Abstract
• Background and Aims Melocacatus paucispinus (Cactaceae) is endemic to the state of Bahia, Brazil, and due to its rarity and desirability to collectors it has been considered threatened with extinction. This species is usually sympatric and inter-fertile with M. concinnus, and morphological evidence for hybridization between them is present in some populations. Levels of genetic and morphological variation and sub-structuring in populations of these species were assessed and an attempt was made to verify the occurrence of natural hybridization between them.
• Methods Genetic variability was surveyed using allozymes (12 loci) and morphological variability using multivariate morphometric analyses (17 vegetative characters) in ten populations of M. paucispinus and three of M. concinnus occurring in the Chapada Diamantina, Bahia.
• Key Results Genetic variability was low in both species (P = 0·0–33·3, A = 1·0–1·6, He = 0·000–0·123 in M. paucispinus; P = 0·0–25·0, A = 1·0–1·4, He = 0·000–0·104 in M. concinnus). Deficit of heterozygotes within the populations was detected in both species, with high values of FIS (0·732 and 0·901 in M. paucispinus and M. concinnus, respectively). Evidence of hybridization was detected by the relative allele frequency in the two diaphorase loci. High levels of genetic (FST = 0·504 in M. paucispinus and 0·349 in M. concinnus) and morphological (A = 0·20 in M. paucispinus and 0·17 in M. concinnus) structuring among populations were found.
• Conclusions The Melocactus spp. displayed levels of genetic variability lower than the values reported for other cactus species. The evidence indicates the occurrence of introgression in both species at two sites. The high FST values cannot be explained by geographical substructuring, but are consistent with hybridization. Conversely, morphological differentiation in M. paucispinus, but not in M. concinnus, is probably due to isolation by distance.
Keywords: Allozymes, Cactaceae, campo rupestre, Chapada Diamantina, genetic diversity, Melocactus concinnus, Melocactus paucispinus, morphological variability, morphometrics
INTRODUCTION
The genus Melocactus consists of 36 species (Anderson, 2001), being common in arid and semi-arid regions of tropical and subtropical zones of the western hemisphere. Although it has a wide distribution, the greatest concentration of taxa and the centre of diversity lie in eastern Brazil, especially in the state of Bahia. Taylor and Zappi (2004) recognized 22 species and subspecies in eastern Brazil, of which 18 are endemic. Plants in the genus are characterized by a small, globose to slightly elongated, unbranched stem, the fertile part differentiated into a terminal cephalium. Flowers are diurnal, small, and embedded within the cephalium with only the perianth segments visible. According to Taylor (1991), most species are self-compatible, but floral adaptations promote hummingbird-mediated cross-pollination. The fruits are small turbinate berries, with small black seeds embedded in a watery pulp; the seeds are locally dispersed by lizards (Taylor, 1991; Fonseca, 2004; Taylor and Zappi, 2004).
Melocactus spp. are collected and sold by local communities because of their ornamental value. Since this occurs indiscriminately, those species with a more restricted distribution, occupying specific areas and having a small number of individuals in the populations, are at risk of becoming extinct as a result of a single collection (Taylor and Zappi, 2004).
Melocacatus paucispinus is endemic to Bahia, and due to its rarity and desirability to collectors, the species has been listed on Appendix I of CITES. It has been listed as Endangered in the IUCN Red List of Threatened Species (IUCN, 2004) due to its erratic distribution and generally small population sizes. In the revision of the genus by Taylor (1991) only five populations were known for M. paucispinus, two of these possessing less than 50 individuals. The species was known to occur in the municipalities of Seabra, Rio de Contas and Abaíra (adjoining to Rio de Contas), with an area of occurrence of 6585 km2, but with an area occupied smaller than 500 km2. More recently the species has been found further north than the previously known populations, in the municipality of Morro do Chapéu (Machado, 1999; Taylor and Zappi, 2004), where it occurs as several small populations, and again further north in Delfino, in the municipality of Umburanas (Machado and Charles, 2004). Melocactus concinnus occurs in the states of Bahia and Minas Gerais, and is widespread geographically.
The distribution of both species is fragmented, with isolated and often widely disjunct populations. Both are elements of the campo rupestre vegetation of the Cadeia do Espinhaço mountain range (Taylor, 1991). Campo rupestre is a vegetation type that occurs in mountainous areas of the north-eastern and south-eastern regions of Brazil, mainly in Bahia and Minas Gerais. It is characterized by open, herbaceous vegetation on sandy, stony soils mixed with herbs and shrubs growing on outcropping islands of quartzite, sandstone, gneiss or ‘canga’ rocks (Giulietti and Pirani, 1988; Borba and Semir, 1998). Because of the discontinuity of these mountain ranges and outcrops, many species, especially the rupicolous ones, are distributed in disjunct populations. It has been suggested that this characteristic is responsible for the differentiation of plant populations, leading to the great diversity and high levels of endemism in the campo rupestre vegetation, one of the highest among the vegetation types of Brazil (Joly, 1970; Giulietti and Pirani, 1988; Harley, 1988; Borba et al., 2001; Jesus et al., 2001).
Melocactus paucispinus and M. concinnus (Fig. 1) may be distinguished by their habit, depressed in M. paucispinus and globose in M. concinnus, by the presence of both glaucousness in the epidermis and a central spine in M. concinnus (never present in M. paucispinus), and by a small depression between the areoles on the ribs, also present only in M. concinnus. These two species are sympatric and inter-fertile (M. A. S. Colaço et al., unpubl. data). Morphological evidence for hybridization between them is present in some of the populations (Fig. 1), already reported by Taylor (1991) and Taylor and Zappi (2004). According to Taylor (1991), M. concinnus is supposed to be the most promiscuous Melocactus sp., frequently hybridizing when sympatric with other species, and usually generates hybrid swarms, e.g. with M. glaucescens, M. zehntneri and M. oreas. However, such statements are conjectural, arising from observation of morphologically intermediate individuals in the field.
Fig. 1.
(A) Melocactus paucispinus, an individual from Seabra population. (B) M. concinnus, an individual from Morro do Chapéu population. (C–D) M. concinnus, individuals from CM03 population in Morro do Chapéu (possible introgressants or hybrids with M. paucispinus).
Hybridization is frequent in plants, with approx. 70 000 distinct interspecific hybrids having been estimated in nature (Stace, 1984), and it has been considered one of the main problems in conservation (Ellstrand, 1992). According to Barton (2001), in the wide scale, hybridization has been very important for evolution, being suggested as one of the processes that contributes to genetic recombination, increasing the levels of variability existing in nature (Stebbins, 1959; Rieseberg, 1995) and consequently acting as an important resource for adaptation and speciation (Lewontin and Birch, 1966; Arnold, 1996). Nevertheless, hybridization can impoverish biodiversity by allowing the fusion of two different species via interspecific gene flow, which Rhymer and Simberloff (1996) considered to be maximized if the species are sympatric and congeneric. Hybridization may promote the extinction of species by inhibiting the population growth and negatively affecting effective reproduction, competitive status and ecological interactions (Levin et al., 1996), which are particularly important in rare species (Rhymer and Simberloff, 1996). In some cases, hybridization is associated with introgression, which may lead to the complete disappearance of populations through formation of hybrid swarms, the parental species being substituted by hybrids (Forbes and Allendorf, 1991). Taylor and Zappi (2004) reported the occurrence of a hybrid swarm between two Melocactus spp. (M. glaucescens and M. concinnus), in which there were no pure individuals of M. glaucescens.
The objective of this study was to assess levels of genetic and morphological variation and sub-structuring within and between populations of M. paucispinus, and to verify the occurrence of natural hybridization with M. concinnus, using allozyme markers and morphometric data. This study is part of a project for the conservation and management of species of Cactacteae and other endangered plant groups in the Chapada Diamantina, Bahia, involving studies of demography, biology, variability, propagation and ethnobotany of the plants; the results obtained here will help in the determination of actions and priority areas for conservation of the species.
MATERIALS AND METHODS
Populations sampled
Samples were taken from ten populations of Melocactus paucispinus (256 individuals) and from three populations of M. concinnus (64 individuals) (Table 1; Fig. 2). Geographic distances between the Morro do Chapéu populations range from 5·5 to 32 km, and those between the Rio de Contas populations range from 5 to 10 km. The complete matrix of geographical distances can be obtained from the first author on request. All individuals sampled were mature, as evidenced by the presence of a well-developed cephalium. Vouchers for each species are deposited at the herbarium of the Universidade Estadual de Feira de Santana (HUEFS) (M. paucispinus–S. M. Lambert et al. 01, S. M. Lambert et al. 03; M. concinnus–S. M. Lambert et al. 02).
Table 1.
Populations of Melocactus paucispinus and M. concinnus occurring in the Chapada Diamantina, Bahia, Brazil, used in this study
| Species | Name | N* | Municipality | Location | |
|---|---|---|---|---|---|
| M. paucispinus | PM01 | 28 | Morro do Chapéu | Morrão | 11°33′51·4″S, 41°10′38·1″W |
| PM02 | 22 | Morro do Chapéu | Tabuleiro dos Tigres | 11°36′02·6″S, 41°09′53·9″W | |
| PM03 | 26 | Morro do Chapéu | Fazenda 2 Irmãs | 11°33′35·2″S, 41°17′50·3″W | |
| PM04 | 24 | Morro do Chapéu | Margens do rio Jacuipe | 11°33′34·1″S, 41°07′35·7″W | |
| PM05 | 26 | Morro do Chapéu | Cachoeira do Ferro Doido | 11°36′56·7″S, 41°00′20·7″W | |
| PD01 | 25 | Umburanas | Delfino | 10°21′58·7″S, 41°11′54·4″W | |
| PR01 | 29 | Rio de Contas | Campo dos Gerais | 13°29′23·2″S, 41°57′18·7″W | |
| PR02 | 21 | Rio de Contas | Campo do Alto da Cruz | 13°25′28·7″S, 41°54′40·4″W | |
| PR03 | 25 | Rio de Contas | Brumadinho | 13°31′00·3″S, 41°54′30·4″W | |
| PS01 | 30 | Seabra | Palmeira dos Mendes | 12°25′40·4″S, 41°59′45·3″W | |
| M. concinnus | CM01 | 24 | Morro do Chapéu | Orchidário | – |
| CM02 | 20 | Morro do Chapéu | Barrigudas | 11°30′23·0″S, 41°18′17·5″W | |
| CM03 | 20 | Morro do Chapéu | Morrão | 11°34′10·9″S, 41°10′36·6″W |
Vouchers are deposited in the herbarium of the Universidade Estadual de Feira de Santana (HUEFS).
N = number of individuals sampled.
Fig. 2.
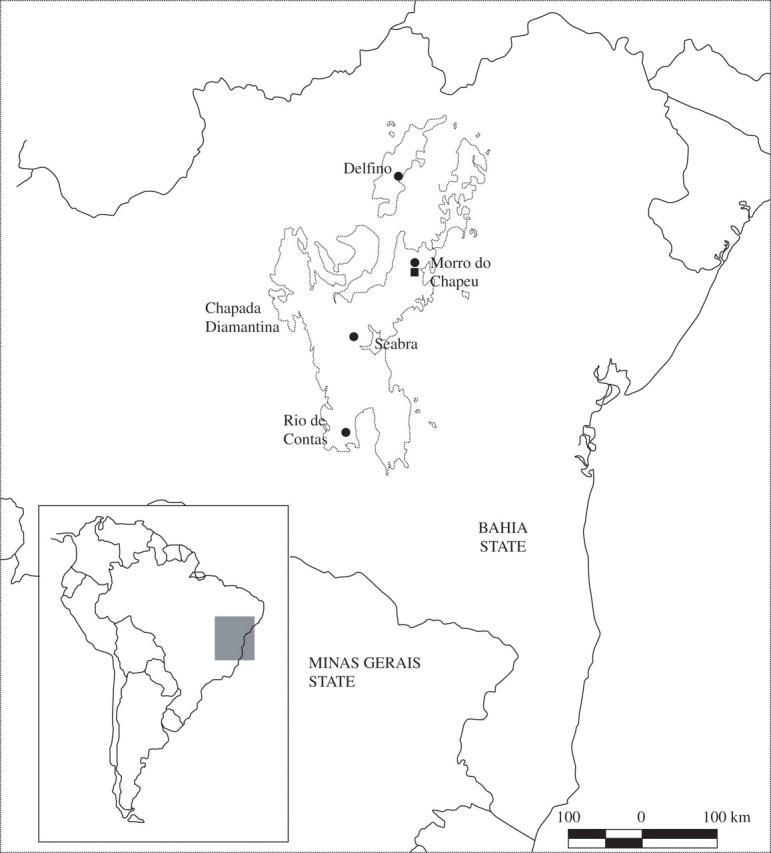
Map of the state of Bahia, north-eastern Brazil, showing the localities of the populations of Melocactus paucispinus (circles) and M. concinnus (squares).
Electrophoretic procedures
Small sections of stem tissue were crushed in 0·5 mL of grinding buffer [100 mL Tris–HCl 0·1 mol L−1 pH 7·0, 6·846 g sucrose, 0·6 g PVP (polyvinylpyrrolidone), 0·0292 g EDTA (ethylenediaminetetraacetic acid), 0·145 g BSA (bovine albumin), 0·13 g DIECA (sodium diethylcarbamate), 0·6 g borax, and 100 μL β-mercaptoethanol; modified from Sun and Ganders, 1990]. Extracts were absorbed in 1·0 × 0·3 cm Whatman number 3 paper wicks, which were loaded into 8·5 % starch gels (Sigma hydrolyzed potato starch).
For the electrodes and gels, four buffer systems were used: (1) electrode: histidine 0·065 mol L−1 adjusted to pH 6·5 with citric acid; gel: electrode buffer diluted 1 : 4; modified from Stuber et al. (1977); (2) electrode: lithium hydroxide 0·05 mol L−1, boric acid 0·0935 mol L−1, EDTA 0·0059 mol L−1, pH 8·0; gel: electrode solution diluted 1 : 10; modified from Ridgway et al. (1970); (3) electrode: citric acid 0·04 m adjusted to pH 6·1 with N-(3-aminopropyl)-morpholine; gel: electrode solution diluted 1 : 20; modified from Clayton and Tretiak (1972); and (4) electrode: boric acid 0·3 mol L−1, NaOH 0·06 mol L−1, pH 8·0; gel: Tris 0·01 mol L−1, pH 8·5; modified from Shaw and Prasad (1970).
Standard horizontal electrophoresis was performed until the inner marker (bromophenol blue) reached 9 cm from the application site using the following running conditions: system 1: 150 V; systems 2, 3 and 4: 25 mA. Nine enzymatic systems gave enough resolution for reading and were used. System 1 was used for malate dehydrogenase locus 1 (MDH; EC 1.1.1.37); system 2 was used for MDH locus 2, phosphoglucomutase (PGM; EC 2.7.5.1), isocitrate dehydrogenase (IDH; EC 1.1.1.42), and shikimate dehydrogenase (SKDH; EC: 1.1.1.25); system 3 was used for diaphorase (DIA; EC 1.8.1.4); and system 4 was used for acid phosphatase (ACPH; EC 3.1.3.2), leucine aminopeptidase (LAP; EC 3.4.11.1), glucose-6-phosphate dehydrogenase (G6PDH; EC 1.1.1.49) and phosphoglucose isomerase (PGI; EC. 5.3.1.9). The staining procedures were similar to but slightly adjusted from Brune et al. (1998; ACPH, LAP, DIA, SKDH, G6PDH), Corrias et al. (1991; IDH, PGI) and Soltis et al. (1983; PGM, MDH). Modifications were mainly in the amounts of the components used; the exact recipes can be obtained on request. Enzymatic systems showing more than one locus were numbered in ascending order from the locus with lowest mobility. The alleles were numbered according their mobility relative to the allele with the highest mobility of a standard individual present in all gels and designated as 100.
Analysis of allozyme data
The allele frequencies were determined by manually counting the banding patterns of the homozygotes and heterozygotes stained in the gels. Genetic variability for every population was estimated by the following parameters: proportion of polymorphic loci (P; 0·95 criterion), mean number of alleles per locus (A), observed (Ho) and expected (He) mean heterozygosity per locus. Deviations from the expected mean heterozygosity under Hardy–Weinberg (HW) equilibrium were tested using χ2 with a correction for small samples according to Levene (1949). A test for linkage disequilibrium was performed using GENEPOP software (Raymond and Rousset, 1995). Default program settings were used: 100 batches of 1000 iterations per batch with 1000 dememorization steps. As multiple tests enhance type 1 errors, the sequential Bonferroni procedure (Rice, 1989) was applied to each population.
Partitioning of genetic diversity among conspecific populations was estimated by F statistics (FIS, the inbreeding coefficient, measures the reduction in heterozygosity due to non-random mating within a population; FST measures the differentiation among populations; Wright, 1978). Two large regions (Morro do Chapéu and Rio de Contas) were sampled for M. paucispinus, which gave an opportunity to test if this species exhibits hierarchical partitioning of genetic variability and if it conforms to the expectations of the ‘stepping stone’ model, where only adjacent populations exchange genes (Kimura and Weiss, 1964). A hierarchical analysis of F was carried out within and among major sampling regions (Morro do Chapéu and Rio de Contas): FSR, variation among populations within regions; FRT, total variation among regions; FST, total variation among the eight populations. Values of fixation indexes and their respective components of variance (σ2) were calculated following Wright (1978).
Gene flow (Nm) among populations was estimated indirectly from the population genetic structure using Wright's (1951) equation as modified by Crow and Aoki (1984): FST = 1/(1 + 4Nmα), where α = [n/(n – 1)]2, where n is the number of populations analysed for each species. This formula establishes two properties of FST for neutral alleles: it is nearly independent of both mutation rate and the number of the demes (Slatkin and Barton, 1989). A second indirect estimate of gene flow was made based on the mean frequency of alleles found exclusively in single populations (‘private alleles’, Barton and Slatkin, 1986), with correction for sample size. The basic rationale underlying the latter method is that private alleles are likely to show a high frequency only when Nm is low. Nm(W) refers to Wright's gene flow estimate, and Nm(S) to Slatkin's estimate.
Matrices of genetic distances (Nei's unbiased genetic distance; 1978) and genetic identities (Nei's unbiased genetic identity; 1978) were calculated for populations and species. Cluster analysis was performed with the genetic distance matrix of the populations with UPGMA as grouping algorithm (Sneath and Sokal, 1973). All analyses were made using the BIOSYS 1.0 software package (Swofford and Selander, 1989), except for the cluster analysis, which was performed in the software package STATISTICA for Windows, Release 5.5 A (StatSoft, 2000), and the linkage disequilibrium and Nm(S) analyses, both performed with GENEPOP software (Raymond and Rousset, 1995).
Morphometric analysis
The individuals sampled for the allozyme analysis were also used in an analysis of morphological variability, in which 17 vegetative morphological characters were used (Table 2). The characters were chosen based on both previous fieldwork and the literature regarding the taxonomy of the group and morphometrics of cacti species (Backer and Pinkava, 1987; Chamberland, 1997; Casas et al., 1999; Backer and Johnson, 2000; Thomson, 2002). All measurements of continuous quantitative characters were taken with the aid of a vernier caliper. The values for the spine characters represent an average of the measurements of four areoles located on different ribs, the measurements always being made on the fourth areole along the rib from the apex of the plant.
Table 2.
Morphological characters used in the morphometric analysis of ten populations of M. paucispinus and three populations of M. concinnus occurring in the Chapada Diamantina, Brazil
| Characters | M. paucispinus | M. concinnus |
|---|---|---|
| Stems | ||
| 01. Length (mm) | 69·21 ± 14·27 (21·00–133·10) | 84·07 ± 14·65 (34·10–107·30) |
| 02. Width (mm) | 155·41 ± 20·15 (16·80–201·10) | 129·96 ± 9·90 (84·10–146·90) |
| Ribs | ||
| 03. Number | 9·64 ± 0·77 (8·0–15·0) | 8·52 ± 0·71 (8·0–11·0) |
| 04. Height (mm) | 15·36 ± 8·44 (0·40–39·30) | 24·70 ± 8·56 (9·60–42·30) |
| 05. Width at 1/4 (mm)* | 43·46 ± 5·79 (27·60–61·40) | 46·40 ± 5·34 (30·0–56·0) |
| 06. Width at 2/4 (mm) | 37·16 ± 4·94 (22·10–50·40) | 37·09 ± 5·58 (19·30–50·30) |
| 07. Width at 3/4 (mm) | 29·99 ± 4·85 (5·0–46·20) | 26·10 ± 4·75 (14·80–35·90) |
| 08. Number of areoles | 5.33 ± 0·75 (3·0–9·0) | 6·66 ± 0·74 (5·0–8·0) |
| Spines (mean of four areoles of different ribs) | ||
| 09. Lower number of radials | 4·21 ± 0·90 (3·0–6·0) | 5·98 ± 0·99 (5·0–7·0) |
| 10. Higher number of radials | 4·52 ± 0·75 (3·0–6·0) | 6·42 ± 0·91 (5·0–7·0) |
| 11. Length of right radial (mm) | 20·26 ± 3·30 (11·36–28·65) | 20·60 ± 2·40 (16·88–26·98) |
| 12. Length of lower radial (mm) | 22·36 ± 3·61 (12·45–33·28) | 22·91 ± 3·38 (15·65–30·30) |
| 13. Length of left radial (mm) | 20·52 ± 4·70 (12·78–60·60) | 21·04 ± 2·45 (16·83–28·20) |
| 14. Diameter of the lower radial (mm) | 1·77 ± 0·99 (1·0–17·0) | 1·43 ± 1·39 (1·0–12·20) |
| 15. Angle between right and left radials | 166·32 ± 14·28 (111·0–182·0) | 160·41 ± 15·73 (130·0–188·0) |
| Cephalium | ||
| 16. Length (mm) | 32·18 ± 15·00 (4·90–78·20) | 33·97 ± 16·40 (8·70–68·10) |
| 17. Width (mm) | 70·75 ± 10·41 (18·60–97·30) | 70·87 ± 16·06 (29·40–102·50) |
Values shown are means ± s.d., with minimum and maximum recorded values in brackets.
Width of ribs at one-, two- and three-quarters along their length.
Patterns of morphological similarity/difference were analysed by multivariate statistical methods using the software package STATISTICA for Windows, Release 5.5A (StatSoft, 2000). The analyses included canonical variate analysis (CVA) and cluster analysis for the calculation of variability parameters and morphological structuring. A basic data matrix was constructed with the morphological characters considered as variables. CVA was performed with the population as the categorical variable (individuals were grouped according to the population to which they belonged). The standardized coefficients for canonical variables resulting from CVA were used to identify the characteristics that contribute most significantly to the resulting patterns observed. The morphological matrix was analysed using discriminant analysis with population as the grouping variable in order to obtain a matrix of squared Mahalanobis distances of individuals to the centroid of the group (D2); the morphological variability of populations was calculated as the median of these distances (D2m) (Goldman et al., 2004). We used the median of the squared Mahalanobis distances instead of an average of these distances because of the non-normal distribution of the data. The non-parametric test of Kruskal–Wallis was applied to verify the occurrence of significant differences between medians of conspecific populations. Cluster analysis was carried out on a matrix of morphological distance among populations calculated using Mahalanobis Generalised Distance as the distance coefficient, and UPGMA was used as the clustering algorithm (Sneath and Sokal, 1973).
A multi-response permutation procedure (MRPP) analysis made with the PC-ORD 4.10 program (McCune and Mefford, 1999) was used to calculate the chance-corrected within-group agreement (A) among populations of every species, and the A-values were compared with the indexes of genetic differentiation among populations (FST) (Borba et al., 2002). The average Euclidian distance (ED) between the individuals of each population resulting from the MRPP analysis was also utilized as a measure of variability within populations (Borba et al., 2002). The two indices of morphological variability are essentially different, as D2m is more affected by form and ED is more affected by size of the characters.
Correlation analyses
For M. paucispinus, the matrix of squared Mahalanobis distances between populations was compared with the matrix of genetic distances (Nei, 1978), and both were also compared with the matrix of geographical distances between populations, using Mantel tests with the randomization (Monte Carlo) method (1000 randomizations) in the PC-ORD 4.10 program (McCune and Mefford, 1999), in order to test for significant correlations between morphological, genetic and geographic distances. This procedure was not used for M. concinnus due to the low number of populations sampled for this species. The pair-wise geographical distances between the populations were computed with geodetic distances on WGS84 earth ellipsoid calculated using the INVERSE 2.0 program (National Geodetic Survey, 2002). A Spearman rank correlation analysis between the morphological (ED and D2m) and genetic (He) variability of populations was also carried out using the software package STATISTICA for Windows, Release 5.5A (StatSoft, 2000).
RESULTS
Intra-populational variability
Using nine enzymatic systems, 12 loci were obtained with good resolution and were used in this study. One locus was monomorphic for all populations studied (IDH). The remaining loci displayed a low degree of polymorphism, with 66·6 % of the loci only having two alleles per locus. SKDH was the most polymorphic locus, with four alleles (Table 3).
Table 3.
Allele frequencies at 12 allozymic loci in ten populations of M. paucispinus and three populations of M. concinnus occurring in the Chapada Diamantina, Brazil
|
Melocactus paucispinus |
Melocactus concinnus |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Allele* | PM01 | PM02 | PM03 | PM04 | PM05 | PD01 | PR01 | PR02 | PR03 | PS01 | CM01 | CM02 | CM03 |
| PGM-1 | 90 | – | – | – | – | – | – | – | – | – | 0·231 | – | – | – |
| 100 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0·769 | 1 | 1 | 1 | |
| N | 24 | 18 | 18 | 21 | 26 | 24 | 23 | 15 | 24 | 26 | 22 | 20 | 20 | |
| PGM-2 | 88 | – | – | – | – | – | – | – | – | – | 0·043 | – | – | – |
| 100 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0·957 | 1 | 1 | 1 | |
| N | 24 | 20 | 16 | 21 | 26 | 24 | 23 | 15 | 24 | 23 | 21 | 19 | 20 | |
| PGI | 100 | 1 | 1 | 0·886 | 0·833 | 1 | 1 | 1 | 1 | 0·833 | 1 | 1 | 1 | 1 |
| 113 | – | – | 0·114 | 0·167 | – | – | – | – | 0·167 | – | – | – | – | |
| N | 18 | 23 | 22 | 24 | 27 | 24 | 24 | 19 | 24 | 22 | 21 | 20 | 20 | |
| LAP | 100 | 0·931 | 1 | 1 | 1 | 1 | 1 | 1 | 0·960 | 1 | 1 | 1 | 1 | 1 |
| 107 | 0·069 | – | – | – | – | – | – | 0·050 | – | – | – | – | – | |
| N | 29 | 17 | 28 | 23 | 26 | 22 | 19 | 20 | 24 | 26 | 22 | 18 | 14 | |
| SKDH | 85 | – | – | – | – | – | – | – | – | – | – | 0·063 | 0·063 | – |
| 100 | 0·977 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0·727 | 0·938 | 0·938 | 1 | |
| 109 | – | – | – | – | – | – | – | – | – | 0·159 | – | – | – | |
| 120 | 0·023 | – | – | – | – | – | – | – | – | 0·114 | – | – | – | |
| N | 22 | 10 | 18 | 23 | 17 | 23 | 19 | 12 | 22 | 22 | 16 | 16 | 13 | |
| IDH | 100 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| N | 25 | 23 | 27 | 23 | 27 | 14 | 19 | 20 | 22 | 27 | 14 | 19 | 14 | |
| G6PD | 100 | 0·895 | 0·656 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 118 | 0·105 | 0·344 | – | – | – | – | – | – | – | – | – | – | – | |
| N | 19 | 16 | 27 | 23 | 19 | 21 | 21 | 21 | 22 | 27 | 20 | 19 | 20 | |
| MDH-1 | 100 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0·971 | 1 |
| 119 | – | – | – | – | – | – | – | – | – | – | – | 0·029 | – | |
| N | 24 | 17 | 26 | 21 | 18 | 18 | 17 | 15 | 21 | 23 | 18 | 17 | 15 | |
| MDH-2 | 80 | – | – | – | – | – | – | – | – | – | – | 0·026 | 0·028 | – |
| 100 | 1 | 1 | 1 | 1 | 1 | 0·955 | 1 | 1 | 1 | 1 | 0·974 | 0·972 | 1 | |
| 111 | – | – | – | – | – | 0·045 | – | – | – | – | – | – | – | |
| N | 28 | 18 | 21 | 23 | 26 | 22 | 24 | 13 | 24 | 18 | 19 | 18 | 19 | |
| ACPH | 83 | 0·036 | – | 0·037 | – | – | – | – | – | – | – | 0·026 | – | – |
| 100 | 0·964 | 1 | 0·963 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0·974 | 1 | 1 | |
| N | 28 | 22 | 27 | 24 | 27 | 22 | 23 | 19 | 24 | 21 | 19 | 17 | 17 | |
| DIA–1 | 80 | – | – | – | – | – | – | – | – | – | 0·103 | – | – | – |
| 96 | – | – | – | – | – | – | – | – | – | 0·069 | 0·667 | 0·714 | – | |
| 100 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0·828 | 0·333 | 0·286 | 1 | |
| N | 24 | 18 | 28 | 23 | 14 | 2 | 20 | 13 | 22 | 29 | 6 | 7 | 6 | |
| DIA-2 | 94 | – | – | – | – | – | 0·857 | – | – | – | 0·167 | 0·5 | 0·714 | – |
| 100 | 1 | 1 | 1 | 1 | 1 | 0·143 | 1 | 1 | 1 | 0·833 | 0·5 | 0·286 | 1 | |
| N | 28 | 17 | 28 | 21 | 25 | 7 | 22 | 15 | 23 | 30 | 8 | 7 | 15 | |
See Table 1 for the names of the populations.
N = sample size.
Some alleles were exclusive to a one species: PGM-1 90, PGM-2 88, PGI 113, LAP 107, SKDH 109, SKDH 120, G6PD 118, MDH-2 111 and DIA-1 80 to M. paucispinus; SKDH 85, MDH-I 119, and MDH-2 80 to M. concinnus. A few of these alleles were exclusive to single populations: PGM-1 90, PGM-2 88, SKDH 109, SKDH 120 and DIA-1 80 (PS01), MDH-2 111 (PD01) and MDH-1 119 (CM02). However, neither of the species was fixed for alternative alleles at any locus, and thus no locus was diagnostic for either species (Table 3).
The percentage of polymorphic loci (P; 0·95 criterion) ranged from 0·0–33·3 %, the mean number of alleles per locus was between 1·0 and 1·6, and mean heterozygosity (He) ranged from 0·0 to 0·123 (Table 4). The populations PM05, PR01 and CM03 did not have any polymorphic loci. The populations PS01 and CM01 possessed the greatest genetic variability.
Table 4.
Genetic variability at 12 allozymic loci and morphological variability (D2m and ED) based on the morphometric analysis of 17 morphological characters in ten populations of M. paucispinus and three populations of M. concinnus occurring in the Chapada Diamantina, Brazil
| Population | N | A | P | Ho | He | D2m* | ED |
|---|---|---|---|---|---|---|---|
| M. paucispinus | |||||||
| PM01 | 24·4 (1·0) | 1·3 (0·1) | 16·7 | 0·004 (0·004) | 0·037 (0·019) | 12·21ac | 3·99 |
| PM02 | 18·3 (1·0) | 1·1 (0·1) | 8·3 | 0·016 (0·016) | 0·039 (0·039) | 12·94ac | 4·07 |
| PM03 | 23·8 (1·3) | 1·2 (0·1) | 8·3 | 0·011 (0·011) | 0·023 (0·018) | 8·54ac | 3·65 |
| PM04 | 22·5 (0·3) | 1·1 (0·1) | 8·3 | 0·014 (0·014) | 0·024 (0·024) | 9·15bc | 3·90 |
| PM05 | 23·2 (1·4) | 1·0 (0·0) | 0·0 | 0·000 (0·000) | 0·000 (0·000) | 8·62bc | 4·69 |
| PD01 | 18·6 (2·1) | 1·2 (0·1) | 8·3 | 0·000 (0·000) | 0·029 (0·023) | 10·33bc | 3·73 |
| PR01 | 21·2 (0·7) | 1·0 (0·0) | 0·0 | 0·000 (0·000) | 0·000 (0·000) | 10·00bc | 3·54 |
| PR02 | 16·4 (0·9) | 1·1 (0·1) | 8·3 | 0·000 (0·000) | 0·008 (0·008) | 10·59ac | 4·64 |
| PR03 | 23·0 (0·3) | 1·1 (0·1) | 8·3 | 0·014 (0·014) | 0·024 (0·024) | 11·89ac | 3·91 |
| PS01 | 24·5 (1·0) | 1·6 (0·2) | 33·3 | 0·022 (0·016) | 0·123 (0·050) | 17·11a | 4·78 |
| Mean | 21·59 | 1·17 | 9·98 | 0·008 | 0·031 | 11·14 | 4·09 |
| M. concinnus | |||||||
| CM01 | 17·2 (1·5) | 1·4 (0·1) | 25·0 | 0·009 (0·006) | 0·104 (0·056) | 13·10ac | 3·78 |
| CM02 | 16·4 (1·3) | 1·4 (0·1) | 25·0 | 0·010 (0·006) | 0·093 (0·048) | 14·54ac | 4·47 |
| CM03 | 16·1 (1·2) | 1·0 (0·0) | 0·0 | 0·000 (0·000) | 0·000 (0·000) | 15·00ac | 4·05 |
| Mean | 16·56 | 1·27 | 16·67 | 0·006 | 0·066 | 14·21 | 4·10 |
See Table 1 for the names of the populations. A locus was considered polymorphic if the frequency of the most common allele did not exceed 0·95.
Different letters in conspecific populations indicate statistically different median values in the Kruskal–Wallis test.
N = mean sample size per locus; A = mean number of alleles per locus; P = percentage of polymorphic loci; Ho = observed and He = expected mean heterozygosity per locus (Nei, 1978; unbiased estimate); D2m = median of the Mahalanobis generalized distance of the individuals to the centroid of the population; ED = mean of the Euclidean distance between the individuals of the population. Numbers in parentheses are s.d.
Of the 13 populations, ten showed significant deviations from the expected values in HW equilibrium for at least one locus; six of these did not have any locus in equilibrium (PM02, PM04, PM06, PR02, PR03 and PS1). Of the 11 polymorphic loci, ten were not in HW equilibrium in at least one population (except MDH-1) and six were not in HW equilibrium in any population in which they were polymorphic (PGM-1, PGM-2, LAP, G6PD, DIA-1, DIA-2). The reason for disequilibrium was a deficit of heterozygotes in all loci except for SKDH, ACPH, MDH-1 and MDH-2 in one population each. The high positive values for FIS (Table 5) reflect the deficit of heterozygotes in the populations.
Table 5.
F statistics (Wright, 1978) and Nm(W) at 12 allozymic loci in ten populations of M. paucispinus and three populations of M. concinnus occurring in the Chapada Diamantina, Brazil
|
M. paucispinus |
M. concinnus |
|||||
|---|---|---|---|---|---|---|
| Locus | M. paucispinusFIS | M. concinnusFIS | FST | Nm(W) | FST | Nm(W) |
| PGM-1 | 0·783 | – | 0·213 | 0·75 | – | |
| PGM-2 | 1·000 | – | 0·039 | 4·99 | – | |
| PGI | 0·38 | – | 0·114 | 1·57 | – | |
| LAP | 1·000 | – | 0·050 | 3·85 | – | |
| SKDH | 0·524 | 1·000 | 0·174 | 0·96 | 0·022 | 4·94 |
| G6PD | 0·707 | – | 0·254 | 0·59 | – | |
| MDH-1 | – | –0·03 | – | 0·02 | 5·44 | |
| MDH-2 | 1·000 | –0·028 | 0·041 | 4·73 | 0·009 | 12·23 |
| ACPH | 1·000 | –0·027 | 0·029 | 6·78 | 0·018 | 6·06 |
| DIA-1 | 1·000 | 1·000 | 0·857 | 0·03 | 0·428 | 0·15 |
| DIA-2 | 1·000 | 1·000 | 0·716 | 0·08 | 0·372 | 0·19 |
| Mean | 0·732 | 0·901 | 0·504 | 0·199 | 0·349 | 0·207 |
| A-value | 0·2 | 0·17 | ||||
A-values of the MRPP analysis of 17 morphological characters in all populations analysed are also presented.
After Bonferroni correction, populations PM01 and PS01 presented significant associations between loci LAP/G6PD (P = 0·006; critical α = 0·008 for six tests) and DIA-1/DIA-2 (P = 0·00001; α = 0·005 for ten tests), respectively. None of the ten linkage disequilibrium tests in M. concinnus were significant after the correction (α = 0·005).
In both morphological analyses (discriminant analysis and MRPP), the population with the highest variability scores was PS01 of M. paucispinus from Seabra, followed by the populations CM01, CM02, CM03 of M. concinnus. The population with the lowest variability was PM03 (Table 4). Among the characters analysed, those that showed the highest variation for both species are the length and width of the stem (variables 1 and 2, respectively), height of the ribs (variable 4), diameter of the radial spine (variable 14) and length and width of the cephalium (variables 16 and 17, respectively). The least variable characters were the numbers of radial spines (variables 9 and 10).
Spearman rank correlation analysis between morphologic and genetic variability resulted in a statistically significant correlation between He and D2m (r = 0·581, P = 0·037), but not between He and ED (r = 0·191, P = 0·532). The population CM03 displayed one of the highest scores for morphological variability in both morphological analyses, but it did not display any genetic variability.
Structuring of the variability
Both species displayed high average values of FST (0·504 for M. paucispinus and 0·349 for M. concinnus), interpreted as a high level of genetic structuring (Table 5). By excluding the populations PD01 of M. paucispinus and CM03 of M. concinnus the average values of FST drop to 0·158 and 0·022, respectively, due to an inversion in the relative frequency of the alleles of DIA-2 (PD01) and DIA-1 (CM03) in these populations.
The hierarchical analysis showed a high FSR among populations (0·338, σ2 = 0·101), an FRT of 0·253 (σ2 = 0·067) and an FST of 0·140 (σ2 = –0·034), showing that there is more genetic structuring within regions than between them or among all populations.
The Nm values estimated from a mean frequency of private alleles of 0·117 in M. paucispinus was 0·571, and the Nm(W) was 0·199 (Table 5). The mean frequency of private alleles was lower in M. concinnus (0·027), resulting in a Nm(S) of 12·17, while Nm(W) was 0·207. These dissimilar values could be attributed to the low number of private alleles in this species, because the Nm(S) method requires that a reasonable number of private alleles be present (Slatkin and Barton, 1989), and only two were found in this species. There is a large variation concerning Nm(W) values among loci, especially in M. concinnus (Table 5).
High levels of morphological structuring were also found in both species (Table 5), with A values of 0·20 and 0·17 for M. paucispinus and M. concinnus, respectively, these values being correlated with the high values of genetic differentiation (FST) found in the species. When the populations PD01 and PS01 are removed from the MRPP analysis the A value still remains high (0·18).
Phenetic relationships
The genetic identities between conspecific populations ranged from 0·842 to 1·000 for M. paucispinus and from 0·914 to 1·000 for M. concinnus. PD01 has the lowest values of genetic identities among the conspecific populations of M. paucispinus (0·842–0·880). This population is genetically more similar to the populations CM01 and CM02 of M. concinnus (0·983 and 0·995, respectively), due to allele frequencies of the DIA-2 (Table 3). If PD01 is removed, the mean genetic identity among the populations of M. paucispinus increases to 0·994, ranging from 0·978 to 1·000. The population CM03 is genetically more similar to the populations of M. paucispinus (0·989–1·000)—except for the population PD01 (0·854)—than to its two conspecific populations (CM01, 0·943; CM02, 0·914), mainly due to the allele frequencies of DIA-1 and DIA-2 (Table 3). The genetic identity between the species ranged from 0·854 to 1·000.
The UPGMA dendrogram obtained from the cluster analysis of Nei (1978) unbiased genetic distances (Fig. 3) reveals the formation of two main groups: one composed of nine of the ten populations of M. paucispinus (except PD01) plus population CM03 of M. concinnus, and the other composed of population PD01 grouped with the remaining two populations of M. concinnus (CM01 and CM02), with a genetic distance between these groups of 0·10.
Fig. 3.
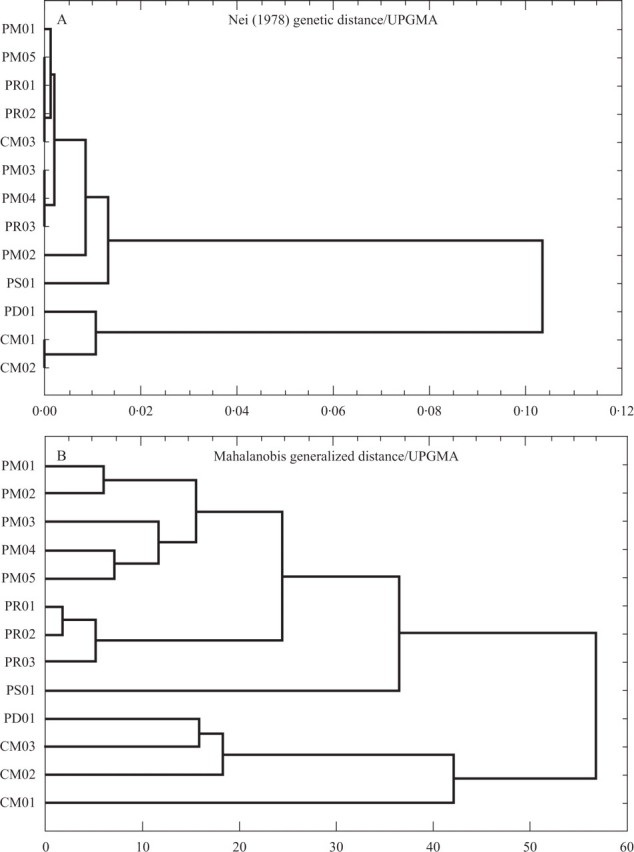
Dendrograms showing the phenetic relationships among ten populations of Melocactus paucispinus and three populations of M. concinnus occurring in the Chapada Diamantina, Brazil, constructed using the matrix of genetic distances (Nei, 1978, unbiased estimate; cophenetic correlation = 0·886) based on 12 allozymic loci (A), and using the matrix of Mahalanobis generalized distance based on 17 morphological characters (B) with UPGMA as the clustering algorithm. See Table 1 for the names of the populations.
Two groups of populations (PM05, PR01, PR02, CM03; and PM03, PM04, PR03) did not display any genetic differentiation. The population from Seabra (PS01) was the most distant within the group due to its higher levels of polymorphism, with four alleles exclusive for this population, some of them at high frequencies (PGM-1 90, SKDH 109, DIA-1 80).
The UPGMA dendrogram obtained from the cluster analysis of morphological distances resulted in the formation of two main groups (Fig. 3): one uniting the populations of M. paucispinus except for PD01 (Delfino), and the other containing the populations of M. concinnus plus PD01. Within the group of M. paucispinus the population PS01 (Seabra) displayed the greatest differentiation, and the remaining populations were divided into two subgroups, one grouping the populations from Morro do Chapéu and the other grouping the populations from Rio de Contas. Smaller genetic distances were found among the populations from Rio de Contas than among the populations from Morro do Chapéu.
Table 6 shows the classification matrix of the individuals analysed. The percentage of correct classifications ranged from 57 to 100 %. The incorrect classifications mostly occurred between conspecific populations, except for one individual each from PM04 and CM02, and three individuals from CM03. Within M. paucispinus the percentage of incorrect classifications was higher among populations from the same locality, with the exception of populations PM02, PR02 and PR03, each of which had one individual incorrectly classified as belonging to a population from a different locality. The populations of M. paucispinus with a higher degree of morphological differentiation (PS01 and PD01) displayed the highest values of correct classifications.
Table 6.
Matrix of classification of the individuals in the discriminant analysis of 17 morphological characters in ten populations of Melocactus paucispinus and three populations of M. concinnus occurring in the Chapada Diamantina, Brazil
|
Melocactus paucispinus |
Melocactus concinnus |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Percent correct | PM1 | PM2 | PM3 | PM4 | PM5 | PD1 | PR1 | PR2 | PR3 | PS1 | CM1 | CM2 | CM3 | |
| PM1 | 71·43 | 20 | 1 | – | 2 | 5 | – | – | – | – | – | – | – | – |
| PM2 | 77·27 | 3 | 17 | – | – | – | – | 1 | – | – | 1 | – | – | – |
| PM3 | 92·31 | – | – | 24 | 2 | – | – | – | – | – | – | – | – | – |
| PM4 | 70·83 | 1 | 1 | 2 | 17 | 2 | – | – | – | – | – | – | – | 1 |
| PM5 | 96·15 | – | – | – | 1 | 25 | – | – | – | – | – | – | – | – |
| PD1 | 100·00 | – | – | – | – | – | 25 | – | – | – | – | – | – | – |
| PR1 | 82·76 | – | – | – | – | – | – | 24 | 3 | 2 | – | – | – | – |
| PR2 | 57·14 | – | 1 | – | – | – | – | 7 | 12 | 1 | – | – | – | – |
| PR3 | 76·00 | – | 1 | – | – | – | – | 5 | – | 19 | – | – | – | – |
| PS1 | 93·33 | – | – | – | – | 2 | – | – | – | – | 28 | – | – | – |
| CM1 | 100·00 | – | – | – | – | – | – | – | – | – | – | 24 | – | – |
| CM2 | 95·00 | – | – | – | – | – | 1 | – | – | – | – | – | 19 | – |
| CM3 | 85·00 | – | – | – | – | – | 3 | – | – | – | – | – | – | 17 |
| Total | 84·69 | 24 | 21 | 26 | 22 | 34 | 29 | 37 | 15 | 22 | 29 | 24 | 19 | 18 |
| P = | 0·087 | 0·068 | 0·081 | 0·075 | 0·081 | 0·078 | 0·090 | 0·065 | 0·078 | 0·093 | 0·075 | 0·062 | 0·062 | |
See Table 1 for the names of the populations.
The scatterplots of the scores of individuals on the first two CVA canonical axes, and on the first and third CVA canonical axes are shown in Fig. 4. The first, second and third canonical axes explained 48·81 %, 27·67 % and 8·82 % of the morphological variation, respectively; in total 85·3 % of the observed variability. On the first canonical axis there is a separation between the populations of M. concinnus plus PD01 from the remaining populations of M. paucispinus, mainly due to the higher height of the ribs (variable 4), higher width at the base of ribs (variable 5) and higher number of areoles per rib (variable 8) in those populations. Within M. paucispinus there is separation of the population PS01 in the same axis due to the higher width of the individuals (variable 2) and the higher number of ribs (variable 3) in this population. Such separation also occurs in the third canonical axes, the greater length of the individuals (variable 1) being the most significant character for this separation. In the second canonical axes there is a slight separation between the populations of Rio de Contas and Morro do Chapéu in the form of a gradient; the individuals from Rio de Contas are mainly distributed in the uppermost region of the scatterplot, with some individuals overlapping with individuals from the Morro do Chapéu populations, which are mostly located in the lower region of the scatterplot.
Fig. 4.
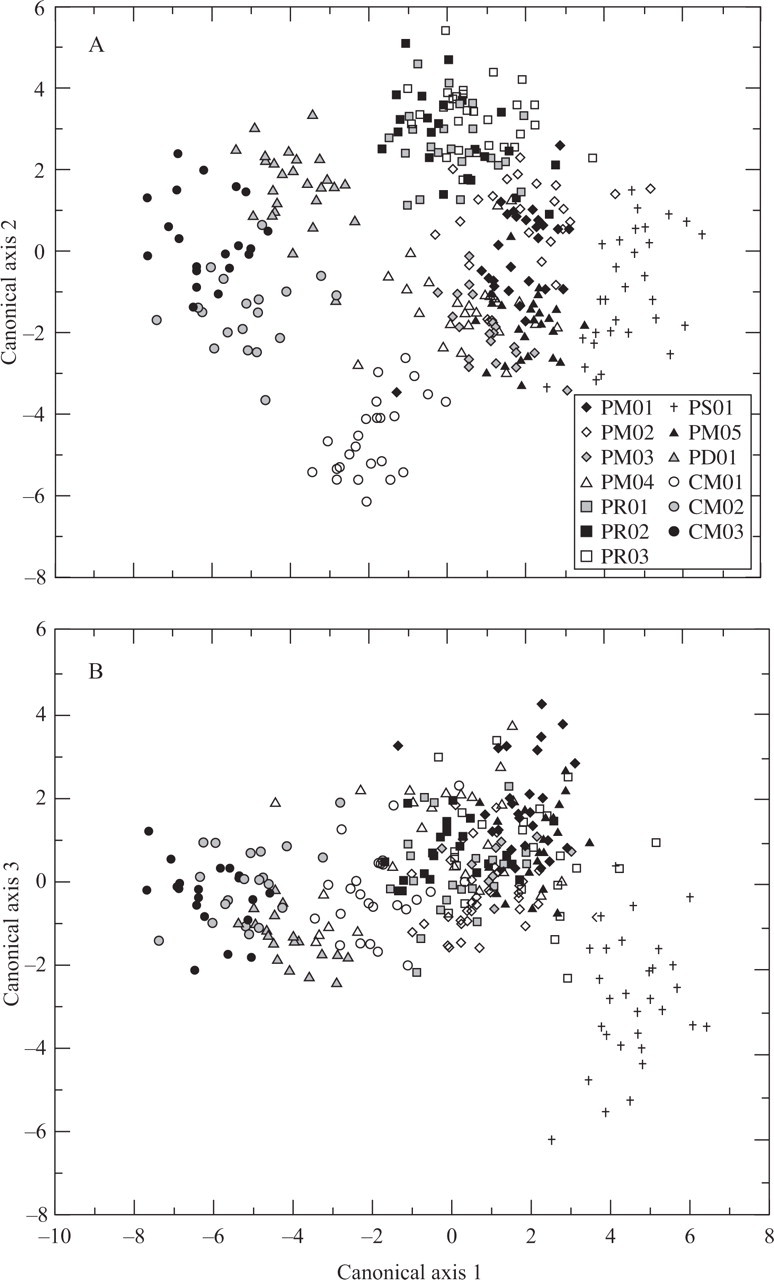
Representation of the scores on the three first canonical axes of the CVA using 17 morphological characters in ten populations of Melocactus paucispinus and three populations of M. concinnus occurring in the Chapada Diamantina, Brazil. (A) Canonical axes 1 and 2. (B) Canonical axes 1 and 3. See Table 1 for the names of the populations.
For M. paucispinus, Mantel tests did not produce statistically significant results for pair-wise correlations between genetic and morphological distances (r = 0·527, P = 0·064), nor between genetic and geographical distances (r = 0·356, P = 0·084) of conspecific populations; however, there was a significant correlation between morphological and geographical distances (r = 0·364, P = 0·022).
DISCUSSION
The Melocactus spp. studied here displayed levels of genetic variability lower than the average values reported for another Melocactus species, M. curvispinus (P = 89·5 %, A = 3·82, He = 0·145; Nassar et al., 2001) and for other cactus species (Parker and Hamrick, 1992; Hamrick et al., 2002; Nassar et al., 2002, 2003; Clark-Tapia and Molina-Freaner, 2003; Moraes et al., 2005) and lower than the average values reported for plant species with similar characteristics, namely woody, long-lived, animal-pollinated plants (Hamrick and Godt, 1992; Colunga-GarcíaMarín et al., 1999; Martínez-Palacios et al., 1999). The low genetic variability displayed by the species surveyed is in sharp contrast with the high levels found in species of the genus Discocactus (Cactaceae) that occur in the same general area, occupy similar habitats, and also have a globose habit (M. C. Machado et al., unpubl. res.). The low variability presented by these Melocactus species may be associated with recent bottleneck effects experienced by the populations, as they occur in disturbed areas, generally next to roads, in agricultural areas or in areas of sand extraction. Moreover, these populations are being exploited by collectors and traders, and this could be adversely affecting the genetic diversity. The same has been observed in populations of Discocactus bahiensis, a species that grows in similar sites and is subject to similar threats (M. C. Machado et al., unpubl. res.).
Studies of levels of intra- and inter-populational morphologic variability are absent in Melocactus and rare in Cactaceae (Cassas et al., 1999; Backer and Johnson, 2000; Schmalzel et al., 2004). The levels of morphological variability in these Melocactus spp. are lower than those observed by M. C. Machado et al. (unpubl. res.) in 17 populations of three Discocactus species (D. bahiensis: D2m = 11·53–17·71; D. catingicola: D2m = 20·39–32·88; D. zehntneri: D2m = 10·64–32·18).
In M. paucispinus, the indirect migration estimates gave values lower than one, suggesting restricted long-term gene flow among populations (Slatkin, 1987; Slatkin and Barton, 1989), possibly due to historical factors, indicating environmental fragmentation. Melocactus concinnus also showed a low value of Nm(W), providing further evidence for genetic drift having played a prominent role in these populations. The population presenting both the highest genetic and morphological diversity was the M. paucispinus population from Seabra (PS01), which possessed exclusive or private alleles, and presented the highest average values for the majority of the morphological characters. In general, allozyme differentiation among populations is a result of genetic drift or directional selection. The presence of four exclusive alleles in this population can be interpreted as a result of its geographic isolation: gene flow between the PS01 population and the remaining M. paucispinus populations is possibly non-existent, and genetic drift could be responsible for the differentiation observed. This kind of process can quickly lead to speciation in small, geographically isolated populations (Levin, 2000). The main issue raised by the distinctness of this population would be taxonomic, since the type specimen of the species comes from this population. However, it cannot be excluded that selection is also occurring, especially since genetic drift affects all loci in the same way, but natural selection does not. The balance of selection and gene flow could be different from the balance between drift and gene flow, where gene flow might be weaker than selection in some loci and stronger than genetic drift at other loci (Slatkin, 1987). This could explain the differences observed in Nm values among the different loci.
The smaller genetic distance between the population CM03 of M. concinnus and populations of M. paucispinus than to its remaining conspecific populations could be explained by the possibility of gene flow occurring between the species, since they are intercompatible (M. A. S. Colaço et al., unpubl. res.). The frequencies of alleles of the two DIA loci in this population strengthens this hypothesis. The population CM03 occurs sympatrically with M. paucispinus in Morro do Chapéu and individuals displaying intermediate characteristics between the two species have been observed in this population, as noted earlier by Taylor (1991) and Taylor and Zappi (2004). Morphologically this population shows characters intermediate between the two species, including the presence of depressed-globose individuals, with diameter of the spines being similar to that found in M. paucispinus individuals. However, they are usually glaucous, have more radial spines per areole than individuals of M. paucispinus and show re-entrance in the ribs: typical characteristics of M. concinnus individuals.
The population CM03 is monomorphic for an allele (DIA-1 100) that is a low-frequency allele in other M. concinnus populations and which is monomorphic in all populations of M. paucispinus (except PS01). This strengthens the hypothesis of gene flow between the two species (see Fig. 1). However, the grouping of CM03 with populations of M. paucispinus could also be merely incidental: CM03 is monomorphic for all loci sampled, as are the populations PM05, PR01 and PR02 of M. paucispinus with which CM03 is grouped. The lack of variation thus cannot be taken uncritically as an indication of close relationship. Since CM03 is monomorphic for all loci, another explanation is that the small size of the CM03 population coupled with genetic drift led to it becoming fixed for an allele that occurs in low frequency in the species as a whole, and that happens to be the most frequent allele in M. paucispinus. The above two explanations are not mutually exclusive, and the most plausible hypothesis is that hybrization and genetic drift have acted together.
The population PD01 groups with populations of M. concinnus in the morphological and genetic analyses. The characteristics that contribute most significantly to the grouping of PD01 with the populations of M. concinnus in the morphological analysis are the stem width (variable 2), rib height (variable 4), rib width at mid-region (variable 6) and number of areoles on the ribs (variable 8), all of them with higher values in PD01. In genetic terms, the grouping of PD01 with populations of M. concinnus is due to the higher frequency of the allele 94 at the locus DIA-2 (Table 3). These results could be a consequence of gene flow between M. paucispinus and M. concinnus at the location where PD01 occurs, since the latter species is widely distributed and possibly occurs in that region (Taylor, 1991; Taylor and Zappi, 2004). Thus, hybridization and/or introgression processes could be generating the genetic and morphological differentiation of the population PD01. Another explanation for grouping of PD01 with populations of M. concinnus is that this population in fact represents a M. concinnus morph that is superficially similar to M. paucispinus. The most important morphological characters in diagnosing it as a M. paucispinus population are the relative lack of glaucousness in the epidermis (plants of M. paucispinus are never glaucous, whereas plants of M. concinnus are glaucous, often intensely so), the depressed habit, and the lack of a central spine in the areoles (characters typical of M. paucispinus). However, other morphological characteristics strongly suggest that the plants from the PD01 population are more akin to M. concinnus. Thus, PD01 can be interpreted either as an M. paucispinus population that has undergone extensive introgression with M. concinnus or as an M. concinnus population that displays morphological convergence with M. paucispinus.
Hybridization and introgression may have significant influence in the conservation of species as they can promote the extinction of pure populations and consequently compromise the survival of rare species (Levin et al., 1996; Rhymer and Symberloff, 1996). Arnold (1997) argued that the greatest barrier to gene flow between some species of plants is the non-formation of F1 hybrids; if these are formed, generally introgression is verified. Thus, rare species could have numerical disadvantages due to the proliferation of these fertile hybrids, which promote a reduction in the proportional representation of pure individuals and inhibit the growth of populations of the rare species (Levin et al., 1996).
The high average values of FIS observed in both species (Table 5) are much higher than those found in M. curvispinus by Nassar et al. (2001). However, several works have demonstrated moderate to high levels of endogamy in Cactaceae, such as in Stenocereus griseus (FIS = 0·145; Nassar et al., 2003), Cereus repandus (FIS = 0·182; Nassar et al., 2003), Pilosocereus lanuginosus (FIS = 0·176; Nassar et al., 2003), Stenocereus gummosus (FIS = 0·608; Clark-Tapia and Molina-Freaner, 2003), Pereskia guamacho (FIS = 0·301; Nassar et al., 2002), and, in Brazil, Pilosocereus machrisii and P. euchlorus (FIS ranged from 0·025 to 0·569 and from −0·276 to 0·529, respectively; Moraes et al., 2005).
The high values of FIS observed indicate a strong heterozygote deficit, which could be the result of endogamy or sub-structuring of the populations. Local subdivision could also explain the significant associations among different loci observed in two populations of M. paucispinus. Hummingbirds are the main pollinators of Melocactus spp. (Taylor, 1991). The most frequent floral visitor to M. paucispinus is the hummingbird Chlorostilbon aureoventris (M. A. S. Colaço et al., unpubl. res.), a territorial species. The behaviour of this pollinator may contribute to genetic subdivision of the populations, since hummingbirds promote gene flow only among the individual plants within their feeding territories. Endogamy could also be a factor responsible for lack of heterozygotes in M. paucispinus because this species is self-compatible and autogamous (M. A. S. Colaço et al., unpubl. res.). In spite of being autogamous, studies of reproductive biology for the species indicate a low level of fruit production in spontaneous auto-pollination experiments, suggesting the possibility of inbreeding depression resulting from recent endogamy within the populations. Endogamy could also occur because of crosses between closely related individuals, since seed dispersal is extremely local, being mediated by lizards (Taylor, 1991; Fonseca, 2004). The limited dispersal ability of the lizards could contribute to an increase both in endogamy and genetic sub-structuring within the Melocactus populations (Nassar et al., 2001). Inbreeding depression is generally associated with a decrease of fitness of the individuals, affecting viability, fecundity, development and susceptibility to environmental stress, therefore increasing the probability of extinction of small populations (Hauser and Loescheke, 1995; Bijlsma et al., 2000). This is especially important for species threatened with extinction, in which the disappearance of one population may affect the survival the whole species.
The genetic identity values found between conspecific populations are similar to those reported for other plant species (Thorpe, 1982; Crawford, 1989; Borba et al., 2001; Jesus et al., 2001). The values of genetic differentiation found for both species are similar to those observed by Nassar et al. (2001) for different populations of M. curvispinus, and this may be conservative in the genus, due to similar pollination and seed dispersal mechanisms. In spite of the lack of correlation between the genetic variability and geographic distance, the latter may be a factor influencing the differentiation of population PD01. This population is geographically isolated and its differentiation is reflected in the high FST value (0·504) observed, indicating a geographic sub-structuring of the species. After removing this population from the analysis, the FST reduces dramatically (to 0·158), a value lower than that found in M. curvispinus (FST = 0·193; Nassar et al., 2001) and close to the values found in for other cacti (Lophocereus schottii: FST = 0·130, Parker and Hamrick, 1992; Pereskia guamacho: FST = 0·112, Nassar et al., 2002; Stenocereus gummosus: FST = 0·10, Clarkia-Trapia and Molina-Freaner, 2003). However, it should be also noticed that there is no evidence of a ‘stepping-stone’ pattern in M. paucispinus and that there is more structuring at a local scale (FSR > FRT > FST) than at a large scale. Limited gene flow among close populations combined with a low level of natural selection (Linhart and Grant, 1996) could explain these results.
Geographic sub-structuring of populations is one of the factors responsible for the high FST values reported for other plant species with disjunct populations occurring in the mountainous regions of the Espinhaço Range, such as Orchidaceae (Borba et al., 2001), Asteraceae (Jesus et al., 2001), Eriocaulaceae (A. C. S. Pereira et al., unpubl. res.) and other Cactaceae (M. C. Machado et al., unpubl. res.). A major contribution to the observed differentiation among populations is probably the restricted gene flow and local dispersion of seeds in these species of cactus, as in some bat-pollinated species (Nassar et al., 2002, 2003). However, as stated earlier, the differentiation of PD01 may be due to hybridization/introgression or it may be a M. concinnus morph. This is clearly the case in M. concinnus, in which the high FST value cannot be explained by geographic sub-structuring, but only by hybrization and/or introgression in CM03.
Besides the population PD01 from Delfino, which clusters with populations of M. concinnus, the remaining populations of M. paucispinus differ morphologically on a geographical basis, with the populations of each municipality grouping together, resulting in the high A-value for this species. However, the lack of correlation between geographical and morphological distances is probably a result of the higher differentiation displayed by the population PS01 from Seabra, mainly due to characters of the stem and ribs. The sets of populations from Morro do Chapéu and Rio de Contas are morphologically more similar to each other. However the population from Seabra is geographically located between Morro do Chapéu and Rio de Contas. As in the FST analysis, the high A-value for M. concinnus cannot be explained by geographic sub-structuring, but only by hybridization and/or introgression.
Acknowledgments
The authors thank Ana C. S. Pereira, Patrícia L. Ribeiro, Delmar L. Alvim, Francisco H. F. Nascimento and Rosineide B. S. Fonseca for helping on field trips, and Luis E. Eguiarte and an anonymous reviewer for suggestions. This work was supported by a grant from Fundo Nacional do Meio Ambiente to ELB (FNMA #75/2001). SML received a scholarship from the CNPq. ELB is supported by a grant (PQ2) from CNPq.
LITERATURE CITED
- Anderson EF. 2001. The cactus family. Portland, OR: Timber Press.
- Arnold ML. 1996. Natural hybridization and introgression. Princeton, NJ: Princeton University Press.
- Arnold ML. 1997. Natural hybridization and evolution. Oxford: Oxford University Press.
- Backer MA, Johnson RA. 2000. Morphometric analysis of Escobaria sneedii var. sneedii, E. sneedii var. leei, and E. guadalupensis (Cactaceae). Systematic Botany 25: 577–587. [Google Scholar]
- Backer MA, Pinkava DJ. 1987. A cytological and morphometric analysis of a triploid apomict, Opuntia × kelvinensis (subgenus Cylindropyntia, Cactaceae). Brittonia 39: 387–401. [Google Scholar]
- Barton NH. 2001. The role of hybridization in evolution. Molecular Ecology 10: 551–568. [DOI] [PubMed] [Google Scholar]
- Barton NH, Slatkin M. 1986. A quasi-equilibrium theory of the distribution of rare alleles in a subdivided population. Heredity 56: 409–415. [DOI] [PubMed] [Google Scholar]
- Bijlsma R, Bundgaard J, Boerema AC. 2000. Does inbreeding affect the extinction risk of small populations? Predictions from Drosophila. Journal of Evolutionary Biology 13: 502–514. [Google Scholar]
- Borba EL, Semir J. 1998. Wind-assisted fly pollination in three Bulbophyllum (Orchidaceae) species occurring in the Brazilian ‘campos rupestres’. Lindleyana 13: 203–218. [Google Scholar]
- Borba EL, Felix JM, Solferini VN, Semir J. 2001. Fly-pollinated Pleurothallis (Orchidaceae) species have high genetic variability: evidence from isozyme markers. American Journal of Botany 88: 419–428. [PubMed] [Google Scholar]
- Borba EL, Shepherd GJ, van den Berg C, Semir J. 2002. Floral and vegetative morphometrics of five Pleurothalis (Orchidaceae) species: correlation with taxonomy, phylogeny, genetic variability and pollination systems. Annals of Botany 90: 219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune W, Alfenas AC, Junghans TG. 1998. Identificações específicas de enzimas em géis. In: Alfenas AC, ed. Eletroforese de isoenzimas e proteinas afins: fundamentos e aplicações em plantas e microorganismos. Viçosa: Universidade Federal de Viçosa, 201–328.
- Casas A, Caballero J, Valiente-Banuet A, Soriano JA, Dávila P. 1999. Morphological variation and the process of domestication of Stenocereus stellatus (Cactaceae) in Central México. American Journal of Botany 86: 522–533. [PubMed] [Google Scholar]
- Chamberland M. 1997. Systematics of the Echinocactus polycephalus complex (Cactaceae). Systematic Botany 22: 303–313. [Google Scholar]
- Clark-Tapia R, Molina-Freaner F. 2003. The genetic structure of a columnar cactus with a disjunct distribution: Stenocereus gummosus in the Sonoran desert. Heredity 90: 443–450. [DOI] [PubMed] [Google Scholar]
- Clayton JW, Tretiak DN. 1972. Amine-citrate buffers for pH control in starch gel eletrophoresis. Journal of the Fisheries Research Board of Canada 29: 1169–1172. [Google Scholar]
- Colunga-GarcíaMarín P, Coello-Coello J, Eguiarte LE, Piñero D. 1999. Isozymatic variation and phylogenetic relationships (Agave fourcroydes) and its wild ancestor A. angustifolia (Agavaceae). American Journal of Botany 86: 115–123. [PubMed] [Google Scholar]
- Corrias B, Rossi W, Arduino P, Cianchi R, Bullini L. 1991. Orchis longicornu Poiret in Sardinina: genetic, morphological and chorological data. Webbia 45: 71–101. [Google Scholar]
- Crawford DJ. 1989. Enzyme eletrophoresis and plant systematics. In: Soltis DE, Soltis PS, eds. Isozymes in plant biology. Portland, OR: Dioscorides Press, 146–164.
- Crow JF, Aoki K. 1984. Group selection for a polygenic behavioral trait: estimating the degree of population subdivision. Proceedings of the National Academy of Sciences USA 81: 6073–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellstrand NC. 1992. Gene flow by pollen: implication for plant conservation genetics. Oikos 63: 77–86. [Google Scholar]
- Fonseca RBS. 2004. Fenologia reprodutiva e dispersão de Melocactus glaucescens Buining & Brederoo e M. paucispinus G. Heiman & R. Paul (Cactaceae) no município de Morro do Chapéu, Chapada Diamantina—Bahia—Brasil. Masters Thesis, Universidade Estadual de Feira de Santana, Brazil.
- Forbes SH, Allendorf FW. 1991. Mitochondrial genotypes have no detectable effects on meristic traits in cutthroat trout hybrid swarms. Evolution 45: 1350–1359. [DOI] [PubMed] [Google Scholar]
- Giulietti AM, Pirani JR. 1988. Patterns of geographic distribution of some plant species from the Espinhaço Range, Minas Gerais and Bahia, Brazil. In: Heyer WR, Vanzolini PE, eds. Proceedings of a workshop on Neotropical distribution patterns. Rio de Janeiro: Academia Brasileira de Ciências, 39–63.
- Goldman DH, van den Berg C, Griffith MP. 2004. Morphometric circumscription of species and infraspecific taxa in Calopogon R.Br. (Orchidaceae). Plant Systematics and Evolution 274: 37–60. [Google Scholar]
- Hamrick JL, Godt MJ. 1992. Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS, eds. Plant population genetics, breeding and genetic resources. Sunderland, MA: Sinauer, 43–63.
- Hamrick JL, Nason JD, Fleming TH, Nassar JM. 2002. Genetic diversity in columnar cacti. In: Fleming TH, Valiente-Banuet A, eds. Columnar cacti and their mutualisms: evolution, ecology and conservation. Tucson, AR: University of Arizona Press, 122–133.
- Harley RM. 1988. Evolution and distribution of Eriope (Labiatae) and its relatives, in Brazil. In: Heyer WR, Vanzolini PE, eds. Proceedings of a workshop on Neotropical distribution patterns. Rio de Janeiro: Academia Brasileira de Ciências, 71–120.
- Hauser TP, Loescheke V. 1995. Inbreeding depression in Lychnis flos-cuculi (Caryophyllaceae): effects of different levels of inbreeding. Journal of Evolutionary Biology 8: 589–600. [Google Scholar]
- IUCN. 2004. 2004 IUCN Red List of Threatened Species. www.redlist.org. 17 Apr. 2005.
- Jesus FF, Solferini VN, Semir J, Prado PI. 2001. Local genetic differentiation in Proteopsis argentea (Asteraceae), a perennial herb endemic in Brazil. Plant Systematics and Evolution 226: 59–68. [Google Scholar]
- Joly AB. 1970. Conheça a vegetação brasileira. São Paulo: EDUSP.
- Kimura M, Weiss GH. 1964. The stepping stone model of population structure and the decrease of genetic correlation with distance. Genetics 49: 561–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene H. 1949. On a matching problem arising in genetics. Annals of Mathematical Statistics 20: 91–94. [Google Scholar]
- Levin DA. 2000. The origin, expansion, and demise of plant species. New York: Oxford University Press.
- Levin DA, Francisco-Ortega J, Jansen RK. 1996. Hybridization and the extinction of rare plant species. Conservation Biology 10: 10–16. [Google Scholar]
- Lewontin RC, Birch LC. 1966. Hybridization as a source of variation for adaptation to new environments. Evolution 20: 315–336. [DOI] [PubMed] [Google Scholar]
- Linhart YB, Grant MC. 1996. Evolutionary significance of local genetic differentiation in plants. Annual Review of Ecology and Systematics 27: 237–277. [Google Scholar]
- McCune B, Mefford MJ. 1999. PCOrd—Multivariate analysis of ecological data, version 4.10. Gleneder Beach: MjM Software.
- Machado MC. 1999. The cacti of Morro do Chapéu, Bahia, Brazil. British Cactus and Succulent Journal 17: 201–213. [Google Scholar]
- Machado MC, Charles, G. 2004. Pilosocereus bohlei Hofacker—a remarkable new species from Brazil. British Cactus and Succulent Journal 22: 188–192. [Google Scholar]
- Martínez-Palacios A, Eguiarte LE, Furnier G. 1999. Genetic diversity of the endangered endemic Agave victoriae-reginae (Agavaceae) in the Chihuahuan desert. American Journal of Botany 86: 1093–1098. [PubMed] [Google Scholar]
- Moraes EM, Abreu AG, Andrade SCS, Sene FM, Solferini VN. 2005. Population genetic structure of two columnar cacti with a patchy distribution in eastern Brazil. Genetica 125: 311–323. [DOI] [PubMed] [Google Scholar]
- Nassar JM, Hamrick JL, Fleming TH. 2001. Genetic variation and population structure of the mixed-mating cactus, Melocactus curvispinus (Cactaceae). Heredity 87: 69–79. [DOI] [PubMed] [Google Scholar]
- Nassar JM, Hamrick JL, Fleming TH. 2002. Allozyme diversity and genetic structure of the leafy cactus (Pereskia guamacho [Cactaceae]). The American Genetic Association 93: 193–200. [DOI] [PubMed] [Google Scholar]
- Nassar JM, Hamrick JL, Fleming TH. 2003. Population genetic structure of Venezuelan chiropterophilous columnar cacti (Cactaceae). American Journal of Botany 90: 1628–1637. [DOI] [PubMed] [Google Scholar]
- National Geodetic Survey. 2002. INVERSE—Version 2.0. Silver Spring: 1315 East-West Highway, Silver Spring, MD 20910–3282. http://ngs.noaa.gov/. 18 Jan. 2005.
- Nei M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker KC, Hamrick JL. 1992. Genetic diversity and clonal structure in a columnar cactus, Lophocereus schotii. American Journal of Botany 79: 86–96. [Google Scholar]
- Raymond M, Rousset F. 1995. GENEPOP (version 1·2): population genetics software for exact tests and ecumenicism. Journal of Heredity 86: 248–249. [Google Scholar]
- Rhymer JM, Simberloff D. 1996. Extinction by hybridization and introgression. Annual Review of Ecology and Systematics 27: 83–109. [Google Scholar]
- Rice WR. 1989. Analyzing tables of statistical tests. Evolution 43: 223–225. [DOI] [PubMed] [Google Scholar]
- Ridgway GJ, Sherburne SW, Lewis RD. 1970. Polymorphism in the esterases of Atlantic herring. Transactions of the American Fisheries Society 99: 147–151. [Google Scholar]
- Rieseberg LH. 1995. The role hybridization in evolution: old wine in new skins. American Journal of Botany 82: 944–953. [Google Scholar]
- Schmalzel RJ, Nixon RT, Best AL, Tress Jr JA. 2004. Morphometric variation in Coryphantha robustipina (Cactaceae). Systematic Botany 29: 553–568. [Google Scholar]
- Shaw CR, Prasad R. 1970. Starch gel eletrophoresis of enzymes—a compilation of recipes. Biochemical Genetics 4: 297–320. [DOI] [PubMed] [Google Scholar]
- Slatkin M. 1987. Gene flow and the geographic structure of natural populations. Science 236: 787–792. [DOI] [PubMed] [Google Scholar]
- Slatkin M, Barton NH. 1989. A comparison of three indirect methods for estimating average levels of gene flow. Evolution 43: 1349–1368. [DOI] [PubMed] [Google Scholar]
- Sneath PHA, Sokal RR. 1973. Numerical taxonomy. San Francisco: Freeman and Co.
- Soltis DE, Haufler CH, Darrow DC, Gastony GJ. 1983. Starch gel eletrophoresis of ferns: a compilation of grinding buffers, gel and electrode buffers, and staining schedule. American Fern Journal 73: 9–27. [Google Scholar]
- Stace CA. 1984. Plant taxonomy and biosystematics. London: Edward Arnold.
- StatSoft INC. 2000. Statistica for windows (computer program manual). Tulsa: Salt Soft Inc.
- Stebbins GL. 1959. The role of hybridization in evolution. Proceedings of the American Philosophical Society 103: 231–251. [Google Scholar]
- Stuber CW, Goodman MM, Johnson FM. 1977. Genetic control and racial variation of β-glucosidase isozymes in maize (Zea mays L.). Biochemical Genetics 15: 383–394. [DOI] [PubMed] [Google Scholar]
- Sun M, Ganders FR. 1990. Outcrossing rates and allozymes variation in rayed and rayless morphs of Bidens pilosa. Heredity 64: 139–143. [Google Scholar]
- Swofford DL, Selander RB. 1989. BIOSYS-1: computer program for the analysis of allelic variation in population genetics and biochemical systematics. Champaign: Illinois Natural History Survey.
- Taylor NP. 1991. The genus Melocactus (Cactaceae) in Central and South America. Bradleya 9: 1–80. [Google Scholar]
- Taylor NP, Zappi DC. 2004. Cacti of Eastern Brazil. Richmond, Surrey, UK: Royal Botanic Gardens, Kew.
- Thomson G. 2002. A re-evaluation of the taxonomic status of the genus Melocactus in Aruba, Netherlands Antilles. Bradleya 20: 29–44. [Google Scholar]
- Thorpe JP. 1982. The molecular clock hypothesis: biochemical evolution, genetic differentiation and systematics. Annual Review of Ecology and Systematics 13: 139–168. [Google Scholar]
- Wright S. 1951. The genetical structure of populations. Annals of Eugenics 15: 323–354. [DOI] [PubMed] [Google Scholar]
- Wright S. 1978. Evolution and the genetics of populations—Variability within and among natural populations. Vol. 4. Chicago: University of Chicago Press.



