Abstract
CYP2D6 plays a major role in the metabolism of tamoxifen, and polymorphism of P-glycoprotein has been associated with resistance of many drug therapies. This study investigates the clinical impact of genetic variants of CYP2D6 and ABCB1 in breast cancer patients treated with tamoxifen. Blood samples from 95 breast cancer patients treated with tamoxifen were collected and genotyped for CYP2D6 and ABCB1 variants using allele-specific PCR method. Recurrence risks were calculated using Kaplan–Meier analysis and compared using the log-rank test. Patients carrying CYP2D6*10/*10 and heterozygous null allele (IM) showed higher risks of developing recurrence and metastasis (OR 13.14; 95% CI 1.57–109.94; P = 0.004) than patients with CYP2D6*1/*1 and *1/*10 genotypes. Patients with homozygous CC genotypes of ABCB1 C3435T showed a shorter time to recurrence. Patients who were CYP2D6 IM and homozygous CC genotype of C3435T have statistically significant higher risks of recurrence (P = 0.002). Similarly, median time to recurrence in these patients was only 12 months (95% CI = 0.79–23.2) compared to those without this combination which was 48 months (95% CI = 14.7–81.2). Patients with CYP2D6 IM and homozygous CC genotype of ABCB1 C3435T have shorter times to recurrence. The results confirmed the findings of previous studies and support FDA recommendation to perform pre-genotyping in patients before the choice of therapy is determined in breast cancer patients.
KEY WORDS: ABCB1, breast cancer, CYP2D6, recurrence and metastasis, tamoxifen
INTRODUCTION
Pharmacogenomics is an area with fast-evolving implication in clinical setting, and thus, its incorporation to pharmaceutical care is believed to enhance patient-centred quality care. The US Food Drug and Administration has encouraged the practice of pharmacogenetics testing prior to drug therapy initiation in some clinically important therapeutics, and tamoxifen is one of them. Tamoxifen has been widely used as the standard adjuvant therapy for hormone receptor-positive breast cancer patients, especially in the high-risk pre- and postmenopausal women. However, 30% to 50% of oestrogen receptor (ER)-positive breast cancer patients do not respond to tamoxifen therapy (1–3). Major challenges to effective tamoxifen therapy include issues of drug resistance and tamoxifen-induced adverse events. Tamoxifen is metabolized by a number of cytochrome P-450 enzymes including CYP2D6, CYP3A4 and CYP2C9. Genetic polymorphisms of CYP2D6 have been reported as the major cause of variation in the metabolism of tamoxifen that leads to adverse effects or lack of therapeutic efficacy (4,5).
Genetic variation in CYP2D6 results in different metabolic phenotypes including extensive (EM), intermediate (IM), ultra-rapid (UM) and poor metabolizer. CYP2D6*10 is responsible for the reduced enzyme activity in IMs whereas CYP2D6*4, CYP2D6*5 and CYP2D6*14 are null alleles which encode no enzyme at all. UM (CYP2D6*xN) has a duplication or multiduplication of functional alleles and leads to increase enzymes activity (6–8). This variation causes variability in patients’ outcomes because CYP2D6 had been reported to biotransform tamoxifen into several active metabolites. Studies on the pharmacokinetics of tamoxifen demonstrated significantly lower steady-state plasma and serum concentrations of 4-hydroxy-N-desmethyltamoxifen and 4-hydroxytamoxifen in CYP2D6*10/*10 than other genotypes (4,9,10).
Active metabolites from primary and secondary metabolism pathways of tamoxifen known as endoxifen (4-hydroxy-N-desmethyltamoxifen) and 4-hydroxytamoxifen were found to be 100-fold more potent than tamoxifen (11). A recent study among the breast cancer patients with CYP2D6*10/*10 in Asians has shown that the steady-state plasma levels of 4-hydroxytamoxifen and endoxifen were significantly lower, and these patients have significant shorter median time to disease progression compared to patients who were wild type or heterozygote of CYP2D6*1/*10 (4,9,12). Furthermore, co-administration of selective serotonin reuptake inhibitors in breast cancer patients who suffer depressive syndrome was found to reduce the metabolite concentrations and thus affect the outcomes of tamoxifen therapy (13,14).
ABCB1 is responsible for multidrug resistance, the principal mechanism by which many patients with cancer disorders develop resistance to chemotherapeutic drugs. This gene encodes P-glycoprotein (P-gp) and functions as an energy-dependent drug efflux pump and transports a variety of toxins, nutrients, environmental carcinogens and drugs (15). Over-expression of this protein in breast cancer tumours was significantly associated with disease relapse and shorter disease-free survival period (16). This gene was found to be highly polymorphic and causes susceptibility to various disease and therapeutic clinical outcomes. A silent mutation in exon 26 namely C3435T has been reported to be associated with therapeutic outcomes in breast cancer treatment and other disease (17–19). Another allele G2677A/T in exon 21 had been shown to be associated with an amino acid change in Ala893Thr and Ala893Ser. Substitution of these nucleotides results in change of a lipophilic residue to a hydrophilic one and affects the geometric precision of the interaction site and the secondary structure (20). Tamoxifen, 4-hydroxytamoxifen and endoxifen are known to bind P-gp and are substrates of P-gp which may acts as a barrier and limits the accessibility of active metabolites of tamoxifen to various critical target tissues and the success of tamoxifen therapy (21,22). P-gp expression was also found to increase from 40–50% to 60–70% after chemotherapy in breast cancer patients and resulted in a shorter overall survival in patients (21).
Our earlier studies reported the heterogeneity of CYP2D6 among the three different ethnic groups in Malaysia (23–26), but no studies have investigated the influence of the polymorphism of CYP2D6 and ABCB1 in tamoxifen therapy among the patients in Malaysia. We therefore investigated the impact of CYP2D6 and ABCB1 genotypes on the outcomes of tamoxifen therapy in a cohort of breast cancer patients.
MATERIALS AND METHOD
Subjects
The study was approved by the Medical Research and Ethics Committee of Ministry of Health Malaysia, institutional review board of both Universiti Teknologi MARA and Universiti Kebangsaan Malaysia Medical Centre. Five millilitres of blood samples was collected from breast cancer patients at Universiti Kebangsaan Malaysia Medical Centre, Selayang Hospital and Tengku Ampuan Afzan Hospital after a written informed consent. Patients recruited comprised of three ethnic groups predominant in Malaysian, namely Malays, Chinese and Indians. The ethnicities of all subjects were confirmed by individual screening and verified against the new National Registry Identification Cards. Ninety-five breast cancer patients (53 Malays, 36 Chinese and 6 Indian) were successfully recruited. Status of ER and progesterone receptor breast cancer tumour was determined by immunohistochemistry, and patients received tamoxifen 20 mg/day.
Sample Preparation and Genotyping
Genomic DNA was extracted using alkaline lysis method as described previously (10), and DNA was stored at −20°C until analysis. Patients’ samples were genotyped for CYP2D6*xN, CYP2D6*4 (rs3892097), CYP2D6*5, CYP2D6*10 (rs1065852), CYP2D6*14 (rs5030862), ABCB1 C3435T (rs1045642) and G2677A/T (rs2032582). Nested PCR approach was used to identify variants of CYP2D6. In the first PCR, 0.2 μM of each primer, fragment A; forward primer—5′ ACCAGGCCCCTCCACCGG 3′, reverse primer—5′ AATGCGCCACGCTCACCCATTGG 3′; fragment B: forward primer—5′ ATGGGTGAGCGTGGCGCATTTCC 3′, reverse primer—5′ TGCACTGTTTCCCAGATGGGCTC 3′, 1.0 U of BioTools Taq (Biotools B&M Labs. S.A) and 100 ng of DNA were used to amplify fragments A and B. Fragment A spanning exon 1 and 2 served as template for CYP2D6*10 while fragment B spanning exon 3 to 6 served as template for detection of CYP2D6*4 and CYP2D6*14. The thermocycling conditions for this PCR were 95°C for 5 min (initial denaturation), followed by 35 cycles of 94°C for 1 min; 64°C for 30 s; 72°C for 1.5 min and a final extension of 72°C for 5 min. Eight microlitres of the PCR product was checked on 1% agarose gel and appropriately diluted product and was used as the template in the second PCR.
Allele-specific primers were used for detection of variant and wild-type alleles in a multiplex PCR. The final reaction was carried out in a total volume of 25 μl PCR mix containing diluted first PCR product (1:100), 0.06 μM of common primer, forward and reverse primer: CYP2D6*4—5′ TTGGAGTGGGTGGTGGATGGT 3′, 5′ AGCCCGACTCCTCCTTCAG 3′; CYP2D6*10—5′ ATTTGGTAGTGAGGCAGGTAT 3′, 5′ AACGGCACTCAGGACTAACT 3′; CYP2D6*14—5′ CAGAGACGAGGTGGGGCAA 3′, 5′ TTGCTCACGGCTTTGTCCA 3′, 0.4 μM of wild-type primers, forward primer: CYP2D6*4—5′ CGCATCTCCCACCCCCCG 3′, CYP2D6*10—5′ GCTGGGCTGCACGCTAAC 3′, CYP2D6*14—5′ GCCTTCGCCAACCACTCAG 3′, 0.2 μM of variant primer, reverse primer: CYP2D6*4—5′ TGGGGCGAAAGGGGCGTGT 3′, CYP2D6*10—5′ TGGCAGGGGGCCTGGTTA 3′, CYP2D6*14—5′ CTTCTGCCCATCACCCATT 3′, 1% of dimethyl sulfoxide (Sigma-Aldrich Co., St. Louis USA) and 12.5 μl of 1× of GoTaq® Green Master Mix (Promega Corporation, Madison, USA) for amplification of CYP2D6*4, CYP2D6*10 and CYP2D6*14. The thermocycling conditions were 94°C for 5 min (initial denaturation), followed by 18 cycles of 94°C for 45 s; 56°C for 45 s; 72°C for 1.0 min and 72°C for 5 min (extra elongation). Eight microlitres of the PCR product was checked on 1.5% agarose gel. The specificity of the PCR amplification was confirmed using a positive control for each allele.
For gene duplication and deletion of CYP2D6, previously reported methods were used with some modification (27,28). Amplification reactions were performed using 1 U Biotools® Taq DNA Polymerase (Biotools B&M Labs. S.A, Madrid, Spain), 0.2 μM of primers for duplication and 0.4 μM of primers for deletion with 12 s denaturing at 94°C and 5 min annealing/extension at 68°C for 35 cycles and final extension at 72°C for 10 min. All reactions were performed in a Takara PCR machine (TP600, Takara Bio Inc., Japan).
Genotyping for ABCB1 C3435T was performed using previously designed primers (13) with modification for common reverse primer: 5′ CAAATAAACAGCATGGGAGCA 3′. Amplification reactions were performed using 12.5 μl of 1× GoTaq® Green Master Mix (Promega Corporation, Madison, USA), 0.1 μM of forward primer, 0.2 μM of reverse primer and 0.3 μM of each allele-specific primers were used. The reaction was split into two tubes for parallel amplification of wild-type and variant allele. The conditions for this PCR were 94°C for 5 min (initial denaturation), followed by 10 cycles of 94°C for 30 s; 61°C for 30 s; 72°C for 20 s; followed by 25 cycles of 94°C for 30 s; 55°C for 30 s and 72°C for 30 s.
The variant and wild-type specific primers of ABCB1 G2667A/T were split into three tubes, and each reaction was internally controlled for amplification failure. 1× GoTaq® Green Master Mix (Promega Corporation, Madison, USA), 0.1 μM of common forward primer: 5′ CTATAG GTTCCAGGCTTGCTG 3′, 0.2 μM of common reverse primer: 5′ GGAAGGAAGAACAGTGTGAAGAC 3′ and 0.3 μM allele-specific primers: 5′ TGAAAGATAAGAAAGAACTAGAAGGCG 3′, 5′ TGAAAGATAAGAAAGAACTAGAAGGCA 3′, 5′ TGAAAGATAAGAAAGAACTAGAAGGCT 3′ were used for each allele. Specificity of the primer sets was confirmed using a positive control for each allele. The PCR conditions were 80°C for 5 min and 94°C for another 5 min (initial denaturation), followed by 8 cycles of 94°C for 30 s; 61°C for 45 s; 72°C for 20 s; and subsequent 25 cycles of 94°C for 45 s; 58°C for 30 s; 72°C for 1 min and 72°C for 5 min. Ten microlitres of the PCR product was checked on 1.5% agarose gel.
The metabolizer status of CYP2D6 was defined as (1) EM, with absence of variant alleles; (2) heterozygous intermediate metabolizer (Het-IM), with presence of one allele of CYP2D6*10 and (3) IM with homozygous CYP2D6*10 or having a combined allele of heterozygous null alleles. For statistical reasons, individuals with duplicated allele without any null alleles were included in the EM group.
Statistical Analysis
The association of the genetic variants of CYP2D6 and ABCB1 with recurrence and metastasis of breast cancer was investigated. Allele frequencies of CYP2D6 and ABCB1 with their respective Hardy–Weinberg equilibrium were calculated. The differences in allele frequencies with different patients’ characteristic were determined using the ϰ2 test or Fisher’s exact test. Odds ratios and corresponding 95% CI were calculated for risk estimation and trend of association between genotypes and recurrence or metastasis. Tukey’s test was used to determine the combination effects of CYP2D6 and ABCB1 based on the number of risk alleles. Recurrence-free rate was analysed using Kaplan–Meier methods. The recurrence-free period was defined as the period between surgery and initiation of tamoxifen therapy to the recurrence of breast cancer or metastasis. Statistical significance analysis of a relationship between outcomes and genetic polymorphism was assessed by the log-rank test. All P values were two sided, and values less than 0.05 were considered statistically significant. Statistical tests were run using SPSS version 18 on an IBM-compatible computer.
RESULT
Allelic Distribution of CYP2D6 and ABCB1
CYP2D6*4, CYP2D6*xN, CYP2D6*5, CYP2D6*10, CYP2D6*14 and C3435T, G2677A/T alleles of ABCB1 were successfully amplified in 95 patients (Table I). Their genotype frequencies are as shown in Table II. The genotype and allele frequencies of CYP2D6 in the breast cancer patients were compared with those among Malaysian healthy populations and Singapore breast cancer patients (7,10–13). The most common allele detected was CYP2D6*10; 47.2% patients were heterozygous and 22.6% were homozygous CYP2D6*10. The genotypes for both CYP2D6 and ABCB1 were in Hardy–Weinberg equilibrium.
Table 1.
Clinical and Pathological Characteristics of 95 Patients Subgrouped According to CYP2D6 and ABCB1 Alleles
| Variant | CYP2D6 | ABCB1 C3435T | ABCB1 G2677 (A/T) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| WT | VT | P (VT vs. WT) | C | T | P (C vs. T) | G | A/T | P (G vs. A/T) | |
| Characteristic (n = 95, alleles = 190) | P value/OR (95% CI) | P value/OR (95% CI) | P value/OR (95% CI) | ||||||
| Age at surgery | |||||||||
| Median (range), 51 (33–79) | |||||||||
| Menopausal status (pre- vs. post-) | |||||||||
| Pre- (n = 33, alleles = 66) | 25 | 41 | 0.13a | 43 | 23 | 0.68a | 38 | 28 | 0.37a |
| Post- (n = 62, alleles = 124) | 61 | 63 | 1.58 (0.86–2.92) | 77 | 47 | 1.14 (0.61–2.13) | 63 | 61 | 1.31 (0.71–2.39) |
| Histology (DIC vs. LIC) | |||||||||
| DIC (n = 82, alleles = 164) | 77 | 87 | 0.29b | 103 | 61 | 0.71b | 90 | 74 | 0.14b |
| LIC (n = 4, alleles = 8) | 2 | 6 | 0.37(0.07–1.92) | 6 | 2 | 0.56 (0.11–2.88) | 2 | 6 | 3.65 (0.72–18.62) |
| Others (n = 9, alleles = 18) | |||||||||
| Tumour grade (2 + 3 vs. 1) | |||||||||
| 1 (n = 34, alleles = 86) | 39 | 47 | 1.0a | 52 | 34 | 0.008a, c | 45 | 41 | 0.84b |
| 2 and 3 (n = 52, alleles = 104) | 47 | 57 | 1.01 (0.57–1.79) | 68 | 18 | 2.47 (1.26–4.86) | 56 | 48 | 1.06 (0.59–1.88) |
| Tumour stage (III + IV vs. 0–II) | |||||||||
| 0–II (n = 70, alleles = 140) | 69 | 69 | 0.05a | 81 | 47 | 0.06a | 75 | 65 | 0.84a |
| III and IV (n = 25, alleles = 50) | 17 | 33 | 1.94 (0.99–3.81) | 39 | 11 | 2.05 (0.96–4.39) | 26 | 24 | 0.94 (0.49–1.79) |
| ER and/or PR status (Pos vs. Neg) | |||||||||
| Pos (n = 78, alleles = 156) | 69 | 87 | 0.54a | 97 | 59 | 0.55a | 85 | 71 | 0.43a |
| Neg (n = 17, alleles = 34) | 17 | 17 | 1.26 (0.59–2.65) | 23 | 11 | 0.78 (0.36–1.73) | 16 | 18 | 1.34 (0.64–2.03) |
Patients with C allele of ABCB1 C3435T were found to have statistically higher odds ratio for tumour grade 2 and 3 compared to tumour grade 1 (OR 2.47; 95% CI 1.26–4.86; P = 0.008)
Abbreviations: WT wild type, VT variant, DIC ductal invasive carcinoma, LIC lobular invasive carcinoma, ER oestrogen receptor, PR progesterone receptor, Pos positive, Neg negative
a ϰ 2 test
bFisher’s exact test
cA significant association
Table II.
The CYP2D6 and ABCB1 Allele Frequencies
| Polymorphism | Allele | Allele frequencies | P value (95% CI) | |||
|---|---|---|---|---|---|---|
| Current study | Malaysian populationa | Singapore breast cancer patientsb | ||||
| A (n = 95) | B (n = 429) | C (n = 165) | A vs. B | A vs. C | ||
| CYP2D6 | * 1 | 41.6 | 51.5 | 36.7 | Refc | |
| * 4 | 2.1 | 1.6 | 0.3 | 1.0 | 0.5 | |
| * 5 | 4.7 | 1.9 | 8.0 | 0.2 | 0.4 | |
| * 10 | 48.9 | 43.1 | 51.0 | 0.3 | 0.4 | |
| * 14 | 1.1 | 2.0 | ||||
| * xN | 1.6 | 1.9 | 2.0 | 1.0 | 1.0 | |
| ABCB1 | ||||||
| C3435T | C | 63.2 | 56.5d | 0.3 | ||
| T | 36.8 | 43.5 | ||||
| G2677A/T | G | 53.2 | ||||
| A | 11.0 | |||||
| T | 35.8 | |||||
Abbreviations: A vs. B current study vs. Malaysian population, A vs. C current study vs. Singapore breast cancer patients
a(1) Teh et al. 2001, (2) Ismail et al. 2001 and (3) Ismail et al. 2003
bLim et al. 2011
c CYP2D6*1 was used as the reference
dTeh et al. 2007
Risk Estimation Between Genotypes of CYP2D6 and ABCB1 Variants
Patients with CYP2D6 IM genotype appeared to be at higher risk of developing recurrence and metastasis compared to CYP2D6 EM genotype (OR 13.14; 95% CI 1.54–109.94; P = 0.004; Table III). Carriers of CYP2D6 Het-IM genotype also showed an increased risk, but the difference was not statistically significant (OR 2.71; 95% CI 0.28–25.78; P = 0.78). On the other hand, patients with combination of CT and TT genotypes of ABCB1 C3435T have lower risk to disease relapse but was not significant (OR 0.46; 95% CI 0.16–1.34; P = 0.15). Patients with genotypes of G/A or G/T and AT of G2677A/T were found to have higher risk of disease relapse, but no statistically significant association was observed (OR 1.41; 0.35–5.71; P = 0.63).
Table III.
Risk Estimation Between Genotype and Recurrences and Metastasis in Breast Cancer Patients
| Variant | Overall | No event | Event | Statistic | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. | Percentage | No. | Percentage | No. | Percentage | OR | 95% CI | P | ||
| CYP2D6 | ||||||||||
| Homo-EM | 24 | 25.3 | 23 | 29.5 | 1 | 5.9 | 1.0 (ref)a | |||
| Het-IM | 38 | 40.0 | 34 | 43.6 | 4 | 23.5 | 2.71 | 0.28–25.78 | 0.78b | |
| IM | 33 | 34.7 | 21 | 26.9 | 12 | 70.6 | 13.14 | 1.54–109.94 | 0.004b, c | |
| ABCB1 | ||||||||||
| C3435T | ||||||||||
| CC | 41 | 43.2 | 31 | 39.7 | 10 | 58.8 | 1.0 (ref)a | |||
| CT | 38 | 40.0 | 32 | 41.0 | 6 | 35.3 | 0.58 | 0.19–1.79 | 0.34d | |
| TT | 16 | 16.8 | 15 | 19.3 | 1 | 5.9 | 0.21 | 0.02–1.76 | 0.15e | |
| CT and TT combined | ||||||||||
| CT + TT | 54 | 56.8 | 47 | 60.3 | 7 | 41.2 | 0.46 | 0.16–1.34 | 0.15d | |
| G2677A/T | ||||||||||
| GG | 32 | 33.7 | 29 | 34.5 | 3 | 27.3 | 1.0 (ref)a | |||
| G/(A or T) | 37 | 38.9 | 32 | 38.1 | 5 | 45.5 | 1.51 | 0.33–6.89 | 0.72e | |
| AT | 26 | 27.4 | 23 | 27.4 | 3 | 1.2 | 1.26 | 0.23–6.84 | 1.00e | |
| G/(A or T) and AT combined | ||||||||||
| G/(A or T) + AT | 63 | 66.3 | 55 | 65.5 | 8 | 72.7 | 1.41 | 0.35–5.71 | 0.63e | |
| No. of risk alleles of CYP2D6 and ABCB1 C3435T | ||||||||||
| 0 | 6 | 6.3 | 6 | 7.7 | 0 | 0 | ||||
| 1 | 15 | 15.8 | 15 | 19.2 | 0 | 0 | ||||
| 2 | 28 | 29.5 | 26 | 33.3 | 2 | 11.8 | 1.0 (ref)f | |||
| 3 | 30 | 31.6 | 20 | 25.6 | 10 | 58.8 | 6.5 | 1.28–33.05 | 0.04b, c | |
| 4 | 16 | 16.8 | 11 | 14.2 | 5 | 29.4 | 5.9 | 0.99–35.21 | 0.15b | |
| No. of risk alleles of CYP2D6 and ABCB1 C3435T combined | ||||||||||
| 0 | 6 | 6.3 | 6 | 7.7 | 0 | 0 | ||||
| 1 + 2 | 43 | 45.3 | 41 | 52.6 | 2 | 11.8 | 1.0 (ref)f | |||
| 3 + 4 | 46 | 48.4 | 31 | 39.7 | 15 | 88.2 | 9.9 | 2.11–46.6 | 0.001b, c | |
No. of risk alleles—patients were classified into five groups according to combination genotypes of CYP2D6 and ABCB1 C3435T (0—Homo-EM of CYP2D6 and homozygous mutant of ABCB1, 1—Het-IM of CYP2D6 and homozygous mutant of ABCB1 or Homo-EM of CYP2D6 and heterozygous ABCB1, 2—Het-IM ofCYP2D6 and heterozygous of ABCB1, 3—Het-IM of CYP2D6 and homozygous wild type of ABCB1 or IM CYP2D6 and heterozygous of ABCB1, 4—IM of CYP2D6 and homozygous wild type for ABCB1)
Abbreviations: Homo-EM homozygous wild-type allele for *xN, *4, *5, *10, *14, HetIM one allele of *10 and IM homozygous of *10 or having a combined allele of heterozygous null alleles; ABCB1 C3435T (CC homozygous wild type, CT heterozygous, TT homozygous mutant) and G2677A/T (GG homozygous wild type, G/(A or T) heterozygous, AT homozygous mutant)
aThe reference category included variant without any risk allele
bTukey’s test
cA significant association
dϰ2 test
eFisher exact test
fThe reference category included ≤2 risk alleles
CYP2D6 and ABCB1 Variants and Onset of Recurrence and Metastasis
Time to develop recurrence and metastasis was evaluated using the Kaplan–Meier analysis. The recurrence-free rate was measured from the date of diagnosis to the date of the first instance of recurrence and metastasis. Patients carrying CYP2D6 IM genotypes had significantly shorter recurrence-free survival compared to patients with CYP2D6 EM and Het-IM genotypes (log-rank; P = 0.031, Fig. 1a). No statistically significant association was detectable for ABCB1 C3435T (log-rank; P = 0.22, Fig. 1b), but patients with CC genotype have earlier onset of relapse compared to CT and TT genotypes. Kaplan–Meier analysis showed a different trend for genotypes of ABCB1 G2677T compared to C3435T, but it was not statistically significant (P = 0.97, Fig. 1c).
Fig. 1.
Kaplan–Meier probabilities of time to develop recurrence and metastasis in patients treated with tamoxifen predicted from CYP2D6 and ABCB1 genotypes: a carrier of CYP2D6, extensive metabolizer (EM), heterozygous intermediate metabolizer and intermediate metabolizer (IM); b carrier of ABCB1 C3435T, homozygous wild type (CC) vs. heterozygous (CT) and homozygous mutant (TT); c carrier of ABCB1 G2677A/T, homozygous wild type (GG) vs. heterozygous {G(A or T)} and homozygous mutant (AT)
Association of Genotypes and Patients’ Clinical Outcomes
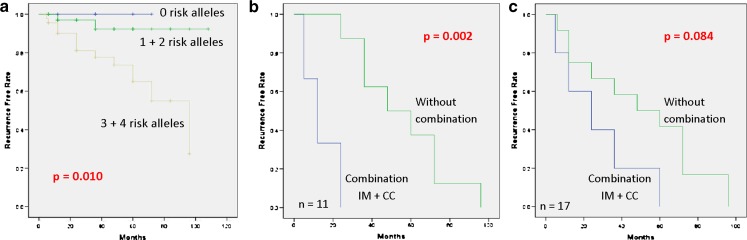
Patients carrying CC genotype of ABCB1 C3435T were found to have shorter recurrence-free survival. This genotype was combined with CYP2D6 IM to investigate the combination effect on the onset of relapse. Patients were classified into five groups according to the number of risk alleles (0, 1, 2, 3, 4 risk alleles group; Table III). Patients carrying three and four risk alleles were found to have higher risk in developing disease compared with other alleles (OR 9.9; 95% CI 2.11–46.6; P = 0.001). Kaplan–Meier analysis revealed that patients have shorter recurrence-free survivals (P = 0.01, Fig. 2a) if they have a combination of risk alleles. Time to recurrence and metastasis were evaluated among relapsing patients with their genotypes (recurrence and metastasis, n = 11; metastasis only, n = 6). Median time to develop recurrence and metastasis in intermediate metabolizers of CYP2D6 and CC group of ABCB1 C3435T was only 12 months (95% CI = 0.79–23.2, n = 3) compared to 48 months (95% CI = 14.7–81.2, n = 8) in patients with genotype of CYP2D6 IM or CC group of ABCB1 C3435T or other genotypes. The difference was found to be statistically significant (log-rank; P = 0.002, Fig. 2b). A similar trend was observed in patients who have both recurrence and metastasis (n = 11) and those with metastasis only (n = 6), but no statistical significance was found (Fig. 2c).
Fig. 2.
Kaplan–Meier probabilities of time to develop recurrence and metastasis in patients treated with tamoxifen. Patients were classified according to combined genotypes of CYP2D6 and ABCB1. a Patients were classified into five groups (0, 1, 2, 3, 4) based on the number of risk alleles, combined for 1 + 2 risk alleles and 3 + 4 risk alleles; b combined effect of CYP2D6 and ABCB1 C3435T genotypes with time to develop recurrence + metastasis (combination of CYP2D6 IM + CC genotypes of ABCB1 C3435T vs. without combination—other CYP2D6 and ABCB1 C3435T genotype combination); c combined effect of CYP2D6 and ABCB1 C3435T with time to develop recurrence and metastasis + metastasis only
DISCUSSION
The clinical benefit of tamoxifen has significantly impacted ER-positive breast cancer patients, but approximately one third of patients have recurrence within 15 years (29). Heterogeneity in patients’ responses to tamoxifen among breast cancer patients is consistently observed across patient populations where administration of the same dose of this drug results in a range of outcomes which include adverse events or therapeutic failure. Many clinical variables have been associated with drug response such as age, diet, menopausal status, lifestyle and tumour biology. Important achievements also have been obtained in optimisation of drug therapy based on classification of diseases using protein expression profiles of the breast cancer tumours. However, the genetic background of a patient with respect to metabolizing enzymes, drug transporters and correlation of patients’ clinical data that determine efficacy and toxicity of the therapy is still lacking.
Lim et al. demonstrated that patients prescribed with tamoxifen and carrying CYP2D6*10/*10 genotype have slower response towards therapy compared patients with CYP2D6*1/*1 or CYP2D6*1/*10 genotypes (4). Furthermore, breast cancer patients who received adjuvant tamoxifen therapy revealed worse therapy outcomes in patients with the CYP2D6*10/*10 genotype and significantly higher incidence of recurrence within 10 years after the operation (10,12). Among Caucasians, tamoxifen-treated patients carrying the heterozygous and homozygous CYP2D6*4 had a significantly increased risk of recurrence of breast cancer, shorter relapse-free periods and worse event-free survival rates compared with carriers of functional alleles (30,31). In contrast, two studies from Japan showed that CYP2D6*10/*10 genotype was unlikely to have any clinical significance on breast cancer patients receiving adjuvant tamoxifen (32,33).
Our result are consistent with the hypothesis that the CYP2D6*10/*10 and heterozygous null alleles metabolize tamoxifen slower and thus produce less pharmacologically active tamoxifen metabolites resulting in a higher risk of recurrence and metastasis. We demonstrated that carrier of CYP2D6*10/*10 with combination heterozygous null allele showed higher incidence of developing recurrence and metastasis than patients with CYP2D6*1/*1 and CYP2D6*1/*10 (log-rank test; P = 0.031). In addition, genetic polymorphism of Malays was found to be different from Chinese and Far Eastern races and their CYP2D6 activity was hypothesised to be intermediate between East Asian and Caucasian (23). This could explain the discrepancy results between this study and the study from Japan.
From our study, the CC genotype for ABCB1 C3435T has a shorter time in developing recurrence and metastasis although the association was not statistically significant (P = 0.22). Previous studies showed that expression of P-gp protein level was twofold higher for homozygous CC genotype compared to TT genotype (34). Thus, the expression of this genotype would cause lower bioavailability of tamoxifen and its metabolites in patients. Genotypes of CT, TT and combination of CT/TT did not show an increased risk in developing relapse. Homozygous TT genotype is associated with lower ABCB1 expression and has been correlated with reduced expression and functional status of P-gp protein compared to CC genotypes (35). Transportation of tamoxifen would be slower and benefited patients with TT genotype. However, accumulation of tamoxifen or active metabolites after several years would increase chances of cells becoming resistant towards therapy. In addition, ABCB1 mRNA levels were higher in high-grade tumours compared to low-grade tumours and this relationship might be an indicator of patient’s clinical outcomes (36). Recent studies in non-small cell lung cancer showed GG and CC genotype of G2677A/T and C3435T were associated with a significantly better response to docetaxel–cisplatin compared with the combined of heterozygous and homozygous mutant of both gene, respectively (37).
In this study, we also performed combination analysis of patients having both CYP2D6 IM and CC genotype of ABCB1 in patients with relapse. This group was compared with similar groups of patients but carrying only either CYP2D6 IM or homozygous wild-type CC or other genotypes of ABCB1. Patients with combination of these genotypes have shorter recurrence-free rates (P = 0.002). To our knowledge, this is the first study to address the correlation between CYP2D6 and ABCB1 variant with clinical outcomes among breast cancer patients treated with tamoxifen.
CONCLUSION
This finding is particularly important in the discovery of the effectiveness and resistance of tamoxifen therapy among breast cancer patients. These lines of evidence imply that pre-genotyping of CYP2D6 intermediate metabolizer and homozygous wild-type C3435T is important before prescribing tamoxifen. It is thus recommended that clinical pharmacists should be aware of this and incorporate the practice of “PharmacoDiagnostics” in the relevant pharmacy practice as routine when appropriate.
ACKNOWLEDGEMENTS
This research is supported by a grant from the Ministry of Science, Technology and Innovation Malaysia (Grant no: 02-01-01-SF0173). We thank the director general of Ministry of Health Malaysia, Suzana Ismail and Khairussaleh Jalaludin for the approval and assistance in collection of patients’ samples.
REFERENCES
- 1.Fisher B, Costantino JP, Wickerham DL, Redmond CK, Kavanah M, Cronin WM, et al. Tamoxifen for prevention of breast cancer: Report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Insts. 1998;90:1371–88. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 2.Osborne CK. Tamoxifen in the treatment of breast cancer. N Engl J Med. 1998;339:1609–1618. doi: 10.1056/NEJM199811263392207. [DOI] [PubMed] [Google Scholar]
- 3.Girault I, Bi'eche I, Lidereau R. Role of estrogen receptor transcriptional. Coregulators in tamoxifen resistance in breast cancer. Maturitas. 2006;54:342–351. doi: 10.1016/j.maturitas.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Lim HS, Ju Lee H, Seok Lee K, Sook Lee E, Jang IJ, Ro J. Clinical implications of CYP2D6 genotypes predictive of tamoxifen pharmacokinetics in metastatic breast cancer. J Clin Oncol. 2007;25:3837–45. doi: 10.1200/JCO.2007.11.4850. [DOI] [PubMed] [Google Scholar]
- 5.Goetz MP, Knox SK, Suman VJ, Rae JM, Safgren SL, Ames MM, et al. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat. 2007;101:113–121. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]
- 6.Eichelbaum M, Mineshita S, Ohnhaus EE, Zekorn C. The influence of enzyme induction on polymorphic sparteine oxidation. Br J Clin Pharmacol. 1986;22:49–53. doi: 10.1111/j.1365-2125.1986.tb02879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steiner E, Bertilsson L, Sawe J, Bertling I, Sjoqvist F. Polymorphic debrisoquin hydroxylation in 757 Swedish subjects. Clin Pharmacol Ther. 1988;44:431–435. doi: 10.1038/clpt.1988.176. [DOI] [PubMed] [Google Scholar]
- 8.Bertilsson L, Dahl ML, Sjoqvist F, Aberg-Wistedt A, Humble M, Johansson I, et al. Molecular basis for rational megaprescribing in ultrarapid hydroxylators of debrisoquine. Lancet. 1993;341:63. doi: 10.1016/0140-6736(93)92546-6. [DOI] [PubMed] [Google Scholar]
- 9.Lim JSL, Chen XA, Singh O, Yap YS, Ng RCH, Wong NS, et al. Impact of CYP2D6, CYP3A5, CYP2C9 and CYP2C19 polymorphism on tamoxifen pharmacokinetics in Asian breast cancer patients. Br J Clin Pharmacol. 2011;71(5):737–750. doi: 10.1111/j.1365-2125.2011.03905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y, Sun Y, Yao L, Shi L, Wu Y, Ouyang T, et al. Association between CYP2D6*10 genotype and survival of breast cancer patients receiving tamoxifen treatment. Ann Oncol. 2008;19:1423–1429. doi: 10.1093/annonc/mdn155. [DOI] [PubMed] [Google Scholar]
- 11.Desta Z, Wars BA, Soukhova NV, Floackhart DA. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther. 2004;310:1062–1075. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- 12.Kiyotani K, Mushiroda T, Sasa M, Bando Y, Sumitomo I, Hosono N, et al. Impact of CYP2D6*10 on recurrence-free survival in breast cancer patients receiving adjuvant tamoxifen therapy. Cancer Sci. 2008;99:995–999. doi: 10.1111/j.1349-7006.2008.00780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loprinzi CL. Venlafaxine in management of hot flashes in survivors of breast cancer: a randomised controlled trial. Lancet. 2000;356:2059–2063. doi: 10.1016/S0140-6736(00)03403-6. [DOI] [PubMed] [Google Scholar]
- 14.Jin Y, Desta Z, Stearns V, Ward B, Ho H, Lee KH, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97:30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 15.Leslie EM, Deeley RG, Cole SP. Multidrug resistance proteins: role of P-glycoprotein, MRP1, MRP2, and BCRP (ABCG2) in tissue defense. Toxicol Appl Pharmacol. 2005;204:216–237. doi: 10.1016/j.taap.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 16.Tsukamoto F, Shiba E, Taguchi T, Sugimoto T, Watanabe T, Kim SJ, et al. Immunohistochemical detection of P-glycoprotein in breast cancer and its significance as a prognostic factor. Breast Cancer. 1997;4:259–263. doi: 10.1007/BF02966518. [DOI] [PubMed] [Google Scholar]
- 17.Kafka A, Sauer G, Jaeger C, Grudmann R, Kreienberg R, Zeillinger R, et al. Polymorphism C3435T of the MDR-1 gene predicts response to preoperative chemotherapy in locally advanced breast cancer. Int J Oncol. 2003;22:1117–1121. [PubMed] [Google Scholar]
- 18.Kelleher D, Farrell R, McManus R. Pharmacogenetics of inflammatory bowel disease. Novatis Found Symp. 2004;263:41–53. doi: 10.1002/0470090480.ch4. [DOI] [PubMed] [Google Scholar]
- 19.Cizmarikova M, Wagnerova M, Schono L, Habalova V, Kohut A, Linkova A, et al. MDR1 (C3435T) polymorphism: relation to the risk of breast cancer and therapeutic outcomes. Pharmacogenetics. 2010;10:62–69. doi: 10.1038/tpj.2009.41. [DOI] [PubMed] [Google Scholar]
- 20.Tanabe M, Ieiri I, Nagata N, Inoue K, Ito S, Kanamori Y, et al. Expression of P-glycoprotein in human placenta: relation to genetic polymorphism of the multidrug resistance (MDR)-1 gene. J Pharmacol Exp Ther. 2001;297:1137–1143. [PubMed] [Google Scholar]
- 21.Vargas-Roig LM, Gago FE, Tello O, Martin De Civetta MT, Ciocca DR. c-erbB-2 (Her-2/neu) protein and drug resistance in breast cancer patients treated with induction chemotherapy. Int J Cancer. 1999;84:129–134. doi: 10.1002/(SICI)1097-0215(19990420)84:2<129::AID-IJC6>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 22.Teft WA, Mansell SE, Kim R. Endoxifen, the active metabolite of tamoxifen, is a substrate of the efflux transporter P-glycoprotein (MDR1) Drug Metab Dispos. 2011;39:588–562. doi: 10.1124/dmd.110.036160. [DOI] [PubMed] [Google Scholar]
- 23.Teh LK, Ismail R, Yusoff R, Hussein A, Isa MN, Rahman AR. Heterogeneity of the CYP2D6 gene among Malays in Malaysia. J Clin Pharm Ther. 2001;26:1–7. doi: 10.1046/j.1365-2710.2001.00347.x. [DOI] [PubMed] [Google Scholar]
- 24.Ismail R, Teh LK. Genetic polymorphism of CYP2D6: Malaysian Indians have the highest frequency for CYP2D6*4 in Asia. Eur J Clin Pharmacol. 2001;57:617–618. doi: 10.1007/s002280100360. [DOI] [PubMed] [Google Scholar]
- 25.Ismail R, Teh LK, Amir J, Alwi Z, Lopez CG. Genetic polymorphism of CYP2D6 in Chinese subjects in Malaysia. J Clin Pharm Ther. 2003;28:279–284. doi: 10.1046/j.1365-2710.2003.00490.x. [DOI] [PubMed] [Google Scholar]
- 26.Teh LK, Lee WL, Amir J, Salleh MZ, Ismail R, Amir J, Alwi Z. Single step PCR for detection of allelic variation of MDR1 gene (P-glycoprotein) among three ethnic groups in Malaysia. J Clin Pharm Ther. 2007;32:313–319. doi: 10.1111/j.1365-2710.2007.00822.x. [DOI] [PubMed] [Google Scholar]
- 27.Lundqvist E, Johansson I, Ingelman-Sundberg M. Genetic mechanisms for duplication and multiduplication of the human CYP2D6 gene and methods for detection of duplicated CYP2D6 genes. Gene. 1999;226:327–338. doi: 10.1016/S0378-1119(98)00567-8. [DOI] [PubMed] [Google Scholar]
- 28.Steen VM, Andreassen OA, Daly AK, Tefre T, Borresen AL, Idle JR, et al. Detection of the poor metabolizer-associated CYP2D6 (D) gene deletion allele by long PCR technology. Pharmacogenetics. 1995;5:215–223. doi: 10.1097/00008571-199508000-00005. [DOI] [PubMed] [Google Scholar]
- 29.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Effects of chemotherapy hormonal therapy for early breast cancer on recurrence 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 30.Schroth W, Antoniadou L, Fritz P, Schwab M, Muerdter T, Zanger UM, et al. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol. 2007;25:5187–5193. doi: 10.1200/JCO.2007.12.2705. [DOI] [PubMed] [Google Scholar]
- 31.Schroth W, Goetz MP, Hamann U, Fasching PA, Schmidt M, Winter S, et al. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA. 2009;302:1429–1436. doi: 10.1001/jama.2009.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toyama T, Yamashita H, Sugiura H, Kondo N, Iwase H, Fujii Y. No association between CYP2D6*10 genotype and survival of node-negative Japanese breast cancer patients receiving adjuvant tamoxifen treatment. Jpn J Clin Oncol. 2009;39(10):651–65621. doi: 10.1093/jjco/hyp076. [DOI] [PubMed] [Google Scholar]
- 33.Okishiro M, Taguchi T, Jin Kim S, Shimazu K, Tamaki Y, Noguchi S. Genetic polymorphisms of CYP2D6 10 and CYP2C19 2, 3 are not associated with prognosis, endometrial thickness, or bone mineral density in Japanese breast cancer patients treated with adjuvant tamoxifen. Cancer. 2009;115:952–961. doi: 10.1002/cncr.24111. [DOI] [PubMed] [Google Scholar]
- 34.Hoffmeyer S, Burk O, Von Richter O, Arnold HP, Brochmoller J, Johne A, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci U S A. 2000;97:3473–3478. doi: 10.1073/pnas.050585397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taheri M, Mahjoubi F, Omranipour R. Effect of MDR1 polymorphism on multidrug resistance expression in breast cancer patients. Genet Mol Res. 2010;9:34–40. doi: 10.4238/vol9-1gmr669. [DOI] [PubMed] [Google Scholar]
- 36.Clifford SC, Thomas DJ, Neal DE, Lunec J. Increased MDR1 gene transcript levels in high-grade carcinoma of the bladder determined by quantitative PCR-based assay. Br J Cancer. 1994;69(4):680–686. doi: 10.1038/bjc.1994.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan JH, Han JX, Wu JM, Huang HN, Yu QZ, Sheng LJ. MDR1 single nucleotide polymorphism G2677T/A and haplotype are correlated with response to docetaxel-cisplatin chemotherapy in patients with non-small-cell lung cancer. Respiration. 2009;78:49–55. doi: 10.1159/000158454. [DOI] [PubMed] [Google Scholar]