Short abstract
The septins are guanine-nucleotide binding proteins that mostly form filaments. They are important in cytokinesis and also have roles in sporulation in yeasts and embryonic development and in the nervous system in animals.
Summary
The septins make up a family of guanine-nucleotide binding proteins, most of which polymerize to form filaments. Septin genes have been found in fungi and animals but not in protozoa or plants; yeasts have seven septin genes and humans have twelve, but Caenorhabditis elegans has only two. Some septin genes generate multiple polypeptides by alternative splicing or alternative translation start sites. Of the five conserved motifs found in other members of the GTPase superfamily, three are highly conserved in septins. Septin filaments are thought to form a cytoskeletal system that organizes higher-order structures by self-assembly and templated assembly. These multifunctional proteins are best known for their role in cytokinesis, but other functions in dividing and non-dividing cells have evolved in different lineages: budding yeast has septins specific for sporulation; nematode septins are implicated in postembryonic morphogenesis of multiple cell lineages; fly septins are associated with the development of germ cells, photoreceptor cells and nervous system; and mammalian septins are implicated in exocytosis, tumorigenesis, apoptosis, synaptogenesis and neurodegeneration.
The septin genes were originally discovered through genetic screening for budding yeast mutants defective in the cell-cycle progression [1]. Mutants of any one of the genetic loci CDC3, CDC10, CDC11 or CDC12 commonly form multinucleated cellular clusters [2-4]. These mutants cannot organize the 'bud neck filaments' that normally encircle and demarcate the cell cortex between a mother cell and the bud (daughter) [5]. From these and other data, the septins have been regarded as the major constituents of the bud-neck filaments, which have essential roles in cytokinesis [2-4]. Molecular genetic studies revealed that the four CDC genes encode similar polypeptides, each with some of the set of conserved motifs found in GTPases. The four encoded proteins, termed septins, thus founded a protein family within the GTPase superfamily [2-4]. The septins that were later found in other fungi, nematodes, flies, and mammals have also been shown to have roles in cytokinesis and other cellular processes.
Gene organization and evolutionary history
Septins have been found in diverse eukaryotes, including animals and fungi but not protozoa and plants. Most septin genes generate one or more polypeptides by alternative splicing and/or multiple translation start sites; the number of variants is not yet established for many of the genes. The septin genes in five organisms, and the largest product of each gene known from the current databases, are shown in Table 1, and a phylogenetic tree illustrating their structural relationships and molecular evolution is shown in Figure 1. It is noteworthy that considerable diversity has been generated within each species; for example, the human septins are 39-63% identical to human Sept2 at the amino-acid level. It may be possible to classify the septins in each species into two to four groups by sequence homology. Orthologs can be found within the fungi (such as Saccharomyces cerevisiae CDC3, Schizosaccharomyces pombe Spn1 and their Candida albicans orthologs, not shown) and within metazoa (such as Drosophila Sep1 and mammalian Sept2), but not between distant lineages (fungi and metazoa). This pattern suggests that there have been independent expansions of the family in different lineages. Thus, any rules and functions found in the fungal septin systems may not necessarily apply to the metazoan ones, and vice versa.
Table 1.
The septin genes and proteins in five representative organisms
| Species | Approved gene name | Chromosomal location* | Size (amino acids) | Mass (kDa) | Isoelectric point (pI) | Charge | Number of predicted coiled coils | Accession number |
| Saccharomyces cerevisiae | Cdc3 | XII:764137 | 520 | 60.0 | 5.3 | A | 2 | L16548 |
| Cdc10 | III:118342 | 322 | 37.0 | 5.6 | A | 0 | L16549 | |
| Cdc11 | X:576294 | 415 | 47.6 | 4.8 | A | 1 | L16550 | |
| Cdc12 | VIII:328038 | 407 | 46.7 | 8.2 | B | 1 | L16551 | |
| Spr3 | VII:607564 | 512 | 59.8 | 7.3 | N | 3 | U24129 | |
| Spr28 | IV:905043 | 423 | 48.2 | 5.8 | A | 1 | NP_010504 | |
| Shs1/Sep7 | IV:52446 | 551 | 62.6 | 5.3 | A | 2 | Z74273 | |
| Schizosaccharomyces pombe | spn1 | I:1067997 | 469 | 53.7 | 5.3 | A | 1 | U31742 |
| spn2 | I:960487 | 331 | 38.1 | 8.0 | B | 0 | U29888 | |
| spn3 | II:2682372 | 412 | 46.6 | 4.7 | A | 1 | U29889 | |
| spn4 | I:1962024 | 380 | 44.7 | 7.0 | N | 1 | U29890 | |
| spn5 | I:3044705 | 464 | 53.1 | 8.2 | B | 1 | U29891 | |
| spn6 | III:421493 | 380 | 44.0 | 6.9 | N | 1 | AL032824 | |
| spn7 | II:161221 | 428 | 49.2 | 5.2 | A | 0 | AF417166 | |
| Caenorhabditis elegans | unc-59 | I:21.15 | 459 | 52.9 | 8.9 | B | 1 | NM_060987 |
| unc-61 | V:6.66 | 530 | 60.7 | 8.9 | B | 1 | NM_182356 | |
| Drosophila melanogaster | Pnut | 44C2 | 539 | 60.2 | 9.0 | B | 1 | NM_165597 |
| Sep1 | 19F5 | 361 | 41.1 | 6.1 | A | 2 | NM_167747 | |
| Sep2 | 92F2 | 419 | 48.5 | 7.4 | N | 1 | NM_079693 | |
| Sep4 | 15A1 | 427 | 49.0 | 6.9 | N | 2 | NM_167530 | |
| Sep5 | 43F8 | 422 | 48.5 | 7.3 | N | 1 | NM_165578 | |
| Homo sapiens | Sept1 | 16p11.1 | 366 | 41.8 | 5.5 | A | 2 | NM_052838 |
| Sept2 | 2q37.3 | 361 | 41.5 | 6.2 | A | 2 | NM_004404 | |
| Sept3 | 22q13.2 | 345 | 39.3 | 6.8 | N | 0 | NM_145733 | |
| Sept4 | 17q23 | 478 | 55.1 | 5.7 | A | 2 | NM_004574 | |
| Sept5 | 22q11.2 | 369 | 42.3 | 6.4 | A | 2 | NM_002688 | |
| Sept6 | Xq24 | 427 | 48.9 | 6.4 | A | 1 | NM_145799 | |
| Sept7 | 7q36.1 | 418 | 48.8 | 9.0 | B | 1 | NM_001788 | |
| Sept8 | 5q31 | 483 | 55.8 | 5.9 | A | 1 | XM_034872 | |
| Sept9 | 17q25.3 | 586 | 65.4 | 9.3 | B | 0 | AF189713 | |
| Sept10 | 8q11.23 | 517 | 60.0 | 6.6 | N | 1 | BC020502 | |
| Sept11 | 4q21.22 | 429 | 49.4 | 6.4 | A | 1 | NM_018243 | |
| Sept12 | 16p13.3 | 358 | 40.8 | 6.7 | N | 0 | NM_144605 |
The data refer to the largest gene products for each gene, deduced from cDNAs on the sequence databases. *The numbers given for the yeast genes refer to the position of the gene along the sequence of the chromosome. A, acidic (pI < 6.5); B, basic (pI > 7.5); N, neutral. The algorithm COILS [56,57] was used to predict coiled coils, and the peaks above an arbitrary threshold (p > 0.8 at a window size of 14) were counted. The mouse genome has counterparts to each of the 12 human septin genes (not shown). For comparative nomenclature of the mouse and human Sept1-Sept10 genes and the products, see [58].
Figure 1.
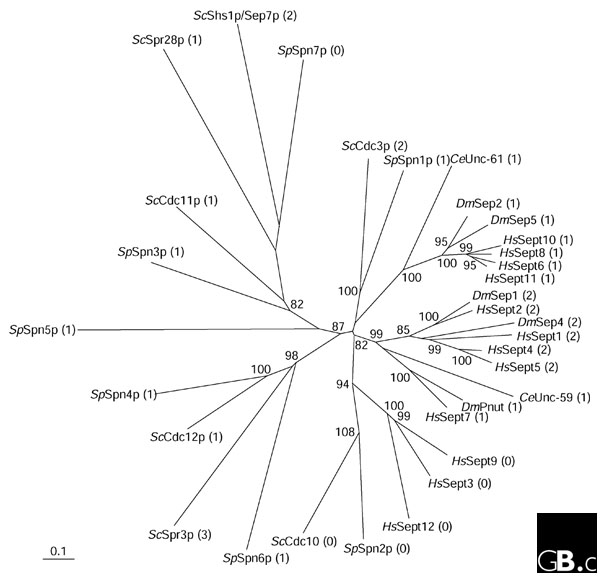
A phylogenetic tree of the septins in Saccharomyces cerevisiae (Sc), Schizosaccharomyces pombe (Sp), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm) and humans (Hs). The longest amino-acid sequence among the putative polypeptides generated by each gene was analyzed with the software Phylip [59] using the default mode with the UPGMA method, 1,000 bootstrap replicates and systematic tie-breaking, and Poisson-corrected distances with proportionally distributed gaps. The numbers of predicted coiled coils are shown in parentheses. The scale bar represents 0.1 substitutions.
Characteristic structural features
The full-length septin cDNAs in the current sequence databases encode polypeptides of 30-65 kDa. Most of these gene products have a set of GTPase motifs, G-1, G-3 and G-4, found in members of the GTPase superfamily, (Figure 2 and not shown). The GTPase motifs of the septins are closer to those of the Ras family than of other members of the GTPase superfamily [6] such as the other cytoskeletal GTPases, tubulins in eukaryotes or FtsZ in bacteria. The G-1 motif (which has a consensus in the superfamily of GxxxxGK[S/T] in the single-letter amino-acid code) is well conserved, and the consensus around the G-1 motif of septins is GESGLGKSTLINTLF (where the bold residues are strictly conserved). The G-3 motif (DxxG) is moderately conserved, with the consensus sequence DTPG; the G-4 motif (xKxD) is strictly conserved with a unique septin consensus of AKAD. The G-2 and G-5 regions cannot be defined in septins; some other classes of GTPases also lack these motifs. GTP-binding and GTP-hydrolyzing activities of the purified and recombinant septin complexes or polypeptides have been demonstrated in vitro ([7-11] and M.K., C.M. Field, M.L. Coughlin and T.J. Mitchison, unpublished observations). The biochemical and biological significance of septin GTPase activity remains a conundrum in the field, however.
Figure 2.
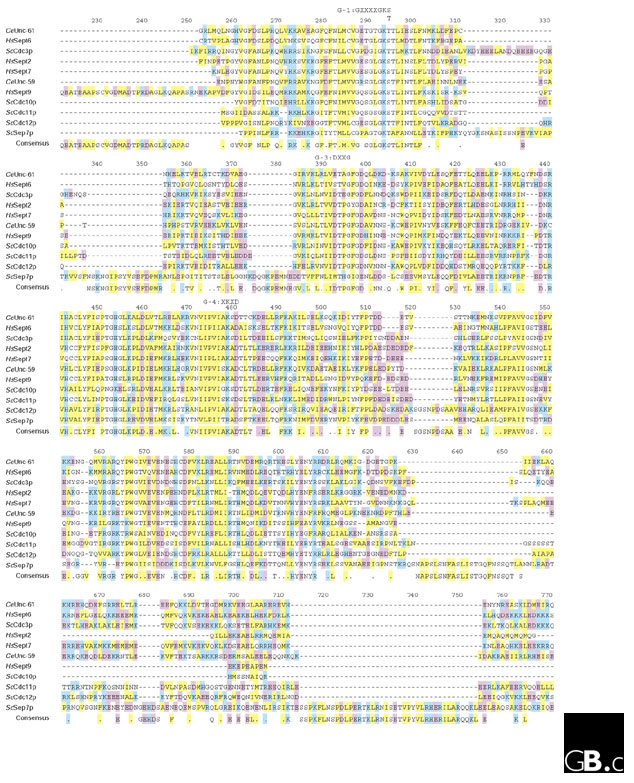
Multiple alignment of the central regions of representative septins. Amino-acid sequences of the representative septins were aligned using MacVector. Acidic, basic and hydrophobic residues are in purple, blue, and yellow respectively. The GTPase motifs that are conserved in this family - G-1, G-3 and G-4 - are indicated above the sequence. A few other conserved stretches of hydrophobic and charged residues are also recognizable. Species abbreviations are as in Figure 1.
Septins polymerize to form rod-shaped hetero-oligomeric complexes, which in turn are arranged in tandem arrays to form filaments that appear by electron microscopy to be 7-9 nm thick. These filaments can assemble in vitro into even higher-order structures by self-assembly and templated assembly. Repeating unit complexes made up of Cdc3p, Cdc10p, Cdc11p, and Cdc12p in budding yeast, Sep1, Sep2 and Pnut in flies, and Sept2, Sept6, and Sept7 in mouse and human have been purified and characterized [4,7,10,12-14]. The majority of septins are predicted to have one or more coiled-coil regions, each spanning about 50-100 amino-acid residues, mostly near the carboxyl termini. In the metazoan septins, proteins that are close on the phylogenetic tree have the same number of coiled coils (Figure 1). Some of the coiled-coil regions are necessary for intermolecular interaction upon septin complex formation [10], whereas others are dispensable ([11,15] and M.K., C.M. Field, M.L. Coughlin and T.J. Mitchison, unpublished observations). Some septins have no predictable coiled-coil region (for example, S. cerevisiae Cdc10p, S. pombe Spn2p and Spn7p, and human Sept3, Sept 9 and Sept12). The mechanism of inter-septin interaction other than through coiled coils is unknown.
The isoelectric points of most septin polypeptides are within the acidic to neutral range, but each organism has one or two septins of basic charge (for example, S. cerevisiae Cdc12p, S. pombe Spn2p and Spn5p, both C. elegans septins, Drosophila Pnut and human Sept7 and Sept9; these are indicated in Table 1). The nematode is exceptional in that it has only two septin genes, both of which encode highly basic proteins. The significance of the isoelectric points of septins is currently unknown. Regardless of the total charge, a short stretch of basic residues preceding the G-1 region is shared by most, but not all, of the septins. Some of these basic residues are critical for interactions with phospholipids in vitro [9,11].
The budding yeast septins Cdc3p, Cdc11p and Shs1p have one or more motifs for sumoylation, [I/V/L]KX[E/D]; the lysine is the attachment site for the ubiquitin-like protein SUMO. Mutating these sites results in loss of bud-neck-associated SUMO and persistent septin rings [16]. Thus, SUMO conjugation is a prerequisite for septin-ring disassembly. This discovery provided a breakthrough towards an understanding of the regulatory mechanism of yeast septin dynamics, and it also suggests that the significance of the sumoylation motifs found in septins from other organisms should be tested.
Localization and function
Expression of the septin genes seems to be regulated according to the cell cycle, cell lineage, and developmental stage. In accordance with a generally accepted notion that the hetero-oligomeric complex is the main functional unit of the septin system [4], the cell-type distributions of different septin proteins largely overlap one another. Paradoxically, however, their subcellular localization is not necessarily identical; this is demonstrated, for example in postmitotic cells in the mouse brain [17]. The differential localization of septin proteins or complexes may reflect their distinct roles in vivo. Besides the best-known functions in cytokinesis, the septin system seems to have evolved to fulfill multiple roles in dividing and non-dividing cells. The normal localization, mutant phenotypes, and possible functions inferred from genetic and cell biological data are summarized for key organisms below.
S. cerevisiae and S. pombe
The 'classical' septins of budding yeast (Cdc3p, Cdc10p, Cdc10p, Cdc12p, and Shs1p/Sep7p) predominantly occur as ring(s) encircling the mother-bud neck, but they also localize at the cell cortices near the presumptive bud site, at the bud scar after cytokinesis, and at the tapering part and the tip of the shmoo, a pheromone-induced protrusion [2,18,19]. As described above, the main phenotype of the original temperature-sensitive mutants (cdc3, cdc10, cdc11 and cdc12) is a lack of bud-neck filaments and cytokinesis defects. The CDC3Δ and CDC12Δ mutants are lethal; the CDC10Δ and CDC11Δ mutants are viable but are unable to organize the bud-neck filaments (the septin ring), and the other septins localize to the bud neck to partially fulfill the functions of the missing septins [12,19].
The septin ring is a multifunctional structure that serves several functions: firstly, as a spatial landmark to establish cell polarity for bud-site selection, in cooperation with other proteins (such as the bud-site selection proteins Bud3p and Bud4p) [20,21]; secondly, as a barrier that prevents bud-specific cortical molecules (Spa2p, Sec3p, Sec5p, Ist2p and others) from diffusing laterally into the mother-cell cortex [22,23]; thirdly, as a scaffold to recruit molecules for cell-wall synthesis (for example, the chitin synthases Chs4p and Chs3p and the scaffold protein Bni4p) [24] and for positioning of the mitotic spindle [25]; and finally, as an apparatus to monitor and control progression of mitosis in conjunction with the cell-cycle regulatory kinases Gin4p, Hsl1p and Kcc4p [26-28], and a component of the mitosis exit network, Tem1p [29,30].
The 'non-classical' S. cerevisiae septins (Spr3p and Spr28p) are expressed in a temporally limited manner during spore formation and are targeted beneath the developing prospore wall [15,31,32]. Deletion of the SPR3 or SPR28 genes causes no obvious phenotype, and a double mutant has minimal defects in sporulation, suggesting that there is compensation by the other septins [15,32].
S. pombe Spn1p, Spn3p, and Spn4p localize to medial ring(s) around the circumference of the dividing cell, where they functionally interact with Mid2p (which is related to the actin-binding protein anillin in animals). The spn1Δ and spn4Δ mutants show mild cytokinetic defects such as delayed cell-cell separation and accumulation of cells with one or more septa [2,33,34].
Animals
The C. elegans UNC-59 and UNC-61 septin proteins localize to the leading edge of the cleavage furrow and the spindle midbody. Mutants of either or both of them exhibit minimal defects in embryonic cytokinesis, but abnormalities in postembryonic morphogenesis occur in multiple organs; these include vulva protrusion, germ-cell defects including gonad extrusion, egg-laying defects, and deformities in the male tail and male sensory neurons. The uncoordinated movement defect through which the mutants were originally isolated also indicates some functional defects in the mutants' nervous systems. Some of these phenotypes are recapitulated by silencing unc-59 and/or unc-61 through siRNA microinjection of small interfering RNAs (siRNAs) [35,36].
In the Drosophila embryo, the Pnut, Sep1, and Sep2 septin proteins have been found in the front of cellularization moving along the early embryo, in the cleavage furrows of dividing cells, and at the leading edges of the epithelium during embryonic dorsal closure. Later in development, they are found in the apical and basal cell cortices of larval imaginal discs, in the cell cortices of the embryonic and larval central nervous system and of photoreceptor cells in the eye imaginal discs [37-39], and in ring canals (stable intercellular bridges formed by incomplete cytokinesis of male and female germ cells) [7,40,41]. The pnut gene was identified as an enhancer of the seven in absentia defect, which results in loss of the R7 photoreceptor cells; pnut-null mutant larvae have severely reduced cell number, with multinucleated cells in the imaginal discs and brain, and they die shortly after pupation [37]. Mutant embryos lacking the Pnut contribution from both the mother and the zygote have abnormal organization of actin rings in the late cellularization stage of embryogenesis and extensive morphological defects during gastrulation and in the formation of cuticle, head, tail, and denticles [39].
Mammalian septins have been found in the cell cortex, contractile ring and midbody of mitotic cells (Sept2, Sept4, Sept6, Sept7, and Sept9) and in the cell cortex, actin stress fibers (Sept2, Sept4, Sept6, Sept7, and Sept9) and microtubules (Sept9) of interphase cells ([8,9,13,14,42-46] and M.K., C.M. Field, M.L. Coughlin and T.J. Mitchison, unpublished observations). In the nervous system, they are seen on the cytoplasmic side of presynaptic membranes (Sept7) and synaptic vesicles (Sept5 and Sept6) and in the endfeet of astroglia (Sept4 and Sept7) [17]. Cytokinesis is perturbed by microinjection of anti-septin antibodies (against Sept2 and Sept9) or transfection of siRNAs (against Sept2, Sept7, Sept9) [8,45,46]. Depletion of Sept2 or Sept7 protein by RNA interference also causes disorganization of actin stress fibers, leading to a flat cell morphology in interphase cells [14]. Although Sept5 is highly expressed in mature nervous systems, no brain abnormality is seen in the Sept5-null mice, probably because of compensation by redundant septin species [47]. Sept5-null mice do, however, aggregate and release granules from blood platelets more readily than do wild-type mice [48].
Frontiers
A number of open questions remain with regard to the septins. Firstly, the fine structures of septins beyond the ultrastructural level are totally unknown. Resolving the atomic structures of septin monomers, oligomers and polymers should help us to address the major questions in septin biochemistry, such as the mechanisms of septin polymer assembly and disassembly and how GTP hydrolysis might be coupled to changes in the structure and activity of the proteins. It will be important to elucidate the mechanisms by which sumoylation and phosphorylation might control septin assembly and disassembly at the structural, biochemical, and cellular levels [16,49].
The interactions of septins with non-septin molecules - such as actin and anillin [8,14,33,34], microtubules [25,45,46], mitosis-associated proteins (see above), and lipids [9,11] - should help to reveal their unknown cellular functions and to clarify the mechanisms underlying the events in which they are involved. Likewise, the discovery of new subcellular localizations of septins may also lead to discoveries of novel roles for the proteins, as is illustrated by a mitochondrial septin variant that has been implicated in apoptosis [50].
Many research groups have found independently that two human septin genes from different groups (Sept6 and Sept9; see Figure 1) have translocated to, and fused in-frame with, the mixed lineage leukemia (MLL) gene, and that a few human and mouse septin genes (Sept2, Sept4, and Sept9) are amplified and/or aberrantly expressed in a variety of malignancies, including leukemia, lymphoma and solid tumors (see, for example, [51,52]). Although the hypothetical oncogenic activities of these septins and the fusion proteins remain to be tested, exploring the involvement of septins in carcinogenesis should bring novel perspectives to cancer research as well as to septin biology.
As mentioned above, a subset of septins are abundantly expressed in metazoan nervous systems, but the biological significance of septins in postmitotic neurons and glial cells has not been understood. Considering the functional redundancy and complexity of the septins, determining their precise roles in neural development and in synaptic or glial functions is challenging, even using systematic genetic analysis including multiple and conditional gene disruption. The septins of nematodes have the potential to lead the field of septin neurobiology, given their relative simplicity.
Finally, in addition to the elusive functions of septins in normal brains, aberrant deposits of septins have been found in neurofibrillary tangles in Alzheimer's disease, in Lewy bodies in Parkinson's disease, and in related pathological aggregates in human brains [53,54]. Exploring the possible linkages between septins and the major players in each disease (such as amyloid-precursor protein, presenilins, and tau proteins in Alzheimer's disease and parkin, the Pael receptor, and synucleins in Parkinson's disease [54,55]) is expected to reveal functions for septins in the brain and help to clarify the unknown pathophysiology underlying these disorders.
References
- Hartwell LH. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. The original report describing the isolation of the septin mutants. [DOI] [PubMed] [Google Scholar]
- Longtine MS, Demarini DJ, Valencik ML, Al-Awar OS, Fares H, De Virgilio C, Pringle JR. The septins, roles in cytokinesis and other processes. Curr Opin Cell Biol. 1996;8:106–119. doi: 10.1016/S0955-0674(96)80054-8. The first comprehensive review on the septin family, including important previously unpublished observations and insights based on original data. [DOI] [PubMed] [Google Scholar]
- Cooper JA, Kiehart DP. Septins may form a ubiquitous family of cytoskeletal filaments. J Cell Biol. 1996;134:1345–1348. doi: 10.1083/jcb.134.6.1345. This article and [4] are reviews of the septin family that focus on their role as components of a novel cytoskeletal system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field CM, Kellogg D. Septins, cytoskeletal polymers or signalling GTPases? Trends Cell Biol. 1999;9:387–394. doi: 10.1016/S0962-8924(99)01632-3. See [3]. [DOI] [PubMed] [Google Scholar]
- Byers B, Goetsch L. A highly ordered ring of membrane-associated filaments in budding yeast. J Cell Biol. 1976;69:717–721. doi: 10.1083/jcb.69.3.717. The historical discovery of the electron-dense striations at the mother-bud neck. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. A classic review on biochemical and structural aspects of the GTP-binding protein families. [DOI] [PubMed] [Google Scholar]
- Field CM, Al-Awar O, Rosenblatt J, Wong ML, Alberts BM, Mitchison TJ. A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J Cell Biol. 1996;133:605–616. doi: 10.1083/jcb.133.3.605. A pioneering study of the biochemistry, ultrastructure, and self-assembly of the fly septin complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M, Kumar S, Mizoguchi A, Ide C, Kinoshita A, Haraguchi T, Hiraoka Y, Noda M. Nedd5, a mammalian septin, is a novel cytoskeletal component interacting with actin-based structures. Genes Dev. 1997;11:1535–1547. doi: 10.1101/gad.11.12.1535. Identification of the mouse and human Sept2 proteins and cell biological studies for their roles in cytokinesis and interphase. [DOI] [PubMed] [Google Scholar]
- Zhang J, Kong C, Xie H, McPherson PS, Grinstein S, Trimble WS. Phosphatidylinositol polyphosphate binding to the mammalian septin H5 is modulated by GTP. Curr Biol. 1999;9:1458–1467. doi: 10.1016/S0960-9822(00)80115-3. The discovery of a septin-phospholipid interaction in vitro. [DOI] [PubMed] [Google Scholar]
- Sheffield PJ, Oliver CJ, Kremer BE, Sheng S, Shao Z, Macara IG. Borg/septin interactions and the assembly of mammalian septin heterodimers, trimers, and filaments. J Biol Chem. 2003;278:3483–3488. doi: 10.1074/jbc.M209701200. The first reconstitution of partial and complete septin complexes using a bacterial expression system. [DOI] [PubMed] [Google Scholar]
- Casamayor A, Snyder M. Molecular dissection of a yeast septin: distinct domains are required for septin interaction, localization, and function. Mol Cell Biol. 2003;23:2762–2777. doi: 10.1128/MCB.23.8.2762-2777.2003. A series of mutant studies to map septins' functional domains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier JA, Wong ML, Longtine MS, Pringle JR, Mann M, Mitchison TJ, Field CM. Polymerization of purified yeast septins, evidence that organized filament arrays may not be required for septin function. J Cell Biol. 1998;143:737–749. doi: 10.1083/jcb.143.3.737. An excellent combination of genetic, biochemical and ultrastructural analyses on the budding yeast septin complexes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joberty G, Perlungher RR, Sheffield PJ, Kinoshita M, Noda M, Haystead T, Macara IG. Borg proteins control septin organisation and are negatively regulated by Cdc42. Nat Cell Biol. 2001;3:861–866. doi: 10.1038/ncb1001-861. The discovery of a family of adapter proteins that link Cdc42 and the mammalian Sept2-Sept6-Sept7 complex. [DOI] [PubMed] [Google Scholar]
- Kinoshita M, Field CM, Coughlin ML, Straight AF, Mitchison TJ. Self- and actin-templated assembly of mammalian septins. Dev Cell. 2002;3:791–802. doi: 10.1016/s1534-5807(02)00366-0. The first reconstitution of the filamentous septin complex and the higher-order structures, also demonstrating a septin-actin interaction mediated by anillin. [DOI] [PubMed] [Google Scholar]
- De Virgilio C, DeMarini DJ, Pringle JR. SPR28, a sixth member of the septin gene family in Saccharomyces cerevisiae that is expressed specifically in sporulating cells. Microbiology. 1996;142:2897–2905. doi: 10.1099/13500872-142-10-2897. This report and [31,32] describe the identification and characterization of the sporulation-specific septins. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147:981–994. doi: 10.1083/jcb.147.5.981. An original study connecting the two distinct fields of 'septinology' and 'sumology'. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A, Noda M, Kinoshita M. Differential localization of septins in the mouse brain. J Comp Neurol. 2000;428:223–239. doi: 10.1002/1096-9861(20001211)428:2<223::AID-CNE3>3.0.CO;2-M. The first ultrastructural mapping of septins in postmitotic neurons and glial cells by immuno-electron microscopy. [DOI] [PubMed] [Google Scholar]
- Gladfelter AS, Pringle JR, Lew DJ. The septin cortex at the yeast mother-bud neck. Curr Opin Microbiol. 2001;4:681–689. doi: 10.1016/S1369-5274(01)00269-7. A comprehensive review of the septins and other molecules localized at the cell cortex. [DOI] [PubMed] [Google Scholar]
- Mino A, Tanaka K, Kamei T, Umikawa M, Fujiwara T, Takai Y. Shs1p: a novel member of septin that interacts with Spa2p, involved in polarized growth in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1998;251:732–736. doi: 10.1006/bbrc.1998.9541. The discovery and characterization of a budding yeast septin that had not been identified in [1]. [DOI] [PubMed] [Google Scholar]
- Chant J, Mischke M, Mitchell E, Herskowitz I, Pringle JR. Role of Bud3p in producing the axial budding pattern of yeast. J Cell Biol. 1995;129:767–778. doi: 10.1083/jcb.129.3.767. This paper and [21] are the initial studies on the relationship between cell polarity-related proteins and the septin ring. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Herskowitz I. The BUD4 protein of yeast, required for axial budding, is localized to the mother/BUD neck in a cell cycle-dependent manner. J Cell Biol. 1996;134:413–427. doi: 10.1083/jcb.134.2.413. See [20]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral Y, Mermall V, Mooseker MS, Snyder M. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol Cell. 2000;5:841–851. doi: 10.1016/S1097-2765(00)80324-X. This paper and [23] are two independent studies that established a concept of the septin ring as a diffusion barrier for bud-specific cortical proteins. [DOI] [PubMed] [Google Scholar]
- Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290:341–344. doi: 10.1126/science.290.5490.341. See [22]. [DOI] [PubMed] [Google Scholar]
- DeMarini DJ, Adams AE, Fares H, De Virgilio C, Valle G, Chuang JS, Pringle JR. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J Cell Biol. 1997;139:75–93. doi: 10.1083/jcb.139.1.75. An elaborate study that established the concept of the septin ring as a scaffold for the cell-wall synthesis machinery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch J, Meyer A, Snyder MP, Barral Y. Microtubule capture by the cleavage apparatus is required for proper spindle positioning in yeast. Genes Dev. 2002;16:1627–1639. doi: 10.1101/gad.222602. A study proposing that the mitotic spindle may physically contact the septin ring for proper positioning against the cell cortex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Altman R, Schieltz D, Yates JR, Kellogg D. The septins are required for the mitosis-specific activation of the Gin4 kinase. J Cell Biol. 1998;143:709–717. doi: 10.1083/jcb.143.3.709. This article and [27,28] report the discoveries that the three similar kinases, Gin4p, Hsl1p and Kcc4p and the septin ring are inter-dependent and cooperatively control mitosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, Fares H, Pringle JR. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J Cell Biol. 1998;143:719–736. doi: 10.1083/jcb.143.3.719. See [26]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral Y, Parra M, Bidlingmaier S, Snyder M. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 1999;13:176–187. doi: 10.1101/gad.13.2.176. See [26]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez J, Cid VJ, Cenamor R, Yuste M, Molero G, Nombela C, Sanchez M. Morphogenesis beyond cytokinetic arrest in Saccharomyces cerevisiae. J Cell Biol. 1998;143:1617–1634. doi: 10.1083/jcb.143.6.1617. This paper and [30] report initial studies indicating septin's functional interaction with components of the mitotic exit network. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J, Shannon KB, Shou W, Deshaies RJ, Li R. The Tem1 small GTPase controls actomyosin and septin dynamics during cytokinesis. J Cell Sci. 2001;114:1379–1386. doi: 10.1242/jcs.114.7.1379. See [29]. [DOI] [PubMed] [Google Scholar]
- Ozsarac N, Bhattacharyya M, Dawes IW, Clancy MJ. The SPR3 gene encodes a sporulation-specific homologue of the yeast CDC3/10/11/12 family of bud neck microfilaments and is regulated by ABFI. Gene. 1995;164:157–162. doi: 10.1016/0378-1119(95)00438-C. See [15]. [DOI] [PubMed] [Google Scholar]
- Fares H, Goetsch L, Pringle JR. Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. J Cell Biol. 1996;132:399–411. doi: 10.1083/jcb.132.3.399. See [15]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin A, Paoletti A, Chang F. Mid2p stabilizes septin rings during cytokinesis in fission yeast. J Cell Biol. 2003;160:1083–1092. doi: 10.1083/jcb.200212016. This paper and [34] report characterization of the septin ring and its dependence on one of the two anillin-related proteins in fission yeast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasto JJ, Morrell JL, Gould KL. An anillin homologue, Mid2p, acts during fission yeast cytokinesis to organize the septin ring and promote cell separation. J Cell Biol. 2003;160:1093–1103. doi: 10.1083/jcb.200211126. See [33]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Horvitz HR, Sulston JE. Neurone differentiation in cell lineage mutants of Caenorhabditis elegans. Nature. 1982;297:584–587. doi: 10.1038/297584a0. This paper and [36] are the original reports describing genetic screening and morphology of the nematode septin mutants. [DOI] [PubMed] [Google Scholar]
- Nguyen TQ, Sawa H, Okano H, White JG. The C. elegans septin genes, unc-59 and unc-61, are required for normal postembryonic cytokineses and morphogenesis but have no essential function in embryogenesis. J Cell Sci. 2000;113:3825–3837. doi: 10.1242/jcs.113.21.3825. See [35]. [DOI] [PubMed] [Google Scholar]
- Neufeld TP, Rubin GM. The Drosophila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell. 1994;77:371–379. doi: 10.1016/0092-8674(94)90152-x. A citation classic describing identification and phenotype of the first metazoan septin mutant lacking Pnut. [DOI] [PubMed] [Google Scholar]
- Fares H, Peifer M, Pringle JR. Localization and possible functions of Drosophila septins. Mol Biol Cell. 1995;6:1843–1859. doi: 10.1091/mbc.6.12.1843. A report on subcellular co-localization and co-immunoprecipitation of Pnut and Sep1, indicating their complex formation in cytokinesis and embryonic development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam JC, Pringle JR, Peifer M. Evidence for functional differentiation among Drosophila septins in cytokinesis and cellularization. Mol Biol Cell. 2000;11:3123–3135. doi: 10.1091/mbc.11.9.3123. A genetic and morphological dataset indicating functional diversity and redundancy of the fly septin system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hime GR, Brill JA, Fuller MT. Assembly of ring canals in the male germ line from structural components of the contractile ring. J Cell Sci. 1996;109:2779–2788. doi: 10.1242/jcs.109.12.2779. A report on similarities and differences in the ring canals between male and female germ cells in Drosophila. [DOI] [PubMed] [Google Scholar]
- Robinson DN, Cooley L. Genetic analysis of the actin cytoskeleton in the Drosophila ovary. Annu Rev Cell Dev Biol. 1997;13:147–170. doi: 10.1146/annurev.cellbio.13.1.147. An insightful review on the ring canals of Drosophila. [DOI] [PubMed] [Google Scholar]
- Hsu SC, Hazuka CD, Roth R, Foletti DL, Heuser J, Scheller RH. Subunit composition, protein interactions, and structures of the mammalian brain Sec6/8 complex and septin filaments. Neuron. 1998;20:1111–1122. doi: 10.1016/s0896-6273(00)80493-6. A report on copurification of the mammalian septin complexes with the Sec6/8 complex from the rat brain. Their direct interaction has not been demonstrated. [DOI] [PubMed] [Google Scholar]
- Xie H, Surka M, Howard J, Trimble WS. Characterization of the mammalian septin H5: distinct patterns of cytoskeletal and membrane association from other septin proteins. Cell Motil Cytoskeleton. 1999;43:52–62. doi: 10.1002/(SICI)1097-0169(1999)43:1<52::AID-CM6>3.3.CO;2-X. A careful study showing the differential subcellular localization of Sept2 and Sept4. [DOI] [PubMed] [Google Scholar]
- Beites CL, Xie H, Bowser R, Trimble WS. The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat Neurosci. 1999;2:434–439. doi: 10.1038/8100. The discovery of a physical and functional interaction between the general secretory machinery and Sept5. [DOI] [PubMed] [Google Scholar]
- Surka MC, Tsang CW, Trimble WS. The mammalian septin MSF localizes with microtubules and is required for completion of cytokinesis. Mol Biol Cell. 2002;13:3532–3545. doi: 10.1091/mbc.E02-01-0042. This paper and [46] are two careful studies demonstrating a Sept9-microtubule interaction and examining its significance in mitosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata KI, Kawajiri A, Matsui S, Takagishi M, Shiromizu T, Saitoh N, Izawa I, Kiyono T, Itoh TJ, Hotani H, Inagaki M. Filament formation of MSF-A, a mammalian septin, in mammary epithelial cells depends on interactions with microtubules. J Biol Chem. 2003;278:18538–18543. doi: 10.1074/jbc.M205246200. See [45]. [DOI] [PubMed] [Google Scholar]
- Peng XR, Jia Z, Zhang Y, Ware J, Trimble WS. The septin CDCrel-1 is dispensable for normal development and neurotransmitter release. Mol Cell Biol. 2002;22:378–387. doi: 10.1128/MCB.22.1.378-387.2002. This paper and [48] are the first attempts to genetically disrupt a mammalian septin that is mainly expressed in the brain and blood platelets. The absence of Sept5 affected platelet functions but was tolerated in the brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent J, Kato K, Peng XR, Martinez C, Cattaneo M, Poujol C, Nurden P, Nurden A, Trimble WS, Ware J. A prototypic platelet septin and its participation in secretion. Proc Natl Acad Sci USA. 2002;99:3064–3069. doi: 10.1073/pnas.052715199. See [47]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J, Gentry MS, Hallberg RL, Barral Y. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev Cell. 2003;4:345–357. doi: 10.1016/s1534-5807(03)00061-3. A breakthrough demonstrating the two kinases and a phosphatase subunit that control the turnover of the septin ring. [DOI] [PubMed] [Google Scholar]
- Larisch S, Yi Y, Lotan R, Kerner H, Eimerl S, Tony Parks W, Gottfried Y, Birkey Reffey S, de Caestecker MP, Danielpour D, et al. A novel mitochondrial septin-like protein, ARTS, mediates apoptosis dependent on its P-loop motif. Nat Cell Biol. 2000;2:915–921. doi: 10.1038/35046566. The discovery of a splice variant of Sept4 with an incomplete set of GTPase motifs. [DOI] [PubMed] [Google Scholar]
- Osaka M, Rowley JD, Zeleznik-Le NJ. MSF (MLL septin-like fusion), a fusion partner gene of MLL, in a therapy-related acute myeloid leukemia with a t(11;17)(q23;q25). Proc Natl Acad Sci USA. 1999;96:6428–6433. doi: 10.1073/pnas.96.11.6428. One of the earliest reports on the Sept9-MLL gene fusion in human malignancies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagna C, Lyu MS, Hunter K, Lukes L, Lowther W, Reppert T, Hissong B, Weaver Z, Ried T. The Septin 9 (MSF) gene is amplified and overexpressed in mouse mammary gland adenocarcinomas and human breast cancer cell lines. Cancer Res. 2003;63:2179–2187. A demonstration of cancer-associated amplification and overexpression of the Sept9 gene in two species. [PubMed] [Google Scholar]
- Kinoshita A, Kinoshita M, Akiyama H, Tomimoto H, Kumar S, Noda M, Kimura J. Identification of septins in the neurofibrillary tangles in Alzheimer's disease. Am J Pathol. 1998;153:1551–1560. doi: 10.1016/S0002-9440(10)65743-4. A serendipitous discovery of a subset of septins in the human pathology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara M, Tomimoto H, Kitayama H, Morioka Y, Akiguchi I, Shibasaki H, Noda M, Kinoshita M. Association of the cytoskeletal GTP-binding protein Sept4/H5 with cytoplasmic inclusions found in Parkinson's disease and other synucleinopathies. J Biol Chem. 2003;278:24095–24102. doi: 10.1074/jbc.M301352200. Histopathological and biochemical analyses of the interaction between Sept4 and α-synuclein. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gao J, Chung KK, Huang H, Dawson VL, Dawson TM. Parkin functions as an E2-dependent ubiquitin-protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc Natl Acad Sci USA. 2000;97:13354–13359. doi: 10.1073/pnas.240347797. Identification of Sept5 as a substrate of an E3 ligase, parkin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. A program for predicting coiled-coil-forming regions (available at [57]). [DOI] [PubMed] [Google Scholar]
- COILS - prediction of coiled coil regions in proteins http://www.ch.embnet.org/software/COILS_form.html See [56].
- Macara IG, Baldarelli R, Field CM, Glotzer M, Hayashi Y, Hsu SC, Kennedy MB, Kinoshita M, Longtine M, Low C, et al. Mammalian septins nomenclature. Mol Biol Cell. 2002;13:4111–4113. doi: 10.1091/mbc.E02-07-0438. The nightmarish confusion of the mammalian septins nomenclature becomes history. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phylip http://evolution.genetics.washington.edu/phylip.html A free package of programs for inferring phylogenies.


