Abstract
Like other energy-dependent proteases, proteasomes, which are found across the three domains of life, are self-compartmentalized and important in the early steps of proteolysis. Proteasomes degrade improperly synthesized, damaged or misfolded proteins and hydrolyse regulatory proteins that must be specifically removed or cleaved for cell signalling. In eukaryotes, proteins are typically targeted for proteasome-mediated destruction through polyubiquitylation, although ubiquitin-independent pathways also exist. Interestingly, actinobacteria and archaea also covalently attach small proteins (prokaryotic ubiquitin-like protein (Pup) and small archaeal modifier proteins (Samps), respectively) to certain proteins, and this may serve to target the modified proteins for degradation by proteasomes.
Proteasomes are large self-compartmentalized, energy-dependent proteases found in eukaryotes, archaea and actinobacteria1. These nanomachines function in protein quality control by degrading misfolded, damaged and inaccurately synthesized proteins2,3. Proteasomes also serve as highly specialized proteases that regulate cell division, DNA repair and other important processes by destroying regulatory proteins at specific times and locations in the cell4–6. Most proteins degraded by proteasomes are hydrolysed processively into small peptides7. However, proteasomes can also cleave precursors to yield biologically active proteins8.
Proteins that are targeted to proteasomes often contain amino acid sequences that act as specific degradation signals, or ‘degrons’. Degrons initiate the process of proteolysis and can vary greatly, ranging from phosphorylated amino acid residues to exposed amino or carboxyl termini9,10. Often, eukaryotic degrons are recognized by the ubiquitylation system, resulting in the covalent attachment of polyubiquitin chains to the substrate protein; these chains are then recognized by proteasomes for proteolysis11.
The degrons that stimulate proteasome-mediated proteolysis in archaea and actinobacteria are not as well defined as those in eukaryotes. However, recent evidence reveals that protein conjugation may serve as an intermediary step in the proteolytic processes of archaea and actinobacteria, similarly to ubiquitylation in eukaryotes. Through groundbreaking work, actinobacteria were shown to modify proteins by the attachment of a small protein modifier termed prokaryotic ubiquitin-like protein (Pup), which can target proteins for degradation by proteasomes12,13. More recently, archaea were found to covalently modify proteins by a mechanism termed sampylation (using small archaeal modifier proteins (Samps))14. Although sampylation is more closely related to ubiquitylation than pupylation14,15, and sampylated proteins accumulate in proteasomal mutants14, a direct connection between sampylation and proteasomes has yet to be demonstrated.
This Review discusses what is known about proteasomes, including their structure and function, across the three domains of life, and describes the three protein conjugation systems (ubiquitylation, pupylation and sampylation) that are used to target proteins for proteasomal degradation.
Proteasome structure and function
All three domains of life use proteasomes to catalyse protein degradation. Below, I describe the structure of protea somes in eukaryotes (using yeast nomenclature for protein names), actinobacteria and archaea, and discuss the mechanism by which they mediate proteolysis.
The proteasome core particle
The central component of all proteasomes is a self-compartmentalized 20S core particle (CP) that harbours the proteolytic active sites16 (FIG. 1a). The CPs are cylindrical, with narrow entry portals on each end that open to a central channel connecting three interior chambers. The central chamber is lined with the proteolytic active sites, which form during CP assembly. Although the active sites are reasonably nonspecific in the types of peptide bonds that they cleave, substrate specificity can be provided by gating at the entry portals on each end of the CP, which also limits substrate access (see below). Regulatory complexes such as ATPases of the AAA+ superfamily can physically interact with CPs and stimulate proteolysis by unfolding the protein substrate, opening the CP gates and translocating the protein substrate into the interior of the CP17.
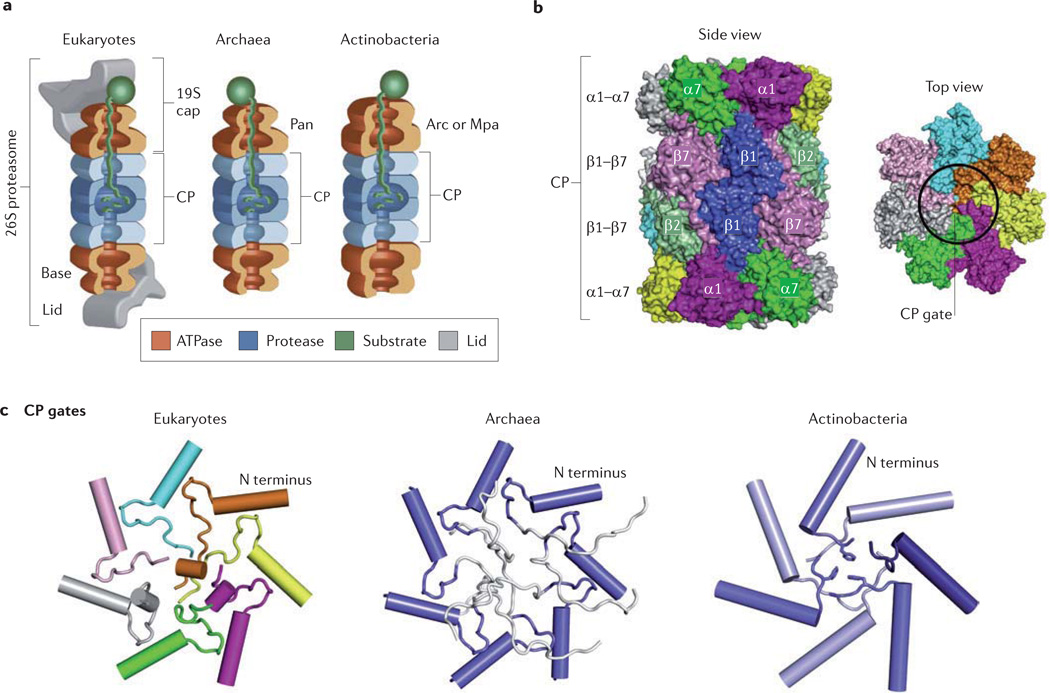
Figure 1. Basic structures of proteasomes across domains of life.
a | All proteasomes are composed of a 20S catalytic core particle (CP) formed from four stacked heptameric rings of α- and β-subunits. The CPs can associate with AAA+ ATPases, which unfold and translocate substrate proteins into the CP by an ATP-dependent mechanism. In eukaryotes (using yeast as an example), six different ATPase subunits (Rpt1–Rpt6) form the hexameric ring of the 19S cap, which associates with CPs to form 26S proteasomes. The 19S cap can be separated into base and lid subcomplexes, with the base harbouring the Rpt1–Rpt6 subunits (which use ATP to fuel CP-mediated degradation of folded proteins), and the lid including the deubiquitylating enzyme Rpn11. The proteasomal AAA+ proteins of archaea (proteasome-activating nucleotidase (Pan)) and of actinobacteria (AAA+ ATPase forming a ring-shaped complex (Arc) or mycobacterial proteasome ATPase (Mpa)) assemble into homohexameric rings and associate with CPs in vitro, but the evidence that these ATPases interact with their cognate CPs in vivo is limited. b | Side and top views of a yeast CP provide a perspective on the basic CP structure. c | Across the domains of life, the α-subunit amino-terminal tails that gate the openings of CPs differ in the extent to which they seal the CP channel from substrate entry. Eukaryotic CPs are gated primarily by the well-ordered N-terminal tails of α2, α3 and α4 subunits, which form numerous hydrogen bonds and van der Waals contacts. Gates of archaeal CPs (Thermoplasma acidophlium CPs synthesized in recombinant Escherichia coli) are disordered (residues in white are highly mobile). In actinobacterial CPs, the seven α-subunits are identical but can adopt three different conformations at their N termini (indicated by different shading) to form an ordered closed gate. Part a is modified, with permission, from REF. 117 © (2009) Elsevier. Parts b and c are reproduced, with permission, from REF. 21 © (2011) Elsevier.
CPs from all three domains of life are similar in overall structure and are formed from structurally related α- and β-subunits; these associate as four stacked heptameric rings16, with the outermost rings made up of α-subunits and the inner two rings made of β-subunits, assembled in an α7β7β7α7 symmetry (FIG. 1b). The proteolytic active sites, sequestered within the central chamber of CPs, are formed by the amino-terminal Thr residues of β-subunits. These active sites are exposed after autocatalytic removal of N-terminal propeptides from β-subunits during CP assembly. Unlike the HslV and ClpP proteases of bacteria, the central proteolytic chamber of proteasomal CPs in all three domains of life is flanked by two antechambers (FIG. 1a). When the rate of substrate translocation is slower than the rate of proteolysis, the antechambers can store substrate proteins before their degradation18; the antechambers can also can maintain substrate proteins in an unfolded state19.
Although proteasomal CPs in the three domains of life have a similar overall structure, they differ in subunit composition (TABLE 1) and the number of active sites16. Eukaryotic CPs are composed of seven different α-subunits (α1–α7) and seven different β-subunits (β1–β7) assembled in dyad symmetry (that is, the subunit organization is repeated after a 180° rotation around a 2-fold axis). Furthermore, in eukaryotes three of the seven different β-subunits are active, leading to a total of six active sites per CP (in housekeeping CPs, β1 catalyses endopeptiase Glu-C-like activity, β2 catalyses tryptic peptidase activity and β5 catalyses chymotryptic peptidase activity). By contrast, CPs of actinobacteria and archaea are simpler in composition, with their heptameric rings assembled from one to two different α-subunits and one to two different β-subunits. Typically, each β-subunit harbours one proteolytic active site that mediates chymotryptic, tryptic and/or endopeptidase Glu-C-like peptidase activities, although inactive β-subunits are predicted for some archaea20. Even with these differences, the overall size distribution of peptide products generated by CPs is not influenced by the number of active sites or their types of peptidase activities7.
Table 1.
Proteasome and protein conjugation systems across the domains of life
| Bacteria | Archaea | Eukarya | |
|---|---|---|---|
| Distribution of proteasomes | Actinobacteria | All organisms* | All organisms |
| Proteasomal CP subunits‡ | One to two different α- and β-subunits | One to two different α- and β-subunits | Seven different α- and β-subunits (α1–α7 and β1–β7) |
| Protein conjugation system | Pupylation | Sampylation (Samp1 and Samp2) | Ubiquitylation, urmylation and other ubiquitin-like systems |
| AAA+ regulators of proteasomal CP function | Arc and Mpa | Pan§ | 19S RP (Rpt1–Rpt6 subunits form the ATPase ring) and Cdc48 |
| Non-ATPase regulators of proteasome function | None identified to date | None identified to date | 11S regulators, Blm10, Lot6–NQO1 family proteins and others |
| Maturation factors involved in proteasome assembly and/or maintenance | None confirmed to date | None confirmed to date | Pac1–Pac2 and Pac3–Pac4 (CP α-ring formation) |
| Ump1 (CP maturation, including assembly of the β-subunits onto α-rings and Pac3–Pac4 displacement) | |||
| Nas2, Nas6, Hsm3 and Rpn14 (ATPase ring assembly) | |||
| Hsp90 (lid formation) | |||
| Not4 E3 ligase and Ecm29 (putatively, 26S proteasome assembly and/or maintenance) |
Arc, AAA+ ATPase forming a ring-shaped complex; Cdc48, cell division cycle 48; CP, core particle; Hsp90, heat shock protein 90;Mpa, mycobacterial proteasome ATPase; Not4, general negative regulator of transcription subunit 4; Pac, proteasome assembly chaperone; Pan, proteasome-activating nucleotidase; RP, regulatory particle; Samp, small archaeal modifier protein.
Includes all archaeal genomes of the phyla Euryarchaeota, Crenarchaeota, Korarchaeota and Thaumarchaeota that are available to date.
All proteasomal CPs are composed of 14 α- and 14 β-subunits organized in an α7β7β7α7 symmetry, with the number of different subunits varying among organisms. In most eukaryotic proteasomes, β1, β2 and β5 harbour the amino-terminal Thr active-site residues. There are eukaryotic CPs with alternative formations, including the thymoproteasome (in which β5 is replaced by β5t) and the immunoproteasome (in which β1, β2 and β5 are replaced by β1i, β2i and β5i)119.
Although not all archaea encode Pan homologues, related AAA+ Cdc48 homologues seem to be present throughout the archaea. Some archaea encode two Pan homologues (for example, haloarchaea and methanosarina).
Proteasomal gates
All proteasomal CPs have an opening on each end of their cylindrical structure that is gated by the N-terminal tails of α-subunits21 (FIG. 1c). Many α-subunits of archaeal and eukaryotic CPs are Nα-acetylated at their initiator Met residue, and this acetylation seems to promote further CP gating22,23. In the absence of regulatory proteins, the gates of CPs from all three domains can be in a closed conformation that minimizes substrate access to the proteolytic active sites (as described below).
The number of different α-subunit N-terminal tails that form the entrance gates, and the degree to which the gates block the CP channel, varies among the CPs from eukaryotes, actinobacteria and archaea (FIG. 1c). Eukaryotic CPs can be purified in a latent state, with little to no peptidase activity, and have gates that are closed by three to four different α-subunit N termini (primarily α2, α3 and α4)24. For example, in the X-ray crystal structures of yeast25 and bovine26 CPs, the gates are fully closed, with no portal for substrate entry. Supporting the role of the CP gate in restricting substrate entry in eukaryotes, deletion of the α3 N-terminal tail that forms the gate derepresses the peptidase activity of CPs in yeast27. By contrast, the CPs of archaea and actinobacteria are purified in an active state that can hydrolyse short peptides (fewer than nine residues), and have gates on each end that seem to fluctuate between open and closed states (even in the absence of regulators). Archaeal CP gates are measured to be dynamic by TROSY NMR, with the α-subunit N-terminal tails interchanging between closed and open conformations that are correlated with slower and faster rates of peptide hydrolysis, respectively28. In addition, archaeal and mycobacterial CPs typically seem to be in an open gate conformation in crystal structures, most probably owing to the partial disorder of α-subunit N-terminal residues in these structures29–31. However, closed gate structures have been detected for archaeal and mycobacterial CPs by cryoelectron microscopy and transmission electron microscopy, suggesting that the method of analysis influences CP gate structure30,32–34. In addition, the crystal structure of a mycobacterial CP active-site variant (β-subunit Thr1Ala) reveals how seven identical α-subunit N-terminal tails can take on three distinct conformations to close CP gates35. Likewise, using a different archaeal species as the source for proteasomal genes has yielded a CP crystal structure with gates that appear to be in the closed conformation36. As for eukaryotic CPs, deletion of the α-subunit N-terminal residues that form the gates in archaeal and mycobacterial CPs stimulates the hydrolysis of peptides and disordered proteins34,37. Furthermore, association with C-terminal peptides of regulatory ATPases stimulates a switch from the closed to the open gate conformation for archaeal CPs32,33,38 (see below).
Proteasome-associated regulators
Proteasomal CPs associate with AAA+ and non-ATPase regulators21. Eukaryotic CPs are often assembled with 19S regulatory particles (RPs) or caps to form 26S proteasomes (FIG. 1a). In yeast, the 19S RP can be separated into lid and base subcomplexes by deletion of the regulatory particle non-ATPase subunit Rpn10 (REF. 39). The base is composed of nine subunits: a hexameric ring of six different regulatory particle triphosphatase AAA+ subunits (Rpt1–Rpt6)39 and three non-ATPase subunits, Rpn1, Rpn2 and Rpn13 (with Rpn13 and Rpn10 binding ubiquitin chains with high affinity40). The base contacts and activates CPs for the ATP-dependent degradation of folded proteins39. The lid harbours nine other Rpn subunits, including the deubiquitylating enzyme (DUB) Rpn11 (REF. 41). Like Rpt1–Rpt6 of 26S proteasomes, other eukaryotic members of the AAA+ subfamily, including yeast cell division cycle 48 (Cdc48; known as p97 in mammals), form hexameric rings that seem to guide ubiquitylated proteins to proteasomes42. Non-ATPase regulators also associate with CPs in eukaryotes, including Blm10 in S. cerevisiae, the Blm10-related protein PA200 in mammals, and 11S regulators (also known as PA28 or REG in mammals and PA26 in trypanosomes), opening the axial CP gates and stimulating peptide hydrolysis21. Archaeal and actinobacterial CPs can be reconstituted in vitro with homohexameric rings of AAA+ proteins (including proteasome-activating nucleotidase (Pan) in archaea, mycobacterial proteasome ATPase (Mpa) in myco bacteria and AAA+ ATPase forming a ring-shaped complex (Arc) in other actinobacteria)43–45 (FIG. 1a). However, archaeal and bacterial CPs have yet to be purified from a native host in association with either ATPase or non-ATPase regulators.
Although CPs alone can degrade denatured or intrinsically disordered proteins46, they require AAA+ proteins and the hydrolysis of ATP to fuel the degradation of folded proteins. Proteasome-associated AAA+ proteins from all three domains of life (the eukaryotic 19S RP with Rpt1–Rpt6, archaeal Pan and actinobacterial Arc and Mpa) seem to interact with the ends of the CP cylinder, selectively bind and unfold substrate proteins, open the CP gate and facilitate the unidirectional, processive translocation of substrate proteins into the CP channel for proteolysis38,47,48. Among these processes, unfolding of substrate proteins and their subsequent translocation into the CP are both coupled to ATP hydrolysis.
Proteasome assembly
The formation of CPs is a complex process involving protein folding, subunit assembly and β-subunit maturation49. Assembly of archaeal and eukaryotic CPs proceeds through the formation of a heptameric ring of α-subunits that provides a scaffold for β-subunits to assemble into half-proteasome intermediates. During the assembly of half proteasomes into active CPs, the propeptides of β-subunits (β-propeptides) are autocatalytically removed to expose the N-terminal active-site Thr residues. In actinobacteria, the α-subunits do not form heptameric rings in the absence of β-subunits. Instead, α- and β-subunits associate as heterodimers and oligomerize to form the half-proteasomes needed for CP maturation. On the basis of CP structures, the inability of actinobacterial α-subunits to independently form rings seems to be due to the small contact regions between α-subunits35,50.
In eukaryotes, numerous maturation factors facilitate 26S proteasome biogenesis39,51. Two heterodimeric chaperones, proteasome assembly chaperone (Pac) complexes Pac1–Pac2 and Pac3–Pac4, work in concert to form the CP α-ring. An additional chaperone, Ump1, promotes β-subunit assembly onto (and Pac3–Pac4 displacement from) the α-rings. During this process, half proteasomes associated with Ump1 retain their β-propeptides and are inactive. Conversion of the two half-proteasomes into mature CPs is associated with autocatalytic processing of the β-propeptides to expose the active sites and results in the degradation of Ump1. Pac1 and Pac2 remain associated with the α-subunit N termini that form the CP gate until regulatory components, such as the 19S RP, are in place. Assembly of the 19S RP, although not fully understood, involves at least four different external proteins (Nas2, Nas6, Hsm3 and Rpn14) for ATPase ring assembly and one, heat shock protein 90 (Hsp90; also known as Hsp82 in yeast), for lid formation. Proteins that promote 26S proteasome assembly and/or maintenance (general negative regulator of transcription subunit 4 (Not4) and Ecm29) have also been reported39,51,52.
It is unclear whether specialized factors are needed for proteasome maturation in actinobacteria and archaea. Actinobacterial and archaeal CPs and their associated AAA+ proteins are simple in composition and often assemble spontaneously49. β-propeptides can even be deleted with little effect on in vitro CP assembly53. However, no CP has yet been isolated with its AAA+ partner from a native archaeal or actinobacterial host, suggesting that factors are needed to stabilize the energy-dependent proteasomal complexes. Factors may also be needed to regulate the populations of CP and ATPase subtypes in these domains. Indeed, in the haloarchaeon Haloferax volcanii, the CP α1, α2 and β-subunits can form three different CP subtypes but accumulate in mixed dimers to heptamers when the ratio of α1 to α2 is genetically perturbed, suggesting that wild-type archaea have mechanisms to maintain appropriate subunit ratios for the proper assembly of CP subtypes54.
Homologues of the eukaryotic chaperone complex Pac1–Pac2 (which is needed for proteasomal α-ring assembly) are also found in actinobacteria and archaea. Archaeal Pac1–Pac2 homologues have been purified (from recombinant Escherichia coli) and shown to bind the α–α intersubunit pockets of α-ring heptamers and immature CP mimics (that is, CPs with intact β-propeptides) but not mature CPs55. Much like the yeast proteins, archaeal Pac1–Pac2 homologues require a conserved C-terminal HbYX (in which Hb represents a hydrophobic residue) motif for binding to the α-ring55. Eukaryotic Pac1–Pac2 chaperones are thought to bind α-rings to prevent the association of activators such as the 19S RP until CPs are mature and/or no longer inhibited at their active site by β-propeptides. Although it remains to be determined in vivo, the demonstration that archaeal Pac1–Pac2 homologues can bind immature CP mimics suggests that, similarly to their eukaryotic counterparts, these proteins function as chaperones in the assembly of proteasomes in archaea.
Proteasome-mediated protein degradation
Molecular details are now available to devise models that explain how proteasomes convert chemical bond energy (ATP) into mechanical work (protein degradation). Recent X-ray crystal structures of subdomains of proteasomal ATPases (archaeal Pan, and actinobacterial Arc and Mpa) have been assembled and docked with proteasomal CPs36,44,56,57. In addition, a new structure of the yeast 26S proteasome has been resolved by cryoelectron microscopy to 9.1 Å58.
Along with these new proteasomal structures, biochemical studies of proteasomes and related bacterial AAA+ ATPases provide evidence to support models that explain how proteasomes degrade proteins36,44,48,56–60. In current models (FIG. 2), substrate proteins are thought to bind the N-terminal coiled-coil domains of the AAA+ ATPases during proteasome-mediated proteolysis. In archaea and bacteria, the N-terminal coiled-coils associate in pairs that protrude like three tentacles from the ATPase face distal to the CP. Although the basic coiled-coil structure is conserved in proteasomal ATPases from all three domains of life, the coiled-coils of the eukaryotic Rpt subunits are embedded within the 26S proteasomes58,61. However, the coiled-coils of the related ATPases from actinobacteria and archaea are not masked, and their location suggests that they interact with substrate proteins early in the degradation pathway36,56,57.
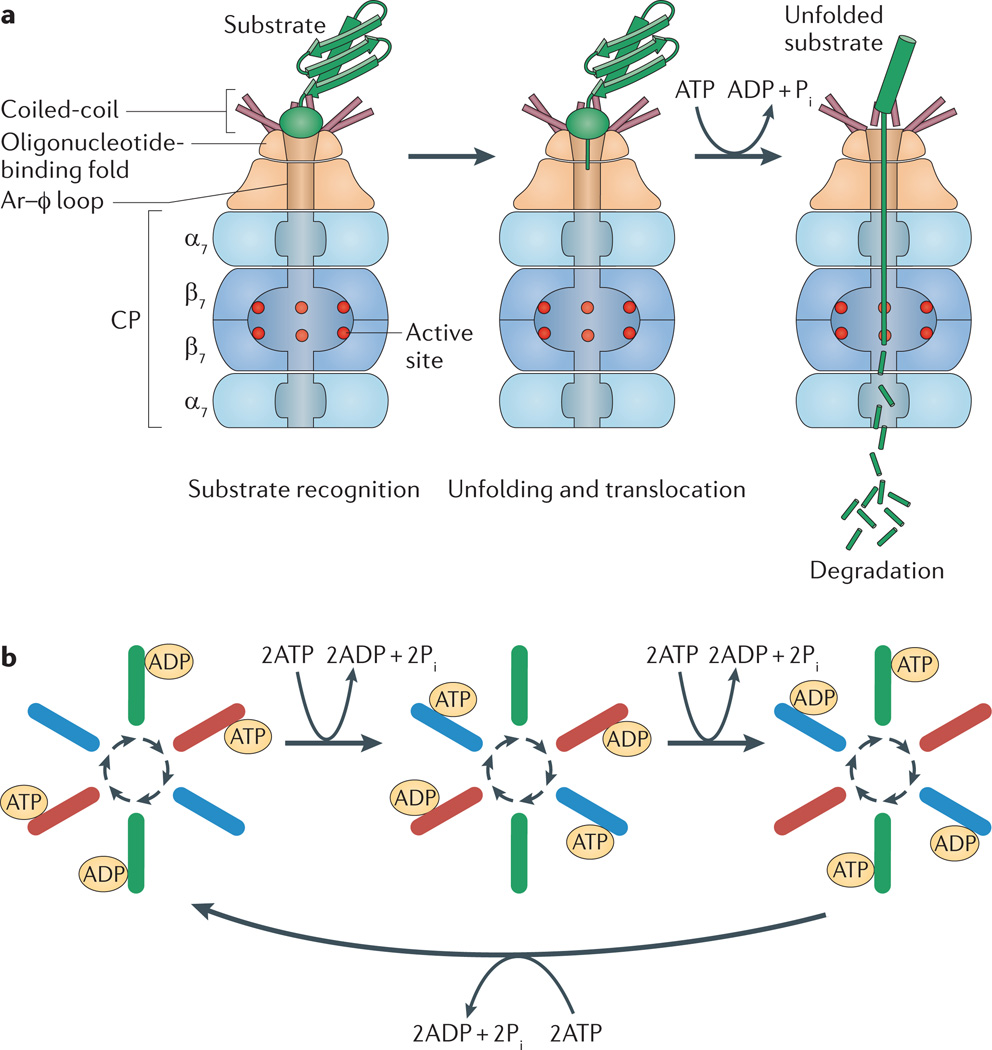
Figure 2. Ordered reaction cycle in protein degradation by proteasomes.
a | Model of proteolysis based on archaeal proteasome-activating nucleotidase (Pan) and core particle (CP) complexes. The amino-terminal coiled-coil domain of each Pan subunit forms a pair with one of its neighbours. The three tentacle-like coiled-coil pairs protrude from the ATPase face most distal to the CP and surround a pore formed by an oligonucleotide-binding fold, which may serve as an entry point for substrate proteins to traverse into the ATPase channel. An aromatic–hydrophobic (Ar–φ) loop within the narrowest region of the ATPase channel may grip and pull down on substrate proteins, a process driven by ATP hydrolysis. Protein unfolding is thought to occur from these repetitive power strokes, with the oligonucleotide-binding fold providing a rigid platform and narrow opening to stimulate this unfolding. Unfolded protein substrates are translocated through the ATPase channel to the CP for degradation. b | Proteasomal ATPases seem to function as para-subunit pairs in ATP binding, ATP hydrolysis and ADP release during protein unfolding and docking to the CP. ATP binding to a para-subunit pair (red) induces conformational changes in adjacent subunit pairs, so that the clockwise pair (blue) becomes nucleotide free and the anticlockwise pair (green) becomes bound to ADP. Following ATP hydrolysis, the ATP-bound partners (red) are converted to an ADP-bound state, thus simulating the clockwise pair to bind ATP and the anticlockwise pair to release ADP. Thus, an ordered reaction cycle is perpetuated with coordinated conformational changes in para-subunit pairs, probably providing the power strokes for pulling and unfolding the substrates. At any given time, only a subset of the carboxy-terminal HbYX motifs in the ATPase (those in para-subunits bound to ATP) may be extended to open the CP gates. Pi, inorganic phosphate. Part b is modified, with permission, from REF. 63 © (2011) Elsevier.
The coiled-coil domains surround a pore at the distal ATPase face. This pore is formed by an oligonucleotide-binding fold, which holds the hexameric ATPase ring together, and it may serve as a narrow entry point for the translocation of substrate proteins into the channel traversing the ATPase ring. A highly conserved aromatic–hydrophobic (Ar–φ) loop, which is crucial for the unfolding and degradation of proteins by AAA+ proteases60, is located at the narrowest region of the ATPase channel formed by the AAA+ domain (based on modelling)36. This loop is thought to grip hydrophobic residues of substrate proteins that extend within the ATPase channel, and to pull down on the substrate, with cycles of ATP hydrolysis driving this motion. As the rigid oligonucleotide-binding fold serves as a narrow opening that resists the entry of folded proteins into this Ar–φ trap, overall protein unfolding may occur from repetitive energy-dependent power strokes of the ATPase. Protein substrates that are unfolded by this process would be translocated through the ATPase channel to the coaxial CP channel for ultimate destruction.
The homohexameric archaeal AAA+ ATPase Pan provides a simple model for understanding how the binding and hydrolysis of ATP facilitates proteasome-mediated proteolysis62. Pan subunits can exist in one of three conformational states, with high, low and no affinity for ATP63. The subunits directly opposite each other in the ATPase ring (known as para-subunits) are proposed to team up as partners in an ordered clockwise reaction cycle (FIG. 2b). In this model, a para-subunit pair binds two ATPs (one per subunit) with high affinity and induces distinct conformational changes in each neighbouring pair. Subunits immediately clockwise to the ATP-bound pair are in a nucleotide-free state (based on a study of the related bacterial protein ClpX60), whereas subunits anticlockwise to the ATP-bound pair are in an ADP-bound state. On ATP hydrolysis, the ATP-bound partners become ADP bound. The subunit pair clockwise to this binds ATP, and the pair anticlockwise releases ADP to take on a nucleotide-free state, thus perpetuating a reaction cycle that facilitates coordinated conformational changes in para-subunit pairs63. As the Ar–φ loop grips the substrate protein, the coordinated ATP-dependent conformational changes in para-subunit pairs of Pan are likely to provide the power strokes needed to pull and unfold the substrate proteins.
Entry of the substrate protein into the proteolytic chamber of the proteasome also requires opening of the CP gates. Proteasome-associated ATPases with conserved C-terminal HbYX motifs can mediate gate opening, and ATP binding stimulates this activity33,38,47. The C termini of the para-subunits within the hexameric ATPase ring are at an atomic distance that is compatible with binding the α–α intersubunit pockets of the heptameric outer CP rings. Indeed, in analogy to the mechanism of gate opening in bacterial protease HslUV64,65, ATP binding to the ATPase subunits is thought to extend the C-terminal residues of the ATPase, which bind the pockets formed between the outer-ring α-subunits of CP, to promote CP gate opening. Interestingly, only three of the six Rpt subunits (Rpt2, Rpt3 and Rpt5) of eukaryotic 26S proteasomes have the HbYX motif required for gate opening. However, each Rpt subunit that does not harbour an HbYX motif (Rpt1, Rpt4 and Rpt6) is paired opposite to one that does66. Thus, ATP binding to Rpt para-subunits in the ATPase ring could still facilitate CP gate opening for protein degradation by 26S proteasomes.
Targeting for degradation
Proteolysis is important for bulk protein turnover (to reclaim amino acids and maintain protein quality) and can also be used to destroy specific proteins at key steps to control cell function. To avoid the widespread and uncontrolled breakdown of proteins (which are synthesized at high energy cost), cells select proteins from their milieu for destruction by energy-dependent proteases such as proteasomes. The mechanisms used by cells to target proteins for degradation vary but often involve changes in protein structure. One of the most notable pathways used by eukaryotes to target proteins for proteasome-mediated proteolysis is ubiquitylation, although ubiquitin-independent mechanisms have also been identified. Mechanisms of protein conjugation that seem to be linked to proteasomes have also been identified in actinobacteria (pupylation) and archaea (sampylation); however, these pathways are not well understood.
Ubiquitylation in eukaryotes
In eukaryotes, proteins that are targeted for proteasome-mediated degradation are often modified by ubiquitylation (FIG. 3). This process is mediated by a group of enzymes that select the target protein, generate the appropriate type of ubiquitin modification on the target, regulate the length of ubiquitin chains and maintain free pools of ubiquitin in the cell11. To initiate ubiquitylation, E1 ubiquitin-activating enzyme adenylates the C-terminal carboxyl group of ubiquitin using ATP (FIG. 3). This activation of ubiquitin then leads to the formation of a thioester intermediate between the C terminus of ubiquitin and a conserved E1 Cys residue. The E1–ubiquitin intermediate transfers ubiquitin to a conserved Cys on an E2 ubiquitin-conjugating enzyme to form a second thioester intermediate (E2–ubiquitin). E3 ubiquitin ligases typically assist the E2 enzymes in selecting the proper substrate protein for ubiquitin transfer. Ultimately, a covalent isopeptide bond is formed between the C-terminal carboxyl group of ubiquitin Gly76 and the ε-amino group of a Lys residue on the substrate protein. Ubiquitylation of Ser, Thr and Cys residues and of the N-terminal α-amino group of proteins has also been observed9,67.
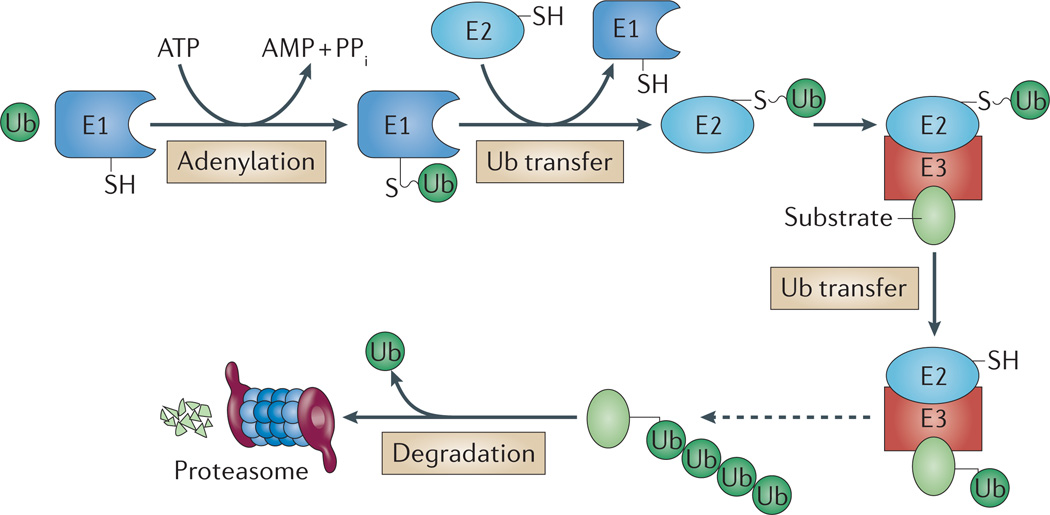
Figure 3. Ubiquitylation as a signal for degradation.
Ubiquitylation is a common signal for eukaryotic 26S proteasomes and involves a cascade of E1 ubiquitin-activating, E2 ubiquitin-conjugating and E3 ubiquitin ligase enzymes. In this cascade, E1 (plus ATP) first adenylates the carboxy-terminal carboxylate of ubiquitin (Ub), forming Ub–AMP, and then forms a Ub thioester intermediate (E1–Ub). Ubiquitin is transferred from E1 to E2, and then to the protein target with assistance from E3 (although ubiquitylation without E3 can occur118). Typically, an isopeptide bond is formed between the ubiquitin C-terminal carboxylate and the ε-amino group of a Lys side chain of the substrate protein or the growing ubiquitin chain (Lys48-linked ubiquitin chains are common signals for 26S proteasomes). Deubiquitylating enzymes within 26S proteasomes release and recycle ubiquitin during substrate protein degradation. PPi, inorganic pyrophosphate.
After a protein is modified with ubiquitin, additional isopeptide bonds can form between the C-terminal Gly76 of another incoming ubiquitin and one of the seven Lys residues of the ubiquitin on the modified protein to generate polyubiquitin chains68. Linear polyubiquitin chains can also form between the C-terminal Gly76 of an incoming ubiquitin and the N-terminal α-amino group of the Met residue in ubiquitin on the modified protein69. Lys48-linked ubiquitin chains are signals for degradation by proteasomes70, and Lys63-linked ubiquitin chains act in non-proteolytic events71,72. The roles of the other ubiquitin chains are only now being elucidated68,69. Interestingly, a protein termed ubiquitin-related modifier 1 (Urm1) has been implicated, along with its E1 enzyme (Uba4), in both sulphur transfer and protein conjugation in eukaryotic cells73 (BOX 1).
Box 1 | Ubiquitin-like systems in protein conjugation and sulphur transfer.
The yeast protein ubiquitin-related modifier 1 (Urm1; one of the most ancestral eukaryotic proteins related to ubiquitin), along with its E1 ubiquitin-activating enzyme, Uba4, provided the first example of a system that functions in both protein conjugation (urmylation) and sulphur transfer (2-thiolation of tRNAs)73. Uba4 adenylates and transfers sulphur to Urm1, resulting in an Urm1 protein that is thiocarboxylated at its carboxyl terminus and is required for tRNA thiolation and, surprisingly, also urmylation115. The Lys residues of protein substrates also seem to be required for urmylation115. Although not all the details of urmylation are clear, it is known that Uba4 belongs to the well-studied E1-like superfamily of proteins, which catalyse ATP-dependent adenylation of the C-terminal carboxylate of β-grasp fold (ubiquitin-like) proteins, such as Urm1 (REFS 91, 116). During sulphur transfer, Cys desulphurase can mobilize sulphur to adenylate ubiquitin-like proteins and generate ubiquitin-like proteins with a C-terminal thiocarboxyl group, which is needed in the formation of sulphurated biomolecules such as molybdenum cofactor (MoCo), thiamine and thiolated tRNA. Thus, ubiquitylation and these sulphur transfer pathways both use ubiquitin-like proteins with a β-grasp fold. However, ubiquitylation contrasts with sulphur transfer in that the adenylated ubiquitin is first converted to an E1–ubiquitin intermediate with a thioester bond between the C-terminal carboxylate of ubiquitin and the catalytic Cys of E1, preceding E2 ubiquitin conjugating- and E3 ubiquitin ligase-mediated transfer of ubiquitin to the substrate protein. By contrast, it is unclear whether a thioester bond is formed between Uba4 and Urm1 in the Urm1 pathway.
Similarly, the E1 homologue in the archaeon Haloferax volcanii, UbaA, was shown to be required not only for sampylation by small archaeal modifier protein 1 (Samp1) and (Samp2), but also for the thiolation of tRNALysUUU and for growth under anaerobic conditions that require sulphur transfer form MoCo15 (Supplementary information S1 (figure)). Furthermore, Samp2 is essential for tRNALysUUU thiolation, and Samp1 seems to be necessary for MoCo biosynthesis15. On the basis of genome neighbourhood analysis95, the respective association of Samp1 and Samp2 orthologues with MoCo biosynthesis and tRNA modification seems to be common among archaea.
Ubiquitin-independent proteolysis in eukaryotes
Although ubiquitylation is typically used to target proteins for proteasome-mediated hydrolysis, proteins can also be degraded by proteasomes through ubiquitin-independent mechanisms. In eukaryotes, intrinsically disordered proteins are thought to be degraded by CPs through a default mechanism unless they are otherwise stabilized during the course of their synthesis (for example, when assembled into an appropriate complex)74,75, at which point they can be targeted for degradation by ubiquitylation. For example, the tumour suppressors p53 and p73 (which are intrinsically disordered) can be degraded by uncapped CPs in the absence of ubiquitylation, through a process regulated by NADH/NAD+ levels and NAD(P)H:quinone oxidoreductase76.
Regulatory proteins can also bind and target proteins for destruction by proteasomes in a ubiquitin-independent manner. A classic example of this type of regulation is in polyamine biosynthesis77. The small protein antizyme binds and targets ornithine de carboxylase (ODC) for ubiquitin-independent proteolysis by proteasomes. Antizyme inhibitor, an inactive ODC homologue, reverses this activity by binding antizyme and rescuing ODC from destruction.
Targeting for degradation in actinobacteria: pupylation
Over the past few years, it has become apparent that the covalent attachment of small protein modifiers to target proteins is not restricted to eukaryotes (TABLE 1). The first discovery of protein conjugation in a non-eukaryotic organism was through the analysis of the DUF797 family of small proteins predicted to be encoded in the vicinity of proteasome genes in actinobacteria12. A member of this protein family, termed Pup, was shown to be covalently attached to target proteins in mycobacteria12. Although this protein conjugation (pupylation) system has only recently been discovered, elegant studies have illuminated its mechanism and biological roles in actinobacteria78.
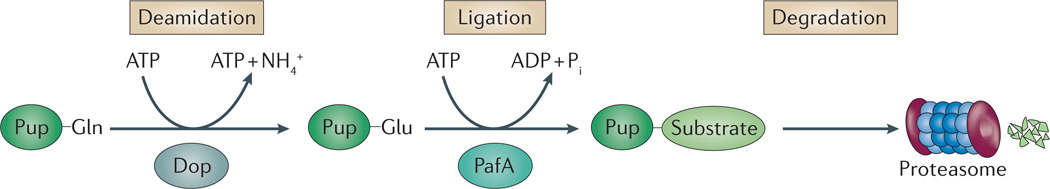
Pupylation has many features that make it distinct from ubiquitylation (FIG. 4). Pup is intrinsically disordered, which is in contrast to the highly ordered β-grasp fold of ubiquitin and ubiquitin-like proteins57,79,80. In addition, the mechanism of Pup activation and attachment to target proteins differs from that of ubiquitylation. During pupylation, the C-terminal Gln64 of Pup is deamidated to glutamate by the Glu synthetase-like protein Dop (which is in contrast to the E1-mediated adenylation of ubiquitin)81–83. Deamidation of Pup exposes a γ-carboxylate that can then be attached to the ε-amino group of Lys residues on substrate proteins through an ATP-dependent reaction catalysed by another Glu synthetase-like protein, PafA84,83. Like ubiquitylation, this process can target protein substrates for degradation by proteasomes. Furthermore, Dop (although not related to DUBs in enzymatic mechanism or structure) catalyses the removal of Pup from substrate proteins and, thus, may function similarly to eukaryotic DUBs in preventing or promoting proteasome-mediated proteolysis83,85,86.
Figure 4. Pupylation as a signal recognized by proteasomes in bacteria.
Pupylation and proteasome-mediated proteolysis in actinobacteria. In pupylation, the carboxy-terminal Gln of prokaryotic ubiquitin-like protein (Pup) is deamidated to Glu by Dop. PafA can then attach Pup to substrates, mediating their proteasomal degradation. Once conjugated to protein substrates, Pup binds to the coiled-coil domain of the proteasomal ATPase (called mycobacterial proteasome ATPase (Mpa) in mycobacteria), and a region of Pup is converted from a disordered state into an α-helix (not shown). Pi, inorganic phosphate.
In mycobacteria, the C-terminal half of Pup is needed for binding to the proteasomal ATPase, and the N-terminal half of Pup is required for the unfolding and degradation of substrate proteins43,87. Importantly, Pup binding to the N-terminal coiled-coil of the proteasomal ATPase, Mpa, converts a small portion of the C-terminal region of Pup from a disordered (randomly coiled) state into an α-helix through a binding-induced folding mechanism57. The randomly coiled state of Pup is thought to facilitate the initial interaction of pupylated proteins with Mpa, whereas the binding-induced folding of Pup may reel pupylated proteins into the Mpa–CP proteasome complex. By contrast, polyubiquitin chains are already highly structured, with a β-grasp fold, and bind distinct receptors within 26S proteasomes of eukaryotes40.
Whether additional factors (beyond Dop and PafA) are required to select proper substrates for pupylation remains to be determined. It also is unclear whether other small disordered proteins that are distinct from DUF797 family proteins function in protein conjugation. Interestingly, synthesis of PafA and Pup in genetically modified E. coli leads to pupylation of the recombinant host proteins, suggesting that a minimal gene set of pup and pafA is needed for the transfer of pupylation within bacteria88.
Targeting for degradation in archaea: sampylation
Previously, the repertoire of enzymes predicted in the genomes of actinobacteria and archaea seemed insufficient for a ubiquitin-like-protein conjugation system. Proteins that are related structurally to ubiquitin, E1 ubiquitin-activating enzymes and JAMM–MPN+ enzymes (which are a type of DUB) are widespread in actinobacteria and archaea89–92. However, bacterial homologues of ubiquitin and E1 enzymes were known to function only in non-protein-conjugating pathways, including the biosynthesis of sulphur-containing biomolecules such as the pterin-based molybdenum cofactor (MoCo), thiamine and thiolated tRNA89–92. Furthermore, although the crystal structure of a JAMM–MPN+ homologue from the archaeon Archaeoglobus fulgidus was determined and used to predict the active-site structure of Rpn11 (a DUB subunit of 26S proteasomes), the archaeal protein has no apparent protease, peptidase or DUB activity93,94. In addition, E2 and E3 homologues have not been identified in most bacterial and archaeal genome sequences92.
Even with this apparent limitation in coding sequence for a ubiquitin-like-protein conjugation pathway in actinobacteria and archaea, a mechanism that has analogies to ubiquitylation, termed sampylation (FIG. 5), was recently identified in the halophilic archaeon H. volcanii14,15. Like most archaea, H. volcanii encodes a single E1 homologue, two DUB (JAMM–MPN+) homologues, multiple ubiquitin-like proteins and no readily apparent E2 or E3 homologues. In a study of H. volcanii, two different ubiquitin-like proteins (denoted Samp1 and Samp2) were found to be attached to protein substrates through covalent (non-thiol) bonds in an apparent E1-type mechanism14,15.
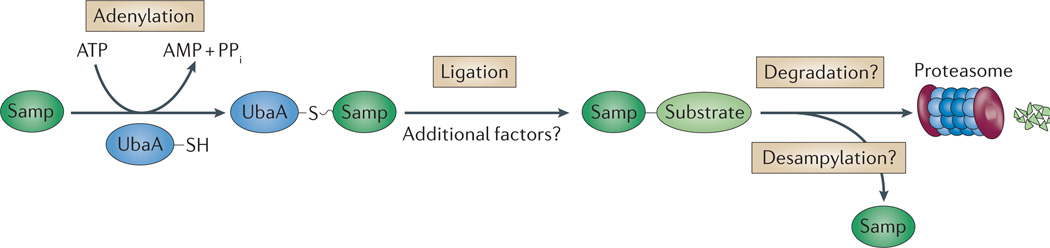
Figure 5. Sampylation and proteasomes in archaea.
Similar to ubiquitylation, evidence suggests that the small archaeal modifier proteins (Samps) are adenylated at their carboxy-terminal carboxylate by an E1 ubiquitin-activating-like enzyme (UbaA) and transferred to Lys side chains of protein substrates. Whether additional factors (other than the E1) are needed to ensure proper selection of protein targets and whether sampylated proteins are degraded by proteasomes remain to be determined. Although Lys58-linked Samp2 chains have been detected, it is unclear whether these chains are anchored to substrate proteins (not shown). PPi, inorganic pyrophosphate.
The proteins subjected to sampylation in H. volcanii have been analysed by tandem mass spectrometry (MS–MS) to determine the identity of the protein substrates, the type of covalent bond formed and the site of protein modification14. Proteins found to be sampylated are associated with a range of functions, including sulphur mobilization, the stress response, metabolism, DNA replication, translation and RNA modification. The proteins that are sampylated differ according to the type of protein modifier (Samp1 versus Samp2) and growth conditions (for example, nitrogen availability). However, some protein targets can be modified by both Samp1 and Samp2, including E1 and methionine-S-sulphoxide reductase (MsrA) homologues. Nitrogen limitation induces sampylation of both Samp1 and Samp2 targets, and both groups of Samp-modified proteins are altered by proteasomal-gene knockouts (the levels of Samp1-modified proteins increase, whereas the levels of Samp2-modified proteins decrease). Thus, at least in H. volcanii, sampylation could target proteins for proteasome-mediated degradation and increase pools of amino acids during nitrogen limitation, but also seems to have non-proteolytic roles.
Although sampylation has yet to be demonstrated in archaea beyond H. volcanii, and the complete sampylation pathway has not been reconstituted in vitro, sampylation is now predicted for all archaea14,95, and components of this system have been investigated using genetic, biochemical and structural approaches (see below). In particular, gene-knockout studies have indicated that an E1 ubiquitin-activating homologue of archaea, termed UbaA, is required for sampylation by both Samp1 and Samp2 in H. volcanii, suggesting that UbaA is the adenylation enzyme for both modifiers15. In addition, a homologue of UbaA (termed Elsa) and homologues of the Samps have been purified from Methanosarcina acetivorans (a methanogenic archaeon). Elsa associates with the Samp homologues in the absence of ATP and adenylates these Samp homologues in the presence of ATP96. Structures of H. volcanii Samp1 and its M. acetivorans homologue have been determined and compared to a three-dimensional model of Samp2 and other ubiquitin-like proteins, such as MoaD and Urm1 (REFS 96,97). Both Samp1 and the model of Samp2 were found to have a β-grasp configuration similar to that of ubiquitin and ubiquitin-like proteins. But, unlike ubiquitin and Samp2, Samp1 has extra α1 and α3 helical segments and is more structurally related to MoaD and Urm1 (both of which are required for sulphur transfer to biomolecules (BOX 1))97.
Further MS–MS-based dissection of the proteins subjected to sampylation in H. volcanii revealed a ‘classical’ ubiquitin-like isopeptide bond between the C-terminal carboxyl group of Samp2 and the ε-amino group of Lys residues within numerous protein targets14. Analogous to ubiquitylation, protein substrates with multiple sites of sampylation by Samp2 were identified, and Lys58-linked poly-Samp2 was detected14. Whether the poly-Samp2 chains are attached to target proteins or are unanchored remains to be determined. Although poly-Samp1 chains have yet to be identified, the single Lys residue (Lys4) of Samp1 aligns structurally with ubiquitin Lys6 and Samp2 Lys58, which are known to form chains14,68. Samp1 and Samp2 also have a region on their surface analogous to the hydrophobic Ile44-centred patch of ubiquitin, which is recognized by more than ten ubiquitin-interacting domains98. As is found for ubiquitin in eukaryotes, these hydrophobic patches of Samps might be involved in non-covalent interactions with other binding proteins in H. volcanii.
Ubiquitin-like protein conjugation in other bacteria and in archaea
In addition to sampylation and pupylation, other types of ubiquitin-like-protein conjugation systems are predicted to exist in bacteria and archaea on the basis of recent DNA sequences. The metagenome of ‘Candidatus Caldiarchaeum subterraneum’ (a free-living archaeon distinct from known archaeal phyla) harbours an apparent operon encoding structural homologues of eukaryotic ubiquitin, E1, E2, E3 and deubiquitylating enzymes of the JAMM–MPN+ family99. Several phylogenetically diverse bacteria (of the phyla Actinobacteria, Planctomycetes and Acidobacteria) also carry related operons92. Overall, these uncharacterized ubiquitylation operons are sporadically dispersed in bacteria and archaea and are often missing in close relatives. On the basis of these in silico findings, the genes seem to be functionally linked, non-essential, highly mobile and disseminated through horizontal transfer. Thus, the origins of ubiquitylation are speculated to be from horizontal gene transfer of operons related to such sequences92.
Proteasome systems in a cellular context
Proteasome-targeting processes such as ubiquitylation regulate many functions that are important for the growth and survival of cells. These have been recently reviewed for eukaryotes and mycobacteria78,100, so below I focus on the functions of proteasomes in archaea.
Function of proteasomes in archaea
Across archaeal phyla, the genes encoding proteasomes are not organized together in operons but are linked with common gene neighbours101,102. The gene neighbours include homologues of proteins mediating 3′-to-5′ mRNA degradation (the exosome), tRNA modification, MoCo binding and other non-proteolytic processes101,102. In H. volcanii, the proteasomal α1 subunit gene is co-transcribed with genes encoding homologues of the RNase P Pop5 subunit and S-adenosylmethionine (SAM)-dependent methyltransferase103. Likewise, the proteasomal α2 subunit gene is co-transcribed with a MoCo-dependent oxidoreductase gene homologue103. These genomic and transcriptional linkages suggest a close physiological association of proteasomes with RNA modification and MoCo biosynthesis in archaea.
Chemical inhibitor and genetic studies have provided experimental insights into the role of proteasomes in archaea. Proteasome-specific inhibitors can partially inhibit CP activity in cells, resulting in reduced growth rates under heat shock conditions for Thermoplasma acidophilum104 and under non-heat-shock conditions for H. volcanii105. Conditional and markerless gene deletion studies in H. volcanii have revealed that the cell must produce at least one CP subtype for viability, whereas genes encoding the sampylation system and Pan AAA+ ATPases are not essential15,106. Furthermore, H. volcanii cells lacking the α1 subunit (one of two CP α-subunits produced in this cell) or PanA (also known as Pan1; one of two Pan proteins produced in this cell) are hypersensitive to nitrogen limitation, low-salt stress and exposure to l-canavanine (an l-Arg analogue that induces protein unfolding). They also show altered responses to thermal stress, but in this case α1 mutants decrease in number, whereas PanA mutants increase in number compared with wild-type cells106. In addition, cells producing ungated CPs are hypersensitive to low-salt stress22. Thus, archaea generally show reduced survival in stressful conditions when proteasomal genes are deleted, and CPs are essential for growth.
Reporter gene constructs and proteomic methods have been used to detect the accumulation of proteins in proteasome-deficient archaeal cells105,107–109. According to these studies, the proteasome seems to be important for controlling the levels of proteins involved in key cellular processes and can destabilize proteins with hydro phobic C termini105,107–109. Furthermore, disruption of PanA results in a marked increase in the number of phosphorylated proteins107. Whether phosphorylation triggers proteolysis or is a stress response caused by the absence of PanA remains to be established. However, several proteins that accumulate in proteasome-deficient cells are also targeted by sampylation14, suggesting a physiological link between the two systems.
Regulation of archaeal proteasomes
Regulation of proteasomes can be at the level of synthesis or posttranslational and co-translational modification of subunits. For example, some archaea synthesize CPs and Pan AAA+ ATPases in a regulated manner depending on the growth conditions. In H. volcanii, three different CP subtypes have been purified, including CPs with a single type of α-subunit (α1β or α2β CPs) and CPs with all three subunits (α1α2β CPs)54,110 (and I. Karadzic, J.M.-F., M. Humbard, P. Singh and D. Goodlett, unpublished observations). Proteasomal ATPases composed of PanA and PanB (also known as Pan2) have also been isolated111. Of these proteins, α1 subunit, β-subunit and PanA levels are relatively high throughout growth, whereas PanB and α2 subunit levels are low and increase during stationary phase112. On the basis of differences in the amino acid residues that are predicted to form the CP α–α interface, the Pan HbYX motif and the Pan coiled-coil domain, these alterations in the levels of proteasomal proteins may influence interactions between the Pan and CP subtypes, and/or substrate recognition. Pyrococcus furiosus also encodes three CP subunits (α, β1 and β2). Of these, β1 subunit transcript levels are upregulated during heat shock, and CPs with the greatest ratio of β1/β2 are the most thermostable113. Thus, P. furiosus might incorporate β1 into CPs to enhance proteasome function during thermal stress.
Archaea modify proteasomal proteins both co-translationally and post-translationally. In H. volcanii, like in eukaryotes, the α-subunits are phosphorylated in the CPs of actively dividing cells111. Substitution of the phosphorylated Thr or Ser to Ala in the α1 subunit results in global changes to the cell, including reduced viability and an apparent reduction in carotenoid levels111. The α-subunits are also Nα-acetylated at their initiator Met residue22,114, and maintaining this modified form of the α1 subunit seems to be important for CP gating. When cells produce an α1 subunit Gln2Ala variant, which enhances the cleavage of the α1 subunit by methionine aminopeptidase and results in Nα-acetylation of the exposed N-terminal Ala, the cells generate CPs with enhanced peptidase activity and are hypersensitive to low-salt stress22. N-terminal α1 subunit residues are also important for maintaining proper levels of α1 subunit in the cell, potentially through an N-end rule pathway of protein degradation22. Cells with unusually high levels of α1 rings (due to alterations in α1 subunit N-terminal residues) display enhanced cell growth and are more tolerant of low-salt and high-temperature stresses than wild-type cells. Thus, altering the post-translational and co-translational modifications of proteasomal subunits can have a global impact on archaeal cell function.
Perspectives
Many new findings have advanced our understanding of the structure and function of proteasomes and protein conjugation; however, there is still a lot to be learned if we are to optimally control these systems in a living cell. For example, it remains unclear how proteasomes couple ATP hydrolysis to the unfolding and degradation of proteins. Although the study of bacterial AAA+ ATPases has provided insights into how ATP energy fuels proteolysis, it remains to be fully understood how proteasomes convert ATP into the mechanical energy used for protein degradation. It is also not clear whether sampylation and pupylation only target proteins to proteasomes or whether these protein modification systems also serve non-proteolytic roles. Interestingly, most archaea encode only one E1-like protein that, at least in H. volcanii, seems to activate multiple ubiquitin-like Samps for protein conjugation and sulphur mobilization. Although the control mechanism is not known, archaea must somehow regulate these multifunctional E1- and ubiquitin-like proteins to ensure that appropriate substrates are sampylated while levels of activated sulphur are maintained as needed for the synthesis of biomolecules such as MoCo and thiolated tRNA. Furthermore, the E1- and ubiquitin-like proteins of H. volcanii seem to stand at the crossroads between sulphur mobilization and protein conjugation, and it will be interesting to determine whether related bacterial systems (previously thought to be involved only in sulphur mobilization) also mediate the covalent modification of proteins.
Experimental advances and developments to address these questions will surely be multidisciplinary and hypothesis driven. An atomic structure of an intact proteasomal ATPase complex will augment the currently available subdomain structures and will be helpful for guiding models that explain how proteasomes couple ATP hydrolysis to the unfolding and degradation of proteins. Approaches that combine genetics with biochemistry and proteomics will continue to be valuable for the identification of new factors and pathways associated with proteasomes and protein conjugation. Combined approaches will also assist in understanding whether proteins tagged by sampylation and pupylation are targeted only for proteolysis or whether they have other biological fates. Functional studies guided by atomic structures should provide insights into how the E1-like proteins UbaA (which is involved in sampylation) and Uba4 (which is involved in urmylation) can catalyse both protein conjugation and sulphur transfer. Likewise, genomics will continue to provide a window for identifying new components of ubiquitin–proteasome pathways. For example, homologues of Pac1–Pac2 and JAMM–MPN+ proteins in archaea are predicted to function in proteasome assembly and desampylation, respectively. Likewise, the newly discovered bacterial and archaeal operons that include genes encoding E2 and E3 homologues have provided insights into the evolution of ubiquitylation and may prove to be functional in protein conjugation. As with any biological process, a great deal of understanding will be gained from comparing proteasomes and associated protein conjugation systems across domains of life.
Supplementary Material
Acknowledgements
The author thanks the reviewers for providing important insights and constructive comments, and apologizes for not citing many important references that have contributed to this Review, owing to space limitations. Work in the author’s laboratory is funded in part by grants from the US National Institutes of Health (GM57498) and the US Department of Energy Office of Basic Energy Sciences (DE-FG02-05ER15650).
Glossary
- Actinobacteria
A group of Gram-positive bacteria with high genomic GC contents, including Mycobacterium, Rhodococcus, Streptomyces and Frankia spp. Actinobateria have been shown to have proteasomes.
- HslV and ClpP proteases
Self-compartmentalized proteins that are located within bacteria and eukaryotic organelles, harbour proteolytic active sites and associate with the hexameric rings of AAA+ ATPases to form HslUV, ClpXP and ClpAP proteases, which mediate the energy-dependent degradation of structured proteins.
- TROSY NMR
A method for analysing large biomolecules such as proteasomes by measuring the cancellation between dipolar coupling and chemical shift anisotropy or between different dipolar couplings.
- E1-like superfamily
A group of conserved proteins that catalyse the adenylation of proteins containing a β-grasp fold, such as ubiquitin. Examples include the E1 enzyme used to activate ubiquitin during ubiquitylation, MoeB (which activates MoaD during sulphur transfer to form molybdenum cofactor (MoCo)) and ThiF (which activates ThiS in sulphur transfer during thiamine biosynthesis)
- JAMM–MPN+ enzymes
A family of proteins that typically coordinate a catalytic zinc ion. Members of this family include the yeast protein Rpn11 (or POH1 in humans), an isopeptidase that is required for the deubiquitylase activity of 26S proteasomes.
Footnotes
Competing interests statement
The author declares no competing financial interests.
FURTHER INFORMATION
Julie Maupin-Furlow’s homepage: http://microcell.ufl.edu/personnel/faculty/maupin.shtml
SUPPLEMENTARY INFORMATION
See online article: S1 (figure)
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Volker C, Lupas AN. Molecular evolution of proteasomes. Curr. Top. Microbiol. Immunol. 2002;268:1–22. doi: 10.1007/978-3-642-59414-4_1. [DOI] [PubMed] [Google Scholar]
- 2.Mehnert M, Sommer T, Jarosch E. ERAD ubiquitin ligases: multifunctional tools for protein quality control and waste disposal in the endoplasmic reticulum. Bioessays. 2010;32:905–913. doi: 10.1002/bies.201000046. [DOI] [PubMed] [Google Scholar]
- 3.Bengtson MH, Joazeiro CA. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467:470–473. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barford D. Structure, function and mechanism of the anaphase promoting complex (APC/C) Q. Rev. Biophys. 2011;44:153–190. doi: 10.1017/S0033583510000259. [DOI] [PubMed] [Google Scholar]
- 5.Bader M, Steller H. Regulation of cell death by the ubiquitin-proteasome system. Curr. Opin. Cell Biol. 2009;21:878–884. doi: 10.1016/j.ceb.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al Hakim A, et al. The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair (Amst.) 2010;9:1229–1240. doi: 10.1016/j.dnarep.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kisselev AF, Akopian TN, Woo KM, Goldberg AL. The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J. Biol. Chem. 1999;274:3363–3371. doi: 10.1074/jbc.274.6.3363. [DOI] [PubMed] [Google Scholar]
- 8.Piwko W, Jentsch S. Proteasome-mediated protein processing by bidirectional degradation initiated from an internal site. Nature Struct. Mol. Biol. 2006;13:691–697. doi: 10.1038/nsmb1122. [DOI] [PubMed] [Google Scholar]
- 9.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nature Rev. Mol. Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20:1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burns KE, Darwin KH. Pupylation versus ubiquitylation: tagging for proteasome-dependent degradation. Cell. Microbiol. 2010;12:424–431. doi: 10.1111/j.1462-5822.2010.01447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humbard MA, et al. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature. 2010;463:54–60. doi: 10.1038/nature08659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miranda HV, et al. E1- and ubiquitin-like proteins provide a direct link between protein conjugation and sulfur transfer in archaea. Proc. Natl Acad. Sci. USA. 2011;108:4417–4422. doi: 10.1073/pnas.1018151108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 17.Smith DM, Benaroudj N, Goldberg A. Proteasomes and their associated ATPases: a destructive combination. J. Struct. Biol. 2006;156:72–83. doi: 10.1016/j.jsb.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 18.Sharon M, et al. 20S proteasomes have the potential to keep substrates in store for continual degradation. J. Biol. Chem. 2006;281:9569–9575. doi: 10.1074/jbc.M511951200. [DOI] [PubMed] [Google Scholar]
- 19.Ruschak AM, Religa TL, Breuer S, Witt S, Kay LE. The proteasome antechamber maintains substrates in an unfolded state. Nature. 2010;467:868–871. doi: 10.1038/nature09444. [DOI] [PubMed] [Google Scholar]
- 20.Maupin-Furlow JA, et al. Proteasomes from structure to function: perspectives from Archaea. Curr. Top. Dev. Biol. 2006;75:125–169. doi: 10.1016/S0070-2153(06)75005-0. [DOI] [PubMed] [Google Scholar]
- 21.Stadtmueller BM, Hill CP. Proteasome activators. Mol. Cell. 2011;41:8–19. doi: 10.1016/j.molcel.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Humbard MA, Zhou G, Maupin-Furlow JA. The N-terminal penultimate residue of 20S proteasome α1 influences its Nα-acetylation and protein levels as well as growth rate and stress responses of Haloferax volcanii. J. Bacteriol. 2009;191:3794–3803. doi: 10.1128/JB.00090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura Y, et al. Nα-acetylation and proteolytic activity of the yeast 20 S proteasome. J. Biol. Chem. 2000;275:4635–4639. doi: 10.1074/jbc.275.7.4635. [DOI] [PubMed] [Google Scholar]
- 24.Forster A, Whitby FG, Hill CP. The pore of activated 20S proteasomes has an ordered 7-fold symmetric conformation. EMBO J. 2003;22:4356–4364. doi: 10.1093/emboj/cdg436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groll M, et al. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 26.Unno M, et al. Structure determination of the constitutive 20S proteasome from bovine liver at 2.75 Å resolution. J. Biochem. 2002;131:171–173. doi: 10.1093/oxfordjournals.jbchem.a003084. [DOI] [PubMed] [Google Scholar]
- 27.Kohler A, et al. The substrate translocation channel of the proteasome. Biochimie. 2001;83:325–332. doi: 10.1016/s0300-9084(01)01242-1. [DOI] [PubMed] [Google Scholar]
- 28.Religa TL, Sprangers R, Kay LE. Dynamic regulation of archaeal proteasome gate opening as studied by TROSY NMR. Science. 2010;328:98–102. doi: 10.1126/science.1184991. [DOI] [PubMed] [Google Scholar]
- 29.Groll M, Brandstetter H, Bartunik H, Bourenkow G, Huber R. Investigations on the maturation and regulation of archaebacterial proteasomes. J. Mol. Biol. 2003;327:75–83. doi: 10.1016/s0022-2836(03)00080-9. [DOI] [PubMed] [Google Scholar]
- 30.Hu G, et al. Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate. Mol. Microbiol. 2006;59:1417–1428. doi: 10.1111/j.1365-2958.2005.05036.x. [DOI] [PubMed] [Google Scholar]
- 31.Lowe J, et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 32.Rabl J, et al. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu Y, et al. Interactions of PAN’s C-termini with archaeal 20S proteasome and implications for the eukaryotic proteasome-ATPase interactions. EMBO J. 2010;29:692–702. doi: 10.1038/emboj.2009.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin G, et al. Mycobacterium tuberculosis prcBA genes encode a gated proteasome with broad oligopeptide specificity. Mol. Microbiol. 2006;59:1405–1416. doi: 10.1111/j.1365-2958.2005.05035.x. [DOI] [PubMed] [Google Scholar]
- 35.Li D, et al. Structural basis for the assembly and gate closure mechanisms of the Mycobacterium tuberculosis 20S proteasome. EMBO J. 2010;29:2037–2047. doi: 10.1038/emboj.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang F, et al. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol. Cell. 2009;34:473–484. doi: 10.1016/j.molcel.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benaroudj N, Zwickl P, Seemuller E, Baumeister W, Goldberg AL. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol. Cell. 2003;11:69–78. doi: 10.1016/s1097-2765(02)00775-x. [DOI] [PubMed] [Google Scholar]
- 38.Smith DM, et al. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s α ring opens the gate for substrate entry. Mol. Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomko RJ, Jr, Hochstrasser M. Order of the proteasomal ATPases and eukaryotic proteasome assembly. Cell Biochem. Biophys. 2011;60:13–20. doi: 10.1007/s12013-011-9178-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peth A, Uchiki T, Goldberg AL. ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Mol. Cell. 2010;40:671–681. doi: 10.1016/j.molcel.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee MJ, Lee BH, Hanna J, King RW, Finley D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.R110.003871. R110.003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stolz A, Hilt W, Buchberger A, Wolf DH. Cdc48: a power machine in protein degradation. Trends Biochem. Sci. 2011;36:515–523. doi: 10.1016/j.tibs.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 43.Striebel F, Hunkeler M, Summer H, Weber-Ban E. The mycobacterial Mpa-proteasome unfolds and degrades pupylated substrates by engaging Pup’s N-terminus. EMBO J. 2010;29:1262–1271. doi: 10.1038/emboj.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang T, et al. Structural insights on the Mycobacterium tuberculosis proteasomal ATPase Mpa. Structure. 2009;17:1377–1385. doi: 10.1016/j.str.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zwickl P, Ng D, Woo KM, Klenk HP, Goldberg AL. An archaebacterial ATPase, homologous to ATPases in the eukaryotic 26 S proteasome, activates protein breakdown by 20 S proteasomes. J. Biol. Chem. 1999;274:26008–26014. doi: 10.1074/jbc.274.37.26008. [DOI] [PubMed] [Google Scholar]
- 46.Tsvetkov P, Reuven N, Shaul Y. The nanny model for IDPs. Nature Chem. Biol. 2009;5:778–781. doi: 10.1038/nchembio.233. [DOI] [PubMed] [Google Scholar]
- 47.Smith DM, et al. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol. Cell. 2005;20:687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 48.Medalia N, et al. Architecture and molecular mechanism of PAN the archaeal proteasome regulatory ATPase. J. Biol. Chem. 2009;284:22952–22960. doi: 10.1074/jbc.M809643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zwickl P, Kleinz J, Baumeister W. Critical elements in proteasome assembly. Nature Struct. Biol. 1994;1:765–770. doi: 10.1038/nsb1194-765. [DOI] [PubMed] [Google Scholar]
- 50.Kwon YD, Nagy I, Adams PD, Baumeister W, Jap BK. Crystal structures of the Rhodococcus proteasome with and without its pro-peptides: implications for the role of the pro-peptide in proteasome assembly. J. Mol. Biol. 2004;335:233–245. doi: 10.1016/j.jmb.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 51.Gallastegui N, Groll M. The 26S proteasome: assembly and function of a destructive machine. Trends Biochem. Sci. 2010;35:634–642. doi: 10.1016/j.tibs.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 52.Panasenko OO, Collart MA. Not4 E3 ligase contributes to proteasome assembly and functional integrity in part through Ecm29. Mol. Cell. Biol. 2011;31:1610–1623. doi: 10.1128/MCB.01210-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maupin-Furlow JA, Aldrich HC, Ferry JG. Biochemical characterization of the 20S proteasome from the methanoarchaeon Methanosarcina thermophila. J. Bacteriol. 1998;180:1480–1487. doi: 10.1128/jb.180.6.1480-1487.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaczowka SJ, Maupin-Furlow JA. Subunit topology of two 20S proteasomes from Haloferax volcanii. J. Bacteriol. 2003;185:165–174. doi: 10.1128/JB.185.1.165-174.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kusmierczyk AR, Kunjappu MJ, Kim RY, Hochstrasser M. A conserved 20S proteasome assembly factor requires a C-terminal HbYX motif for proteasomal precursor binding. Nature Struct. Mol. Biol. 2011;18:622–629. doi: 10.1038/nsmb.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Djuranovic S, et al. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol. Cell. 2009;34:580–590. doi: 10.1016/j.molcel.2009.04.030. [DOI] [PubMed] [Google Scholar]
- 57.Wang T, Darwin KH, Li H. Binding-induced folding of prokaryotic ubiquitin-like protein on the Mycobacterium proteasomal ATPase targets substrates for degradation. Nature Struct. Mol. Biol. 2010;17:1352–1357. doi: 10.1038/nsmb.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bohn S, et al. Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc. Natl Acad. Sci. USA. 2010;107:20992–20997. doi: 10.1073/pnas.1015530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang F, et al. Mechanism of substrate unfolding and translocation by the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol. Cell. 2009;34:485–496. doi: 10.1016/j.molcel.2009.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 61.Nickell S, et al. Insights into the molecular architecture of the 26S proteasome. Proc. Natl Acad. Sci. USA. 2009;106:11943–11947. doi: 10.1073/pnas.0905081106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bar-Nun S, Glickman MH. Proteasomal AAA-ATPases: structure and function. Biochim. Biophys. Acta. 2011 Jul 23; doi: 10.1016/j.bbamcr.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 63.Smith DM, Fraga H, Reis C, Kafri G, Goldberg AL. ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell. 2011;144:526–538. doi: 10.1016/j.cell.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seong IS, et al. The C-terminal tails of HslU ATPase act as a molecular switch for activation of HslV peptidase. J. Biol. Chem. 2002;277:25976–25982. doi: 10.1074/jbc.M202793200. [DOI] [PubMed] [Google Scholar]
- 65.Wang J, et al. Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU. Structure. 2001;9:1107–1116. doi: 10.1016/s0969-2126(01)00670-0. [DOI] [PubMed] [Google Scholar]
- 66.Tomko RJ, Jr, Funakoshi M, Schneider K, Wang J, Hochstrasser M. Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: implications for proteasome structure and assembly. Mol. Cell. 2010;38:393–403. doi: 10.1016/j.molcel.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bloom J, Amador V, Bartolini F, DeMartino G, Pagano M. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell. 2003;115:71–82. doi: 10.1016/s0092-8674(03)00755-4. [DOI] [PubMed] [Google Scholar]
- 68.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iwai K, Tokunaga F. Linear polyubiquitination: a new regulator of NF-㮫 activation. EMBO Rep. 2009;10:706–713. doi: 10.1038/embor.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pickart CM. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 71.Spence J, Sadis S, Haas AL, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell. Biol. 1995;15:1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 73.Wang F, Liu M, Qiu R, Ji C. The dual role of ubiquitin-like protein Urm1 as a protein modifier and sulfur carrier. Protein Cell. 2011;2:612–619. doi: 10.1007/s13238-011-1074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsvetkov P, et al. Operational definition of intrinsically unstructured protein sequences based on susceptibility to the 20S proteasome. Proteins. 2008;70:1357–1366. doi: 10.1002/prot.21614. [DOI] [PubMed] [Google Scholar]
- 75.Baugh JM, Viktorova EG, Pilipenko EV. Proteasomes can degrade a significant proportion of cellular proteins independent of ubiquitination. J. Mol. Biol. 2009;386:814–827. doi: 10.1016/j.jmb.2008.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Asher G, Tsvetkov P, Kahana C, Shaul Y. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 2005;19:316–321. doi: 10.1101/gad.319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kahana C. Identification, assay, and functional analysis of the antizyme inhibitor family. Methods Mol. Biol. 2011;720:269–278. doi: 10.1007/978-1-61779-034-8_16. [DOI] [PubMed] [Google Scholar]
- 78.Darwin KH. Prokaryotic ubiquitin-like protein (Pup), proteasomes and pathogenesis. Nature Rev. Microbiol. 2009;7:485–491. doi: 10.1038/nrmicro2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liao S, et al. Pup, a prokaryotic ubiquitin-like protein, is an intrinsically disordered protein. Biochem. J. 2009;422:207–215. doi: 10.1042/BJ20090738. [DOI] [PubMed] [Google Scholar]
- 80.Chen X, et al. Prokaryotic ubiquitin-like protein pup is intrinsically disordered. J. Mol. Biol. 2009;392:208–217. doi: 10.1016/j.jmb.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Imkamp F, et al. Deletion of dop in Mycobacterium smegmatis abolishes pupylation of protein substrates in vivo. Mol. Microbiol. 2010;75:744–754. doi: 10.1111/j.1365-2958.2009.07013.x. [DOI] [PubMed] [Google Scholar]
- 82.Striebel F, et al. Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nature Struct. Mol. Biol. 2009;16:647–651. doi: 10.1038/nsmb.1597. [DOI] [PubMed] [Google Scholar]
- 83.Cerda-Maira FA, et al. Molecular analysis of the prokaryotic ubiquitin-like protein (Pup) conjugation pathway in Mycobacterium tuberculosis. Mol. Microbiol. 2010;77:1123–1135. doi: 10.1111/j.1365-2958.2010.07276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sutter M, Damberger FF, Imkamp F, Allain FH, Weber-Ban E. Prokaryotic ubiquitin-like protein (Pup) is coupled to substrates via the side chain of its C-terminal glutamate. J. Am. Chem. Soc. 2010;132:5610–5612. doi: 10.1021/ja910546x. [DOI] [PubMed] [Google Scholar]
- 85.Burns KE, et al. “Depupylation” of prokaryotic ubiquitin-like protein from mycobacterial proteasome substrates. Mol. Cell. 2010;39:821–827. doi: 10.1016/j.molcel.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Imkamp F, et al. Dop functions as a depupylase in the prokaryotic ubiquitin-like modification pathway. EMBO Rep. 2010;11:791–797. doi: 10.1038/embor.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Burns KE, Pearce MJ, Darwin KH. Prokaryotic ubiquitin-like protein provides a two-part degron to Mycobacterium proteasome substrates. J. Bacteriol. 2010;192:2933–2935. doi: 10.1128/JB.01639-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cerda-Maira FA, et al. Reconstitution of the Mycobacterium tuberculosis pupylation pathway in Escherichia coli. EMBO Rep. 2011;12:863–870. doi: 10.1038/embor.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Iyer LM, Burroughs AM, Aravind L. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like β-grasp domains. Genome Biol. 2006;7:R60. doi: 10.1186/gb-2006-7-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Burroughs AM, Balaji S, Iyer LM, Aravind L. Small but versatile: the extraordinary functional and structural diversity of the β-grasp fold. Biol. Direct. 2007;2:18. doi: 10.1186/1745-6150-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Burroughs AM, Iyer LM, Aravind L. Natural history of the E1-like superfamily: implication for adenylation, sulfur transfer, and ubiquitin conjugation. Proteins. 2009;75:895–910. doi: 10.1002/prot.22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Burroughs AM, Iyer LM, Aravind L. Functional diversification of the RING finger and other binuclear treble clef domains in prokaryotes and the early evolution of the ubiquitin system. Mol. Biosyst. 2011;7:2261–2277. doi: 10.1039/c1mb05061c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ambroggio XI, Rees DC, Deshaies RJ. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2004;2:e2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tran HJ, Allen MD, Löwe J, Bycroft M. Structure of the Jab1/MPN domain and its implications for proteasome function. Biochemistry. 2003;42:11460–11465. doi: 10.1021/bi035033g. [DOI] [PubMed] [Google Scholar]
- 95.Makarova KS, Koonin EV. Archaeal ubiquitin-like proteins: functional versatility and putative ancestral involvement in tRNA modification revealed by comparative genomic analysis. Archaea. 2010;2010:710303. doi: 10.1155/2010/710303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ranjan N, Damberger FF, Sutter M, Allain FH, Weber-Ban E. Solution structure and activation mechanism of ubiquitin-like small archaeal modifier proteins. J. Mol. Biol. 2010;405:1040–1055. doi: 10.1016/j.jmb.2010.11.040. [DOI] [PubMed] [Google Scholar]
- 97.Jeong YJ, Jeong BC, Song HK. Crystal structure of ubiquitin-like small archaeal modifier protein 1 (SAMP1) from Haloferax volcanii. Biochem. Biophys. Res. Commun. 2011;405:112–117. doi: 10.1016/j.bbrc.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 98.Raiborg C, Slagsvold T, Stenmark H. A new side to ubiquitin. Trends Biochem. Sci. 2006;31:541–544. doi: 10.1016/j.tibs.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 99.Nunoura T, et al. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 2011;39:3204–3223. doi: 10.1093/nar/gkq1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 101.Maupin-Furlow JA, Wilson HL, Kaczowka SJ, Ou MS. Proteasomes in the archaea: from structure to function. Front. Biosci. 2000;5:D837–D865. doi: 10.2741/furlow. [DOI] [PubMed] [Google Scholar]
- 102.Koonin EV, Wolf YI, Aravind L. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 2001;11:240–252. doi: 10.1101/gr.162001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gil MA, Sherwood KE, Maupin-Furlow JA. Transcriptional linkage of Haloferax volcanii proteasomal genes with non-proteasomal gene neighbours including RNase P, MOSC domain and SAM-methyltransferase homologues. Microbiology. 2007;153:3009–3022. doi: 10.1099/mic.0.2007/008177-0. [DOI] [PubMed] [Google Scholar]
- 104.Ruepp A, Eckerskorn C, Bogyo M, Baumeister W. Proteasome function is dispensable under normal but not under heat shock conditions in Thermoplasma acidophilum. FEBS Lett. 1998;425:87–90. doi: 10.1016/s0014-5793(98)00205-1. [DOI] [PubMed] [Google Scholar]
- 105.Reuter CJ, Maupin-Furlow JA. Analysis of proteasome-dependent proteolysis in Haloferax volcanii cells, using short-lived green fluorescent proteins. Appl. Environ. Microbiol. 2004;70:7530–7538. doi: 10.1128/AEM.70.12.7530-7538.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhou G, Kowalczyk D, Humbard MA, Rohatgi S, Maupin-Furlow JA. Proteasomal components required for cell growth and stress responses in the haloarchaeon Haloferax volcanii. J. Bacteriol. 2008;190:8096–8105. doi: 10.1128/JB.01180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kirkland PA, Gil MA, Karadzic IM, Maupin-Furlow JA. Genetic and proteomic analyses of a proteasome-activating nucleotidase A mutant of the haloarchaeon Haloferax volcanii. J. Bacteriol. 2008;190:193–205. doi: 10.1128/JB.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kirkland PA, Reuter CJ, Maupin-Furlow JA. Effect of proteasome inhibitor clasto-lactacystin-β-lactone on the proteome of the haloarchaeon Haloferax volcanii. Microbiology. 2007;153:2271–2280. doi: 10.1099/mic.0.2007/005769-0. [DOI] [PubMed] [Google Scholar]
- 109.Kirkland PA, Maupin-Furlow JA. Stabilization of an archaeal DNA-sliding clamp protein, PCNA, by proteasome-activating nucleotidase gene knockout in Haloferax volcanii. FEMS Microbiol. Lett. 2009;294:32–36. doi: 10.1111/j.1574-6968.2009.01547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wilson HL, Aldrich HC, Maupin-Furlow J. Halophilic 20S proteasomes of the archaeon Haloferax volcanii: purification, characterization, and gene sequence analysis. J. Bacteriol. 1999;181:5814–5824. doi: 10.1128/jb.181.18.5814-5824.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Humbard MA, Reuter CJ, Zuobi-Hasona K, Zhou G, Maupin-Furlow JA. Phosphorylation and methylation of proteasomal proteins of the haloarcheon Haloferax volcanii. Archaea. 2010;2010:481725. doi: 10.1155/2010/481725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Reuter CJ, Kaczowka SJ, Maupin-Furlow JA. Differential regulation of the PanA and PanB proteasome-activating nucleotidase and 20S proteasomal proteins of the haloarchaeon Haloferax volcanii. J. Bacteriol. 2004;186:7763–7772. doi: 10.1128/JB.186.22.7763-7772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Madding LS, et al. Role of the β1 subunit in the function and stability of the 20S proteasome in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 2007;189:583–590. doi: 10.1128/JB.01382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Humbard MA, Stevens SM, Jr, Maupin-Furlow JA. Posttranslational modification of the 20S proteasomal proteins of the archaeon Haloferax volcanii. J. Bacteriol. 2006;188:7521–7530. doi: 10.1128/JB.00943-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Van der Veen AG, et al. Role of the ubiquitin-like protein Urm1 as a noncanonical lysine-directed protein modifier. Proc. Natl Acad. Sci. USA. 2011;108:1763–1770. doi: 10.1073/pnas.1014402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kessler D. Enzymatic activation of sulfur for incorporation into biomolecules in prokaryotes. FEMS Microbiol. Rev. 2006;30:825–840. doi: 10.1111/j.1574-6976.2006.00036.x. [DOI] [PubMed] [Google Scholar]
- 117.Striebel F, Kress W, Weber-Ban E. Controlled destruction: AAA+ ATPases in protein degradation from bacteria to eukaryotes. Curr. Opin. Struct. Biol. 2009;19:209–217. doi: 10.1016/j.sbi.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 118.Sorkin A. Ubiquitination without E3. Mol. Cell. 2007;26:771–773. doi: 10.1016/j.molcel.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 119.Murata S, Takahama Y, Tanaka K. Thymoproteasome: probable role in generating positively selecting peptides. Curr. Opin. Immunol. 2008;20:192–196. doi: 10.1016/j.coi.2008.03.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.