Abstract
In mammalian cells, flat Golgi cisternae closely arrange together to form stacks. During mitosis, the stacked structure undergoes a continuous fragmentation process. The generated mitotic Golgi fragments are distributed into the daughter cells, where they are reassembled into new Golgi stacks. In this study, an in vitro assay has been developed using purified proteins and Golgi membranes to reconstitute the Golgi disassembly and reassembly processes. This technique provides a useful tool to delineate the mechanisms underlying the morphological change. There are two processes during Golgi disassembly: unstacking and vesiculation. Unstacking is mediated by two mitotic kinases, cdc2 and plk, which phosphorylate the Golgi stacking protein GRASP65 and thus disrupt the oligomer of this protein. Vesiculation is mediated by the COPI budding machinery ARF1 and the coatomer complex. When treated with a combination of purified kinases, ARF1 and coatomer, the Golgi membranes were completely fragmented into vesicles. After mitosis, there are also two processes in Golgi reassembly: formation of single cisternae by membrane fusion, and restacking. Cisternal membrane fusion requires two AAA ATPases, p97 and NSF (N-ethylmaleimide-sensitive fusion protein), each of which functions together with specific adaptor proteins. Restacking of the newly formed Golgi cisternae requires dephosphorylation of Golgi stacking proteins by the protein phosphatase PP2A. This systematic study revealed the minimal machinery that controls the mitotic Golgi disassembly and reassembly processes.
The interphase Golgi apparatus, as seen by light or fluorescence microscopy, is a compact juxta-nuclear reticulum, located most often in the peri-centriolar region of the cell (1). Electron microscopy (EM)2 shows that it is comprised of discrete Golgi stacks linked together by tubules that connect equivalent cisternae in adjacent stacks (2). At the onset of mitosis, the characteristic stacked organization of the Golgi apparatus undergoes extensive fragmentation (3, 4). The mitotic Golgi fragments generated by this disassembly process are subsequently distributed to daughter cells, where they are reassembled into new Golgi stacks after mitosis. So far, the mechanism that controls the Golgi disassembly and reassembly processes is not well understood.
Biochemical reconstitution experiments have provided powerful tools with which to dissect biological processes. Two basic experimental approaches have been taken to reconstitute mitotic Golgi disassembly and reassembly. One involves semi-permeabilized cells, in which cells are permeabilized gently with detergent (e.g. digitonin), washed with 1 M KCl to remove endogenous cytosol and peripheral membrane proteins, and then incubated in cytosol prepared from mitotic (or interphase) cells (5, 6). Cells can then be processed directly for immunofluorescence or electron microscopy, or biochemical analysis of proteins. This approach has been used to test the mitotic kinases that regulate the Golgi disassembly process (5, 6).
The second method involves purified Golgi membranes to which mitotic or interphase cytosol is added as above (7–9). After incubation, membranes are separated from cytosol by centrifugation through a sucrose cushion. Membranes can then be processed for biochemical analysis of proteins and morphological analysis of the membranes. This approach has contributed to the discovery and examination of much of the currently identified proteins that mediate Golgi membrane tethering (10, 11), fusion (8, 12–14), and Golgi cisternal stacking (15–17). Although the discovery of these proteins that are involved in regulation of Golgi membrane dynamics has contributed much to our understanding of the biogenesis of the Golgi apparatus, it is unclear whether these proteins are sufficient to control mitotic Golgi disassembly and reassembly, as all these studies used cytosol, or cell extract, in the reconstitution assays. Because cytosol contains many kinds of proteins, it is difficult to identify the minimal machinery or the key components that control Golgi disassembly during mitosis and reassembly afterward. This study describes for the first time an in vitro assay that reconstitutes the entire mitotic Golgi disassembly and reassembly processes using biochemically purified components. Our results show that the disassembly process is mediated by two independent but interactive processes: cisternal membrane unstacking mediated by mitotic kinases, and membrane vesiculation mediated by the COPI vesicle budding machinery. Post-mitotic Golgi reassembly also consists of two processes: membrane fusion mediated by two AAA ATPases, p97 and NSF, and cisternal membrane restacking, mediated by dephosphorylation of the Golgi stacking proteins by protein phosphatase PP2A. Our method provides a powerful tool to further dissect the molecular mechanism that regulates Golgi membrane dynamics during the cell cycle.
EXPERIMENTAL PROCEDURES
Reagents
All reagents were from Sigma, Roche Applied Sciences, or Calbiochem, unless otherwise stated. The following antibodies were used: monoclonal antibodies against ARF1 (1D9, Abcam), Bet1 (BD Transduction Laboratories), β-COP (M3A5, T. Kreis), GM130 and Gos28 (BD Transduction Laboratories), GRASP65 (F. Barr), and α-tubulin (K. Gull); polyclonal antibodies against ARF1 (D. Shields and D. Sheff), β-COP (EAGE, T. Kreis), GM130 (MLO7, M. Lowe), GRASP55 (J. Seemann), GRASP65 (17), α-Mannosidase I (J. Seemann) and II (K. Moremen), Golgin-84 (A. Satoh), rat serum albumin (G. Warren), syntaxin 5 (A. Price), and TGN38 (18). The polyclonal antibody for phosphorylated GM130 was generously provided by M. Lowe. Secondary antibodies for immunofluorescence and for Western blotting were from Molecular Probes and Jackson Immunoresearch Laboratories, respectively.
Protein Expression and Purification
Golgi membranes were purified from rat liver (19). Interphase (IC) and mitotic (MC) cytosol were prepared from HeLa S3 cells (7, 17). The histone kinase activity of mitotic cytosol was 20–25-fold higher than that of interphase cytosol (20). Cdc2 kinase (complexed with cyclin B1) and plk kinases were expressed and purified as described, and kinase activity was measured (17). Coatomer complex was purified from rabbit cytosol (21). Myristoylated ARF1 was co-expressed in bacteria with a second plasmid encoding the yeast N-myristoyltransferase in BL21 bacterial cells and purified as previously described (22, 23). Recombinant His-tagged NSF was purified as previously described (24). Recombinant His-tagged α-SNAP (soluble NSF attachment protein) and γ-SNAP were expressed in bacteria and purified with nickel beads (13). Recombinant p97 and p47 were expressed in bacteria and purified by chromatography using nickel beads (25). p97 was also purified by chromatography from rat liver cytosol (12, 25). p115 was purified from rat liver cytosol by chromatography as previously described (10, 26).
GRASP65 Dephosphorylation
Purified protein serine/threonine phosphatases PP1, PP2A, PP2B, PP2C, and protein tyrosine phosphatase (PTP) were purchased from Upstate Biotech. Purified PP1 originated from skeletal muscle and contained multiple PP1 isoforms. PP2A was purified from human red blood cells as a heterodimer of A and C subunits; the trimeric form of PP2A, called PP2A1, was purified from rabbit skeletal muscles and contained ACBα subunits. PP2B/calcineurin was purified from bovine brain. Recombinant PP2Cα and protein tyrosine phosphatase 1B (PTP-1B) were expressed in bacteria and purified by chromatography. GRASP65 phosphorylation was achieved by treating Golgi membranes with mitotic cytosol, as in the Golgi disassembly assay. Briefly, 5 μg of purified rat liver Golgi membranes were mixed with 500 μg of mitotic cytosol in MEB buffer (50 mM Tris-HCl, pH 7.4, 0.2 M sucrose, 50 mM KCl, 20 mM β-glycerophosphate, 15 mM EGTA, 10 mM MgCl2, 2 mM ATP, 1 mM GTP, 1 mM glutathione, and protease inhibitors) and an ATP regeneration system (10 mM creatine phosphate, 1 mM ATP, 20 μg/ml creatine kinase, 20 ng/ml cytochalasin B) in a 50-μl reaction. After 20 min of incubation at 37 °C, the membranes were pelleted through a 0.4 M sucrose layer at 55,000 rpm for 30 min in a TLA55 rotor (Beckman). To dephosphorylate GRASP65, the membrane pellets were directly resuspended in KHM buffer (20 mM Hepes-KOH, pH7, 0.2 M sucrose, 60 mM KCl, 5 mM Mg(OAc)2, 2 mM ATP, 1 mM GTP, 1 mM glutathione, protease inhibitors) containing 100 μg interphase cytosol, or 0.5 units of purified phosphatases (except for PP2A1, for which 0.125 milliunits was used), and then incubated at 30 °C for 60 min. The membranes were solubilized in SDS buffer and analyzed by Western blotting for GRASP65. GRASP65 phosphorylation and dephosphorylation were analyzed by the migration shift of GRASP65 on SDS-PAGE (17).
To express isoform-specific PP2A, sf9 cells were co-infected by baculoviruses (generously provided by Gary Thomas, the Vollum Institute, Oregon) encoding the A, Bα, B′α, and C subunits. Cell lysates of the infected cells were prepared as described (27). Each lysate exhibited similar activity toward phosphorylase a (28), and Western blotting confirmed that the appropriate subunits were expressed at similar levels by the recombinant baculoviruses in each combinatorial infection. Relative equal amounts of the lysate (20 μl each) with AC, ACBα, ACB′α subunits expressed were used to treat 5 μg of mitotic Golgi fragments prepared as described above. The same amount of lysate from non-infected cells was used as a control. Okadaic acid was purchased from Calbiochem, inhibitor-2 from Upstate Biotech, and microcystin LR from Sigma.
Golgi Disassembly and Reassembly Assay
All recombinant proteins used in this study were wild type. Although the non-hydrolyzable GTP analogue GTPγS could enhance the Golgi disassembly, the resulting Golgi fragments could not be reassembled; thus GTPγS was not used in this study. Mitotic Golgi disassembly assay was performed as previously described (9, 17, 23, 29). Briefly, Golgi membranes (200 μg) were mixed with 10 mg of mitotic cytosol, or with purified coatomer (100 μg), recombinant myristoylated ARF1 (50 μg), 1 mM GTP, and an ATP-regenerating system in MEB buffer, in a final volume of 1000 μl. After incubation for 20 min at 37 °C, mitotic Golgi fragments (MGFs) were isolated and soluble factors were removed by centrifugation (55,000 rpm for 30 min in a TLA55 rotor) through a 0.4 M sucrose cushion in KHM buffer onto a 6-μl 2 M sucrose cushion. The membranes were resuspended in KHM buffer, and aliquots were succeeded to fixation and EM processing, or to reassembly reactions described below.
For Golgi reassembly, 20 μg of MGFs were treated with the following combinations of proteins in 30-μl reactions: 1) 400 μl of interphase cytosol; 2) 3 μg of NSF (100 ng/μl), 0.75 μg of α-SNAP (25 ng/μl), 0.75 μg of γ-SNAP (25 ng/μl) (8), 0.9 μg of p115 (30 ng/μl) (10), and 2 units of PP2A (0.07 units/μl); 3) 3 μg of p97 (100 ng/μl), 0.75 μg of p47 (14), 0.9 μg of p115 (30 ng/μl), and 2 units of PP2A (0.07 units/μl). Reactions were incubated at 37 °C, and membranes were pelleted and processed for EM. The percentage of membranes in cisternae or in vesicles was determined by the intersection method (10, 17). Cisternae were defined as long membrane profiles with a length more than four times their width, with the width no more than 80 nm. Vesicles were defined as round profiles of ~70 nm in diameter. The average size of the vesicles was determined by measuring the diameter of 50 vesicles. Results were quantitated from three independent experiments and the statistical significance was assessed by Student’s t test.
Analysis of the Contents in Golgi Remnants and Vesicles
To determine which proteins were sorted into the vesicles, Golgi disassembly reactions described above using 200 μg of Golgi treated with ARF1 and coatomer in the presence or absence of mitotic kinases were supplemented with 250 mM KCl in cold assay buffer to stop the reaction and release the vesicles. The reactions were directly loaded onto a step gradient comprised of 1.0 ml 0.5 M, 1.5 ml 0.8 M, 2 ml 1.2 M, 2 ml 1.4 M, and 2 ml 1.6 M sucrose in assay buffer containing 250 mM KCl (23). Membranes were centrifuged to equilibrium at 200,000 × g, 4 °C for 3 h in a near vertical rotor (NVT65, Beckman Instruments, 50,000 rpm). COPI-coated vesicles typically peaked at 1.2–1.3 M sucrose, while uncoated Golgi remnants peaked at about 0.8 M sucrose (23). Fractions were diluted 3-fold with a buffer without sucrose and membranes in each fraction were pelleted by centrifugation in a Beckman TLA55 rotor at 55,000 rpm for 60 min followed by Western blotting analysis. The soluble proteins, most of which were found in the top two fractions, were not pelleted after this centrifugation. This allowed us to determine the distribution of only the proteins that were associated with Golgi remnants and vesicles. Six categories of proteins were analyzed: 1) COPI coat proteins ARF1 and β-COP; 2) Golgi matrix proteins GM130, GRASP65, GRASP55, and Golgin-84; 3) Golgi enzymes α-mannosidase I and II; 4) Golgi SNAREs Syntaxin 5, Gos28 and Bet1; 5) the trans-Golgi protein TGN38; and 6) a secretory cargo protein rat serum albumin (RSA). For morphological analysis, fractions containing the Golgi remnants (no. 1–3) and vesicles (no. 7–9) were pooled, diluted 3-fold using a buffer without sucrose, and pelleted by centrifugation. Membrane samples were fixed and processed for EM.
RESULTS
Defined Golgi Disassembly
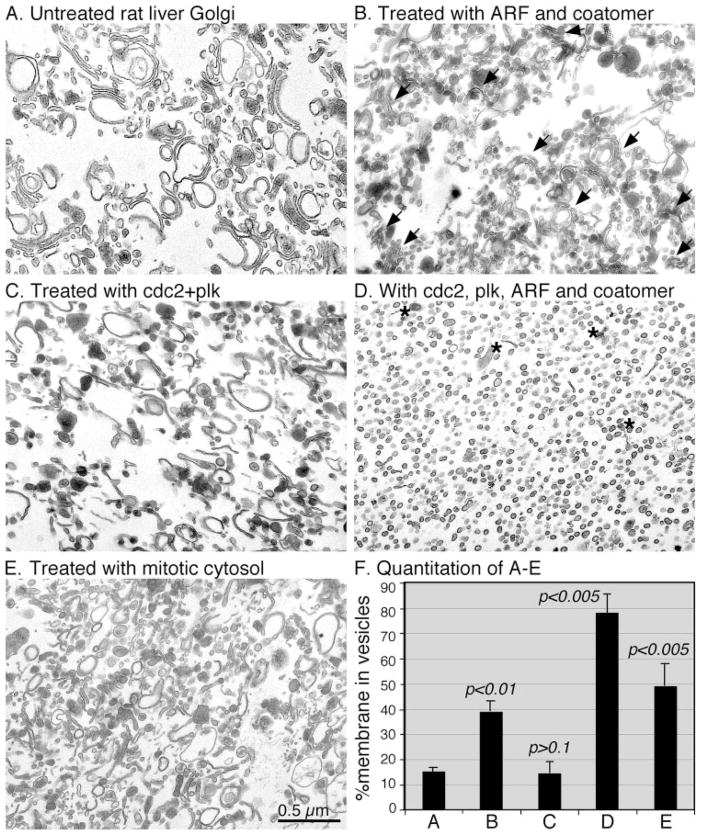
During mitosis, the Golgi apparatus undergoes continuous fragmentation, which requires ARF1 and COPI budding (23). To test whether COPI vesicle budding activity is sufficient to fragment the entire Golgi stack, we treated purified Golgi membranes with purified ARF1 and coatomer (Fig. 1B). This treatment led to fragmentation of a large fraction (40%, Fig. 1F) of the Golgi membranes into vesicles. However, a significant amount of the membranes remained as short cisternae concentrated in ministacks (Fig. 1B, arrows). These ministacks did not diminish even when increased amounts of ARF1 and coatomer were used (not shown), suggesting that vesicle budding alone is not sufficient to complete the fragmentation process of the entire Golgi stack. This result indicates that fragmentation of the Golgi during mitosis may require a COPI-independent fragmentation process, as previously proposed (7). However, since most of the observed cisternae were found in ministacks, it is possible that unstacking of the cisternae may help to improve the fragmentation process. Previous work showed that Golgi membrane stacking is mediated by GRASP65, whose oligomerization functions as a glue to hold the cisternal membrane together (17, 30). Oligomerization of GRASP65 is regulated by phosphorylation, which is mediated by two mitotic kinases, cdc2 (in complex with cyclin B1) and plk. GRASP65 phosphorylation disrupts its oligomerization and thus causes cisternal membrane unstacking (17). In addition, cdc2 and plk also mimic mitotic cytosol to phosphorylate GM130, another Golgi matrix protein whose phosphorylation during mitosis is involved in mitotic Golgi disassembly (20). In this study, when the Golgi membranes were treated with these mitotic kinases, the Golgi membrane became unstacked and thus generated a large amount of single cisternae. However, this treatment did not vesiculate the Golgi cisternal membranes (Fig. 1C). Quantitation results showed that about 15% of the membranes were in vesicles when the Golgi membranes were treated with both mitotic kinases; thus, there is no change compared with that of untreated membranes (Fig. 1F). To test whether unstacking and vesiculation have a synergistic effect in terms of Golgi fragmentation, both sets of components, ARF1/coatomer and kinases, were combined and used to treat Golgi membranes. As expected, the result showed that fragmentation was complete; essentially all membranes were fragmented into vesicles except the elements of the trans-Golgi network (TGN) indicated by the electron-dense contents of lipoproteins (Fig. 1D, asterisks). The size of the vesicles generated by purified components was 70 ± 9 nm (Fig. 1D); no significant change was seen compared with an average diameter of 72 ± 6 nm for those generated by mitotic cytosol (Fig. 1E). Quantitation of the results showed that about 79% of membranes were vesiculated by the treatment with ARF1/coatomer in the presence of mitotic kinases cdc2 and plk, a significant increase compared with treatment with ARF1 and coatomer alone (40%), or with only the two mitotic kinases (15%) (Fig. 1F), suggesting that vesiculation and unstacking are independent but synergistic. The Golgi disassembly after this treatment was also slightly higher than with mitotic cytosol, which had ~50% of membranes in vesicles (Fig. 1, E and F). Detailed examination of EM images at higher magnification indicated that most of the vesicles, generated by treatment with either mitotic cytosol or with ARF1 and coatomer in the presence or absence of mitotic kinases, were coated (data not shown). These results suggest that the mitotic Golgi fragmentation consists of two independent but interactive processes: unstacking, mediated by mitotic kinases that phosphorylate Golgi stacking proteins, and vesiculation, mediated by ARF1 and the COPI coat protein complex coatomer.
FIGURE 1. Golgi disassembly using purified kinases, ARF1 and coatomer.
EM photographs showing fragmentation of purified Golgi membranes using purified ARF1, coatomer, and mitotic kinases. Purified rat liver Golgi membranes (A) were treated with ARF1 and coatomer (B), purified cdc2 and plk kinases (C), or both ARF1/coatomer and kinases (D), at 37 °C for 20 min. Mitotic cytosol treatment was used as a control (E). Membranes were fixed and processed for EM. Bar, 0.5 μm. Note that treatment with purified ARF1 and coatomer generated vesicles and mini-Golgi stacks (arrows in B), treatment with mitotic kinases led to Golgi unstacking but not vesiculation (C) and fragmentation was enhanced when both budding machinery and mitotic kinases were present (D). Asterisks in D indicate electron-dense lipoprotein-enriched TGN elements. The average diameter of the vesicles is 70 ± 9 nm (mean ± S.D.) in D and 72 ± 6 nm in E. F, quantitation of A–E by the intersection method to estimate the percentage of membrane in vesicles. Results represent the mean of three independent experiments ± S.D. To assess the statistical significance, the result for treatment with purified components was compared with that of no treatment.
Golgi Enzymes and SNARE Proteins Are Enriched in Vesicles
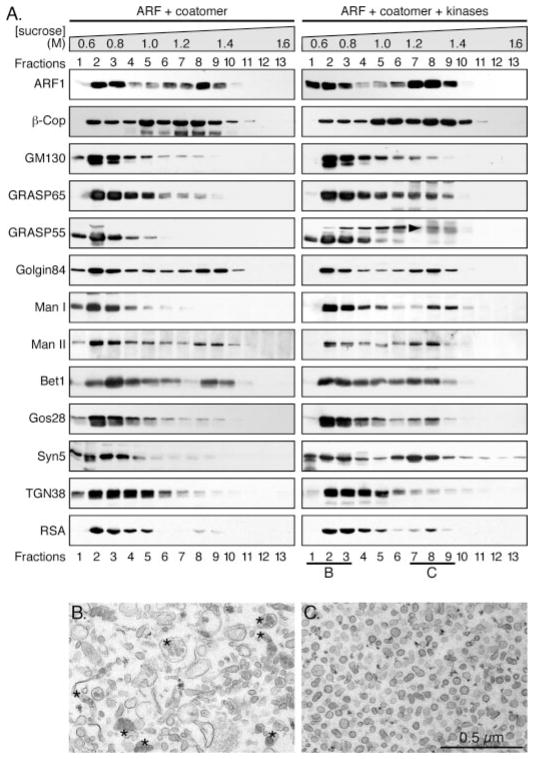
To determine which proteins are enriched in the vesicles generated by treatment with ARF1, coatomer and mitotic kinases, we fractionated the fragmented Golgi membranes on a 0.5–1.6 M sucrose gradient, as described earlier (23). After centrifugation to equilibrium, membrane-bound proteins in each fraction were collected by high-speed centrifugation. Previous work showed that coated vesicles are enriched in a fraction equivalent to the density of 1.2 M sucrose (29, 31), which is fraction 8 (23) in this gradient, while the Golgi remnants remained on the top 3 fractions of the gradient with the density of about 0.8 M sucrose, which is consistent with the density of the Golgi membranes (23, 32). To determine which proteins had been recruited into the vesicles, a number of proteins were tested by Western blotting for their distributions in the gradient. Both ARF1 and the β-COP subunit of the coatomer were found in fractions 7–9, indicating the location of the coated vesicles in the gradient, although a subpopulation of both proteins were found on the top of the gradient where the Golgi remnants were enriched (Fig. 2A). Perhaps these were membranes undergoing a budding process.
FIGURE 2. Distribution of Golgi proteins in vesicles and Golgi remnants after mitotic Golgi disassembly.
Purified rat liver Golgi membranes were treated with purified ARF1 and coatomer in the absence or presence of mitotic kinases at 37 °C for 20 min. Reactions were fractionated by equilibrium centrifugation on sucrose gradients (0.5–1.6 M, indicated on the top). Membranes in each fraction were pelleted and equal fractions (by volume) were analyzed by Western blotting for coat proteins ARF1 and coatomer; Golgi matrix proteins GM130, GRASP65, GRASP55, and Golgin-84; Golgi enzymes α-mannosidase I and II (Man I and II); Golgi SNAREs syntaxin 5 (syn5), Gos28, and Bet1; TGN38 and a secretory cargo protein rat serum albumin (RSA) (A). The upper band for GRASP55 marked by an arrowhead indicates the molecules that were phosphorylated. Membranes in fractions 1–3, which contained 0.6 – 0.8 M sucrose and Golgi remnants, and in fractions 7–9, which contained 1.3–1.4 M sucrose and COPI vesicles, were collected and processed for EM. The EM images are shown in B and C, respectively. Asterisks in B indicate lipoprotein-enriched TGN elements. The diameter of the vesicles in C is 70 ± 6 nm (mean ± S.D.). Bar, 0.5 μm.
There are indications that the Golgi matrix proteins, which include the Golgin and GRASP families of cisternal stacking proteins (33), play a critical role in Golgi structure formation as well as in Golgi inheritance (34, 35). Two Golgi matrix proteins, GM130 and GRASP65, were found mainly in the Golgi remnants; only a minor peak for GRASP65 in the vesicle fractions was observed in the presence of mitotic kinases (Fig. 2A), suggesting that the majority of these proteins associate with the Golgi remnants during mitosis. In the absence of mitotic kinases, GRASP55 was enriched in the Golgi remnants. In the presence of mitotic kinases, GRASP55 was partially phosphorylated. Unlike the unphosphorylated GRASP55 that was enriched in the Golgi remnants, the phosphorylated molecules of GRASP55, indicated by the upshift of the band (Fig. 2A, arrowhead), were partially recruited into the vesicle fractions, with a minor peak in fraction 8 (Fig. 2A). In contrast with the three Golgi matrix proteins described above, another Golgi matrix protein, Golgin-84, was found to be enriched in the vesicle fractions, consistent with its role as a tethering factor to hold COPI vesicles to the Golgi membranes (29). The different behavior of these Golgi matrix proteins might reflect different roles for these proteins in Golgi structure formation and biogenesis.
Two Golgi enzymes, α-mannosidase I (Man I) and II (Man II) are localized on the cis- and medial-Golgi subcompartments, respectively. In the absence of mitotic kinases, Man I was enriched in the top fractions (no. 1–3) that contained the Golgi remnants, while Man II had an additional peak in the fractions (no. 7–9) that contained the vesicles, suggesting that under interphase conditions Man II, rather than Man I, is recruited into COPI vesicles. This observation is consistent with the cisternal maturation model in which COPI vesicles mediate retrograde transport of Golgi enzymes, while cargo proteins move across the stack by maturation of the cisternae (see “Discussion”). In the presence of mitotic kinases, both Man I and Man II were found in the vesicle fractions, suggesting that both enzymes are recruited into vesicles during mitosis when the entire Golgi is vesiculated, indicating the different degrees of vesiculation between interphase and mitotic conditions. The pronounced increase of Man I in the vesicles in the presence of mitotic kinases, compared with that in the absence of mitotic kinases, suggests that Golgi fragmentation in the presence of mitotic kinases is much more extensive than the vesicle formation under interphase conditions.
SNARE proteins behave differently in the gradient upon different treatments. For example, Bet1 was found to be enriched in the vesicle fractions, regardless of the presence of mitotic kinases, consistent with the previously described data that it is a recycling SNARE protein in the cell (36, 37). Gos28, a Golgi t-SNARE, was not found in the vesicles under interphase conditions, but it was slightly enriched in the vesicles in the presence of mitotic kinases. Similar to Gos28, Syntaxin 5, a major t-SNARE on the Golgi that forms a complex with Gos28 (36, 38) and is involved in intra-Golgi transport, was highly enriched in the vesicle fractions in the presence of mitotic kinases.
TGN-38, a transmembrane glycoprotein enriched in the TGN, was found on the top of the gradient regardless of the presence of mitotic kinases, which is consistent with the observation that the TGN membranes do not provide membranes for COPI vesicle budding; instead, they generate clathrin-coated vesicles for trafficking to the endosomal/lysosomal system or to the plasma membrane (39, 40). Rat serum albumin (RSA), a cargo protein secreted by the liver, was found to be enriched in the Golgi remnants, although a small amount was also found in the vesicles when the Golgi membranes were fragmented by ARF1/coatomer and mitotic kinases (Fig. 2A).
For morphological analysis of the membrane components in the fractions enriched with Golgi remnants or vesicles, we pooled fractions 1–3 and 7–9 from the gradient of the membranes that were treated with both sets of proteins. Membranes in each pool were collected by centrifugation and analyzed by EM. As shown in Fig. 2B, fractions 1–3 contained the Golgi remnants, including TGN membranes (asterisks indicate the dense lipoproteins), and other vesicular-tubular structures. Fractions 7–9 contained essentially pure COPI vesicles (Fig. 2C), with a uniform size of about 70 nm in diameter, with coat proteins visible on the surface.
Reassembly of Mitotic Golgi Fragments Using Interphase Cytosol
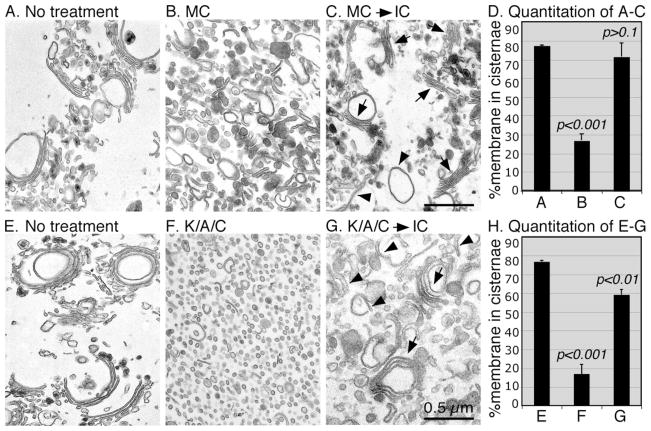
To determine whether the disassembled Golgi membranes can be reassembled into new Golgi cisternae and stacks, the mitotic Golgi fragments were re-isolated by centrifugation through a 0.4 M sucrose cushion to separate the membranes from the soluble proteins of the mitotic cytosol, or from the mitotic kinases. The mitotic Golgi fragments were then treated with interphase HeLa cell cytosol (Fig. 3). When the Golgi was fragmented by mitotic cytosol treatment, 71% of the membranes were reassembled into cisternal membranes, similar to the level of untreated Golgi membranes (77%), which is significantly higher than the 26% observed in mitotic Golgi fragments (Fig. 3, A–D). This result showed the reversibility of the mitotic Golgi disassembly and reassembly processes.
FIGURE 3. Reassembly of mitotic Golgi fragments into Golgi stacks when treated with interphase cytosol.
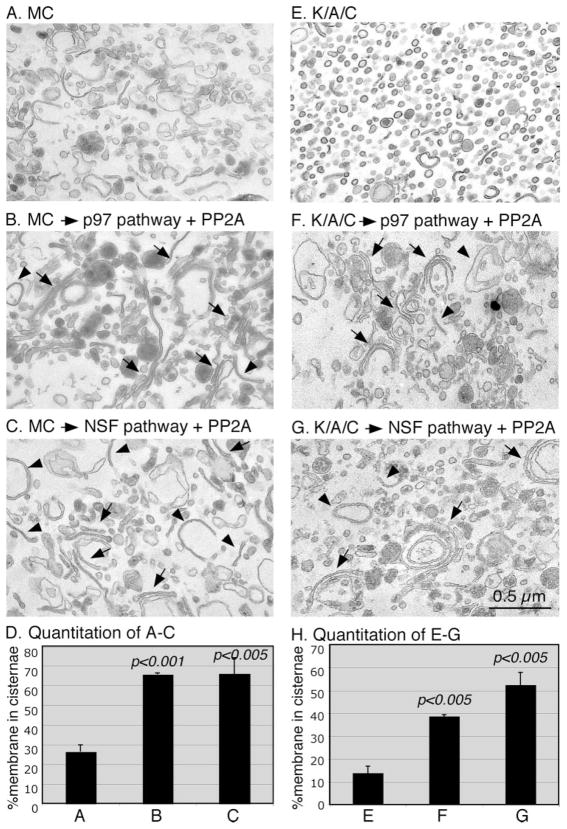
Purified rat liver Golgi membranes (A and E) were treated with MC (B) or a combination of kinases, ARF1 and coatomer (K/A/C, F), at 37 °C for 20 min. Membranes were re-isolated and further treated with IC (C and G) at 37 °C for 60 min. Membranes were fixed and processed for EM. Bar, 0.5 μm. D and H, quantitation of A–C and E–G, respectively, by the intersection method (9), to estimate the percentage of membrane in cisternae. Results represent the mean of three independent experiments ± S.D. To assess the statistical significance, all the results were compared with that of no treatment. Note the newly formed Golgi stacks (arrows) and single cisternae (arrowheads) after reassembly.
When the Golgi membranes were disassembled by treatment with ARF1/coatomer and mitotic kinases, the disassembly was more complete (Fig. 3F), resulting mostly in vesicles and with only about 16% of membranes in cisternae. Most of these cisternae are TGN elements that are indicated by the electron-dense lipoproteins (Fig. 3F). Previous work showed that at least parts of the TGN elements are fragmented during mitosis (41). When these membranes were further treated with interphase cytosol, about 60% of the membranes were reassembled into cisternae and stacks (Fig. 3G), which is greater than three times of that of the disassembled membranes (Fig. 3H). This result showed that the membranes disassembled by the defined disassembly assay are active and can be reassembled into new Golgi stacks, indicating that the conditions used in this study were physiological.
Phosphatases That Dephosphorylate GRASP65 after Mitosis
We then further analyzed the processes that mediate Golgi reassembly. Since we have shown that mitotic Golgi disassembly is mediated by two processes, unstacking and vesiculation, it would be interesting to test whether reassembly is mediated by the reverse processes: membrane fusion and cisternal restacking. Golgi cisternal stacking requires the formation of trans-oligomers of the Golgi stacking protein GRASP65 (17). Oligomerization of GRASP65 is regulated by protein phosphorylation; phosphorylation of GRASP65 by cdc2 and plk during mitosis disrupts the oligomers, while dephosphorylation of GRASP65 leads to reformation of the oligomers (17, 30). Thus restacking of the newly formed Golgi cisternae requires dephosphorylation of the Golgi-stacking protein GRASP65. So far the phosphatase that dephosphorylates mitotic GRASP65 in telophase is unknown. Four major classes of protein serine/threonine phosphatases have been identified in mammalian cells: PP1, PP2A, PP2B, and PP2C (for reviews see Refs. 42, 43). Each of these phosphatases has a different substrate specificity, divalent cation requirement, and sensitivity to various inhibitors (44), which can be used to distinguish the one specific to GRASP65. Previous work showed that GRASP65 was dephosphorylated when the mitotic Golgi fragments were treated with interphase cytosol, e.g. in the Golgi reassembly assay, indicating a phosphatase activity in the interphase cytosol (17). This activity was sensitive to okadaic acid, a strong inhibitor of PP2A and weak inhibitor of PP1, and was insensitive to inhibitor-2, a specific peptide inhibitor of PP1 (17), suggesting a role for protein phosphatase PP2A in post-mitotic GRASP65 dephosphorylation.
PP2A exists as a trimeric complex in vivo, with a constant dimeric core comprised of a 36-kDa catalytic (PP2Ac or C) subunit and a 65-kDa (PR65 or A) subunit bound to a third, variable B subunit (45). Three classes of the B subunit have so far been identified as Bα, B′α and B″α. The B subunits have been shown to act as positive regulators to enhance catalytic activity toward particular substrates (45, 46). Furthermore, the B subunits may target isoforms of PP2A to particular subcellular locations (46); the Bα regulatory subunit has been shown to be associated with the Golgi apparatus (28).
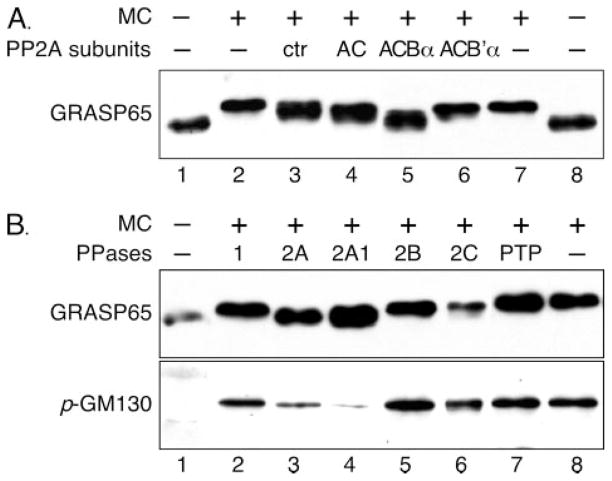
To test which of the B subunits of PP2A plays a role in modulating the activity of PP2A toward GRASP65, sf9 cells were co-infected with baculovirus recombinants expressing combinations of the C, A, and one of the variable B subunits (27), and lysates were assayed for their ability to dephosphorylate GRASP65 in vitro. Golgi membranes were first treated with mitotic cytosol followed by treatment with baculoviral lysate with different combinations of the PP2A subunits expressed. Phosphorylation of GRASP65 in the membranes was determined by the shift of the band on Western blots, shown in Fig. 4. Some GRASP65 phosphatase activity was observed in the lysate prepared from uninfected cells (Fig. 4A, lane 3), most likely due to endogenous PP2A. This was slightly increased by expression of the AC dimer (Fig. 4A, lane 4). Co-expression of the Bα subunit with AC dramatically increased the GRASP65 phosphatase activity (Fig. 4A, lane 5); this treatment shifted GRASP65 down to the level of untreated Golgi membranes. In contrast, co-expression of the B′α subunit with AC did not increase GRASP65 phosphatase activity. In fact, the B′α subunit slightly inhibited basal activity toward GRASP65 (Fig. 4A, lanes 6 versus 4), most likely by displacing endogenous sf9 cell Bα subunits from the holoenzyme complexes (47). These results strongly suggest that PP2A with ACBα subunits is the GRASP65 phosphatase.
FIGURE 4. GRASP65 is dephosphorylated by protein phosphatase PP2A.
A, dephosphorylation of GRASP65 by isoform-specific PP2A. PP2A subunits as indicated were expressed using a baculoviral system in insect cells. Cells were harvested after 64 –72 h infection and the same amounts of the lysates, each of which had similar phosphatase activity assayed using phosphorylase a as the substrate, were used to treat mitotic Golgi fragments (lanes 3– 6). Uninfected cells were used as control (ctr, lane 3). Untreated Golgi membranes (lanes 1 and 8) and mitotic Golgi fragments (lanes 2 and 7) were also loaded on the gel. Shown is a Western blot of GRASP65 after treatment. Dephosphorylation of GRASP65 is detected by downshift of the band (17) compared with that of mitotic Golgi fragments (lanes 2 and 7). Note that uninfected insect cells (none) had a detectable level of GRASP65 phosphatase activity, which is slightly increased by expression of the AC subunits alone or in combination with the B′α subunit. In contrast, the Bα subunit significantly increased the GRASP65 dephosphorylation activity, as GRASP65 is shifted down after treatment to a level that is close to that of untreated membranes (lanes 5 versus 1 and 8). B, dephosphorylation of GRASP65 and GM130 by purified PP2A. Purified Golgi membranes (lane 1) were treated with mitotic cytosol (lanes 2– 8) followed by re-isolation of the mitotic Golgi fragments, which were further treated with 0.5 units of purified PP1 (lane 2), PP2A (purified from human red blood cells, lane 3), calcineurin/PP2B (lane 5), PP2Ca (lane 6), and protein-tyrosine phosphatase (PTP, lane 7), or with 0.125 milliunits of PP2A1 (PP2A purified from rabbit skeletal muscle, lane 4). Membranes were solubilized in SDS buffer and analyzed by Western blotting for GRASP65 and phosphorylated GM130 (p-GM130). Note that GRASP65 is shifted down only when the mitotic Golgi membranes were treated with PP2A (lanes 3– 4), while GM130 dephosphorylation is indicated by the reduction of the band intensity.
To further confirm that dephosphorylation of GRASP65 is mediated by PP2A, mitotic Golgi fragments were further treated with purified phosphatases. Five microgram mitotic Golgi fragments were treated with 0.5 units each of PP1 (Fig. 4B, lane 2), PP2A AC subunit complex (lane 3), PP2B/calcineurin (lane 5), PP2C (lane 6), protein-tyrosine phosphatase (PTP, lane 7), or 0.125 milliunits of PP2A1, the holoenzyme of PP2A with ACBα subunits (Fig. 4B, lane 4). As shown in Fig. 4B, only the PP2A treatments led to downshift/dephosphorylation of GRASP65 to the level comparable to untreated Golgi membranes (lanes 3 and 4 versus lane 1). Notably, the ACBα trimer of PP2A, PP2A1, exhibited much higher activity, with as little as 0.125 milliunits needed to dephosphorylate GRASP65 efficiently (lane 4). A similar result was observed using an antibody that specifically recognizes GM130 phosphorylated on serine 25, consistent with the previous report that GM130 is also dephosphorylated by PP2A after mitosis (28). Taken together, we have identified PP2A as the GRASP65 phosphatase, which is sufficient to dephosphorylate GRASP65 after mitosis.
Defined Golgi Membrane Reassembly
Reassembly of mitotic Golgi fragments into stacks requires membrane fusion and thus generation of single cisternae. Previous work has suggested that post-mitotic Golgi membrane fusion requires two AAA ATPases, p97 and NSF, each of which functions together with specific adaptor proteins (8, 12–14, 48, 49). The activity of p97 in membrane fusion requires its cofactors p47 and p135, the latter of which is a deubiquitinating enzyme associated with the Golgi membranes (14, 50). NSF works together with two adaptor proteins, α- and γ-SNAP. In addition, the Golgi tethering factor p115 has been implicated in post-mitotic Golgi reassembly (10). Therefore, we treated mitotic Golgi fragments with the two sets of proteins: 1) p97, p47, p115, and PP2A, and 2) NSF, α/γ-SNAPs, p115 and PP2A. When the Golgi membranes were disassembled by treatment with mitotic cytosol, reassembly using proteins in the p97 pathway generated long cisternae, some of which formed stacks (Fig. 5B). The same Golgi fragments were also reassembled into cisternae and stacks when treated with the proteins in the NSF pathway, although the long cisternae were seen less often (Fig. 5C). Quantitation of the EM images showed that the percentage of the total membrane in cisternae increased significantly after the reassembly reactions, from 26% of the mitotic Golgi fragments to 66% after reassembly with both p97 and NSF treatments (Fig. 5D). This is consistent with the previous report that both ATPases are sufficient to mediate membrane fusion (8, 13, 48, 49).
FIGURE 5. Reassembly of mitotic Golgi fragments into Golgi stacks using defined components.
Purified rat liver Golgi membranes were treated with MC (A) or a combination of cdc2 and plk kinases, ARF1 and coatomer (K/A/C, E), at 37 °C for 20 min. Membrane fragments were re-isolated and further treated with PP2A and purified proteins in the p97 pathway (p97, p47, p115 and PP2A, B and F), or in the NSF pathway (NSF, α/γ-SNAP, p115 and PP2A, C and G) at 37 °C for 60 min. Membranes were fixed and processed for EM. Bar, 0.5 μm. D and H, quantitation of A–C and E–G, respectively, by the intersection method to estimate the percentage of membrane in cisternae. Results represent the mean of three independent experiments ± S.D. Statistical significance was assessed by comparison of results after reassembly with that of mitotic Golgi fragments used for the reassembly reactions. Note the newly formed Golgi stacks (arrows) and single cisternae (arrowheads) after reassembly.
We then treated the mitotic Golgi fragments disassembled with purified ARF1/coatomer and the mitotic kinases cdc2 and plk (Fig. 5, E–H). Treatment with the proteins in the p97 pathway (in the presence of PP2A) significantly increased the percentage of the total membrane in cisternae. This was raised to 38% after the reassembly by p97 from 13% of the mitotic Golgi fragments (Fig. 5H), with single cisternae and stacks observed (Fig. 5F). The degree of reassembly after NSF treatment is even higher, with 52% of membranes in cisternae, most of which were found in stacks (Fig. 5G). This result showed that the two ATPases were sufficient for postmitotic Golgi membrane fusion to generate single cisternae, and the single cisternae were then assembled into stacks by dephosphorylation of the Golgi stacking proteins mediated by PP2A. To summarize, we have successfully reconstituted the mitotic Golgi disassembly and reassembly processes using biochemically purified components.
In addition, our results revealed the minimal machinery that controls Golgi membrane dynamics during the cell cycle. There are two processes in Golgi disassembly: unstacking, mediated by phosphorylation of Golgi stacking proteins by mitotic kinases, and vesiculation, mediated by the COPI budding machinery ARF1 and coatomer. When treated with a combination of purified kinases, ARF1 and coatomer, the Golgi membranes were completely broken into vesicles. After mitosis, Golgi reassembly is also mediated by two processes: formation of single cisternae by membrane fusion, mediated by the two AAA ATPases p97 and NSF, and restacking, mediated by dephosphorylation of Golgi stacking proteins by the protein phosphatase PP2A. These results are summarized in the model described in Fig. 6.
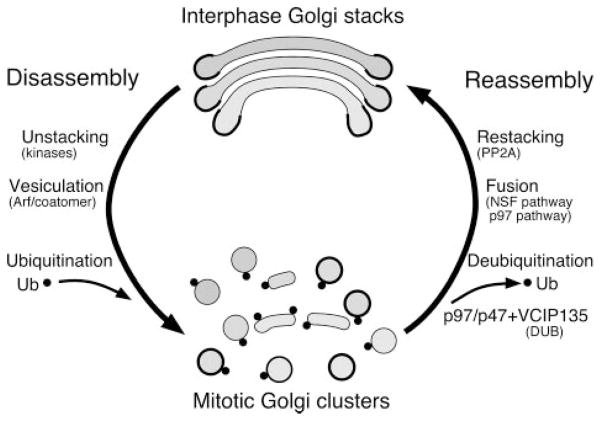
FIGURE 6. Illustration of the processes and proteins involved in Golgi disassembly and reassembly.
There are two processes in Golgi disassembly: unstacking and vesiculation. Two mitotic kinases, cdc2 and plk, phosphorylate GRASP65 and thus disrupt the GRASP65 oligomer and unstack the Golgi. ARF1 and the coatomer complex are the minimal components needed for budding of COPI vesicles and thus vesiculation of the Golgi membranes. A combination of these two sets of proteins is sufficient to fragment the Golgi membranes completely. After mitosis, there are also two processes involved in Golgi reassembly: formation of single cisternae by membrane fusion and restacking. Cisternal membrane fusion requires two AAA ATPases, p97 and NSF, each of which functions together with specific adaptor proteins. Membrane fusion mediated by the p97 pathway requires mono-ubiquitination of Golgi membrane proteins that occurs during mitosis. Restacking of the newly formed Golgi cisternae requires dephosphorylation of Golgi-stacking proteins by the protein phosphatase PP2A.
DISCUSSION
Pathways That Mediate Mitotic Golgi Disassembly
It is known that COPI vesicles can be generated when purified Golgi membranes are treated with purified ARF1 and coatomer (23, 29, 31). The fact that active ARF1 is required for mitotic Golgi fragmentation suggests that COPI vesicle formation plays a major role in mitotic Golgi fragmentation (23). However, it has been proposed that there is an additional COPI-independent pathway during mitotic Golgi disassembly (7). In the current study, fragmentation of the Golgi membranes was not complete upon treatment with purified ARF1 and coatomer, with about 40% of the total membrane in vesicles. The cores of the stacks could not be vesiculated, and were left as ministacks. This, however, does not mean there is another mechanism that shatters these parts of the membranes into fragments. Instead, when this vesiculation process was coupled with the cisternal unstacking machinery, phosphorylation of Golgi stacking proteins by mitotic kinases cdc2 and plk, the Golgi membranes were completely fragmented into vesicles. Consistent with this observation, the disassembly intermediates generated by the previously described COPI-independent Golgi fragmentation pathway can also be fragmented by the COPI-dependent pathway (51). Some cisternae remained after the disassembly reaction, which is consistent with the observations that some Golgi membrane remain as clusters during mitosis (52), most of which are associated with the mitosis spindle (53). Some of the remaining cister-nae are TGN elements, marked by the presence of electron-dense lipoproteins. Previous studies using immuno-EM showed that the TGN elements, marked by the trans-Golgi enzyme galactosyltransferase, remain at least partially in large structures in mitotic cells (41). In addition, the trans-Golgi protein TGN38 does not seem to enter the vesicles after disassembly using both sets of proteins (Fig. 2A). This is consistent with the previous report that the galactosyltransferase (GalT) containing TGN elements remain in the Golgi remnants when mitotic cytosol was used for the disassembly reaction (51). In addition, the size of the vesicles generated by purified components was the same as those generated by mitotic cytosol (Figs. 1 and 2). These results strongly suggest that this assay faithfully represents the condition in mitotic cells. Taken together, our results showed that a combination of mitotic kinases and the COPI budding machinery is sufficient to fragment the Golgi to the same level as in mitotic cells.
Models for Intra-Golgi Trafficking
The flow pattern of materials within the Golgi stack is governed by COPI vesicles. In one view, proposed by the vesicular transport model, anter-ograde vesicles deliver newly synthesized protein and lipid cargo to successive cisternae, where they can undergo post-translational modifications. Retrograde vesicles would then salvage components of the fusion machinery as well as those Golgi enzymes that have strayed beyond the cisterna(e) in which they function. In another view, proposed by the cisternal maturation model, the protein and lipid cargo do not move forward in vesicles; rather, the retrograde COPI vesicles carry Golgi enzymes and fusion machinery back to the preceding cisterna, which matures into the next one (54–56). A recent modification to this view suggests that tubular continuities mediate the retrograde transport of Golgi enzymes, leaving COPI vesicles to carry the fusion machinery (57). These views are not mutually exclusive, but they do require considerable coordination of the COPI vesicle flow pattern (58).
In this study, we have analyzed the proteins that entered the vesicles when the Golgi membranes were treated with ARF1/coatomer in the absence or presence of mitotic kinases. Our results showing Bet1 and Golgin-84 to be enriched in the vesicle fractions under interphase conditions suggest that these proteins are recruited into COPI vesicles and perhaps are recycled between different cisternae or between the Golgi and the ER in interphase cells. One interesting observation is the pattern of the two Golgi enzymes Man I and Man II, which reside in the cis and medial Golgi, respectively. Although both enzymes were recruited into vesicles in the presence of mitotic kinases, there was little Man I in the vesicle fractions compared with a relatively large amount of Man II in the absence of mitotic kinases (Fig. 2A), suggesting that late Golgi enzymes (e.g. Man II), but not early Golgi enzymes (e.g. Man I), were transported by COPI vesicles, presumably from later to earlier cisternae. TGN38, a trans-Golgi protein, was not found in the vesicle fractions even in the presence of mitotic kinases (Fig. 2A). In addition, a very little amount of rat serum albumin, a soluble cargo protein secreted by liver cells, was found in the vesicle fractions in the absence of mitotic kinases (Fig. 2A), suggesting that its transport through the Golgi might not be mediated by COPI vesicles. In summary, our results support the Golgi cisternal maturation model.
Pathways That Mediate Mitotic Golgi Reassembly
Mitotic Golgi fragments achieved by treatment with mitotic cytosol or with the defined components were converted to cisternae and stacks when further treated with interphase cytosol, indicating that the disassembly process is physiological and reversible (Fig. 3). Previous work has shown that post-mitotic Golgi membrane fusion requires two AAA ATPases, NSF and p97, each of which functions together with its cofactors. In this study, when the Golgi membranes were fragmented by mitotic cytosol, reassembly with either p97 or NSF was achieved to the same level, with the majority (66%) of the membranes in cisternae (Fig. 5, A–D). However, when the membranes were disassembled with purified components, reassembly with p97 was less efficient than with NSF (Fig. 5, E–H). It has been shown that a mono-ubiquitination process occurs during Golgi disassembly and that this is required for the subsequent reassembly mediated by p97 (14). In this experiment, as the ubiquitination machinery (such as the ubiquitin E3 ligase) was missing, the reassembly process with p97 was not as efficient. Thus this assay can be used to test the function of ubiquitination in regulation of the Golgi membrane dynamics during the cell cycle.
The reversibility of the disassembly and reassembly reactions provides strong evidence that our assay represents a physiological condition. However, the efficiency of the Golgi reassembly is lower with purified components than with mitotic and inter-phase cytosols (Fig. 5), suggesting that there might be additional factors that modulate these reactions. For example, GTP exchange and hydrolysis are important for ARF activity. The fact that additional ARFGAP is not absolutely required for the disassembly and reassembly indicates that there is some GTP hydrolysis due to the intrinsic GTPase activity from ARF1, or that some ARFGAP proteins are associated with the Golgi membranes. The role of ARFGAP and/or GEF and other proteins in Golgi biogenesis can be easily tested using this assay. To summarize, this study detailed the major pathways involved in mitotic Golgi disassembly: membrane unstacking and vesiculation, and in post-mitotic Golgi membrane reassembly: membrane fusion mediated by p97 and NSF and cisternal restacking. This technique also provided a useful tool to further study the mechanism of Golgi disassembly and reassembly during the mammalian cell cycle.
Acknowledgments
We thank F. Barr, K. Gull, T. Kreis, M. Lowe, K. Moremen, A. Price, A. Satoh, J. Seemann, D. Sheff, D. Shields, T. Taguchi, G. Warren for antibodies, J. Rothman for ARF1, NSF, α- and γ-SNAP cDNAs, J. Malsam for technical help with ARF1 and coatomer purification, H. Meyer for p97 and p47 cDNAs and for p97 purification, M. Jackman for cdc2 and cyclin B1 baculoviruses and purification, and G. Thomas for PP2A baculoviruses. We also thank other members of the Wang Laboratory for suggestions and reagents.
Footnotes
The abbreviations used are: EM, electron microscopy; ARF1, ADP-ribosylation factor-1; IC, interphase cytosol; MC, mitotic cytosol; plk, Polo-like kinase-1; NSF, N-ethylmaleimide-sensitive fusion protein; PP2A, protein serine/threonine phosphatase type 2A; PTP, protein-tyrosine phosphatase; SNAP, soluble NSF attachment protein; TGN, trans-Golgi network.
References
- 1.Louvard D, Reggio H, Warren G. J Cell Biol. 1982;92:92–107. doi: 10.1083/jcb.92.1.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rambourg A, Clermont Y. Eur J Cell Biol. 1990;51:189–200. [PubMed] [Google Scholar]
- 3.Robbins E, Gonatas NK. J Histochem Cytochem. 1964;12:704–711. doi: 10.1177/12.9.704. [DOI] [PubMed] [Google Scholar]
- 4.Lucocq JM, Pryde JG, Berger EG, Warren G. J Cell Biol. 1987;104:865–874. doi: 10.1083/jcb.104.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kano F, Takenaka K, Yamamoto A, Nagayama K, Nishida E, Murata M. J Cell Biol. 2000;149:357–368. doi: 10.1083/jcb.149.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colanzi A, Deerinck TJ, Ellisman MH, Malhotra V. J Cell Biol. 2000;149:331–339. doi: 10.1083/jcb.149.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misteli T, Warren G. J Cell Biol. 1994;125:269–282. doi: 10.1083/jcb.125.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rabouille C, Kondo H, Newman R, Hui N, Freemont P, Warren G. Cell. 1998;92:603–610. doi: 10.1016/s0092-8674(00)81128-9. [DOI] [PubMed] [Google Scholar]
- 9.Rabouille C, Misteli T, Watson R, Warren G. J Cell Biol. 1995;129:605–618. doi: 10.1083/jcb.129.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shorter J, Warren G. J Cell Biol. 1999;146:57–70. doi: 10.1083/jcb.146.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Satoh A, Wang Y, Malsam J, Beard MB, Warren G. Traffic. 2003;4:153–161. doi: 10.1034/j.1600-0854.2003.00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondo H, Rabouille C, Newman R, Levine TP, Pappin D, Freemont P, Warren G. Nature. 1997;388:75–78. doi: 10.1038/40411. [DOI] [PubMed] [Google Scholar]
- 13.Rabouille C, Levine TP, Peters JM, Warren G. Cell. 1995;82:905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Satoh A, Warren G, Meyer HH. J Cell Biol. 2004;164:973–978. doi: 10.1083/jcb.200401010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barr FA, Puype M, Vandekerckhove J, Warren G. Cell. 1997;91:253–262. doi: 10.1016/s0092-8674(00)80407-9. [DOI] [PubMed] [Google Scholar]
- 16.Shorter J, Watson R, Giannakou ME, Clarke M, Warren G, Barr FA. EMBO J. 1999;18:4949–4960. doi: 10.1093/emboj/18.18.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Seemann J, Pypaert M, Shorter J, Warren G. EMBO J. 2003;22:3279–3290. doi: 10.1093/emboj/cdg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taguchi T, Pypaert M, Warren G. Traffic. 2003;4:344–352. doi: 10.1034/j.1600-0854.2003.00091.x. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Taguchi T, Warren G. In: Cell Biology: A Laboratory Handbook. 3. Celis J, editor. Elsevier Science; San Diego: 2006. [Google Scholar]
- 20.Lowe M, Rabouille C, Nakamura N, Watson R, Jackman M, Jamsa E, Rahman D, Pappin DJ, Warren G. Cell. 1998;94:783–793. doi: 10.1016/s0092-8674(00)81737-7. [DOI] [PubMed] [Google Scholar]
- 21.Pavel J, Harter C, Wieland FT. Proc Natl Acad Sci U S A. 1998;95:2140–2145. doi: 10.1073/pnas.95.5.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Randazzo PA, Weiss O, Kahn RA. Methods Enzymol. 1995;257:128–135. doi: 10.1016/s0076-6879(95)57018-7. [DOI] [PubMed] [Google Scholar]
- 23.Xiang Y, Seemann J, Bisel B, Punthambaker S, Wang Y. J Biol Chem. 2007;282:21829–21837. doi: 10.1074/jbc.M611716200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whiteheart SW, Rossnagel K, Buhrow SA, Brunner M, Jaenicke R, Rothman JE. J Cell Biol. 1994;126:945–954. doi: 10.1083/jcb.126.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer HH, Wang Y, Warren G. EMBO J. 2002;21:5645–5652. doi: 10.1093/emboj/cdf579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine TP, Rabouille C, Kieckbusch RH, Warren G. J Biol Chem. 1996;271:17304–17311. doi: 10.1074/jbc.271.29.17304. [DOI] [PubMed] [Google Scholar]
- 27.Molloy SS, Thomas L, Kamibayashi C, Mumby MC, Thomas G. J Cell Biol. 1998;142:1399–1411. doi: 10.1083/jcb.142.6.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowe M, Gonatas NK, Warren G. J Cell Biol. 2000;149:341–356. doi: 10.1083/jcb.149.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malsam J, Satoh A, Pelletier L, Warren G. Science. 2005;307:1095–1098. doi: 10.1126/science.1108061. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Satoh A, Warren G. J Biol Chem. 2005;280:4921–4928. doi: 10.1074/jbc.M412407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nickel W, Malsam J, Gorgas K, Ravazzola M, Jenne N, Helms JB, Wieland FT. J Cell Sci. 1998;111:3081–3090. doi: 10.1242/jcs.111.20.3081. [DOI] [PubMed] [Google Scholar]
- 32.Serafini T, Rothman JE. Methods Enzymol. 1992;219:286–299. doi: 10.1016/0076-6879(92)19029-6. [DOI] [PubMed] [Google Scholar]
- 33.Pfeffer SR. J Cell Biol. 2001;155:873–875. doi: 10.1083/jcb.200109095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shorter J, Warren G. Annu Rev Cell Dev Biol. 2002;18:379–420. doi: 10.1146/annurev.cellbio.18.030602.133733. [DOI] [PubMed] [Google Scholar]
- 35.Seemann J, Pypaert M, Taguchi T, Malsam J, Warren G. Science. 2002;295:848–851. doi: 10.1126/science.1068064. [DOI] [PubMed] [Google Scholar]
- 36.Tai G, Lu L, Wang TL, Tang BL, Goud B, Johannes L, Hong W. Mol Biol Cell. 2004;15:4011–4022. doi: 10.1091/mbc.E03-12-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cosson P, Ravazzola M, Varlamov O, Sollner TH, Di Liberto M, Volchuk A, Rothman JE, Orci L. Proc Natl Acad Sci U S A. 2005;102:14647–14652. doi: 10.1073/pnas.0507394102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Y, Martin S, James DE, Hong W. Mol Biol Cell. 2002;13:3493–3507. doi: 10.1091/mbc.E02-01-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McMahon HT, Mills IG. Curr Opin Cell Biol. 2004;16:379–391. doi: 10.1016/j.ceb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 40.Farquhar MG, Palade GE. Trends Cell Biol. 1998;8:2–10. doi: 10.1016/S0962-8924(97)01187-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lucocq JM, Pryde JG, Berger EG, Warren G. Prog Clin Biol Res. 1988;270:431–440. [PubMed] [Google Scholar]
- 42.Cohen P. Annu Rev Biochem. 1989;58:453–508. doi: 10.1146/annurev.bi.58.070189.002321. [DOI] [PubMed] [Google Scholar]
- 43.Shenolikar S. Annu Rev Cell Biol. 1994;10:55–86. doi: 10.1146/annurev.cb.10.110194.000415. [DOI] [PubMed] [Google Scholar]
- 44.Cohen P. Methods Enzymol. 1991;201:389–398. doi: 10.1016/0076-6879(91)01035-z. [DOI] [PubMed] [Google Scholar]
- 45.Mayer-Jaekel RE, Hemmings BA. Trends Cell Biol. 1994;4:287–291. doi: 10.1016/0962-8924(94)90219-4. [DOI] [PubMed] [Google Scholar]
- 46.Turowski P, Myles T, Hemmings BA, Fernandez A, Lamb NJ. Mol Biol Cell. 1999;10:1997–2015. doi: 10.1091/mbc.10.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tehrani MA, Mumby MC, Kamibayashi C. J Biol Chem. 1996;271:5164–5170. doi: 10.1074/jbc.271.9.5164. [DOI] [PubMed] [Google Scholar]
- 48.Otter-Nilsson M, Hendriks R, Pecheur-Huet EI, Hoekstra D, Nilsson T. EMBO J. 1999;18:2074–2083. doi: 10.1093/emboj/18.8.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Acharya U, Jacobs R, Peters JM, Watson N, Farquhar MG, Malhotra V. Cell. 1995;82:895–904. doi: 10.1016/0092-8674(95)90269-4. [DOI] [PubMed] [Google Scholar]
- 50.Uchiyama K, Jokitalo E, Kano F, Murata M, Zhang X, Canas B, Newman R, Rabouille C, Pappin D, Freemont P, Kondo H. J Cell Biol. 2002;159:855–866. doi: 10.1083/jcb.200208112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levine TP, Misteli T, Rabouille C, Warren G. Cold Spring Harb Symp Quant Biol. 1995;60:549–557. doi: 10.1101/sqb.1995.060.01.058. [DOI] [PubMed] [Google Scholar]
- 52.Misteli T, Warren G. J Cell Sci. 1995;108:2715–2727. doi: 10.1242/jcs.108.7.2715. [DOI] [PubMed] [Google Scholar]
- 53.Shima DT, Cabrera-Poch N, Pepperkok R, Warren G. J Cell Biol. 1998;141:955–966. doi: 10.1083/jcb.141.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glick BS, Malhotra V. Cell. 1998;95:883–889. doi: 10.1016/s0092-8674(00)81713-4. [DOI] [PubMed] [Google Scholar]
- 55.Losev E, Reinke CA, Jellen J, Strongin DE, Bevis BJ, Glick BS. Nature. 2006;441:1002–1006. doi: 10.1038/nature04717. [DOI] [PubMed] [Google Scholar]
- 56.Matsuura-Tokita K, Takeuchi M, Ichihara A, Mikuriya K, Nakano A. Nature. 2006;441:1007–1010. doi: 10.1038/nature04737. [DOI] [PubMed] [Google Scholar]
- 57.Trucco A, Polishchuk RS, Martella O, Di Pentima A, Fusella A, Di Giandomenico D, San Pietro E, Beznoussenko GV, Polishchuk EV, Baldassarre M, Buccione R, Geerts WJ, Koster AJ, Burger KN, Mironov AA, Luini A. Nat Cell Biol. 2004;6:1071–1081. doi: 10.1038/ncb1180. [DOI] [PubMed] [Google Scholar]
- 58.Pelham HR, Rothman JE. Cell. 2000;102:713–719. doi: 10.1016/s0092-8674(00)00060-x. [DOI] [PubMed] [Google Scholar]