-195 | 7↓ | 0.039 |
| miRNA-181a-1 | 7↓ | 0.005 |
| miRNA-16-1 | 7↓ | 0.011 |
| miRNA-16-2 | 7↓ | 0.002 |
| miRNA-181c | 7↓ | 0.004 |
| miRNA-330 | 7↓ | 0.019 |
| miRNA-10b | 7↓ | <0.001 |
| miRNA-656 | 6↓ | 0.007 |
| miRNA-539 | 6↓ | 0.049 |
| miRNA-20a | 6↓ | 0.008 |
| miRNA-584 | 5↓ | 0.025 |
| miRNA-181a-2 | 5↓ | 0.047 |
| miRNA-140 | 5↓ | 0.048 |
| miRNA-668 | 5↓ | <0.001 |
| miRNA-889 | 5↓ | 0.01 |
| miRNA-625 | 5↓ | <0.001 |
| miRNA-1226 | 5↓ | 0.009 |
| miRNA-425 | 5↓ | 0.008 |
| miRNA-655 | 5↓ | 0.03 |
| miRNA-362 | 4↓ | <0.001 |
| miRNA-340 | 4↓ | 0.011 |
| miRNA-455 | 4↓ | 0.019 |
| miRNA-34a | 4↓ | 0.04 |
| miRNA-21 | 4↓ | 0.021 |
| miRNA-502 | 4↓ | 0.04 |
| miRNA-10a | 4↓ | 0.029 |
| miRNA-500b | 4↓ | 0.025 |
| miRNA-125a | 3↓ | 0.031 |
| miRNA-497 | 3↓ | 0.004 |
| miRNA-374c | 3↓ | 0.033 |
| miRNA-103-2-as | 3↓ | 0.002 |
| miRNA-151 | 3↓ | 0.016 |
| miRNA-24-2 | 3↓ | 0.037 |
| miRNA-365-2 | 3↓ | <0.001 |
| miRNA-1285-2 | 3↓ | 0.01 |
| miRNA-146a | 3↓ | 0.003 |
| miRNA-186 | 3↓ | 0.012 |
| miRNA-181b-2 | 3↓ | <0.001 |
| miRNA-3074 | 3↓ | 0.018 |
| miRNA-146b | 3↓ | 0.042 |
| miRNA-222 | 3↓ | <0.001 |
| miRNA-23b | 3↓ | 0.023 |
| miRNA-331 | 3↓ | 0.003 |
| miRNA-720 | 3↓ | 0.038 |
| miRNA-342 | 2↓ | 0.039 |
| miRNA-23a | 2↓ | 0.048 |
| miRNA-24-1 | 2↓ | 0.032 |
| miRNA-199a-1 | 2↓ | 0.021 |
| miRNA-30c-2 | 2↓ | 0.004 |
4. Discussion
To investigate whether MP contain specific miRNAs delivered from their maternal cells, NGS of THP-1 cells and HUVECs as well as qRT–PCR of platelet MP and of their corresponding maternal cells were performed. NGS is a promising method for miRNA studies as it has major advantages in comparison to microarrays, including higher sensitivity, wider miRNA spectrum that can be analysed and the ability of de novo sequencing of so far unknown miRNAs.22,23 We could demonstrate that MP contain a significantly distinct miRNA pattern compared with their corresponding stimulated and non-stimulated maternal cells, suggesting a selective miRNA transport into MP. Several of the miRNAs up-regulated in MP are involved in cardiovascular diseases, indicating that MP may act as transport vehicles delivering specific miRNAs (Table 4). Since MP are major constituents of typical plasma preparations, ‘plasma’ miRNA profiles attributed to specific diseases may in fact originate from circulating MP. Our findings, which show that plasma deprived of MP demonstrates a significant reduction in miRNAs, support this notion.
Table 4.
miRNA detected in MP, which are reported to be associated with cardiovascular diseases
| miRNA | Platelet microparticles | THP microparticles | HUVEC microparticles | Disease |
|---|---|---|---|---|
| miRNA-let-7d | x | x | CAD16 | |
| miRNA-17 | x | x | CAD16 | |
| miRNA-19 | x | x | x | Cardiomyopathy47 |
| miRNA-20a | x | x | CAD16 | |
| miRNA-21 | x | x | x | CAD,16 CF48 |
| miRNA-27a | x | x | CAD16 | |
| miRNA-92a-1 | x | x | CAD16 | |
| miRNA-126 | x | x | x | CAD,16 diabetic angiopathy49 |
| miRNA-130 | x | x | CAD16 | |
| miRNA-133 | x | AMI33 | ||
| miRNA-143 | x | x | CAD16 | |
| miRNA-146a | x | x | x | Myocarditis50 |
| miRNA-146b | x | Myocarditis,50 CAD51 | ||
| miRNA-155 | x | x | CAD,16 myocarditis50 | |
| miRNA-199 | x | CAD16 | ||
| miRNA-221 | x | x | CAD16 | |
| miRNA-223 | x | x | AMI + myocarditis,50 P2Y12R,52 atrial fibrillation53 | |
| miRNA-423 | x | x | Cardiac failure40 | |
| miRNA-451 | x | x | x | Macrophage function45 |
MP represent a novel mechanism of intercellular communication mediating inflammation and coagulation.24,25 Their effect on target cells was long accredited to their specific lipid composition, as well as to intravesicular chemokines.1 However, recent data indicate that MP can also affect protein expression of their target cells by delivering RNAs.7,26 As it is evident that MP shedding is a highly structured process, the question arises whether miRNAs are specifically packed from maternal cells into MP, thereby protecting their miRNAs from degradation by plasma RNase.27,28 In a pioneering study by Hunter et al.,29 it was found that blood MP contain some miRNAs that may be released from peripheral blood mononuclear cells. However, as the mixture of MP circulating in blood may originate from different cell types, no conclusion could be drawn on whether the profile of miRNAs in MP reflect the profile seen in their maternal cells.
Here, we show that plasma contains ∼5000 MP/µL and that the majority of plasma miRNAs are associated with MP, both in patients with stable CAD as well as with ACS and that only a small amount of miRNA is located in MP-free plasma, underlining the importance of MP as biological vectors for miRNAs (Figure 3A). Interestingly, several miRNAs that were detected in ‘plasma’ of clinical studies were also found in plasma MP (Figure 3). In addition, ACS patients had significantly higher levels of some MP-associated pro-inflammatory miRNAs than stable CAD patients. Some of these plasma miRNAs were also found to be significantly up-regulated in isolated platelet MP, whereas others were higher expressed in platelets, indicating a specific miRNA transport into platelet MP (Figure 2). Recent data indicate that miRNA-133, which was found in platelet MP, is involved in different cardiovascular diseases, such as myocardial infarction.30 Villar et al.31 demonstrated elegantly that miRNA-133 functions as a surrogate marker for reverse remodelling in cardiac hypertrophy, a disease that is strongly influenced by activated platelets.32 Moreover, miRNA-133 was found to be increased in plasma of patients with myocardial infarction, who are also known to have elevated levels of circulating MP.33 Also, miRNA-19, which has been shown to be involved in cardiac hypertrophy and angiogenesis, was significantly up-regulated in platelet MP (Figure 2).34,35 In conclusion, MP contain the majority of plasma miRNAs. Furthermore, platelet MP have a different miRNA pattern than platelets themselves and contain several miRNAs that were found in plasma of cardiovascular diseases.
To our knowledge, this is the first study showing that THP-1 MP contain a significantly distinct miRNA profile compared with their stimulated and non-stimulated maternal cells (Figure 4). Nineteen miRNAs were found to be significantly increased in THP-1 MP in comparison to stimulated THP-1 cells, and 58 miRNAs discriminated THP-1 MP from their non-stimulated cells of origin. miRNA-130a, miRNA-320, and miRNA-423 were significantly up-regulated in THP-1 MP. Chen et al.36 found that miRNA-130a has strong pro-angiogenetic properties by down-regulation of GAX, an anti-angiogenic homeobox gene. This pro-angiogenic effect of miRNA-130a might be the underlying molecular mechanism for the observation that MP of plaque macrophages induce in vivo intra-plaque neovessel formation, therefore contributing to plaque instability.37 miRNA-320 is known to play a pivotal role in ischaemic reperfusion injuries,38,39 a disease that is associated with enhanced monocytotic MP, suggesting that THP-1 MP directly influence pro-inflammatory gene expression in cardiac tissue by transferring miRNAs, such as miRNA-320. Tijsen et al.40 investigated miRNA in serum of patients with cardiac failure and detected six miRNAs that were significantly increased in comparison to controls. Among these, miRNA-423, which was found to be significantly increased in THP-1 MP, correlated most strongly with the clinical diagnosis of cardiac failure. Recent evidence is suggestive of miRNA-423 being used as a marker for cardiac failure.41 As it is known that the stimulus inducing MP release influences miRNA packaging from the corresponding maternal cell, expression levels of inflammatory miRNA were assessed in stimulated and non-stimulated THP-1 MP (Figure 5). Interestingly, many pro-inflammatory miRNAs were significantly increased in stimulated THP-1 MP (Figure 5A). miRNA-21 is one of the highest up-regulated miRNAs in stimulated vs. non-stimulated THP-MP and is described to be up-regulated under hypoxic conditions in vascular smooth muscle cells inducing endothelial inflammation (e.g. VCAM-1 expression) and to play a pivotal role in the transition of endothelial cells to fibroblasts during inflammation.42–44 Consecutively, stimulated THP-1 MP had a stronger inflammatory effect on HUVECs when compared with MP of non-stimulated THP-1 cells, both on the mRNA level as well as on surface receptor expression (Figure 5B and C).
Intact endothelial cells are essential for maintenance of vascular integrity and haemostasis. However, stimulated endothelium releases endothelial MP, which, besides functioning as surrogate markers for early endothelial dysfunction, are biological vectors mediating vascular inflammation and coagulation at a distant site from the initially damaged endothelial cells.24 Aiming to investigate whether biological effects of endothelial MP may also be attributed to miRNA transfer, miRNA patterns of HUVECs and HUVEC MP were investigated. We found that HUVEC MP had a significantly different miRNA profile than stimulated and non-stimulated HUVECs (Figure 6). Sixty-six miRNAs were differently expressed between HUVEC MP and their stimulated maternal cells, from which 11 were up-regulated in HUVEC MP. Among the up-regulated miRNAs in HUVEC MP were miRNA-451 and miRNA-486. miRNA-486 is a cardiac enriched, highly conserved miRNA that was found to be 3–11 times up-regulated in HUVEC and THP-1 MP in comparison to their stimulated maternal cells (Tables 2 and 3). Our findings are supported by a study of Hunter et al.,29 who found miRNA-486 to be the highest differentially expressed miRNA in plasma microvesicles compared with peripheral blood monocytic cells. miRNA-451 was significantly up-regulated in HUVEC MP and strongly inhibits expression of macrophage migration factor (MIF), a pluripotent pro-inflammatory cardiac depressant.45 Garner et al. showed that MIF impaired left ventricular function, an effect that was reversible after anti-MIF antibody treatment.46 Overall, this indicates that miRNA-451 may possess cardioprotective effects by inhibiting MIF, which might be an attractive therapeutic approach warranting interesting future studies.
In conclusion, using NGS, we could show that THP-1, HUVEC, and platelet MP contain numerous miRNAs, which significantly differ from their maternal cells, both in stimulated and non-stimulated states. These data indicate a specific ‘packaging’ or delivery mechanism of miRNAs from maternal cells into MP, a topic which warrants further studies. Most interestingly, for the use of miRNAs as biomarkers, miRNAs detected in conventional plasma preparations seem to be localized mainly in MP.
Funding
This work was supported by the National Health and Medical Research Council (NHMRC) of Australia. P.D. was supported by the German Research Foundation (DFG) and Monash University, Melbourne. K.P. holds a fellowship of the Australian Research Council. L.S. and A.F. were supported by the German Heart Foundation. A.E.-O. holds a fellowship of the Juvenile Diabetes Research Foundation International (JDRF), the Diabetes Australia Research Trust (DART), the National Health and Medical Research Council (NHMRC), and the National Heart Foundation of Australia (NHF). Funding to pay the Open Access publication charges for this article was provided by Baker IDI, Heart & Diabetes Institute, Melbourne, Australia.
Acknowledgements
We thank Ross Lazarus for statistical advice and Antony Kaspi for statistical assistance.
Conflict of interest: none declared.
References
- 1.Hugel B, Martinez MC, Kunzelmann C, Freyssinet JM. Membrane microparticles: two sides of the coin. Physiology (Bethesda) 2005;20:22–27. doi: 10.1152/physiol.00029.2004. [DOI] [PubMed] [Google Scholar]
- 2.Prokopi M, Pula G, Mayr U, Devue C, Gallagher J, Xiao Q, et al. Proteomic analysis reveals presence of platelet microparticles in endothelial progenitor cell cultures. Blood. 2009;114:723–732. doi: 10.1182/blood-2009-02-205930. [DOI] [PubMed] [Google Scholar]
- 3.Mause SF, Weber C. Microparticles: protagonists of a novel communication network for intercellular information exchange. Circ Res. 2010;107:1047–1057. doi: 10.1161/CIRCRESAHA.110.226456. [DOI] [PubMed] [Google Scholar]
- 4.Distler JH, Pisetsky DS, Huber LC, Kalden JR, Gay S, Distler O. Microparticles as regulators of inflammation: novel players of cellular crosstalk in the rheumatic diseases. Arthritis Rheum. 2005;52:3337–3348. doi: 10.1002/art.21350. [DOI] [PubMed] [Google Scholar]
- 5.Shantsila E, Kamphuisen PW, Lip GY. Circulating microparticles in cardiovascular disease: implications for atherogenesis and atherothrombosis. J Thromb Haemost. 2010;8:2358–2368. doi: 10.1111/j.1538-7836.2010.04007.x. [DOI] [PubMed] [Google Scholar]
- 6.VanWijk MJ, VanBavel E, Sturk A, Nieuwland R. Microparticles in cardiovascular diseases. Cardiovasc Res. 2003;59:277–287. doi: 10.1016/s0008-6363(03)00367-5. [DOI] [PubMed] [Google Scholar]
- 7.Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847–856. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 8.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. Rna. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 10.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 11.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79:581–588. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 12.Gupta SK, Bang C, Thum T. Circulating microRNAs as biomarkers and potential paracrine mediators of cardiovascular disease. Circ Cardiovasc Genet. 2010;3:484–488. doi: 10.1161/CIRCGENETICS.110.958363. [DOI] [PubMed] [Google Scholar]
- 13.Bauersachs J, Thum T. Biogenesis and regulation of cardiovascular microRNAs. Circ Res. 2011;109:334–347. doi: 10.1161/CIRCRESAHA.110.228676. [DOI] [PubMed] [Google Scholar]
- 14.Fichtlscherer S, Zeiher AM, Dimmeler S. Circulating microRNAs: biomarkers or mediators of cardiovascular diseases? Arterioscler Thromb Vasc Biol. 2011;31:2383–2390. doi: 10.1161/ATVBAHA.111.226696. [DOI] [PubMed] [Google Scholar]
- 15.Hartmann D, Thum T. MicroRNAs and vascular (dys)function. Vascul Pharmacol. 2011;55:92–105. doi: 10.1016/j.vph.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, et al. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107:677–684. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- 17.Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31:659–666. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 18.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mar JC, Kimura Y, Schroder K, Irvine KM, Hayashizaki Y, Suzuki H, et al. Data-driven normalization strategies for high-throughput quantitative RT-PCR. BMC Bioinformatics. 2009;10:110. doi: 10.1186/1471-2105-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuana Y, Bertina RM, Osanto S. Pre-analytical and analytical issues in the analysis of blood microparticles. Thromb Haemost. 2011;105:396–408. doi: 10.1160/TH10-09-0595. [DOI] [PubMed] [Google Scholar]
- 21.Morel O, Hugel B, Jesel L, Mallat Z, Lanza F, Douchet MP, et al. Circulating procoagulant microparticles and soluble GPV in myocardial infarction treated by primary percutaneous transluminal coronary angioplasty. A possible role for GPIIb-IIIa antagonists. J Thromb Haemost. 2004;2:1118–1126. doi: 10.1111/j.1538-7836.2004.00805.x. [DOI] [PubMed] [Google Scholar]
- 22.Hurd PJ, Nelson CJ. Advantages of next-generation sequencing versus the microarray in epigenetic research. Brief Funct Genomic Proteomic. 2009;8:174–183. doi: 10.1093/bfgp/elp013. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Chiodini R, Badr A, Zhang G. The impact of next-generation sequencing on genomics. J Genet Genomics. 2011;38:95–109. doi: 10.1016/j.jgg.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bakouboula B, Morel O, Faure A, Zobairi F, Jesel L, Trinh A, et al. Procoagulant membrane microparticles correlate with the severity of pulmonary arterial hypertension. Am J Respir Crit Care Med. 2008;177:536–543. doi: 10.1164/rccm.200706-840OC. [DOI] [PubMed] [Google Scholar]
- 25.Lynch SF, Ludlam CA. Plasma microparticles and vascular disorders. Br J Haematol. 2007;137:36–48. doi: 10.1111/j.1365-2141.2007.06514.x. [DOI] [PubMed] [Google Scholar]
- 26.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 27.Chen TS, Lai RC, Lee MM, Choo AB, Lee CN, Lim SK. Mesenchymal stem cell secretes microparticles enriched in pre-microRNAs. Nucleic Acids Res. 2010;38:215–224. doi: 10.1093/nar/gkp857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stein JM, Luzio JP. Ectocytosis caused by sublytic autologous complement attack on human neutrophils. The sorting of endogenous plasma-membrane proteins and lipids into shed vesicles. Biochem J. 1991;274:381–386. doi: 10.1042/bj2740381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Rosa S, Fichtlscherer S, Lehmann R, Assmus B, Dimmeler S, Zeiher AM. Transcoronary concentration gradients of circulating microRNAs. Circulation. 2011;124:1936–1944. doi: 10.1161/CIRCULATIONAHA.111.037572. [DOI] [PubMed] [Google Scholar]
- 31.Villar AV, Merino D, Wenner M, Llano M, Cobo M, Montalvo C, et al. Myocardial gene expression of microRNA-133a and myosin heavy and light chains, in conjunction with clinical parameters, predict regression of left ventricular hypertrophy after valve replacement in patients with aortic stenosis. Heart. 2011;97:1132–1137. doi: 10.1136/hrt.2010.220418. [DOI] [PubMed] [Google Scholar]
- 32.Gabbasov Z, Parfyonova Y, Popov E, Gavrilov I, Anuchin V, Dubov P, et al. Association of platelet function in hypertensive patients with left ventricular hypertrophy, transient myocardial ischemia, and coronary artery disease. Platelets. 1998;9:191–195. doi: 10.1080/09537109876672. [DOI] [PubMed] [Google Scholar]
- 33.D'Alessandra Y, Devanna P, Limana F, Straino S, Di Carlo A, Brambilla PG, et al. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J. 2010;31:2765–2773. doi: 10.1093/eurheartj/ehq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 35.Doebele C, Bonauer A, Fischer A, Scholz A, Reiss Y, Urbich C, et al. Members of the microRNA-17–92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood. 2010;115:4944–4950. doi: 10.1182/blood-2010-01-264812. [DOI] [PubMed] [Google Scholar]
- 36.Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111:1217–1226. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leroyer AS, Rautou PE, Silvestre JS, Castier Y, Leseche G, Devue C, et al. CD40 ligand + microparticles from human atherosclerotic plaques stimulate endothelial proliferation and angiogenesis a potential mechanism for intraplaque neovascularization. J Am Coll Cardiol. 2008;52:1302–1311. doi: 10.1016/j.jacc.2008.07.032. [DOI] [PubMed] [Google Scholar]
- 38.Ren XP, Wu J, Wang X, Sartor MA, Qian J, Jones K, et al. MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation. 2009;119:2357–2366. doi: 10.1161/CIRCULATIONAHA.108.814145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 40.Tijsen AJ, Creemers EE, Moerland PD, de Windt LJ, van der Wal AC, Kok WE, et al. MiR423–5p as a circulating biomarker for heart failure. Circ Res. 2010;106:1035–1039. doi: 10.1161/CIRCRESAHA.110.218297. [DOI] [PubMed] [Google Scholar]
- 41.Kumarswamy R, Anker SD, Thum T. MicroRNAs as circulating biomarkers for heart failure: questions about MiR-423–5p. Circ Res. 2010;106:e8. doi: 10.1161/CIRCRESAHA.110.220616. author reply e9. [DOI] [PubMed] [Google Scholar]
- 42.Sarkar J, Gou D, Turaka P, Viktorova E, Ramchandran R, Raj JU. MicroRNA-21 plays a role in hypoxia-mediated pulmonary artery smooth muscle cell proliferation and migration. Am J Physiol Lung Cell Mol Physiol. 2010;299:L861–871. doi: 10.1152/ajplung.00201.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou J, Wang KC, Wu W, Subramaniam S, Shyy JY, Chiu JJ, et al. MicroRNA-21 targets peroxisome proliferators-activated receptor-alpha in an autoregulatory loop to modulate flow-induced endothelial inflammation. Proc Natl Acad Sci USA. 2011;108:10355–10360. doi: 10.1073/pnas.1107052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumarswamy R, Volkmann I, Jazbutyte V, Dangwal S, Park DH, Thum T. Transforming Growth Factor-beta-Induced Endothelial-to-Mesenchymal Transition Is Partly Mediated by MicroRNA-21. Arterioscler Thromb Vasc Biol. 2012;32:361–369. doi: 10.1161/ATVBAHA.111.234286. [DOI] [PubMed] [Google Scholar]
- 45.Bandres E, Bitarte N, Arias F, Agorreta J, Fortes P, Agirre X, et al. microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin Cancer Res. 2009;15:2281–2290. doi: 10.1158/1078-0432.CCR-08-1818. [DOI] [PubMed] [Google Scholar]
- 46.Garner LB, Willis MS, Carlson DL, DiMaio JM, White MD, White DJ, et al. Macrophage migration inhibitory factor is a cardiac-derived myocardial depressant factor. Am J Physiol Heart Circ Physiol. 2003;285:H2500–H2509. doi: 10.1152/ajpheart.00432.2003. [DOI] [PubMed] [Google Scholar]
- 47.Ikeda S, Kong SW, Lu J, Bisping E, Zhang H, Allen PD, et al. Altered microRNA expression in human heart disease. Physiol Genomics. 2007;31:367–373. doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- 48.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 49.Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107:810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 50.Corsten MF, Dennert R, Jochems S, Kuznetsova T, Devaux Y, Hofstra L, et al. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet. 2010;3:499–506. doi: 10.1161/CIRCGENETICS.110.957415. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi Y, Satoh M, Minami Y, Tabuchi T, Itoh T, Nakamura M. Expression of miR-146a/b is associated with the Toll-like receptor 4 signal in coronary artery disease: effect of renin-angiotensin system blockade and statins on miRNA-146a/b and Toll-like receptor 4 levels. Clin Sci (Lond) 2010;119:395–405. doi: 10.1042/CS20100003. [DOI] [PubMed] [Google Scholar]
- 52.Landry P, Plante I, Ouellet DL, Perron MP, Rousseau G, Provost P. Existence of a microRNA pathway in anucleate platelets. Nat Struct Mol Biol. 2009;16:961–966. doi: 10.1038/nsmb.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu Y, Zhang Y, Wang N, Pan Z, Gao X, Zhang F, et al. MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation. 2010;122:2378–2387. doi: 10.1161/CIRCULATIONAHA.110.958967. [DOI] [PubMed] [Google Scholar]