Abstract
Circadian clocks generate rhythms in molecular, cellular, physiological, and behavioral processes. Recent studies suggest that disruption of the clock mechanism accelerates organismal senescence and age-related pathologies in mammals. Impaired circadian rhythms are observed in many neurological diseases; however, it is not clear whether loss of rhythms is the cause or result of neurodegeneration, or both. To address this important question, we examined the effects of circadian disruption in Drosophila melanogaster mutants that display clock-unrelated neurodegenerative phenotypes. We combined a null mutation in the clock gene period (per01) that abolishes circadian rhythms, with a hypomorphic mutation in the carbonyl reductase gene sniffer (sni1), which displays oxidative stress induced neurodegeneration. We report that disruption of circadian rhythms in sni1 mutants significantly reduces their lifespan compared to single mutants. Shortened lifespan in double mutants was coupled with accelerated neuronal degeneration evidenced by vacuolization in the adult brain. In addition, per01 sni1 flies showed drastically impaired vertical mobility and increased accumulation of carbonylated proteins compared to age-matched single mutant flies. Loss of per function does not affect sni mRNA expression, suggesting that these genes act via independent pathways producing additive effects. Finally, we show that per01 mutation accelerates the onset of brain pathologies when combined with neurodegeneration-prone mutation in another gene, swiss cheese (sws1), which does not operate through the oxidative stress pathway. Taken together, our data suggest that the period gene may be causally involved in neuroprotective pathways in aging Drosophila.
Keywords: Biological clock, Circadian rhythms, Neuronal health, Protein carbonyls, RING assay
Introduction
Circadian clocks are endogenous timekeeping mechanisms that generate rhythms with circa-24 h periodicity. At the molecular level, circadian clocks consist of cell autonomous networks of core clock genes and proteins engaged in transcriptional–translational feedback loops, which are largely conserved between Drosophila and mammals (Yu and Hardin, 2006). Rhythmic activities of clock genes generate daily fluctuations in the expression level of many target genes that underlie cellular, physiological and behavioral rhythms (Allada and Chung, 2010; Schibler, 2007). Disruption of circadian rhythms by environmental manipulations or mutations in specific clock genes leads to various age-related pathologies and may reduce lifespan in mice (Antoch et al., 2008; Davidson et al., 2006; Kondratov et al., 2006; Lee, 2006).
Functional links between circadian rhythms and aging are supported by observations that an impaired circadian system may predispose organisms to neurodegenerative diseases (Gibson et al., 2009). However, the evidence linking disruption of circadian rhythms to premature neurodegeneration is of correlative nature and the mechanisms involved are not yet understood. Studies in the model organism, Drosophila melanogaster, showed that a null mutation in the clock gene period (per01) is associated with increased susceptibility to oxidative challenge (Krishnan et al., 2008). Furthermore, exposure of aging per01 flies to mild oxidative stress increased their mortality risk, accelerated functional senescence, and increased signs of neurodegeneration compared to the age-matched controls (Krishnan et al., 2009). Together, these data suggest that the clock gene period may protect the health of the nervous system in aging animals.
Neurodegeneration is a detrimental aging phenotype affecting homeostasis, motor performance, and cognitive functions. Several mutants uncovered in Drosophila show these phenotypes (Kretzschmar, 2005); one of them affects the gene sniffer (sni) that encodes for a carbonyl reductase in fruitflies. Carbonyl reductases catalyze the detoxification of lipid peroxides generated by reactive oxygen species (ROS) and help to prevent protein carbonylation (Maser, 2006). Loss of sni function leads to a progressive neurodegenerative phenotype with the formation of spongiform lesions in the brain neuropil, and apoptotic cell death of glia and neurons (Botella et al., 2004). Similar to sni, mutation in the swiss cheese (sws) gene produces age-dependent lesions in the neuropil that are accompanied by apoptotic neuronal death (Kretzschmar et al., 1997). However, the sws gene encodes a phospholipase that interacts with Protein Kinase A (PKA) and it has not been connected with oxidative stress (Muhlig-Versen et al., 2005).
Neurodegeneration is often associated with accumulated oxidative damage in the nervous system (Sayre et al., 2001). We previously reported that arrhythmic per01 flies show significantly increased levels of lipid peroxidation and protein carbonylation during aging (Krishnan et al., 2009). We therefore hypothesized that the circadian system may contribute to cellular homeostasis by curtailing oxidative damage in the nervous system. To test this hypothesis, we examined aging phenotypes in flies carrying mutations in the clock gene per and carbonyl reductase encoded by sni. We report that such double mutants show significantly shortened lifespan, accelerated neurodegeneration, and a decline in climbing ability. Interestingly, these effects were not restricted to the sni gene alone, because arrhythmia due to loss of per function also accelerated neurodegeneration in the sws mutant. Together, our data suggest that the core clock gene period, functions in neuroprotective pathways that may delay the progression of brain pathologies during aging.
Materials and methods
Fly rearing and creation of double mutants
D. melanogaster were reared on 1% agar, 6.25% cornmeal, 6.25% molasses, and 3.5% Red Star yeast at 25 °C in 12-hour light:dark (LD,12:12) cycles (with an average light intensity of ~2000 lx). All experiments were performed between 4 and 8 h after lights-on (or equivalent time in constant light (LL)) in male flies of different ages, as specified in results. To determine lifespan, 3–4 cohorts of 100 mated males of a given genotype were housed in 8 oz round bottom polypropylene bottles (Genesee Scientific) inverted over 60 mm Falcon Primaria Tissue culture dishes (Becton Dickinson Labware) containing 15 ml of diet. Diet was replaced on alternate days without anesthesia after tapping flies to the bottom of the bottle, and mortality was recorded at this time. The per01 mutants were previously backcrossed to the Canton S (CS) for 8 generations and sni1 mutants were backcrossed to yellow white (y w). The per01 sni1 double mutants were created by recombination using per01 w crossed to y w sni1 and selecting flies that were per01 w sni1 (sni1 was detected by the orange eye color). y is localized at 1A5, per at 3B1, w at 3B6 and sni at 7D22. Similarly, the per01 sws1 double mutants were created by recombination with per01 w and y w sws1 Appl-GAL4 (as a visible marker proximal of sws, which is localized at 7D1, detectable by the orange eye color) and selecting flies that were per01 w sws1 Appl-GAL4. The correct genotype was confirmed by external markers, mutant phenotype, and PCR. To determine circadian rhythmicity for each genotype, locomotor activity patterns were monitored in 2–3 independent experiments using the Trikinetics monitor (Waltham, MA). Flies were entrained to LD for 3 days and then recorded for 7 days in constant darkness. Fast Fourier Transform (FFT) analysis was conducted using the ClockLab software (Actimetrics, Coulbourn Instruments). Flies with FFT values>0.04, which showed a single well-defined peak in the periodogram, were classified as rhythmic and included in the calculation of free-running period using the ClockLab software. The y w flies served as control for sni1 and sws1 single mutants and double mutants carrying per01allele.
Neuronal degeneration
Paraffin-embedded sections of heads were processed as previously described (Bettencourt da Cruz et al., 2005; Tschape et al., 2002). Briefly, heads were cut in 7 μm serial sections, the paraffin was removed in SafeClear (Fisher Scientific), sections were embedded in Permount, and analyzed with a Zeiss Axioscope 2 microscope using the auto-fluorescence caused by the eye pigment (no staining was used). Experimental and control flies were put next to each other in the same paraffin block, cut, and processed together. Microscopic pictures were taken at the same level of the brain, the vacuoles (identified by being unstained and exceeding 50 pixels in size) were counted and vacuolized area was calculated using our established methods (Bettencourt da Cruz et al., 2005; Tschape et al., 2002). For sws, the pictures were taken at the level of the great commissure (z=−1; http://web.neurobio.arizona.edu/Flybrain/html/atlas/silver/horiz/index.html) and the holes in the deutocerebral neuropil were measured as described (Bettencourt Da Cruz et al., 2008). For sni, the pictures were taken from sections that contained the ventral deutocerebral neuropil (z=−6; http://web.neurobio.arizona.edu/Flybrain/html/atlas/silver/horiz/index.html), and the vacuoles in all four optic neuropils (lamina, medulla, lobula, and lobula plate) were counted and measured. In both cases, each side of the brain was scored independently (the number of brain hemispheres analyzed for each genotype is indicated in the figures). For a double blind analyses, pictures were taken and numbered, vacuoles were counted, and the area of vacuoles was measured in pixels in Photoshop and subsequently converted into μm2 (Bettencourt da Cruz et al., 2005). Statistical analysis was done using one-way ANOVA.
Rapid iterative negative geotaxis (RING) and oxidative damage assays
Vertical mobility was tested using the RING assay as described (Gargano et al., 2005). Briefly, 2 groups of 25 flies of each genotype were transferred into empty vials without anesthesia, and the vials were loaded into the RING apparatus. The apparatus was rapped three times in rapid succession to initiate a negative geotaxis response. The flies’ movements in tubes were videotaped and digital images captured 4 s after initiating the behavior. The climbed distance was calculated for each fly and expressed as average height climbed in the 4 s interval. The performance of flies in a single vial was calculated as the average of 5 consecutive trials (interspersed with a 30 s rest). To assess oxidative damage, protein carbonyls were measured in male head homogenates of the various genotypes at 370 nm after reaction with 2,4-dinitrophenylhydrazine (DNPH) using a BioTek Synergy 2 plate reader, as described previously (Krishnan et al., 2008). Results were expressed as nmol mg−1 protein using an extinction coefficient of 22,000 M−1 cm−1.
Gene expression by qRT-PCR
The expression of sni gene was measured in per01 mutants and CS control flies collected at 4 h intervals around the clock in LD. Total RNA was extracted from fly heads using TriReagent (Sigma). The samples were purified using the RNeasy mini kit (Qiagen) with on-column DNAse digestion (Qiagen), and cDNA was synthesized with iScript (Bio-Rad). Real-time PCR (qRT-PCR) was performed on the StepOnePlus (Applied Biosystems) under default thermal cycling conditions with a dissociation curve step. Every reaction contained iTaq SYBR Green Supermix with ROX (Bio-Rad), 0.6 ng cDNA, 80 nM primers. Primer sequences are available upon request. Data were analyzed using the 2−ΔΔCT method with mRNA levels normalized to the gene rp49. Relative mRNA levels were calculated with respect to the trough levels set as 1 for control flies.
Statistical analyses
Lifespan and survival curves were plotted using Kaplan Meier survival curves and statistical significance of curves assessed using the Log-Rank (Mantel–Cox) and Gehan–Breslow–Wilcoxon tests (GraphPad Prism v5.0; GraphPad Software Inc. San Diego CA). For statistical analysis of biochemical and gene expression results, one-way ANOVA with post-hoc tests were conducted (GraphPad Instat v3.0).
Results
Loss of circadian rhythms shortens the lifespan of flies mutant for carbonyl reductase
To determine the effects of per on longevity of sni-deficient flies, we created per01 sni1 double mutants. Since both genes are localized on the X chromosome (per at 3B and sni at 7D), we achieved this by recombination using per01 w and y w sni1and selecting flies that lost the y marker but were orange due to the P-element in sni1. The double mutants were confirmed by phenotype and by PCR using primers within the P-element and sni. Several double mutant lines were generated and two lines (referred to as per01 sni1 line 1 and 2) were selected for further analysis. As expected, both double mutant lines exhibited loss of circadian rhythms due to per01 mutation, while single mutants were mostly rhythmic in DD indicating that they had a functional circadian clock (Table 1).
Table 1.
Table showing the percentage of rhythmic flies and mean period length in flies of indicated genotypes. Locomotor activity was monitored in 2–3 independent experiments and the total number of flies (n) analyzed for each genotype is indicated.
| Genotype | n | % rhythmic | Period |
|---|---|---|---|
| y w | 30 | 93 | 23.87 |
| sni1 | 30 | 70 | 23.77 |
| per01 | 30 | 0 | – |
| sws1 | 31 | 97 | 23.70 |
| per01 sni1 (1) | 14 | 0 | – |
| per01 sni1 (2) | 21 | 0 | – |
| per01 sws1 | 23 | 13 | 24.11 |
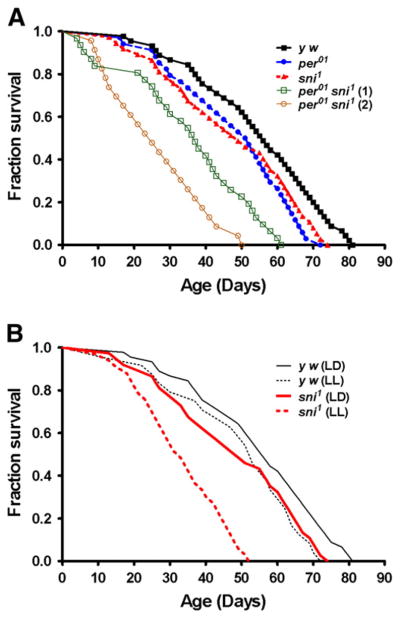
The lifespan of per01 sni1 flies was compared to the sni1 and per01 single mutants, as well as y w controls. There was no difference in mean lifespan between both the single mutants (per01 and sni1) and the y w control (Fig. 1A, Table 2). In contrast, per01 sni1 double mutants showed very significant (p<0.001) reduction of their mean lifespan compared to single mutants. While both double mutant lines were short-lived, we observed significant difference in their lifespan with 32% reduction in recombinant line 1, and 50% reduction in line 2 (Fig. 1A, Table 2).
Fig. 1.
Loss of circadian rhythms dramatically shortens the lifespan of sni1 mutants. A) Survival curves for y w, per01, sni1, and per01 sni1 double mutant lines. B) Lifespan of sni1 and y w in 12 h light: dark (LD) cycles and constant light (LL), which disrupts the circadian clock function.
Table 2.
Disruption of circadian rhythms shortens lifespan in sni1 mutants. Median and mean±SEM lifespan (days) is shown for indicated genotypes with n=sample size. Statistical comparison was conducted using one-way ANOVA with Tukey–Kramer multiple comparison’s test. Values with different superscripts are significantly different at p<0.001.
| Genotype | Regime | n | Median | Mean±SEM |
|---|---|---|---|---|
| y w | LD | 300 | 55 | 55.2±0.7a |
| y w | LL | 300 | 52 | 53.8±0.3a |
| per01 | LD | 295 | 51.5 | 55.7±0.4a |
| sni1 | LD | 197 | 49 | 55.1±0.9a |
| sni1 | LL | 303 | 37 | 37.1±0.4b |
| per01 sni1 (1) | LD | 194 | 37 | 35.7±1.0b |
| per01 sni1 (2) | LD | 268 | 25 | 27.1±0.5c |
We next tested whether disruption of circadian rhythms by non-genetic interventions affect longevity in sni1 single mutants. Adult sni1 flies were reared in constant light (LL) which interferes with the circadian clock mechanism and causes behavioral arrhythmia (Price et al., 1995). The lifespan of sni1 flies maintained in LL was significantly shortened (p<0.0001) compared to sni1 flies reared in LD 12:12, while y w flies showed similar lifespan in both LD and LL (Fig. 1B, Table 2).
Double per01 sni1 mutants show increased neurodegeneration and reduced climbing ability
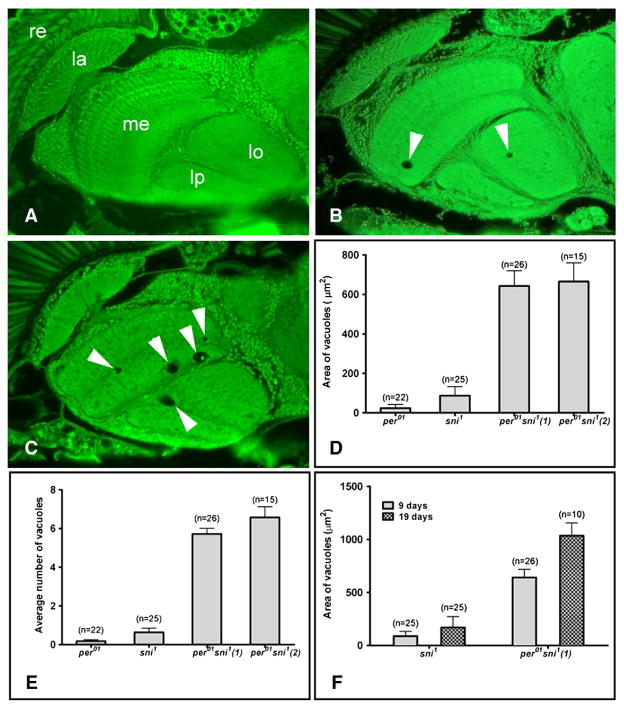
Due to its neuroprotective role, loss of carbonyl reductase in sni1 mutants results in progressive degeneration with small vacuoles appearing in the first 7–9 days of adult life, and becoming larger and more numerous with progressing age (Botella et al., 2004). We demonstrate that neurodegeneration is dramatically increased in per01 sni1 double mutants (Fig. 2). Vacuoles were rarely detected in 9 day-old per01 fly brains (Fig. 2A), while we consistently observed a few small vacuoles in sni1 mutants (Fig. 2B). The vacuolization of the brain was markedly exacerbated in the age-matched per01 sni1 double mutant flies (Fig. 2C). Both, the area taken up by vacuoles and average vacuole number increased very significantly in each of the two per01 sni1 lines examined at 9 days of age (Figs. 2D–E). More pronounced vacuolization was also maintained in 19 day-old per01 sni1 compared to sni1 alone (Fig. 2F). In the next experiment, we asked whether disruption of circadian rhythms by LL, which shortens lifespan of sni1 mutants, affects the levels of neurodegeneration. Brains of 10 day-old sni1 males maintained in LD or LL were sectioned and examined for vacuole formation. Disrupting the circadian clock by constant light significantly increased brain vacuolization in sni1 (Fig. 3), although the effects were less severe than in per01 sni1 double mutants.
Fig. 2.
Interfering with the clock increases neurodegeneration in sni1 mutants. A–C) Paraffin head sections from 9 day-old males (scale bar=25 μm, re=retina, la=lamina, me=medulla, lo=lobula, lp=lobula plate). A) No vacuoles are detectable in the brain of a per01 fly. B) A sni1 fly brain shows a few vacuoles (arrows). C) Brains of per01 sni1 double mutant show increase in the size and number of vacuoles. D) Bar graph showing the mean±SEM area of all vacuoles/brain hemisphere. There is a significant difference between the sni1 and per01 sni1 line 1 (p=2.9×10−6), and sni1 and per01 sni1 line 2 (p=1.6×10−5). E) The mean number of vacuoles/brain hemisphere is increased in per01 sni1 compared to sni1 alone [sni1 to per01 sni1 (1): p=8.42×10−12; sni1 to per01 sni1 (2): p=6.5×10−8]. F) Comparison of the vacuolization between 9 and 19 day-old flies shows that the phenotype is progressive with age for both sni1 (p=0.036) and per01 sni1 line 1 (p=0.03). D–F) The number of brain hemispheres (n) examined to calculate the average values are indicated on the top of each bar.
Fig. 3.
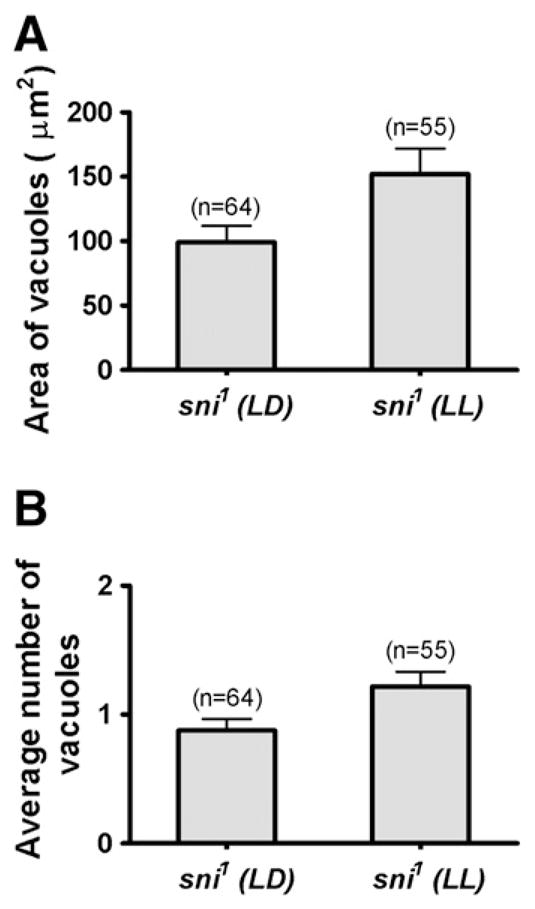
Disrupting the circadian clock by constant light increases vacuolization in sni1 mutant. A) 9 day-old sni1 flies maintained in constant light (LL) show significant increase in the mean area of vacuoles/brain hemisphere compared to 9 day-old sni1 flies in 12 h light:dark (LD, 12:12) cycles (p=0.024). B) The mean number of vacuoles is also significantly higher in sni1 mutant kept in LL (p=0.018). A–B) The number of brain hemispheres (n) examined to calculate the average values is indicated on the top of each bar.
To test whether increased neurodegeneration is associated with altered motor abilities, we conducted the RING assay on 10 day-old single and double mutants along with their controls. The climbing ability of per01 flies did not differ significantly from their CS control, while sni1 mutants showed modest but significant (p<0.05) impairment of climbing ability compared to their y w control. Importantly, the average climbing distance was dramatically reduced (p<0.001) in both per01 sni1 double mutant lines compared to the single sni1 mutants (Fig. 4).
Fig. 4.
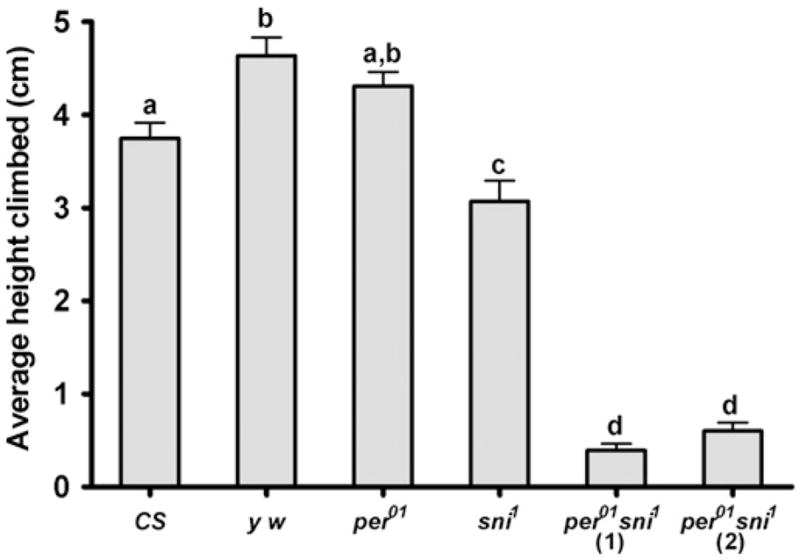
per01 sni1 double mutants show accelerated mobility impairment. Vertical mobility was measured by the RING assay in 10 day-old males of the indicated genotypes. Bars represent mean height climbed (±SEM), based on testing 2 vials per genotype, each containing 25 flies. Bars with different superscripts are significantly different at p<0.01.
Protein carbonyl levels are elevated in per01 sni1 double mutants
Since mutation in the sni gene severely attenuates carbonyl reductase expression (Botella et al., 2004), we tested the levels of oxidatively damaged proteins in heads of per01 and sni1 single mutants as well as per01 sni1 double mutants by measuring protein carbonyls. Levels of protein carbonyls were significantly increased (p<0.01) in heads of both per01 and sni1 single mutants compared to CS and y w controls. Importantly, a significant increase in the protein carbonyl accumulation (p<0.01) was detected in both recombinant lines of the per01 sni1 double mutants compared to the controls and single mutants (Fig. 5).
Fig. 5.
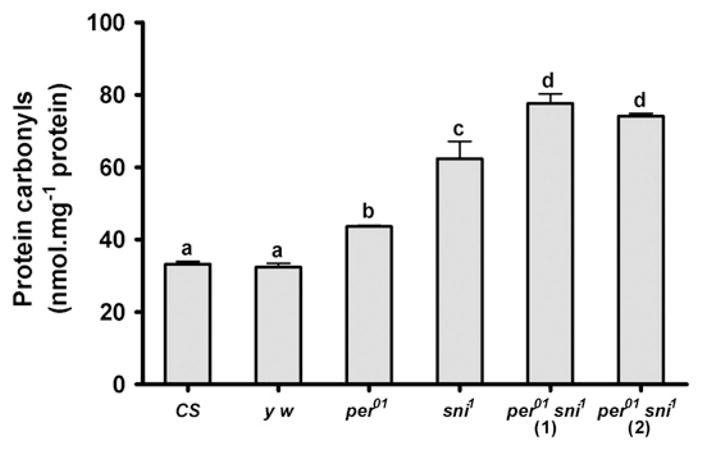
Oxidative damage in the form of protein carbonyls accumulates to higher levels in per01 sni1 flies. Protein carbonyl levels were measured in heads of 10 day-old males of the indicated genotypes. Both per01 and sni1 single mutants had higher protein carbonyls than their respective CS and y w controls. Protein carbonyls were further elevated in per01 sni1 double mutants, compared to per01 or sni1 single mutants. Bars represent mean carbonyl levels (±SEM), based on testing 3 independent sets of flies each containing 75 flies in 3 technical repeats of 25 flies each. Bars with different superscripts are significantly different at p<0.01.
Expression of sni is not affected in per01 mutants
The gene per encodes transcriptional co-regulators that may affect the expression of downstream target genes (Claridge-Chang et al., 2001). Because per01 alone increases protein carbonylation (Krishnan et al., 2008; Fig. 5), we tested whether sni expression might be clock-controlled and therefore altered by the loss of per function. We measured the levels of sni mRNA around the clock in CS and per01 flies by qRT-PCR. The expression of sni did not show a daily rhythm in CS flies, and was not significantly reduced in per01 mutants compared to controls (Fig. 6). These data suggest that sni is not a downstream target of per, rather both mutants appear to act through independent pathways causing additive effects.
Fig. 6.
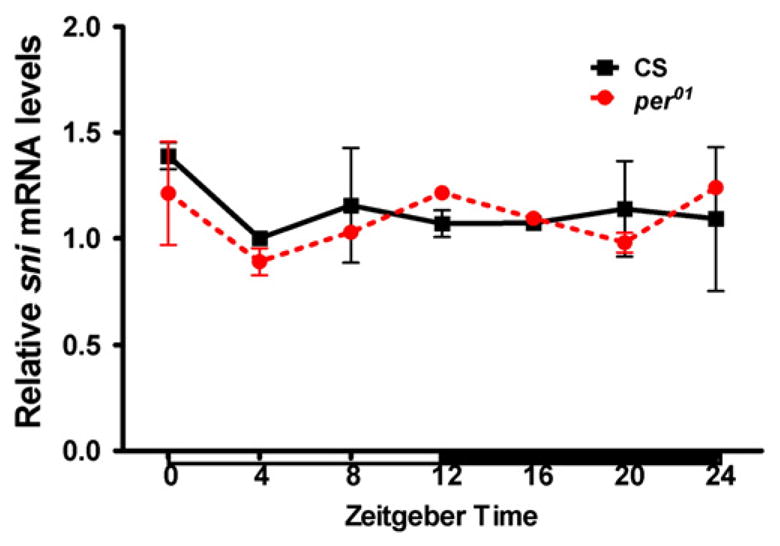
Relative sni mRNA levels are not significantly different between CS and per01 mutants. Expression profile of sni was analyzed by qRT-PCR in heads of flies collected at 4 h intervals in LD, 12:12 cycles. White and black horizontal bars indicate periods of light and darkness, respectively.
Neurodegeneration in sws mutant is substantially increased in a per01 background
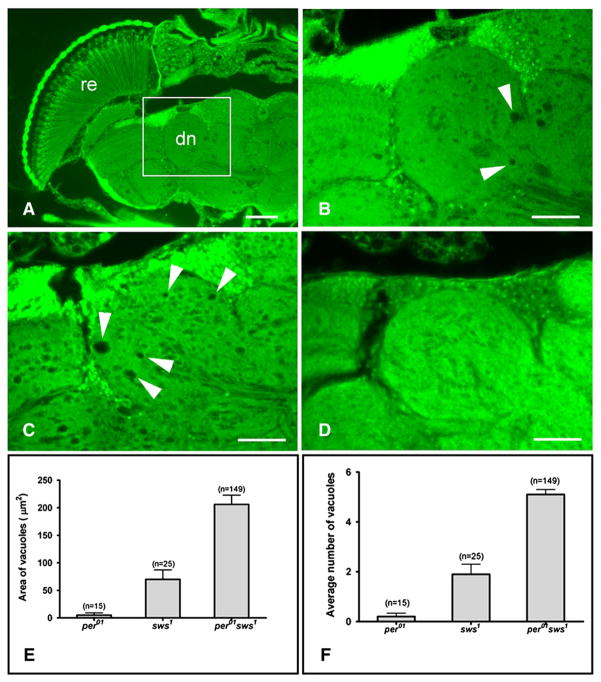
To investigate whether arrhythmicity may affect another neurodegeneration-prone mutant, we compared the neurodegenerative changes caused by a mutation in the swiss cheese (sws) gene in the wild-type and per01 background. The per01 sws1 double mutants were created by recombination between per01 w and y w sws1 Appl-GAL4 (see Materials and methods). Locomotor activity assays indicated that 97% of sws1 single mutants showed rhythmicity with a free-running period similar as in control y w flies, but rhythmicity was mostly lost in per01 sws1 (Table 1). As reported previously (Kretzschmar et al., 1997), 14 day-old sws1 mutant displayed characteristic symptoms of neurodegeneration evidenced by vacuoles in the dorsal neuropil (Figs. 7A, B), which does not occur in y w control flies at this age (Fig. 7D). Importantly, the age-matched per01 sws1 double mutants showed marked increase in the size and number of vacuoles (Fig. 7C arrows) compared to the sws1 single mutants. The three-fold increase in vacuolization was highly significant (Figs. 7E, F).
Fig. 7.
Loss of per function increases neurodegeneration in sws mutants. A) Paraffin head sections from a 14 day-old sws1 fly show widespread degeneration (arrows) characteristic for this mutant (scale bar=50 μm, re=retina, ol=optic lobes, dn=deutocerebral neuropil). B) Magnification from A (box), showing the deutocerebral neuropil that was used for measurements. C) Age-matched per01 sws1 double mutants show increase in the size and number of vacuoles compared to sws1 single mutants. D) Age-matched y w control does not show vacuoles in this area. B–D) Scale bar=25 μm. E) Bar graph showing significant difference in the mean area of all vacuoles in the deutocerebral neuropil between sws1 and per01 sws1 (p=1.9×10−8). F) The mean number of vacuoles/brain hemisphere is also significantly higher in the double mutant compared to sws1 alone (p=0.0033). E–F) The number of brain hemispheres (n) examined to calculate average values is indicated on the top of each bar.
Discussion
Our study demonstrates that disruption of circadian rhythms accelerates aging in two independent mutants that display neurodegenerative phenotypes. We show that lifespan of sni1 flies is reduced by 32–50% in a per01 background, which abolishes molecular and behavioral rhythms. Significant lifespan shortening was also observed in sni1 flies reared in LL, which is known to disrupt circadian systems (Price et al., 1995). Lifespan reduction resulting from the disruption of the clock by either genetic or environmental manipulations strongly suggests that this phenotype is caused by the loss of rhythmicity. However, we cannot exclude that clock-unrelated pleiotropic effects of per may be involved in accelerated aging, since PER protein is unstable in LL (Price et al., 1995). Interestingly, studies in mammals have also shown that interfering with the circadian clock mechanism by the knock-out of specific clock genes may lead to shortened lifespan (Davidson et al., 2006; Yu and Weaver, 2011). Premature aging was observed in mice with mutant core clock genes Bmal1 or Clock, which together form the positive feedback loop of the circadian clock (Antoch et al., 2008; Kondratov et al., 2006). Genetic ablation of per gene homologs in mice resulted in some aging phenotypes and a significant increase in cancer incidence after gamma-radiation challenge (Lee, 2006). Our previous study showed that exposure of per01 flies to external oxidative challenge, significantly increased their mortality risk (Krishnan et al., 2009). Here we show that lifespan is compromised even further when loss of per function is combined with an internal oxidative stress caused by carbonyl reductase deficiency. Taken together, these data suggest that intact circadian clocks promote longevity under various homeostatic challenges.
We show that that per-null related arrhythmia is associated with premature loss of neuronal integrity. A significant increase in the number and size of vacuoles was observed in the brains of per01 sni1 double mutants in LD, or the sni1 mutant in LL compared to sni1 males kept in LD. The increased deterioration of the nervous system might be the cause of the shortened lifespan in these flies because it has been shown that several neurodegenerative mutants are short-lived (Kretzschmar et al., 1997; Tschape et al., 2002). Consistent with the neuronal damage, per01 sni1 flies showed precipitous loss of climbing ability at the age of 10 days, whereas sni1 flies with a functional clock showed only modest (albeit significant) climbing impairment at this age. These data provide experimental evidence suggesting that the disruption of circadian rhythms, which is also observed in human neurodegenerative diseases, may be a causative factor contributing to these pathologies. Indeed, we showed previously that the loss of the clock by itself can lead to neurodegenerative symptoms in per01 mutants later in life (Krishnan et al., 2009). Interestingly, there is also evidence for a reverse relationship such that progressive neurodegeneration may contribute to the loss of clock function in both flies and mice (Morton et al., 2005; Rezaval et al., 2008).
Carbonyl reductase encoded by sni, acts as a neuroprotective enzyme against oxidative stress (Botella et al., 2004); therefore, we tested the levels of oxidatively damaged proteins in heads of single and per01 sni1 double mutants. Consistent with our previous report (Krishnan et al., 2009), there was a significant increase in the protein carbonyl levels in per01 mutants, and these levels were even higher in sni1 mutants (Fig. 4). While protein carbonyl levels were further elevated in per01 sni1 flies, this was not as dramatic as the increase in brain damage observed in these double mutants at the same age (Fig. 2). It is possible that other oxidatively damaged species might accumulate to higher levels in per01 sni1 flies since deficiency in carbonyl reductase activity may also contribute to increased lipid peroxidation (Martin et al., 2011; Sgraja et al., 2004).
To address the nature of per01 and sni1 interactions, we tested daily profiles of sni expression in heads of wild-type CS flies and clock-deficient per01 mutants. The levels of sni mRNA did not change significantly across circadian time points in control flies and neither were they different in per01 mutants. These data suggest that per does not regulate sni expression, consistent with the exacerbated neurodegeneration observed in double mutants. The effects of per mutation appear indirect and additive suggesting more general protective functions of this clock gene. This is further supported by the fact that loss of per function resulted in accelerated neuronal damage in sws mutant, which increases neurodegeneration via different mechanisms. This gene encodes a phospholipase, thereby interfering with the phospholipid homeostasis (Muhlig-Versen et al., 2005). These data show that detrimental effects of per-null allele are not specific to the sni1 mutant with increased oxidative stress, but also extend to the sws1 mutant, which does not appear to act via the oxidative stress pathway. However, the increase in vacuolization was greater in sni1 than in sws1 mutant (Figs. 2, 7), suggesting that per mutation may affect sni via multiple pathways that may or may not be related to oxidative damage.
While our results suggest that a functional circadian system improves the performance of neurodegeneration-prone mutants, the mechanisms involved remain to be investigated. We hypothesize that the circadian clocks slow down the accumulation of neuronal damage in aging organisms by synchronizing the activities of enzymes involved in cellular homeostasis. Indeed, microarray studies of daily gene expression profiles suggested synchronous circadian fluctuations in the expression of some protective enzymes such as glutathione-S-transferases, in fly heads (Wijnen and Young, 2006). Our previous data demonstrated daily fluctuations in the levels of mitochondrial ROS and carbonylated proteins in flies with a functional clock, indicative of a daily rhythm in the removal of oxidative damage (Krishnan et al., 2008). In the absence of the circadian clock, enzymes working in a specific pathway may become dysregulated, leading to the impaired removal of oxidative damage. Consistent with this idea, we reported increased levels of oxidatively damaged lipids and proteins in per01 mutants during aging (Krishnan et al., 2009). It was also shown that oxidative stress may impair the clock function in Drosophila (Zheng et al., 2007).
Circadian clocks are involved in the regulation of response to genotoxic stress and xenobiotics in both mice and fly (Antoch et al., 2008; Beaver et al., 2010; Gachon and Firsov, 2011). Therefore, increased neurodegeneration in arrhythmic sni1 and sws1 mutants may involve additional pathways beyond ROS homeostasis. In mice, the loss of an essential clock component encoded by Bmal1 causes a variety of premature aging phenotypes (Kondratov, 2007). Interestingly, treatment with antioxidants reversed some aging symptoms, but had no effect on other age-related pathologies such as sarcopenia (Kondratov et al., 2009). This suggests that both ROS-dependent and independent mechanisms may contribute to neuroprotection in organisms with functional clocks.
In summary, we show that disrupting clocks in flies has a profound impact on neurodegeneration-prone mutants. Circadian rhythm disturbances such as sleep disorders, which are commonly observed during aging, are exacerbated in neurodegenerative diseases such as Alzheimer’s, Parkinson’s, and Huntington disease (Sterniczuk et al., 2010; Wu and Swaab, 2007). Our functional study, which involved the manipulation of a clock gene to assess its neuroprotective role, substantiates the possibility that arrhythmia is not a mere correlation, but may actually contribute to the onset of neurodegenerative disorders. Conversely, intact circadian clocks appear to promote the health of the nervous system during aging.
Acknowledgments
We thank the anonymous reviewers for insightful comments. This research was supported in part by NIH R21 AG038989 and R21 NS075500 grants to JMG and R01 NS047663 to DK. KR is supported by NSF IGERT in Aging Sciences Fellowship at OSU (DGE 0965820).
References
- Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoch MP, et al. Disruption of the circadian clock due to the Clock mutation has discrete effects on aging and carcinogenesis. Cell Cycle. 2008;7:1197–1204. doi: 10.4161/cc.7.9.5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaver LM, et al. Circadian clock regulates response to pesticides in Drosophila via conserved Pdp1 pathway. Toxicol Sci. 2010;115:513–520. doi: 10.1093/toxsci/kfq083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt da Cruz A, et al. Disruption of the MAP1B-related protein FUTSCH leads to changes in the neuronal cytoskeleton, axonal transport defects, and progressive neurodegeneration in Drosophila. Mol Biol Cell. 2005;16:2433–2442. doi: 10.1091/mbc.E04-11-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt Da Cruz A, et al. Swiss Cheese, a protein involved in progressive neurodegeneration, acts as a noncanonical regulatory subunit for PKA-C3. J Neurosci. 2008;28:10885–10892. doi: 10.1523/JNEUROSCI.3015-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella JA, et al. The Drosophila carbonyl reductase sniffer prevents oxidative stress-induced neurodegeneration. Curr Biol. 2004;14:782–786. doi: 10.1016/j.cub.2004.04.036. [DOI] [PubMed] [Google Scholar]
- Claridge-Chang A, et al. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–671. doi: 10.1016/s0896-6273(01)00515-3. [DOI] [PubMed] [Google Scholar]
- Davidson AJ, et al. Chronic jet-lag increases mortality in aged mice. Curr Biol. 2006;16:R914–R916. doi: 10.1016/j.cub.2006.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon F, Firsov D. The role of circadian timing system on drug metabolism and detoxification. Expert Opin Drug Metab Toxicol. 2011;7:147–158. doi: 10.1517/17425255.2011.544251. [DOI] [PubMed] [Google Scholar]
- Gargano JW, et al. Rapid iterative negative geotaxis (RING): a new method for assessing age-related locomotor decline in Drosophila. Exp Gerontol. 2005;40:386–395. doi: 10.1016/j.exger.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Gibson EM, et al. Aging in the circadian system: considerations for health, disease prevention and longevity. Exp Gerontol. 2009;44:51–56. doi: 10.1016/j.exger.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV. A role of the circadian system and circadian proteins in aging. Ageing Res Rev. 2007;6:12–27. doi: 10.1016/j.arr.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Kondratov RV, et al. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, et al. Antioxidant N-acetyl-L-cysteine ameliorates symptoms of premature aging associated with the deficiency of the circadian protein BMAL1. Aging (Albany NY) 2009;1:979–987. doi: 10.18632/aging.100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar D. Neurodegenerative mutants in Drosophila: a means to identify genes and mechanisms involved in human diseases? Invert Neurosci. 2005;5:97–109. doi: 10.1007/s10158-005-0005-8. [DOI] [PubMed] [Google Scholar]
- Kretzschmar D, et al. The swiss cheese mutant causes glial hyperwrapping and brain degeneration in Drosophila. J Neurosci. 1997;17:7425–7432. doi: 10.1523/JNEUROSCI.17-19-07425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N, et al. Circadian regulation of response to oxidative stress in Drosophila melanogaster. Biochem Biophys Res Commun. 2008;374:299–303. doi: 10.1016/j.bbrc.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N, et al. The circadian clock gene period extends healthspan in aging Drosophila melanogaster. Aging (Albany, NY) 2009;1:937–948. doi: 10.18632/aging.100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC. Tumor suppression by the mammalian Period genes. Cancer Causes Control. 2006;17:525–530. doi: 10.1007/s10552-005-9003-8. [DOI] [PubMed] [Google Scholar]
- Martin HJ, et al. The Drosophila carbonyl reductase sniffer is an efficient 4-oxonon-2-enal (4ONE) reductase. Chem Biol Interact. 2011;191:48–54. doi: 10.1016/j.cbi.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Maser E. Neuroprotective role for carbonyl reductase? Biochem Biophys Res Commun. 2006;340:1019–1022. doi: 10.1016/j.bbrc.2005.12.113. [DOI] [PubMed] [Google Scholar]
- Morton AJ, et al. Disintegration of the sleep-wake cycle and circadian timing in Huntington’s disease. J Neurosci. 2005;25:157–163. doi: 10.1523/JNEUROSCI.3842-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlig-Versen M, et al. Loss of Swiss cheese/neuropathy target esterase activity causes disruption of phosphatidylcholine homeostasis and neuronal and glial death in adult Drosophila. J Neurosci. 2005;25:2865–2873. doi: 10.1523/JNEUROSCI.5097-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, et al. Suppresion of PERIOD protein abundance and circadian cycling by the Drosophila clock mutation timeless. EMBO J. 1995;14:4044–4049. doi: 10.1002/j.1460-2075.1995.tb00075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaval C, et al. A functional misexpression screen uncovers a role for enabled in progressive neurodegeneration. PLoS One. 2008;3:e3332. doi: 10.1371/journal.pone.0003332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayre LM, et al. Chemistry and biochemistry of oxidative stress in neurodegenerative disease. Curr Med Chem. 2001;8:721–738. doi: 10.2174/0929867013372922. [DOI] [PubMed] [Google Scholar]
- Schibler U. The daily timing of gene expresion and physiology in mammals. Dialogues Clin Neurosci. 2007;9:257–272. doi: 10.31887/DCNS.2007.9.3/uschibler. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgraja T, et al. Structural insights into the neuroprotective-acting carbonyl reductase Sniffer of Drosophila melanogaster. J Mol Biol. 2004;342:1613–1624. doi: 10.1016/j.jmb.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Sterniczuk R, et al. Characterization of the 3xTg-AD mouse model of Alzheimer’s disease: part 1. Circadian changes Brain Res. 2010;1348:139–148. doi: 10.1016/j.brainres.2010.05.013. [DOI] [PubMed] [Google Scholar]
- Tschape JA, et al. The neurodegeneration mutant lochrig interferes with cholesterol homeostasis and Appl processing. EMBO J. 2002;21:6367–6376. doi: 10.1093/emboj/cdf636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet. 2006;40:409–448. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- Wu YH, Swaab DF. Disturbance and strategies for reactivation of the circadian rhythm system in aging and Alzheimer’s disease. Sleep Med. 2007;8:623–636. doi: 10.1016/j.sleep.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Yu W, Hardin PE. Circadian oscillators of Drosophila and mammals. J Cell Sci. 2006;119:4793–4795. doi: 10.1242/jcs.03174. [DOI] [PubMed] [Google Scholar]
- Yu EA, Weaver DR. Disrupting the circadian clock: gene-specific effects on aging, cancer, and other phenotypes. Aging (Albany NY) 2011;3:479–493. doi: 10.18632/aging.100323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, et al. FOXO and insulin signaling regulate sensitivity of the circadian clock to oxidative stress. Proc Natl Acad Sci U S A. 2007;104:15899–15904. doi: 10.1073/pnas.0701599104. [DOI] [PMC free article] [PubMed] [Google Scholar]